Abstract
Viral infection or lipopolysaccharide (LPS) treatment induces expression of a large array of genes, the products of which play a critical role in host antipathogen immunity and inflammation. We have previously reported that the expression of ubiquitin-specific protease 25 (USP25) is significantly up-regulated after viral infection or LPS treatment, and this is essential for innate immune signaling. However, the mechanism behind this phenomenon is unclear. In this study, we found that viral infection-induced up-regulation of Usp25 is diminished in cells lacking interferon regulatory factor 7 (IRF7) or interferon α receptor 1 (IFNAR1) but not p65. Sendai virus- or type I interferon-induced up-regulation of Usp25 requires de novo protein synthesis of IRF7. Furthermore, IRF7 directly binds to the two conserved IRF binding sites on the USP25 promoter to drive transcription of Usp25, and mutation of these two sites abolished Sendai virus-induced IRF7-mediated activation of the USP25 promoter. Our study has uncovered a previously unknown mechanism by which viral infection or LPS induces up-regulation of USP25.
Keywords: cell signaling, gene regulation, immunology, interferon, interferon regulatory factor (IRF), transcription, USP25
Introduction
Host pattern recognition receptors recognize pathogen-associated molecular patterns and initiate a series of signaling cascades that lead to activation of transcription factors including NF-κB and interferon regulatory factor 3 (IRF3)2 (1–3). It has been well documented that activation of NF-κB (p65/p50 heterodimer) is dependent on inhibitors of κB kinase (IKK) complex (IKKα/β/γ)-mediated phosphorylation and degradation of IκBα, whereas activation of IRF3 requires phosphorylation by TBK1 or IKKϵ (4–9). The activated NF-κB and IRF3 enter into nucleus, bind to the conserved κB or IRF binding sites of promoters, and recruit co-activators to activate the transcription of target genes.
Viral nucleic acid and lipopolysaccharide (LPS) of Gram-negative bacteria are two common pathogen-associated molecular patterns that trigger signaling cascades to activate NF-κB and IRF3 and induce the production of type I interferons (IFNs) (3, 10). Type I IFNs further induce the expression of hundreds of downstream genes in an autocrine or paracrine manner, and the products of these genes including interferon-induced GTP-binding protein (Mx), 2′-5′-oligoadenylate synthase (OAS), double-stranded RNA-activated protein kinase (PKR), ISG56, and ISG15 orchestrate inhibition of pathogen replication and spread and promote apoptosis and clearance of the infected cells (11). In addition to the direct effect on innate immune cells for antipathogen responses, type I IFNs also regulate adaptive immunity including T cell activation and differentiation and antitumor immunity (12, 13).
The type I IFN family is composed of 13 functional IFNA genes in humans (14 in mice), a single IFNB gene, and others. The IFNα family shares 80% sequence homology among them, whereas the homology between various IFNα and IFNβ is 30% (14, 15). However, all the type I IFNs bind to the same receptors, IFNAR1 and IFNAR2, with affinities varying from picomolar to micromolar orders to recruit tyrosine kinase 2 (TYK2) and Janus kinase 1 (JAK1) for signal transduction, respectively. TYK2 and JAK1 are cross-phosphorylated and activated to phosphorylate several conserved tyrosine residues on IFNAR1 and IFNAR2, which provides docking sites for the downstream effector proteins including STAT1 (16, 17). It has been shown that STAT2 interacts with IFNAR2 constitutively, whereas STAT1 is recruited to IFNAR2-IFNAR1 receptor complex in both STAT2-dependent and -independent manners (18–20). TYK2 and JAK1 further phosphorylate Tyr-701 of STAT1 and Tyr-690 of STAT2, which form the ISGF3 transcription factor complex together with IRF9 to bind to the IFN-stimulated response elements on the promoters of and activate the transcription of ISGs (16). Type I IFN treatment also results in Tyr(P)-STAT1 homodimers that are responsible for the regulation of IFNγ-activated sequence elements (21, 22). In addition to phosphorylation of STAT1 at Tyr-701, the phosphorylation of STAT1 at Ser-708 by IKKϵ accounts for transcriptional activation of about 30% of the ISGF3 target genes (23). Thus, it is conceivable that type I IFN-triggered transcription of ISGs is regulated at multiple steps ranging from the ligand subtypes to the modifications of transcription factors.
IRF7 is strongly induced by type I IFN-mediated signaling in a manner that is dependent on the TYK2-mediated phosphorylation of Tyr-701 of STAT1 but independent of IKKϵ-mediated phosphorylation of Ser-708 of STAT1 (23, 24). Although IRF3 and IRF7 share a similar structure to bind the conserved IRF binding sites and are activated by TBK1- or IKKϵ-mediated phosphorylation, studies with Irf3−/− and Irf7−/− mice or cells suggest that IRF3 is required for early induction of IFNβ and IFNα4, whereas IRF7 is a master transcription factor essential for later induction of IFNα subsets (25–30). Whether and how IRF3 and IRF7 differentially regulate transcription of other genes are of great interest.
We have previously observed that LPS or viral infection substantially up-regulates the expression of Usp25 gene (31, 32). In this study, we found that virus- or LPS-induced expression of Usp25 was significantly abolished in cells lacking IRF7 or IFNAR1. Importantly, type I IFN-triggered signaling indirectly induces up-regulation of Usp25 by inducing expression of IRF7. Furthermore, we have identified two conserved IRF7 binding sites on the promoter of Usp25 gene, and mutation of these two sites impaired SeV-induced or IRF7-mediated activation of the USP25 promoter. Our study has uncovered the type I IFN-IRF7 axis-mediated expression of Usp25 gene.
Experimental Procedures
Mice
Ifnar1+/− mice were purchased from The Jackson Laboratory and maintained and crossed to obtain Ifnar1+/+ and Ifnar1−/− littermates in the specific pathogen-free facility of Wuhan University. Age- and sex-matched Ifnar1+/+ and Ifnar1−/− littermates were used for all experiments. All animal experiments were in accordance with protocols approved by the Institutional Animal Care and Use Committee of Wuhan University.
Cells
Ifnar1+/+ and Ifnar1−/− MEFs were isolated from E14.5 embryos. Bone marrow from Ifnar1+/+ and Ifnar1−/− mice was isolated and differentiated into bone marrow-derived dendritic cells (BMDCs) with GM-CSF (20 ng/ml). MLFs were isolated as described previously (31). Irf3−/−Irf7−/− MEFs were kindly provided by Dr. Pinghui Feng (University of Southern California). p65+/+ and p65−/− MEFs were kind gifts from Dr. Tom Maniatis (Columbia University). Tbk1−/− MEFs were described previously (33, 34) and provided by Dr. Wen-Chen Yeh (University of Toronto). The cells were cultured in DMEM containing 10% FBS, 1% streptomycin-penicillin, and 10 μm mercaptoethanol.
