Abstract
Here we report the crystal structure of M. tuberculosis AldR (Rv2779c) showing that the N-terminal DNA-binding domains are swapped, forming a dimer, and four dimers are assembled into an octamer through crystal symmetry. The C-terminal domain is involved in oligomeric interactions that stabilize the oligomer, and it contains the effector-binding sites. The latter sites are 30–60% larger compared with homologs like MtbFFRP (Rv3291c) and can consequently accommodate larger molecules. MtbAldR binds to the region upstream to the ald gene that is highly up-regulated in nutrient-starved tuberculosis models and codes for l-alanine dehydrogenase (MtbAld; Rv2780). Further, the MtbAldR-DNA complex is inhibited upon binding of Ala, Tyr, Trp and Asp to the protein. Studies involving a ligand-binding site G131T mutant show that the mutant forms a DNA complex that cannot be inhibited by adding the amino acids. Comparative studies suggest that binding of the amino acids changes the relative spatial disposition of the DNA-binding domains and thereby disrupt the protein-DNA complex. Finally, we identified small molecules, including a tetrahydroquinoline carbonitrile derivative (S010-0261), that inhibit the MtbAldR-DNA complex. The latter molecules represent the very first inhibitors of a feast/famine regulatory protein from any source and set the stage for exploring MtbAldR as a potential anti-tuberculosis target.
Keywords: crystal structure, DNA binding protein, DNA-protein interaction, enzyme inhibitor, Mycobacterium tuberculosis, AldR, feast/famine regulatory proteins, Lrp/Asnc, oligomeric association, regulator
Introduction
Feast/famine regulatory proteins (FFRPs),5 also known as Lrp/AsnC family proteins, bind to a variety of effectors like amino acids that modulate the respective regulatory functions. They are involved in the formation of globular nucleoprotein structures, chromosome structure organization, and other regulatory functions (1). Directly or indirectly, they globally regulate a variety of metabolic processes in response to amino acids and nitrogen bases present in the environment. The general understanding is that FFRPs adopt a variety of quaternary structures upon binding/release of effectors. This presumably allows for binding to target promoter regions or for disrupting the nucleoprotein structures formed by them (2). Fine tuning/selection of the target promoter-FFRP interactions is also generally thought to occur due to binding of the effector molecules and in some cases can elicit subtle structural changes as opposed to changes to the oligomeric association itself (3, 4). Escherichia coli Lrp, a better studied member of the family, is known to be important for changes that occur from a “feasting” to a “famine” state, and it controls ∼10% of gene expression (5, 6). The latter protein binds to a variety of amino acids like Leu, Ala, Pro, and Val, and that in turn elicits either positive or negative regulation of the target genes (7).
Crystal structures of several FFRPs/Lrp/AsnC-type proteins have been reported from bacterial and archaeal sources. These include structures of the E. coli Lrp, E. coli AsnC, Mycobacterium tuberculosis FFRP (also called LrpA), Lrp proteins from Pyrococcus furiosus, and Pyrococcus sp. OT3 FL11. Complexes of some of them with several amino acids, DNA, etc. have also been reported (8–10). The studies reveal that the basic functional unit of FFRPs is a dimer where each chain folds into two domains. The N-terminal domain normally faces outside and contains the DNA-binding helix-turn-helix motif and a C-terminal domain that is involved in effector binding and in oligomerization. Both of the domains are connected by a rather long linker region. Some FFRPs like the M. tuberculosis FFRP and E. coli Lrp have been shown to adopt the rare “open” quaternary structure that seems to be an operating principle in these proteins. This presumably allows them to bind to non-symmetrical target sites (10–12). In general, instances of deviations from oligomeric symmetry in proteins are rare, and wherever they are observed, it is attributed to strong functional reasons (13). Helical cylindrical arrangements are the other kind of assembly observed in P. furiosus sp. OT3 Lrp. FFRPs may have one or more types of effector binding sites (e.g. E. coli AsnC has one binding site, whereas those like M. tuberculosis FFRP have at least two types of binding sites, each with different hypothesized roles) (8). The type I binding site is a common site in the FFRP family that occurs at the interdimer interface at the C-terminal oligomerization domain, whereas the type II site was identified in M. tuberculosis FFRP at the intradimer interface and consists of residues from the C-terminal domain also (8). However, both sites are located distal to the DNA-binding helix-turn-helix motif. It has been suggested that binding of ligands to the type I site can affect the formation of higher order oligomers, whereas binding of ligands to the type II site can lead to relative changes to the spatial disposition of the DNA-binding motif, and this in turn can lead to modulation of binding affinity to target DNA (10, 12).
M. tuberculosis harbors five genes that have been annotated as either Lrp- or AsnC-like transcription factors in the databases. These have been drawing attention as potential drug targets due to their important roles in persistence (14). The present work involves structural and functional studies of M. tuberculosis AldR (Rv2779c). In Mycobacterium smegmatis, it has been shown that the ald gene that encodes for alanine dehydrogenase is regulated by its AldR (15). ald is highly up-regulated in tuberculosis nutrient-starved and stress models designed to mimic persistence. In fact one analysis suggested that lysine-ϵ-aminotransferase (16) and alanine dehydrogenase (17) are in the top 3 among over 200 targets considered in the analysis against tuberculosis persistence (18). In M. smegmatis, AldR reportedly senses the level of alanine in cells and acts as an activator or as a repressor of the ald gene, depending upon the alanine levels (15). Ald itself has been shown to function as a dual function enzyme and exhibits both alanine dehydrogenase and glycine dehydrogenase activities (19). In the present study, we report the crystal structure of M. tuberculosis AldR. We also report studies involving its interactions with a variety of amino acids. We have demonstrated through EMSA that MtbAldR binds to the upstream region of the M. tuberculosis ald gene. Amino acids not only bind to MtbAldR but also elicit diverse effects on its interactions with the promoter region. Using EMSAs, supported by in silico docking studies, we have identified small molecule compounds that inhibit the formation of the nucleoprotein complex. To the best of our knowledge, these compounds represent the first inhibitors that act by disrupting FFRP-DNA interactions.
Experimental Procedures
Cloning, Overexpression, and Purification
MtbAldR was cloned, expressed, and purified as described previously (20). In brief, the 540-bp nucleotide sequence encoding MtbAldR was PCR-amplified, with M. tuberculosis H37Rv genomic DNA as a template and cloned into pET21d. The resultant plasmid was transformed into the E. coli C41 (DE3) strain. The C-terminal hexahistidine-tagged protein was purified by standard procedures. The protein was finally concentrated to 7 mg ml−1 in 50 mm HEPES, pH 7.0, 250 mm NaCl, 5 mm EDTA, and 10% glycerol after size exclusion chromatography using a Superdex-200 HR 10/300 column (GE Healthcare). The MtbAldR mutant (G131T) was generated using the following primer pair: F131, 5′-CGCAAACGACAGGTGATGGATGTGTACTTTCTTGCAACCGCT-3′; R131, 5′-GTAGCGAGCGGAGACGTGCAAGATGAAATAGTAGGCGGTGGC-3′. The integrity of the clones was verified by sequencing. G131T was also purified using a similar protocol as that of the wild-type protein. Protein concentration was determined as per the Bradford method (21). Proteins were concentrated using Amicon ultra-10 concentrator (Millipore).
Structure Determination, Refinement, and Analysis
Crystals of the apoprotein diffracted to 2.95 Å. Crystallization and data collection statistics were reported in a preliminary report (20). The molecular replacement method was used to solve the crystal structure of MtbAldR, using the coordinates of Mtb FFRP (Rv3291c; Protein Data Bank code 2IVM) (8) as the search model after removing water molecules and heteroatoms. An unambiguous solution was found by PHASER (22) implemented in the CCP4 program suite (23). Refinements were carried out by Phenix.refine (24) after setting aside 5% of the total reflections for the calculation of Rfree (25). The protein model was iteratively built using COOT (26). The final model was refined to the Rfactor and Rfree of 26 and 29% (Table 1). The quality of the model was checked using Molprobity (27). The geometric parameters as well as the Ramachandran plot (28) fall into acceptable ranges. Analysis of intersubunit contacts and accessible surface area was carried out using the Protein Interaction Calculator (PIC) (29) and other tools implemented in the CCP4 program suite (23), whereas structural superposition was carried out using SUPERPOSE (23). Figures were generated using PyMOL (DeLano Scientific) and UCSF Chimera (30).
TABLE 1.
Data collection and refinement statistics of MtbAldR
| Parameters | Values |
|---|---|
| Data collection | |
| Space group | P21212 |
| Unit cell (Å) | 99.73, 146.09, 49.96 |
| Resolution (Å) | 27.19–2.95 (3.02–2.95)a |
| Total no. of reflections | 17,732 (847) |
| Unique reflections | 3094 (170) |
| Completeness (%) | 99.5 (93.1) |
| Redundancy | 5.7 |
| Rmerge (%) | 6.5 (43.4) |
| I/σI | 25.6 (3.4) |
| Refinement statistics | |
| No. of reflections | 15,581 |
| Rwork/Rfree (%) | 26.3/29.2 |
| No. of atoms | |
| Protein | 4233 |
| Water | 33 |
| B-Factor | |
| Protein | 44.5 |
| Water | 25.14 |
| Root mean square deviation | |
| Bond lengths (Å) | 0.005 |
| Bond angles (degrees) | 0.953 |
a Values in parentheses correspond to the highest resolution shell.
b Rmerge = ΣhklΣi Ii(hkl) − (I(hkl))|/ΣhklΣiIi(hkl), where Ii(hkl) is the intensity of the ith observation of reflection hkl and (I(hkl)) is the average intensity of the i observations.
EMSAs
EMSAs were performed to probe the DNA binding property of the purified MtbAldR to the ald promoter region. A 400-bp upstream region of the ald gene was PCR-amplified using the 6-carboxyfluorescein (FAM)-tagged forward primer 5′-CGAGCTGACGACTGCAAATTGACCGAGATAAC-3′ and reverse primer 5′-GATTACCCCTCTCGATACTGTACGCTTCAGAATGAA-3′. This 400-bp upstream region of the ald gene was further subdivided into four regions of 100 bp each by PCR-amplifying each site with FAM-tagged forward primer and a reverse primer, respectively. The following primers were used (Integrated DNA Technologies): for site S1, 5′-CGAGCTGACTGCAAATTGACCGAG-3′ (forward) and 5′-GGGGTTCTACATCGTCCTGGTCG-3′ (reverse); for site S2, 5′-CGGATCACGCAAAGTCAACCAA-3′ (forward) and 5′-ATGCCAGAATGCCCAACAACGCG-3′ (reverse); for site S3, 5′-CGCCATGCAGCAAGCTCAGGATT-3′ (forward) and 5′-TTCAACGGTACACAAGCTCGACGG-3′ (reverse); and for site S4, 5′-TTATCACGCATCTGGCCTCGAAAA-3′ (forward) and 5′-GATTATCCCTCTCGCTACTGTACGCT-3′ (reverse). DNA protein binding was initiated by mixing increasing concentrations of purified MtbAldR with 30 pmol of FAM-labeled DNA in the binding buffer: 20 mm HEPES (pH 7.5), 500 mm NaCl, 4 mm DTT, 2% glycerol, 2 mm MgCl2. The whole reaction mixture was then incubated for 40 min at 37 °C. Following incubation, the DNA-protein mixture was run on a 8% non-denaturing polyacrylamide gel, and the image was captured by an ImageQuant LAS 4000 system (GE Healthcare). To check the effect of different cofactors on the binding of MtbAldR to the ald promoter region, 650 nm MtbAldR was incubated with 10 mm respective l-amino acids in the same reaction condition. For the inhibition of DNA-protein binding, MtbAldR was incubated with the increasing concentrations of different compounds before the addition of DNA probe. For the determination of the equilibrium binding constant (Ka), densitometry of the EMSA gels was performed by measuring the pixel integrated density of the bands by IQTl (GE Healthcare). The measured pixel intensity was plotted against the concentration of the protein. The equilibrium constant was determined using the non-linear regression analysis implemented in the GraphPad Prism software.