Constructs, Antibodies, and Reagents
Mouse IRF3 or IRF7 was cloned into phage-6tag vector via standard molecular methods. USP25 promoter (−5000 to −1) was cloned into the pGL3-Basic vector (Promega). Site-directed mutagenesis was performed with a kit (Life Technologies) and sequenced for confirmation. FLAG-tagged RelB, p52, and p50 were kindly provided by Dr. Jin Jin (Zhejiang University). IFNα, IFNβ, anti-IFNβ, anti-IFNα (PBL Assay Science), actinomycin D (Sigma), IMD0354, ZM449829, amlexanox, and p38 mitogen-activated protein kinase inhibitor (Abcam) were purchased from the indicated manufacturers. Mouse anti-FLAG (KM8002), mouse anti-β-actin (KM9001), and HRP-conjugated goat anti-mouse or -rabbit IgG (Thermo Scientific, PA1-86717 and SA1-9510) were from the indicated manufacturers. Rabbit anti-USP25 was described previously (35) and kindly provided by Gemma Marfany (University de Barcelona, Barcelona, Spain).
Real Time Quantitative PCR
Cells treated with various stimuli were harvested in TRIzol (Invitrogen), and first strand cDNA was synthesized with a reverse transcription kit (Biotool). Gene expression was examined with a Bio-Rad CFX Connect system with a SYBR Green One Step Real-Time PCR kit (Biotool). Data were normalized to the expression of β-actin. Real time quantitative PCR primers were described previously (31) and are as follows: Irf3: forward, CGG AAA GAA GTG TTG CGG TT; reverse, TTT TCC TGG GAG TGA GGC AG; Irf7: forward, AGA GGG CGT TTT ATC TTG CG; reverse, TGG AGC CCA GCA TTT TCT CT; and Ifnan: forward, TCA AAG GAC TCA TCT GCT GC; reverse, GGT TCC TGC ACC CCC ACC TG.
Viral Infection
Cells were seeded into 24-well plates (2 × 105 cells/well) or 6-well plates (106–107 cells/well). Twenty-four hours later, cells were treated with LPS or infected with SeV or HSV-1 for the indicated time points. The cells were collected for quantitative PCR (qPCR) or immunoblotting assays.
Virus-mediated Gene Transfer
For lentivirus-mediated gene transfer, phage-6tag-IRF3, phage-6tag-IRF7, phage-6tag-rTBK1, phage-6tag-rIKKϵ, or the empty vector was cotransfected with the packaging vectors pSPAX2 and pMD2G into HEK293T cells. Eight hours after transfection, the medium was changed with fresh full medium (10% FBS, 1% streptomycin-penicillin, and 10 μm β-mercaptoethanol). Forty hours later, the supernatants were harvested to infect Irf3−/−Irf7−/− MEFs, Tbk1−/− MEFs, or wild-type MEFs followed by puromycin (1 μg/ml) selection for 2 weeks.
siRNA
siRNA targeting mouse IRF3 or IRF7 was synthesized and transfected into cells by Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol. The sequences of siRNA are as follows: IRF3-siRNA1, 5′-GGA AAG AAG UGU UGC GGU UTT-3′; IRF3-siRNA2, 5′-GGC UAU UGU UUC UGA UCC UTT-3′; IRF3-siRNA3, 5′-GGU UGU UCC UAC AUG UCU UTT-3′; IRF7-siRNA1, 5′-CUU GCG CCA AGA CAA UUC ATT-3′; IRF7-siRNA2, 5′-CU GGA UGU GAC CAU CAU GUTT-3′; IRF7-siRNA3, 5′-GCA CUU UCU UCC GAG AAC UTT-3′; siTBK1, 5′-CCU CUC UCC UGU AGU CUU UTT-3′; siIKKϵ, 5′-CCC ACA ACA CGA UUG CCA UTT-3′, and control siRNA, 5′-GAU GAC GGG AAC UAC AAG ATT-3′.
Reporter Gene Assays
HEK293 cells (4 × 104 cells/well) cultured in 24-well plates were transfected with the reporter plasmid (100 ng) and an internal control vector, phRL-TK-Renilla luciferase (Promega) (2.5 ng). The pGL3-Basic vector served as a negative control, and empty vector was used to equalize the total amount of DNA. Twenty-four hours after transfection, cells were lysed in passive lysis buffer, and the firefly and Renilla luciferase activities were determined using a Dual-Luciferase reporter assay kit (Promega). The firefly luciferase activity was normalized by Renilla luciferase activity and expressed as the -fold stimulation relative to the activity in vector-transfected cells.
Chromatin Immunoprecipitation Assays
Briefly, 5 × 106 cells were fixed with 1% formaldehyde and quenched by glycine. The cells were washed three times with PBS and then harvested in chromatin immunoprecipitation (ChIP) lysis buffer (50 mm Tris·HCl, pH 8.0, 1% SDS, 5 mm EDTA) followed by sonication until the sizes of DNA were 400–600 bp. The lysate was centrifuged at 4 °C for 15 min, and ChIP dilution buffer (20 mm Tris·HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100) was added to the supernatant (4:1 volume). The resulted lysate was then incubated with anti-FLAG at 4 °C overnight. The protein G beads were added into the lysate on the next morning and incubated at 4 °C for 3 h. DNA was eluted using ChIP elution buffer (0.1 m NaHCO3, 1% SDS, 30 μg/ml proteinase K) through incubation at 65 °C overnight, and DNA was purified with a DNA purification kit (TIANGEN). The purified DNA was assayed by quantitative PCR with an CFX Connect system with a SYBR Green One Step Real-Time PCR kit.
Statistical Analysis
Differences between experimental and control groups were determined by Prism software with two-way analysis of variance and Bonferroni test. p values less than 0.05 were considered statistically significant.