Binding Studies Using Competitive 1-Anilino-8-naphthalenesulfonate (ANS) Displacement Assays
ANS, an extrinsic flourophore, binds to pockets and hydrophobic patches of proteins. It has been used in several studies to monitor ligand binding that exploits the displacement of the flourophore upon binding of the ligand (31). A PerkinElmer Life Sciences LS 50B fluorescence spectrophotometer was used in the experiments carried out at 25 °C. The excitation wavelength used was 380 nm, and the emission was monitored between 400 and 600 nm. All samples were incubated for 2 h under specified conditions before recording the spectra. The purified protein (7 μm) and ANS (5 μm) mixture was titrated against 0–25 mm concentrations of the respective amino acids. All of the spectra were recorded in the correct spectrum mode with excitation and emission band passes of 8 and 6 nm. For inhibitor binding studies, the protein-ANS complex was titrated against 0–20 μm concentrations of the respective compounds, and the spectra were recorded in a similar manner as above.
Circular Dichroism Spectroscopy
Far-UV CD spectra were used to quantify the secondary structure content of MtbAldR in the presence of different ligands. The spectra were acquired on a Jasco J810 polarimeter. The spectra were measured from 5 μm protein samples in 10 mm Tris-HCl (pH 7.5) and 10 mm NaCl and titrated with increasing concentrations of ligands. Typically, spectra were recorded from 200 to 260 nm at a scan speed of 10 nm/min, with each spectrum representing an average of three accumulations. The residue ellipticity, [θ]MRW (mean residual weight) was calculated from the measured θ (in degrees) as follows,
where θ is the measured ellipticity in degrees, c is the protein concentration in mg/ml, l is the path length in cm, and Mr and MRW are the protein molecular weight and mean residue weight, respectively. NA is the number of amino acids per protein. [θ]MRW has units of degrees × cm2 × dmol−1. The values obtained were corrected by subtracting the baseline recorded for the buffer.
Size Exclusion Chromatography
Changes to the quaternary association of MtbAldR in the presence of respective amino acids were probed using size exclusion chromatography. A Superdex S-200 HR10/300 (GE Healthcare) column calibrated with low and high molecular weight range markers was mounted on an AKTA-FPLC system (GE Healthcare) for the experiments. The partition coefficient (Kav) was plotted as a function of log Mr of the standard protein according to the equation,
where Ve is the elution volume of the protein, V0 is the void volume of the column, Vt is the total volume of the column, and Mr is the molecular weight of the particular protein. The column was equilibrated with buffer containing 20 mm HEPES (pH 7.0), 500 mm NaCl, 5 mm EDTA, 4 mm DTT and supplemented with a 5 mm concentration of the respective amino acids. 0.5 mg/ml of protein was pre-equilibrated with the required amino acid concentration for 1 h before injection into the column. Also increasing concentrations of MtbAldR alone and in the presence of the corresponding concentration of alanine were subjected to size exclusion chromatography experiments in order to probe for effects of concentration-dependent oligomerization and the effect of alanine on the oligomeric status of MtbAldR.
Dynamic Light Scattering (DLS) Studies
Protein fractions after elution from the size exclusion chromatography column were subjected to DLS analysis. A Zetasizer instrument (Malvern) and the company-provided Malvern DLS version 5.03 software were used in the analysis. The instrument was calibrated using a standard set of protein markers. The hydrodynamic diameter of the protein particles was calculated as per the recommendations of the manufacturer. Protein samples were filtered using a 0.22-μm filter (Millipore) prior to the DLS studies.
Docking and Modeling Studies
AUTODOCK version 3.0.5 (32) was used to probe binding modes of compounds to MtbAldR. The crystal structure of MtbAldR was used as the model to generate the target binding site. The corresponding type I and type II binding sites identified by us earlier in MtbFFRP (8) were used as the target. Three-dimensional structures of the amino acids and the tested compound were built and optimized using the BUILDER module in Insight II (Accelrys) and Sybyl version 8.0 (Tripos Associates, Inc., St. Louis, MO). Crystallographic water atoms and heteroatoms were removed from the docking template. Polar hydrogen was added, and Kollman charges were assigned to all atoms. Ligands were prepared for calculations by adding Gasteiger charges. For calculations involving the type I binding site, a 65 × 69 × 69-Å affinity grid was used, whereas for the type II binding site, a 46 × 52 × 46-Å affinity grid with 0.375-Å spacing was used. The remaining parameters were set to default values. A maximum of 20 poses were sorted based on the scoring function and fitness score implemented in the program. The most stable conformation of the protein-ligand complex was then selected for further analysis. Modeling of the open and closed structures of MtbFFRP and MtbAldR onto the promoter binding region of the ald gene was carried out in an analogous manner as reported by us earlier for MtbFFRP (8).
Results
Crystal Structure of MtbAldR
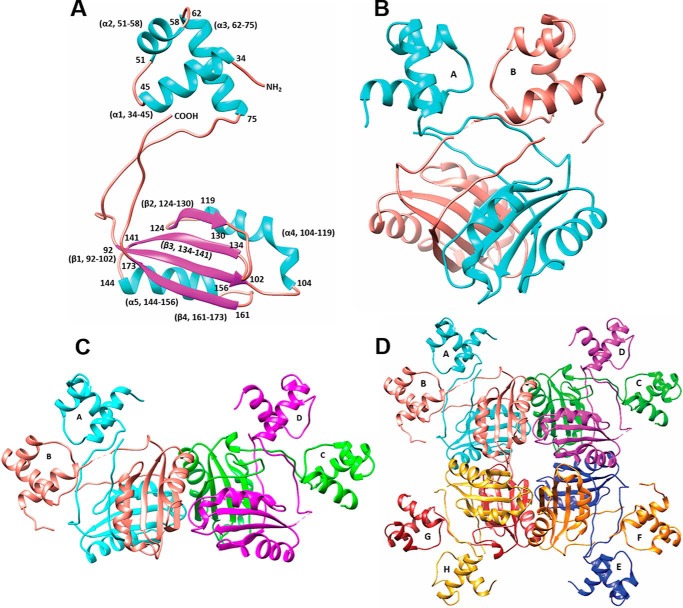
The protein was purified as reported earlier (20). The size of the protein was consistent with octameric association in size exclusion chromatography experiments (20). Molecular replacement calculations were carried out to solve the structure of MtbAldR. There are four independent subunits in the asymmetric unit, and each subunit comprises two domains. MtbAldR belongs to the α/β family of proteins. Each polypeptide chain folds into an N-terminal DNA-binding domain, which initially forms a helix (residues 34–45) and subsequently folds into a helix-turn helix DNA-binding motif (residues 51–75) typical for this protein family (Fig. 1A). A rather long flexible linker (residues 76–93) joins the N-terminal domain to the C-terminal effector-binding/oligomerization domain. The latter domain contains four β-strands and two α-helices arranged in a βαββαβ topology also called the RAM (regulator of amino acid metabolism) domain (residues 94–173). The C terminus ends at residues 174–178.
FIGURE 1.
Crystal structure of MtbAldR. A, schematic representation of the MtbAldR monomer; helices are colored in cyan and sheets in magenta. The helix-turn-helix motif is formed by residues Asn51–Ala75. The C-terminal domain consists of residues Leu94–Leu173. B, MtbAldR dimer. The two chains have been colored cyan and salmon, respectively. C, schematic representation of the two dimers in the asymmetric unit. D, schematic representation of the MtbAldR octamer. Each subunit is labeled A–H, respectively, and colored as follows: A (cyan), B (salmon), C (green), D (magenta), E (blue), F (orange), G (red), and H (yellow).
The flexibility of the linker region allows for variation in positioning of the relatively rigid N- and C-terminal domains with respect to each other. In fact, the dimerization in the protein proceeds with the N-terminal domains being swapped (Fig. 1B). The domain swapping places the C-terminal domain of each monomer below the N-terminal domain of the other monomer of the dimer. Each asymmetric unit of MtbAldR contains two dimers, and four such dimers associate through crystal symmetry to form the octamer (Fig. 1, C and D). The respective DNA-binding domains of each subunit of the octamer face the outside of the packed oligomeric structure. The C-terminal domain is involved in oligomeric interactions and is located inside. There are 47 polar interactions and 18 hydrophobic interactions that stabilize the octameric association.
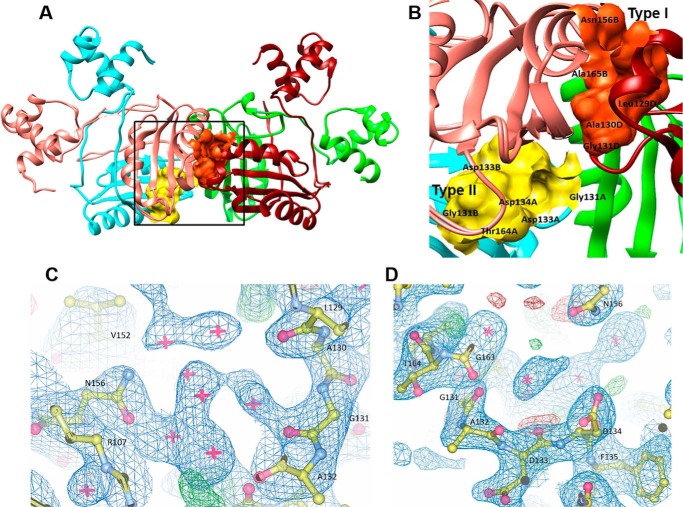
We had earlier (8) identified that a new binding site, called type II, exists in this protein family in the intradimer interface of the protein. This is in addition to the type I binding site that exists in the interdimer interface. Incidentally, the conserved Gly131, essential for binding amino acids/ligands, can form a part of both types of sites (Fig. 2, A and B). An examination of the type I and type II binding sites in MtbAldR shows clusters of water molecules at both of the binding sites in the apo-MtbAldR structure (Fig. 2, C and D). When the volumes of type I and type II binding sites in the MtbFFRP and MtbAldR structures were analyzed using the CASTp server (33), it was found that the volume of the type II binding site is greater than that of type I in both structures. This suggests that the type II site can accommodate larger ligand molecules. Further, the respective volumes of the type I and II sites of MtbAldR (405 and 462 Å3) were about 60 and 30% more than that of MtbFFRP (246 and 344 Å3).
FIGURE 2.
Ligand binding sites of MtbAldR. A, type I binding site at the interdimer interface and type II binding site at the intradimer interface of MtbAldR are shown in orange and yellow, respectively. The region marked by the box has been expanded and shown in B, which shows details of the residues that make up the respective binding sites. Gly131 is common to both sites. 2|Fo| − |Fc| electron density maps contoured at 1.0σ at the type I binding site (C) and the type II binding site (D) are also shown.
The solvent-accessible surface areas of subunits A and B are similar at 9672.8 and 9707.3 Å2, respectively, and a total of 4560 Å2 of surface area was buried upon dimer formation (i.e. ∼23% (2280 Å2) of the accessible surface area of each subunit is buried). Likewise, the individual solvent-accessible surface areas of subunits C and D are 9839.3 and 10,183.0 Å2, respectively, and the interface area of the CD dimer is ∼2467 Å2 (i.e. ∼25% of total surface area). Formation of the octamer and associated additional intersubunit interactions compared with the dimer results in ∼32% of the surface area being buried (i.e. an increase of ∼8% buried surface of each subunit compared with the dimer).
A comparison of the available crystal structures of Lrp/AsnC type proteins shows that the respective proteins share the basic fold, although the proteins exhibit relatively low sequence similarities. The results of structural superimpositions suggested that MtbAldR is relatively distinct as compared with other members and exhibits higher root mean square deviation values upon superposition. MtbAldR is closer to M. tuberculosis FFRP (2.1 Å root mean square deviation) and to a lesser extent to that of E. coli AsnC (3.0 Å root mean square deviation) and B. subtilis LrpC (3.1 Å root mean square deviation) proteins.