Results
IRF7 Plays an Essential Role of LPS- or Virus-induced Expression of Usp25
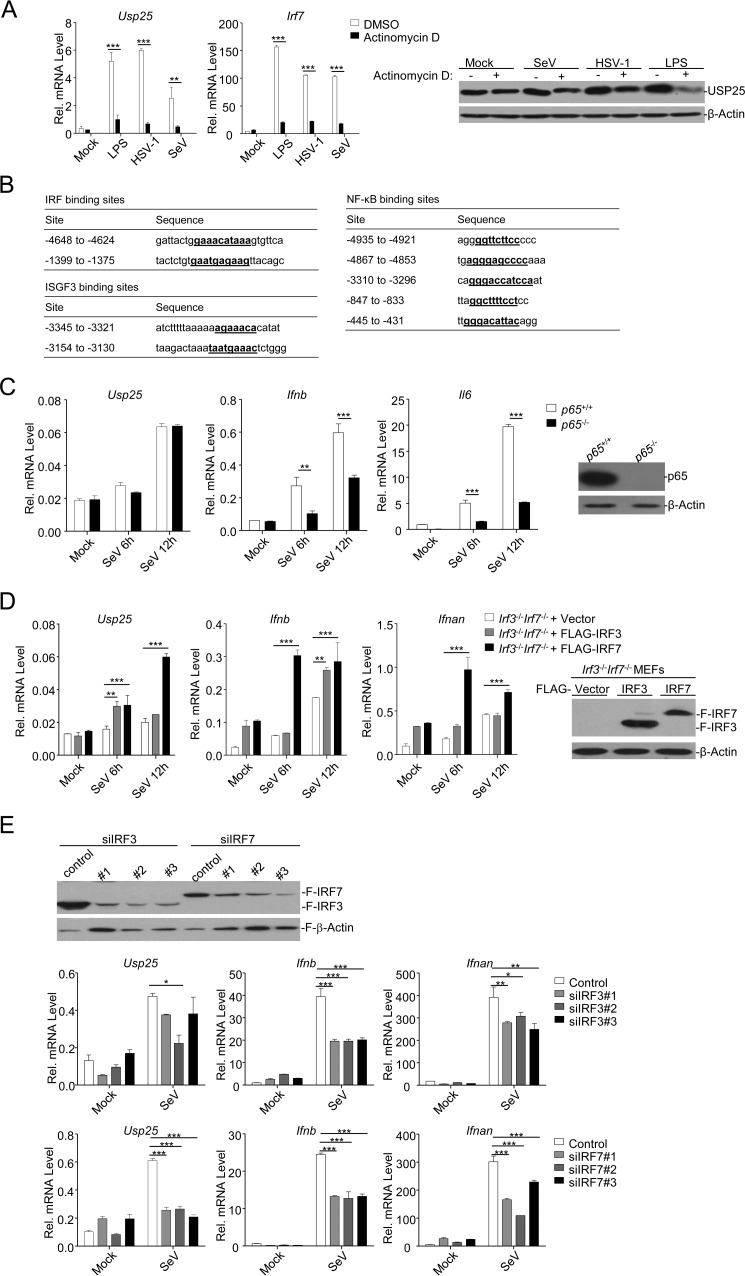
In our previous studies, we have observed that the expression of Usp25 is up-regulated by LPS treatment or viral infection in various types of cells (31, 32). Interestingly, treatment with actinomycin D, a compound that inhibits transcription, almost abolished the up-regulation of Usp25 and the increase of USP25 protein by LPS or SeV or HSV-1 infection in BMDCs (Fig. 1A). Sequence analysis of the promoter of mouse Usp25 gene identified two IRF binding sites (−1399 to −1375 and −4648 to −4624), two ISGF3 binding sites (−3154 to −3130 and −3345 to −3321), and at least five NF-κB binding sites (Fig. 1B). However, p65 deficiency did not affect SeV-induced expression of Usp25 but did inhibit SeV-induced expression of Ifnb and Il6 (Fig. 1C). We reconstituted either IRF3 or IRF7 into Irf3−/−Irf7−/− MEFs and examined SeV-induced expression of Usp25. We found that reconstitution of IRF7 into Irf3−/−Irf7−/− MEFs promoted SeV-induced up-regulation of Usp25 more robustly than did reconstitution of IRF3 (Fig. 1D). In addition, SeV-induced up-regulation of Usp25 was substantially inhibited by knockdown of IRF7 and to a lesser extent by knockdown of IRF3 in MEFs (Fig. 1E). These data suggest that IRF7 and IRF3 (to a lesser extent) but not p65 are essential transcription factors for virus-induced up-regulation of USP25.
FIGURE 1.
IRF7 is the essential transcription factor of LPS- or virus-induced expression of Usp25. A, BMDCs were left untreated, treated with LPS (1 μg/ml), or infected with HSV-1 or SeV in the presence or absence of actinomycin D (2 μg/ml). Twelve hours later, cells were harvested for real time qPCR (left graphs) or immunoblotting analysis (right panels). B, potential IRF, NF-κB, and ISGF3 binding sites and sequences in USP25 promoter. C, p65+/+ and p65−/− MEFs were left uninfected or infected with SeV for 6–12 h followed by qPCR analysis (graphs). The expression of p65 in p65+/+ and p65−/− MEFs was examined by immunoblotting analysis (right panels). D, Irf3−/−Irf7−/− MEFs were reconstituted with the empty vector (Irf3−/−Irf7−/− + Vector), IRF3 (Irf3−/−Irf7−/− + FLAG-IRF3), or IRF7 (Irf3−/−Irf7−/− + FLAG-IRF7) through lentivirus-mediated gene transfer. The expression levels of reconstituted IRF3 and IRF7 were examined by immunoblotting analysis (right panels). The cells were infected with SeV for 6–12 h followed by qPCR analysis (graphs). E, HEK293 cells were transfected with FLAG-IRF3 or FLAG-IRF7 and siRNAs targeting IRF3 or IRF7, respectively. Twenty-four hours later, cells were harvested for immunoblotting analysis (upper panels). MEFs were transfected with siRNAs targeting IRF3 or IRF7. Twenty-four hours later, cells were left untreated or infected with SeV for 12 h followed by qPCR analysis (lower graphs). Data shown are representatives of two (A) or three (C–E) independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars represent S.D. Rel., relative.