MtbAldR Binds to the Upstream Region of the ald Gene
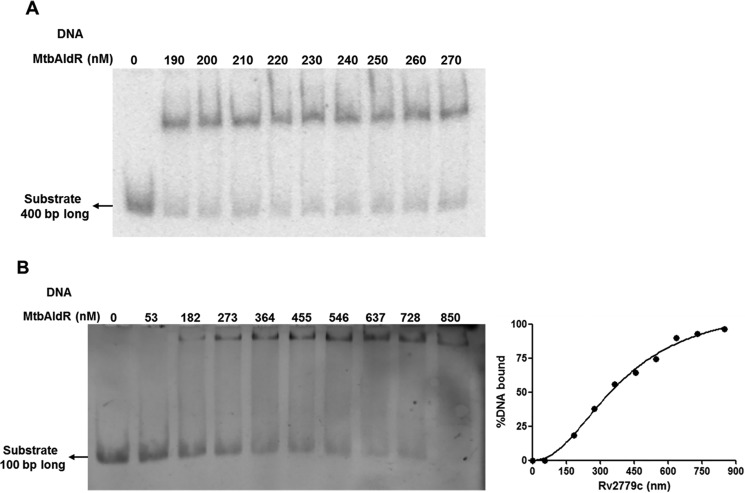
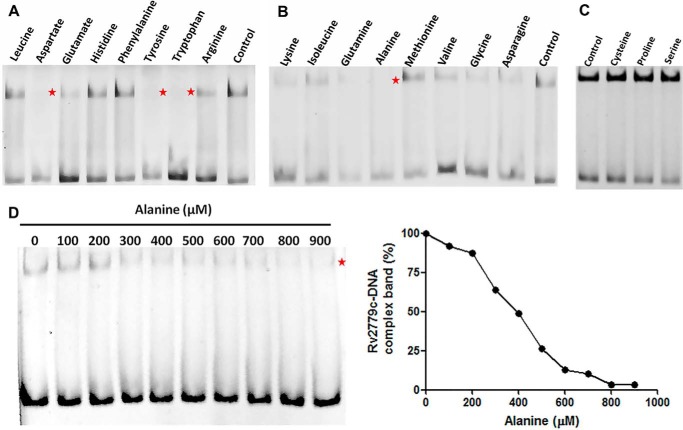
DNA binding and specificity of MtbAldR was investigated by EMSAs. As mentioned earlier (20), MtbAldR lies upstream to the ald gene (Rv2780), which codes for Ald (alanine dehydrogenase; EC 1.4.1.1). Initially, when the extended upstream ald sequence (−400 to +1) was incubated with purified MtbAldR, a single protein-DNA complex was observed (Fig. 3A). To identify the exact site where the protein binds, this 400-bp region was further subdivided into four regions of 100 bp each, and EMSA was performed individually with each of these regions. A single protein-DNA complex was observed when the extended upstream ald sequence S4 (−100 to +1) was incubated with purified MtbAldR, with an apparent equilibrium dissociation constant of 391 ± 32 nm (Fig. 3B). In contrast, no protein-DNA complex was observed when the −200 to −100 (S3) region and −300 to −200 (S2) region of ald was used in EMSA, respectively (data not shown), suggesting that MtbAldR binds more proximally to the upstream region of ald.
FIGURE 3.
EMSAs of MtbAldR with respective FAM-labeled substrates. A, EMSA corresponding to the 400-bp upstream region of Rv2780 (l-alanine dehydrogenase) used as the substrate. B, EMSA corresponding to the 100-bp upstream region (S4) of Rv2780 used as the substrate. Both of the substrates exhibit binding with MtbAldR. The apparent equilibrium dissociation constant for the binding of MtbAldR with the S4 region is 391 ± 32 nm.
Binding of Amino Acids to MtbAldR Probed through Qualitative Competitive ANS Displacement Assays
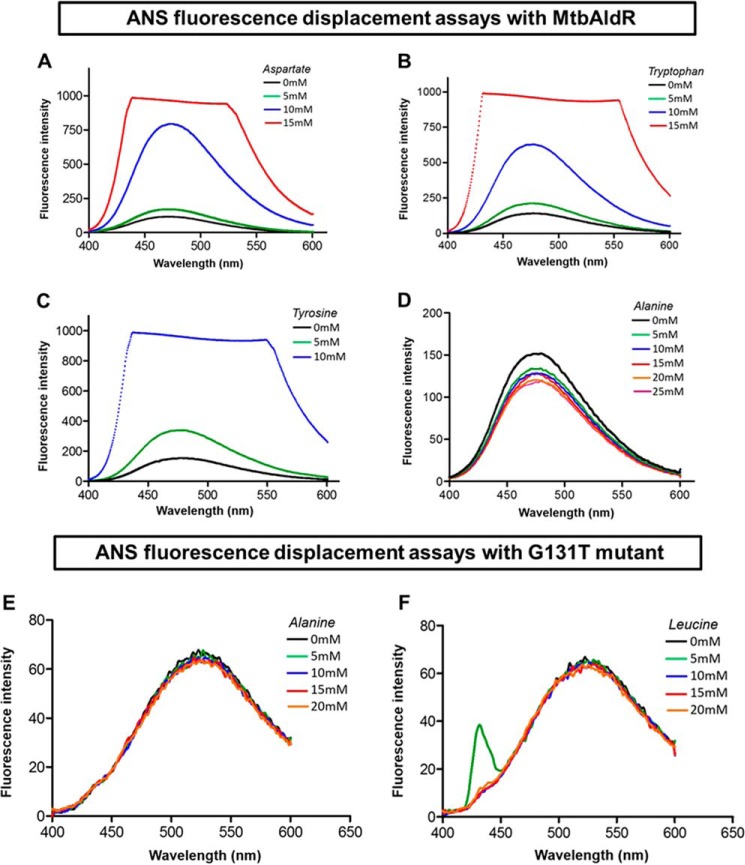
In this part of the study, we examined the binding of various amino acids to MtbAldR. Competitive displacement of ANS (8) was used to probe binding of MtbAldR and its G131T mutant to various amino acids. Gly131 is a conserved Gly that is known to be essential for binding ligands in related Lrp/AsnC proteins. Consequently, this mutant should lose the ability to bind to ligands and was used as a control. Binding of an amino acid/ligand may lead to either an increase or a decrease in the observed ANS fluorescence. The experiments clearly show changes in fluorescence intensity upon respective titration with Asp, Trp, Tyr, and Ala (Fig. 4, A–D, respectively). With His, Phe, Leu, Asn, Gln, Ala, Lys, Arg, Gly, Met, Pro, Ile, Ser, and Thr change in the fluorescence intensity was also observed, but in the case of Cys and Val, no change in intensity was observed (data not shown). The spectroscopy results clearly suggest that various amino acids bind to the wild-type protein. However, no such fluorescence change was observed in the case of the G131T mutant upon the addition of various amino acids, indicating that Gly131 is an essential residue for ligand binding in MtbAldR (Fig. 4, E and F).
FIGURE 4.
Amino acid binding studies using ANS fluorescence displacement assays against MtbAldR and its G131T mutant. Concentrations of the amino acid used in each curve are denoted in the insets with different colors. The change in fluorescence intensity was monitored by titrating the MtbAldR-ANS premix against increasing concentrations of aspartate (A), tryptophan (B), tyrosine (C), and alanine (D). The experiments indicate that these amino acids bind to MtbAldR. Binding studies against the G131T mutant were carried out by monitoring the change in the fluorescence intensity after titrating the G131T-ANS premix against increasing concentrations of alanine (E) and leucine (F), respectively. Clearly, the mutant protein cannot bind to the amino acids.
Binding of Amino Acids Elicits Diverse Responses on the MtbAldR-DNA Complex
Above, we showed that MtbAldR binds to diverse amino acids, including Ala, and also that it binds to the promoter region of ald. Next, we probed whether binding of amino acids to MtbAldR affects its ability to bind to the ald promoter region. Accordingly, we carried out EMSAs of MtbAldR binding to the ald promoter region in the presence of various amino acids. The results show that Ala, Trp, Tyr, and Asp completely abolish the DNA binding ability of MtbAldR. On the other hand, Glu and Asn reduce MtbAldR binding to DNA but do not completely abolish it (Fig. 5, A–C). We probed in more detail the effects of increasing concentration of Ala on the DNA binding activity of the protein, because it was earlier reported that Ala can act as a repressor or an activator for M. smegmatis AldR (15). We found that the addition of increasing concentrations of Ala correspondingly inhibits the formation of the MtbAldR nucleoprotein complex with an IC50 value of ∼400 μm (Fig. 5D).
FIGURE 5.
EMSA to probe for the effects of adding different amino acids to the nucleoprotein complex involving MtbAldR and a FAM-labeled 100-bp DNA fragment derived from the region upstream to Rv2780. The results are shown in A–C, respectively. EMSA was performed in the presence of a 10 mm concentration of each respective amino acid. The control experiment was carried out with only the protein and no amino acid. The protein and DNA concentrations used were 650 nm and 30 pm, respectively. Asp, Tyr, Trp, and Ala could disrupt the nucleoprotein complex and are marked by a red star in the gels. D, the effects of adding l-alanine were analyzed in more detail, and it was found that it inhibits the MtbAldR nucleoprotein complex with an IC50 value of ∼400 μm.
Disruption of the MtbAldR-DNA Complex by Small Molecule Inhibitors
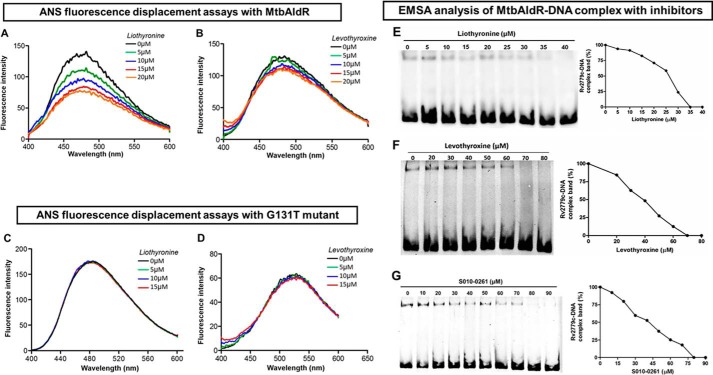
Conceptually, an important step to exploit the potential of AldR as a potential target is the identification of inhibitors that can disrupt the AldR nucleoprotein complex. As part of a long term program, we used an in-house database of small molecule compounds to identify potential binders to MtbAldR using in silico docking. We identified a tetrahydroquinoline carbonitrile derivative (S010-0261), levothyroxine, and liothyronine as potential inhibitors of AldR in this initial exercise. ANS-based fluorescence displacement assays showed that the compounds bind to MtbAldR (Fig. 6, A and B). Gel shift assays were used to probe the ability of the compounds to actually disrupt the MtbAldR-DNA complex. The compounds were individually added in a dose-dependent manner to purified MtbAldR in the DNA binding buffer before the addition of the labeled DNA probe. Fig. 6, E–G, shows that the compounds are able to disrupt/inhibit the formation of the nucleoprotein complex in the gel shift assays. Additionally, the IC50 values of liothyronine, levothyroxine, and S010-0261 were found to be 25, 40, and 42 μm. Indeed, compared with Ala, the compounds exhibit about ∼10–15-fold better inhibition in the assay system (Fig. 7).
FIGURE 6.
Inhibitor/ligand binding studies using ANS fluorescence displacement assays against MtbAldR and its G131T mutant. A and B correspond to the experiments with liothyronine and levothyroxine with MtbAldR, whereas C and D correspond to the experiments involving the inactive G131T mutant. Clearly, liothyronine and levothyroxine exhibit binding to the native protein, but they are unable to bind to the G131T mutant. The y axis represents the fluorescence intensity in arbitrary units, and the x axis corresponds to the scanned wavelength. In the experiments, the MtbAldR-ANS premix or the G131T-ANS premix, was titrated against increasing concentration of the respective compounds, and the resultant change in the fluorescence intensity was monitored. The concentrations of the compound used in each curve are shown as insets in the respective panels. Subsequently, the ability of liothyronine (E), levothyroxine (F), and a tetrahydroquinoline carbonitrile derivative (S010-0261) (G) to inhibit the MtbAldR-DNA complex was probed using EMSA. The first lane in the panels corresponds to the control experiments containing only protein and no compound. The subsequent lanes correspond to increasing concentrations of the added compounds. The IC50 values of liothyronine, levothyroxine, and S010-0261 were found to be ∼25, 40, and 42 μm.