LPS- and Virus-induced Up-regulation of Usp25 Depends on Type I IFN-triggered Signaling
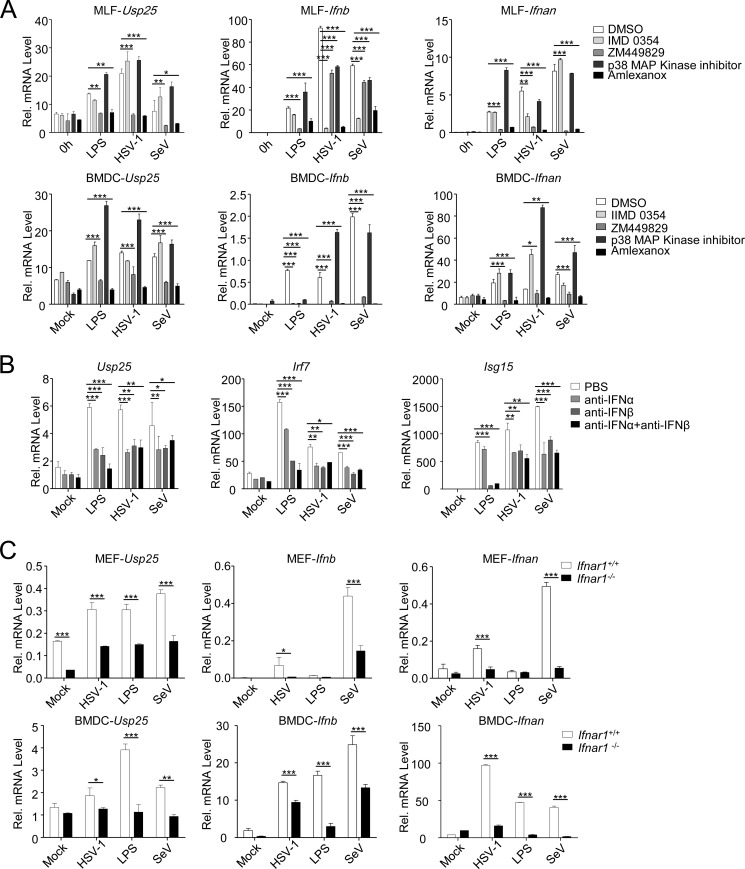
We next examined the effects of various kinase inhibitors on virus- or LPS-induced up-regulation of Usp25 in BMDCs or MLFs. Consistent with the notion that IRF7 and IRF3 are essential for transcriptional up-regulation of Usp25, inhibition of the upstream kinases TBK1 and IKKϵ by amlexanox but not IMD0354 (inhibitor for IKKβ) or the p38 kinase inhibitor impaired LPS- or virus-induced expression of Usp25 (Fig. 2A). Interestingly, we also found that ZM449828 (a JAK1 inhibitor) strongly inhibited up-regulation of Usp25 induced by LPS or viral infection, indicating that JAK1-mediated signaling is critical for the induction of USP25.
FIGURE 2.
LPS- and virus-induced up-regulation of Usp25 depends on type I IFN-triggered signaling. A, BMDCs (lower graphs) or MLFs (upper graphs) were left untreated, treated with LPS (1 μg/ml), or infected with HSV-1 or SeV in the presence or absence of IMD0345 (IKKβ inhibitor), ZM449829 (JAK1 inhibitor), p38 MAPK inhibitor, or amlexanox (TBK1 and IKKϵ inhibitor). Twelve hours later, cells were harvested for qPCR analysis. B, BMDCs were left untreated, treated with LPS (1 μg/ml), or infected with HSV-1 or SeV in the presence of anti-IFNα (3 μg/ml), anti-IFNβ (3 μg/ml), or both. Twelve hours later, cells were harvested for qPCR analysis. C, Ifnar1+/+ and Ifnar1−/− MEFs or BMDCs were left untreated, treated with LPS (1 μg/ml), or infected with HSV-1 or SeV for 12 h followed by qPCR analysis. Data shown are representatives of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars represent S.D. Rel., relative.
Because JAK1 is critical for type I IFN-triggered signaling, we reasoned that LPS or virus up-regulates the expression of Usp25 through type I IFN-triggered signaling. To test this hypothesis, we treated BMDCs with anti-IFNα, anti-IFNβ, or both followed by LPS stimulation or viral infection. As shown in Fig. 2B, blocking IFNα, IFNβ, or both strongly inhibited LPS- or virus-triggered induction of Usp25 and Irf7. Furthermore, LPS- or virus-induced up-regulation of Usp25 was substantially diminished in Ifnar1−/− MEFs and almost completely abolished in Ifnar1−/− BMDCs compared with the wild-type controls (Fig. 2C). These data together suggest that LPS- or virus-induced expression mainly depends on type I IFN-triggered signaling.
Type I IFN-induced Expression of Usp25 Is Dependent on TBK1/IKKϵ and de Novo Synthesized IRF7
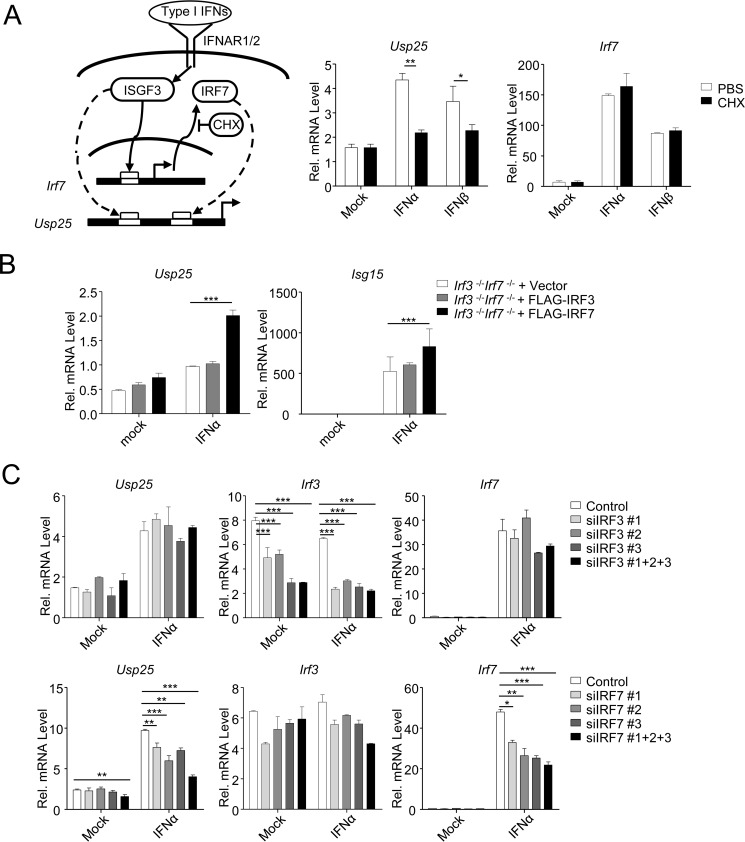
Considering that Usp25 gene promoter contains two ISGF3 binding sites and that type I IFN-triggered signaling is critical for the up-regulation of Usp25, we hypothesized that type I IFNs activate transcription of Usp25 though ISGF3. However, treatment with cycloheximide, a compound that inhibits mRNA translation, impaired IFNα- or IFNβ-induced expression of Usp25. In contrast, IFNα- or IFNβ-induced expression of Irf7, a direct target of ISGF3, was not affected by cycloheximide treatment (Fig. 3A), indicating that virus-triggered type I IFN-mediated up-regulation of Usp25 requires de novo protein synthesis. In addition, IFNα-triggered up-regulation of Usp25 was impaired by knockdown of IRF7 but not IRF3 in MEFs and restored by reconstitution of IRF7 but not IRF3 into Irf3−/−Irf7−/− MEFs (Fig. 3, B and C), indicating that the de novo synthesized IRF7 is required for type I IFN-induced up-regulation of Usp25.
FIGURE 3.
Type I IFN-induced expression of Usp25 is dependent on and the synthesis of IRF7 protein. A, a model of type I IFN-triggered signaling (left). BMDCs were left untreated or treated with IFNα (20 units/ml) or IFNβ (100 ng/ml) in the presence or absence of cycloheximide (CHX) (100 μg/ml) for 12 h followed by qPCR analysis (right graphs). B, Irf3−/−Irf7−/− MEFs were reconstituted with the empty vector (Irf3−/−Irf7−/− + Vector), IRF3 (Irf3−/−Irf7−/− + FLAG-IRF3), or IRF7 (Irf3−/−Irf7−/− + FLAG-IRF7) through lentivirus-mediated gene transfer. Cells were left untreated or treated with IFNα (20 units/ml) for 12 h followed by qPCR analysis. C, MEFs were transfected with control, individual, or combined siRNAs targeting IRF3 or IRF7. Twenty-four hours later, cells were left untreated or treated with IFNα (20 units/ml) for 12 h followed by qPCR analysis. Data shown are representatives of two (A) or at least three independent experiments (B and C). *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars represent S.D. Rel., relative.
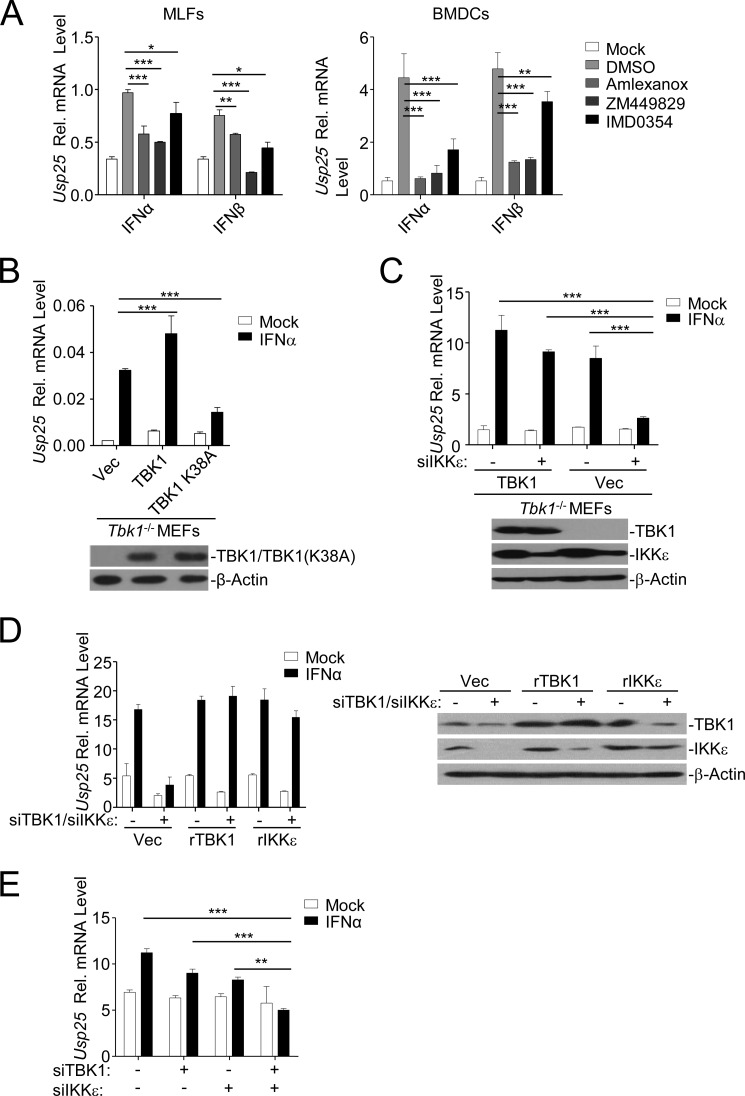
It has been recognized that TBK1- or IKKϵ-mediated phosphorylation of IRF7 is critical for its transcriptional activity. Consistent with this notion, we found that inhibition of TBK1 or IKKϵ impaired type I IFN-triggered up-regulation of Usp25 (Fig. 4A). To further characterize the role of TBK1 and IKKϵ in type I IFN-triggered induction of Usp25, we reconstituted empty vector (Vec), TBK1, or TBK1(K38A) into Tbk1−/− MEFs and examined IFNα-triggered up-regulation of Usp25. As shown in Fig. 4B, reconstitution of TBK1 or TBK1(K38A) substantially promoted or inhibited IFNα-induced up-regulation of Usp25 compared with reconstitution of the empty vector, respectively, indicating that TBK1(K38A) functions as a dominant negative mutant regulating type I IFN-induced up-regulation of Usp25. In addition, knockdown of IKKϵ in Tbk1−/− MEFs reconstituted with the empty vector but not in those reconstituted with TBK1 significantly impaired IFNα-induced expression of Usp25 (Fig. 4C). We further transfected siRNA-resistant TBK1 (rTBK1) or IKKϵ (rIKKϵ) into wild-type MEFs followed by simultaneous knockdown of endogenous TBK1 and IKKϵ. Interestingly, IFNα-induced up-regulation of Usp25 was not affected in MEFs transfected with either rTBK1 or rIKKϵ (Fig. 4D). Furthermore, simultaneous knockdown of TBK1 and IKKϵ by siRNA significantly inhibited IFNα-induced expression of Usp25 in BMDCs (Fig. 4E). Taken together, these data suggest that TBK1 and IKKϵ function redundantly to regulate type I IFN-induced expression of Usp25.