FIGURE 7.
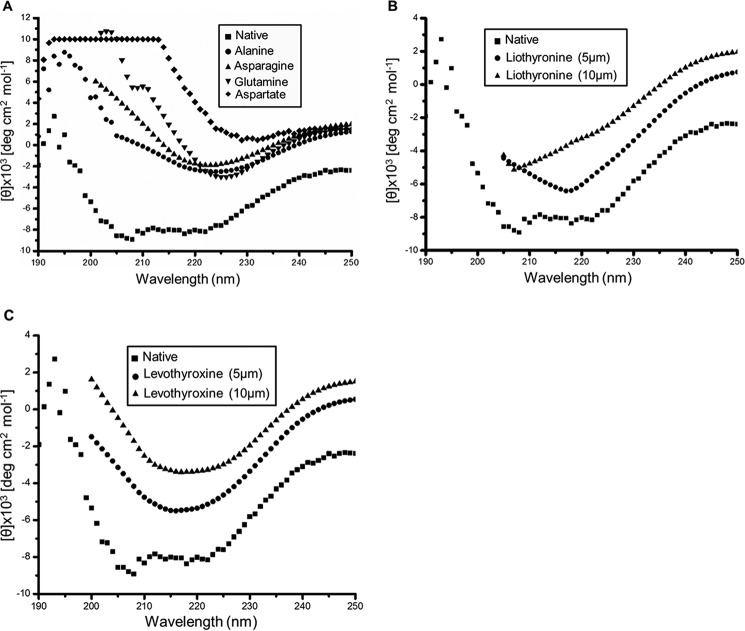
Changes to the far-UV CD spectra of MtbAldR were monitored after the addition of respective compounds. A, changes to the far-UV CD spectra monitored after the addition of amino acids. The inset shows the names of the amino acids corresponding to the spectra. The native spectrum corresponds to MtbAldR alone. B and C show far-UV CD spectral changes monitored upon the addition of liothyronine and levothyroxine to MtbAldR. The experiments suggest that MtbAldR exhibits conformational changes upon the addition of the compounds.
We tried to co-crystallize the protein with amino acids and inhibitors, but this resulted in either no crystals or small poorly diffracting crystals. We then used AUTODOCK to examine the potential binding modes of the compounds. Binding of the inhibitors to the type I site of MtbAldR was found to be unfavorable due to steric interactions of the compounds with the protein and relatively low binding energies. On the other hand, the compounds bind to the type II site with better predicted affinity as compared with the type I site and also do not exhibit unwanted steric clashes. Amino acids like Ala can, however, bind to the type I site also (data not shown).
With the help of qualitative ANS displacement assays, we studied the binding of these inhibitors to the Gly131 mutant, which, as shown above, does not bind to ligands/inhibitors due to the mutation of the essential Gly131 residue (Fig. 6, C and D). Interestingly, the mutant retains the ability to bind to the target DNA but the binding cannot be inhibited by the above compounds. This confirms that the compounds probably bind to the type II binding site as predicted. Further, it also suggests that binding of the inhibitors to the wild-type MtbAldR induces relative changes between the DNA-binding domains/other larger changes that prevent the protein from binding to the promoter region of ald. This was further confirmed using the circular dichroism spectroscopy experiments detailed below.
Circular Dichroism and Size Exclusion Chromatography Experiments
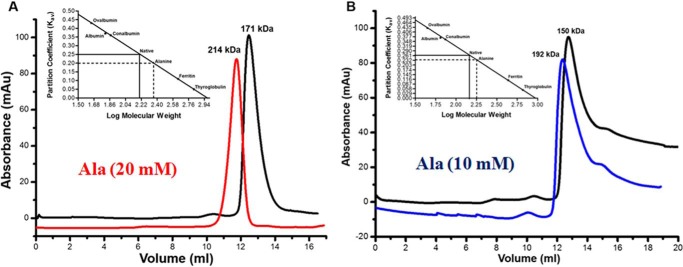
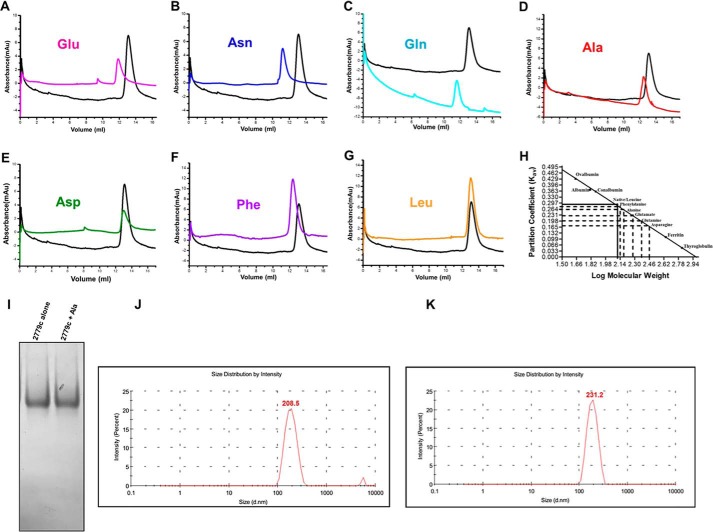
Far-UV CD spectra of both purified apo-MtbAldR and apo-MtbAldR in the presence of 3 mm concentrations of respective amino acids were collected. In the presence of alanine, aspartate, glutamine, and asparagine, significant changes to the respective CD spectra were observed (Fig. 7A). Far-UV CD spectra were also collected in the presence of liothyronine and levothyroxine. In the presence of increasing concentrations of the latter compounds, considerable changes to the CD spectra of the inhibited protein were observed compared with those of the apoprotein (Fig. 7, B and C). The results clearly suggest larger changes to the secondary structure/structural perturbations in the presence of the respective amino acids and compounds. To further probe the nature of the structural changes in MtbAldR in the presence of the amino acids, we used size exclusion chromatography. This protein family exhibits changes to the oligomeric association in the presence of effectors (e.g. amino acids). These changes may be in the form of a different order oligomeric association or even the adoption of an open quaternary structure (10, 11). The size exclusion chromatography experiments show that at 2 mg/ml concentration, MtbAldR elutes as an octamer and further that it elutes as a decamer in the presence of 20 mm Ala (Fig. 8A). At 1 mg/ml concentration, MtbAldR elutes at a position corresponding to a heptamer, and in the presence of 10 mm Ala (Fig. 8B), it apparently elutes as a nonamer. At 0.5 mg/ml concentration, the elution profile is a single peak corresponding to a hexamer. When 5 mm Glu, Asn, and Gln were added to the protein at this concentration, it adopted octameric and higher order oligomeric forms (Fig. 9, A–C). On the other hand, at 5 mm Ala, Asp, and Phe, we also observed a slight left shift in the elution peak of the protein, which can be attributed to either the “opening” of the closed structure or changes in the oligomeric associations (Fig. 9, D–F). No change in the oligomeric association was observed in the presence of Leu in analogous experiments (Fig. 9G).