FIGURE 4.
TBK1 and IKKϵ function redundantly for type I IFN-triggered induction of Usp25. A, MLFs (left graph) or BMDCs (right graph) were left untreated or treated with IFNα (20 units/ml) or IFNβ (100 ng/ml) in the presence or absence of various kinase inhibitors for 12 h followed by qPCR analysis. B, Tbk1−/− MEFs were reconstituted with Vec, TBK1, or TBK1(K38A) through lentivirus-mediated gene transfer. The reconstituted TBK1 or TBK1(K38A) in the cells was examined by immunoblotting analysis (lower panels). Cells were left untreated or treated with IFNα (20 units/ml) for 12 h followed by qPCR analysis (upper graph). C, Tbk1−/− MEFs were reconstituted with Vec or TBK1 through lentivirus-mediated gene transfer. Cells were further transfected with control or siRNA targeting IKKϵ followed by immunoblotting analysis (lower panels) or qPCR analysis after IFNα treatment (20 units/ml) (upper graph). D, wild-type MEFs were stably transfected with Vec, rTBK1, or rIKKϵ followed by transfection of control or siRNAs targeting endogenous TBK1 and IKKϵ. Cells were subjected to immunoblotting analysis (right panels) or qPCR analysis after IFNα treatment (20 units/ml) (left graph). E, wild-type BMDCs were transfected control, siTBK1, or siIKKϵ. Twenty-four hours later, cells were stimulated with IFNα (20 units/ml) for 8 h followed by qPCR analysis. Data shown are representatives of at least three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars represent S.D. Rel., relative.
IRF7 Binds to the USP25 Promoter
To further confirm that IRF7 drives transcription of Usp25, we cloned the upstream 5000 bp starting from the transcription start site of Usp25 into the pGL3-Basic luciferase vector (USP25 promoter), made constructs with various mutations in the IRF or NF-κB binding sites, and performed luciferase reporter assays (Fig. 5A). Interestingly, IRF7 or SeV potently activated the luciferase activity of USP25 promoter, which was substantially impaired by mutation of either the proximal or distal IRF binding site (IB) (USP25 promoter ΔIB1 or USP25 promoter ΔIB2) of USP25 promoter and abolished by simultaneous mutation of both IRF binding sites (USP25 promoter ΔIB1+2), indicating that the two IRF binding sites cooperatively mediate transcription of Usp25 (Fig. 5B). In addition, IRF7 but not IRF3 was sufficient to activate the IB1-, IB2-, or IB1+2-driven reporters in a dose-dependent manner (Fig. 5C). In contrast, IRF3 (at a high dosage) but not p65, p50, or p52-RelB complex activated the USP25 promoter ∼2–4-fold, and mutation of all five NF-κB binding sites (Δ5κB) did not affect SeV-, IRF7-, or IRF3-mediated activation of USP25 promoter (Fig. 5B), indicating that the NF-κB binding sites are dispensable for USP25 transcription after viral infection and that sequences other than the κB sites in USP25 promoter may facilitate IRF3-mediated activation of USP25 promoter. Results from ChIP analysis demonstrated that IRF7 directly bound to the two IRF binding sites (Usp25 IB1 and Usp25 IB2) but not a nonspecific site (Usp25 NS) of USP25 promoter (Fig. 5D and Table 1). Together, these data suggest that IRF7 directly binds to the distal and proximal IRF binding sites in the USP25 promoter and drives transcriptional activation of Usp25 gene.
FIGURE 5.
IRF7 binds to USP25 promoter. A, a schematic model of USP25 promoter and its mutations. B, HEK293 cells were transfected with the indicated luciferase (Luc.) reporter (100 ng) together with Vec, IRF7 (0.1 μg), IRF3 (0.5 μg) (left graph), p65, p50, or p52-RelB (0.2 μg) (middle graph). Twenty hours later, luciferase reporter assays were performed. HEK293 cells were transfected with the indicated luciferase reporter (100 ng). Twenty hours later, cells were infected with SeV for 8 h followed by luciferase reporter assays (right graph). C, HEK293 cells were transfected with the indicated luciferase reporter (100 ng) together with Vec, IRF7 (0.02–0.1 μg), or IRF3 (0.1–0.5 μg) (left graph). Twenty hours later, luciferase reporter assays were performed. Immunoblotting analysis was performed to examine the expression of transfected plasmids (right panels). D, Irf3−/−Irf7−/− MEFs were reconstituted with the empty vector (Irf3−/−Irf7−/− + Vector), IRF3 (Irf3−/−Irf7−/− + FLAG-IRF3), or IRF7 (Irf3−/−Irf7−/− + FLAG-IRF7) through lentivirus-mediated gene transfer. Cells were left untreated or infected with SeV for 12 h followed by ChIP analysis. TSS, transcription start site. Data shown are representatives of four (B) or three independent experiments (C and D). *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars represent S.D. Rel., relative.
TABLE 1.
qPCR primers of USP25 promoter and Ifnb promoter for ChIP analysis
NS, nonspecific site.
| Primer name | Primer sequence |
|---|---|
| USP25 IB1 (−1384 to −1319) | AAGTTACAGCGCTGAGGTCT |
| AGCACGTGTCTGAGAATGGA | |
| USP25 IB2 (−4706 to −4615) | TTGGAGAGATCGAGAGGCTG |
| TCTGCCACCTTTGAGACTGT | |
| USP25 NS (−1018 to −907) | CTGCTTTTCTTGCCGTGGAT |
| GAGTAAGACCGAGACCCAGG | |
| Ifnb promoter | ATTCCTCTGAGGCAGAAAGGACC |
| GCAAGATGAGGCAAAGGCTGTCA |
Discussion
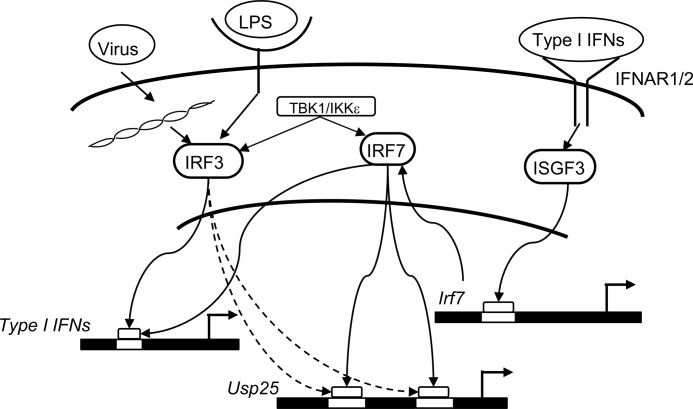
We have previously shown that LPS and viral infection strongly induce up-regulation of USP25. In this study, we further confirmed that LPS and viral infection activated transcription of Usp25 through type I IFN-triggered signaling. In addition, type I IFN-triggered signaling induced expression and protein synthesis of IRF7, which was activated by TBK1 and IKKϵ and bound to the USP25 promoter to activate transcription of Usp25 gene (Fig. 6).
FIGURE 6.
A model on transcriptional regulation of Usp25. LPS or viral infection induces expression of type I IFNs. Type I IFNs bind to IRNAR1 and IRNAR2 to trigger activation of ISGF3, which binds to the promoter of IRF7 and activates expression of IRF7. The de novo synthesized IRF7 is phosphorylated by TBK1 or IKKϵ and binds to the two IRF binding sites in USP25 promoter to drive transcription of IRF7.
Sequence analysis of USP25 promoter indicated that multiple NF-κB binding sites exist in the USP25 promoter. In our study, p65 deficiency did not affect virus-induced expression of Usp25. Mutation of the five NF-κB binding sites did not affect basal, SeV-triggered, or IRF7/3-medaited activation of USP25 promoter. In addition, overexpression of p65, p50, or p52-RelB complex did not activate USP25 promoter, indicating that the NF-κB sites on USP25 promoter are dispensable for virus- or LPS-induced up-regulation of Usp25. However, whether the NF-κB sites are involved in the induction of Usp25 by other stimuli is unknown. The USP25 promoter also contains two potential ISGF3 binding sites. We found that cycloheximide treatment impaired type I IFN-triggered induction of Usp25, indicating that ISGF3 does not directly regulate transcription of Usp25 but instead activates de novo synthesis of other transcription factor(s) to mediate transcription of Usp25.
IRF3 and IRF7 are two structurally related transcription factors that bind to the conserved IRF binding site (5′-GAAANNGAAA-3′) on the promoters and are essential for induction of hundreds of genes involved in innate immunity and inflammation. IRF3 exhibits more restricted DNA binding site specificity compared with IRF7. Mutation of a single nucleotide in either of the two GAAA core sequences impairs IRF3 binding and transcription activity, whereas the G and the third A in the GAAA core sequence are variable for IRF7 binding activity (36). According to this standard, IB1 “gaaacataaa” and IB2 “gaatgagaag” in USP25 promoter are preferentially recognized and bound by IRF7 but not IRF3. Consistent with this notion, we observed that (i) IRF7 but not IRF3 was sufficient to activate the IB1- or IB2-driven reporters and required for virus-triggered type I IFN-mediated up-regulation of Usp25, (ii) IRF7 activated USP25 promoter more potently than did IRF3 in luciferase reporter assays, and (iii) IRF7 bound to the USP25 promoter more potently than did IRF3. However, it should be noted that IRF3 activated USP25 promoter (∼2–4-fold) when transfected at a high dosage and partially rescued USP25 induction in Irf3−/−Irf7−/− MEFs after viral infection. In addition, we observed that IFNAR1 deficiency in MEFs partially inhibited virus-triggered up-regulation of Usp25, whereas IRFAR1 deficiency in BMDCs completely abolished up-regulation of Usp25 after viral infection, indicating that virus-induced expression of Usp25 might be differentially regulated by IRF3 and IRF7 in distinct types of cells. Taken together, it is likely that IRF3 is responsible for minimal expression of Usp25 in MEFs after viral infection, whereas the de novo synthesized IRF7 induced by type I IFNs is a master transcription factor for USP25 expression in MEFs and BMDCs.
Unlike IRF3, which is constitutively expressed and resides in the cytosol, IRF7 is expressed at a low level and strongly induced by type I IFN-triggered signaling. Both IRF3 and IRF7 undergo TBK1- or IKKϵ-mediated phosphorylation, dimerization, and nuclear translocation after LPS treatment or viral infection. We found that treatment with TBK1 and IKKϵ inhibitor severely abolished type I IFN-triggered induction of Usp25. Furthermore, reconstitution of TBK1(K38A) into Tbk1−/− MEFs inhibited IFNα-induced up-regulation of Usp25, and knockdown of IKKϵ in Tbk1−/− + Vec MEFs but not in Tbk1−/− + TBK1 MEFs substantially impaired IFNα-induced up-regulation of Usp25, indicating that TBK1 and IKKϵ function redundantly for USP25 induction downstream of type I IFN stimulation. Further investigations are required to fully address how TBK1 and IKKϵ are involved in type I IFN-triggered signaling. Nonetheless, our data have clearly demonstrated that the type I IFN-IRF7 axis critically regulates viral infection- or LPS-induced transcription of Usp25 and contribute to our understanding of positive feedback regulation of cellular antiviral responses.
Author Contributions
Y. R. and B. Z. conceived and coordinated the study and wrote the paper. Y. R., Y. Z., and D. L. designed, performed, and analyzed experiments. X. X., Q. Z., J. Y., and H. B. S. provided reagents. H. B. S. and B. Z. analyzed data. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Drs. P. Feng (University of Southern California); T. Maniatis (Columbia University); W.-C. Yeh (University of Toronto); J. Jin (Zhejiang University); and D.-Y. Guo, Z.-Y. Song, and M. Wu (Wuhan University) for reagents and members of the Zhong laboratory and the core facilities of College of Life Sciences for technical help.
This work was supported by Ministry of Science and Technology of China Grants 2014CB542601 and 2012CB910201, National Natural Science Foundation of China Grants 31371427 and 31271209, Ministry of Education of China Grant 201427, State Key Laboratory of Veterinary Etiological Biology Grant SKLVEB2015KFKT001, Natural Science Foundation of Hubei Province Grant 2015CFA095, and the Large-scale Instrument and Equipment Sharing Foundation of Wuhan University. The authors declare that they have no conflicts of interest with the contents of this article.
- IRF
- interferon regulatory factor
- ISGF3
- IFN-stimulated gene factor 3
- USP25
- ubiquitin-specific protease 25
- IFNAR
- interferon α receptor
- ISG
- interferon-stimulated gene
- MEF
- mouse embryonic fibroblast
- BMDC
- bone marrow-derived dendritic cell
- TYK2
- tyrosine kinase 2
- IKK
- inhibitor of κB kinase
- SeV
- Sendai virus
- HSV-1
- herpes simplex virus 1
- qPCR
- quantitative PCR
- Vec
- empty vector
- rTBK1
- siRNA-resistant TBK1
- rIKKϵ
- siRNA-resistant IKKϵ
- MLF
- mouse lung fibroblast
- IB
- IRF binding site.
References
- 1. Akira S., Uematsu S., and Takeuchi O. (2006) Pathogen recognition and innate immunity. Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 2. Takeuchi O., and Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 3. Wu J., and Chen Z. J. (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 [DOI] [PubMed] [Google Scholar]
- 4. Baeuerle P. A., and Henkel T. (1994) Function and activation of NF-κB in the immune system. Annu. Rev. Immunol. 12, 141–179 [DOI] [PubMed] [Google Scholar]
- 5. Sharma S., tenOever B. R., Grandvaux N., Zhou G. P., Lin R., and Hiscott J. (2003) Triggering the interferon antiviral response through an IKK-related pathway. Science 300, 1148–1151 [DOI] [PubMed] [Google Scholar]
- 6. Perry A. K., Chow E. K., Goodnough J. B., Yeh W. C., and Cheng G. (2004) Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J. Exp. Med. 199, 1651–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hemmi H., Takeuchi O., Sato S., Yamamoto M., Kaisho T., Sanjo H., Kawai T., Hoshino K., Takeda K., and Akira S. (2004) The roles of two IκB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199, 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., and Maniatis T. (2003) IKKϵ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 9. McWhirter S. M., Fitzgerald K. A., Rosains J., Rowe D. C., Golenbock D. T., and Maniatis T. (2004) IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 101, 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takeda K., and Akira S. (2015) Toll-like receptors. Curr. Protoc. Immunol. 109, 14.12.1–14.12.10 [DOI] [PubMed] [Google Scholar]
- 11. Stifter S. A., and Feng C. G. (2015) Interfering with immunity: detrimental role of type I IFNs during infection. J. Immunol. 194, 2455–2465 [DOI] [PubMed] [Google Scholar]
- 12. Crouse J., Kalinke U., and Oxenius A. (2015) Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 15, 231–242 [DOI] [PubMed] [Google Scholar]
- 13. Gajewski T. F., and Corrales L. (2015) New perspectives on type I IFNs in cancer. Cytokine Growth Factor Rev. 26, 175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoffmann H. H., Schneider W. M., and Rice C. M. (2015) Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 36, 124–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schreiber G., and Piehler J. (2015) The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 36, 139–149 [DOI] [PubMed] [Google Scholar]
- 16. Stark G. R., and Darnell J. E. Jr. (2012) The JAK-STAT pathway at twenty. Immunity 36, 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Platanias L. C. (2005) Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 18. Löchte S., Waichman S., Beutel O., You C., and Piehler J. (2014) Live cell micropatterning reveals the dynamics of signaling complexes at the plasma membrane. J. Cell Biol. 207, 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nguyen V. P., Saleh A. Z., Arch A. E., Yan H., Piazza F., Kim J., and Krolewski J. J. (2002) Stat2 binding to the interferon-α receptor 2 subunit is not required for interferon-α signaling. J. Biol. Chem. 277, 9713–9721 [DOI] [PubMed] [Google Scholar]
- 20. Yang J., and Stark G. R. (2008) Roles of unphosphorylated STATs in signaling. Cell Res. 18, 443–451 [DOI] [PubMed] [Google Scholar]
- 21. Ivashkiv L. B., and Donlin L. T. (2014) Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levy D. E., and Marié I. J. (2012) STATus report on tetramers. Immunity 36, 553–555 [DOI] [PubMed] [Google Scholar]
- 23. Tenoever B. R., Ng S. L., Chua M. A., McWhirter S. M., García-Sastre A., and Maniatis T. (2007) Multiple functions of the IKK-related kinase IKKϵ in interferon-mediated antiviral immunity. Science 315, 1274–1278 [DOI] [PubMed] [Google Scholar]
- 24. Sato M., Hata N., Asagiri M., Nakaya T., Taniguchi T., and Tanaka N. (1998) Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441, 106–110 [DOI] [PubMed] [Google Scholar]
- 25. Taniguchi T., Ogasawara K., Takaoka A., and Tanaka N. (2001) IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19, 623–655 [DOI] [PubMed] [Google Scholar]
- 26. Honda K., Takaoka A., and Taniguchi T. (2006) Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 [DOI] [PubMed] [Google Scholar]
- 27. Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., and Taniguchi T. (2000) Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13, 539–548 [DOI] [PubMed] [Google Scholar]
- 28. Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., and Taniguchi T. (2005) IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777 [DOI] [PubMed] [Google Scholar]
- 29. Prakash A., and Levy D. E. (2006) Regulation of IRF7 through cell type-specific protein stability. Biochem. Biophys. Res. Commun. 342, 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marié I., Durbin J. E., and Levy D. E. (1998) Differential viral induction of distinct interferon-α genes by positive feedback through interferon regulatory factor-7. EMBO J. 17, 6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin D., Zhang M., Zhang M. X., Ren Y., Jin J., Zhao Q., Pan Z., Wu M., Shu H. B., Dong C., and Zhong B. (2015) Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6. Proc. Natl. Acad. Sci. U.S.A. 112, 11324–11329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhong B., Liu X., Wang X., Liu X., Li H., Darnay B. G., Lin X., Sun S. C., and Dong C. (2013) Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitination of the adaptor protein TRAF3. Sci. Signal. 6, ra35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhong B., Yang Y., Li S., Wang Y. Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., and Shu H. B. (2008) The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 [DOI] [PubMed] [Google Scholar]
- 34. Bonnard M., Mirtsos C., Suzuki S., Graham K., Huang J., Ng M., Itié A., Wakeham A., Shahinian A., Henzel W. J., Elia A. J., Shillinglaw W., Mak T. W., Cao Z., and Yeh W. C. (2000) Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-κB-dependent gene transcription. EMBO J. 19, 4976–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bosch-Comas A., Lindsten K., Gonzàlez-Duarte R., Masucci M. G., and Marfany G. (2006) The ubiquitin-specific protease USP25 interacts with three sarcomeric proteins. Cell. Mol. Life Sci. 63, 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin R., Génin P., Mamane Y., and Hiscott J. (2000) Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of α/β interferon genes by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 20, 6342–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]