FIGURE 8.
Analytical gel filtration profiles of MtbAldR carried out under different conditions. The protein concentration used in A was 2 mg/ml, and the profile of the apoprotein is shown in black. The profile of the protein in the presence of 20 mm alanine is shown in red. The elution profiles in B correspond to the experiment involving protein at a concentration of 1 mg/ml. The blue curve corresponds to MtbAldR in the presence of 10 mm alanine. The inset in both panels shows the plot between the partition coefficient (Kav) and log of molecular weight for standard proteins of known molecular weight. This was carried out to calibrate the column. The Kav of the eluted species was calculated and extrapolated in the same plot to determine the log of molecular weight. The anti-log value was calculated to determine the actual molecular weight of the eluted species and is indicated above the curves. The results suggest that MtbAldR apparently adopts a higher order oligomeric form in the presence of alanine. Alternatively, we have hypothesized (see “Discussion”) that the protein forms an open quaternary structure in the presence of alanine, leading to a larger particle size and faster column elution.
FIGURE 9.
Analytical gel filtration experiments involving MtbAldR. The concentration of the protein was 0.5 mg/ml, and the elution profile is shown in black in all of the panels. A–G correspond to the elution profiles of MtbAldR in the presence of a 5 mm concentration of the amino acid indicated in the respective panels. H represents the plot between the partition coefficient (Kav) and log of molecular weight for standard proteins of known molecular weight, used to calibrate the column. I, native PAGE analysis of apo-MtbAldR alone and in the presence of 5 mm alanine. No difference in the molecular weights of the two species could be discerned in the experiments. J and K, DLS experiments carried out on apo-MtbAldR (0.5 mg/ml) alone and in the presence of 5 mm alanine, respectively. The hydrodynamic diameter is indicated. The size of MtbAldR in the presence of alanine exhibits an increase of ∼10% compared with that of the apoprotein.
Additionally, we subjected the fractions corresponding to the elution peaks of the apo- and Ala-bound MtbAldR to native PAGE analysis and DLS studies (Fig. 9). No change in the molecular weights could be discerned in the native PAGE analysis between the apo- and Ala-bound MtbAldR. On the other hand, the z-average size/hydrodynamic diameter of the protein particle in DLS experiments was found to be 231.2 nm for Ala-bound MtbAldR and 208.5 nm for the apoprotein (Fig. 9, J and K). This corresponds to an increase of ∼9.8% in the hydrodynamic diameter of Ala-bound MtbAldR compared with the apoprotein. Overall, the above experiments show that MtbAldR exhibits both concentration-dependent oligomerization changes and effector-mediated changes to the quaternary association.
Discussion
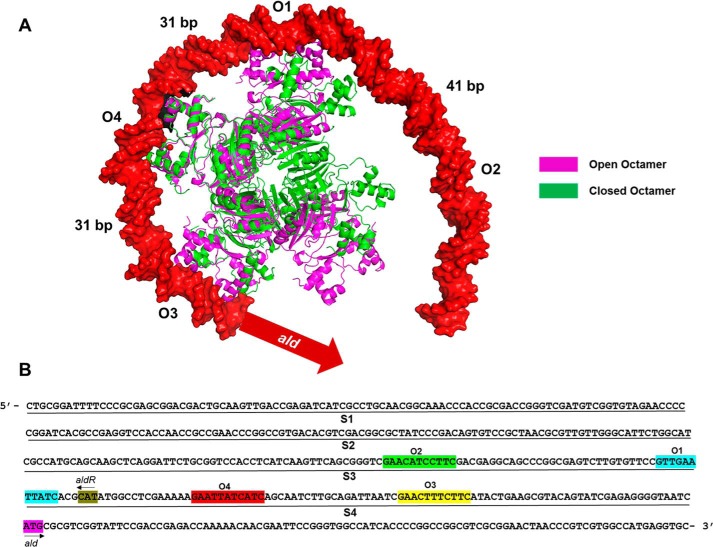
Structurally, MtbAldR exhibits the typical fold observed in this protein family, although it has low sequence homology with other FFRP homologs. The basic functional unit is the dimer that is stabilized, among other things, through “N-terminal domain-swapping” interactions. It exhibits both concentration-dependent and ligand-binding dependent changes to the quaternary structure and association. Deviation from quaternary structure symmetry is a rare event, and where observed, it is attributed to strong functional reasons. The open quaternary structures observed in the feast/famine regulatory family of proteins apparently enable them to bind to target DNA sites separated by ∼20–40 bases between them. The proteins can adopt it in the complex with DNA, as observed in the E. coli Lrp-DNA complex (11). Alternatively, it can be triggered by the binding of a ligand to the effector site, as suggested by the open structure adopted by the G102T mutant of MtbFFRP in its crystal structure (10). Very recently, transmission electron microscopy studies involving M. smegmatis AldR in the presence of Ala showed that this protein adopts an open quaternary structure in the presence of the effector Ala. This was attributed to the necessity of binding to multiple target sites located upstream to the ald gene in M. smegmatis (34). Thus, there appears to be interplay between the interactions of FFRP with the substrate and the effector that results in changes to the quaternary structural assembly and/or adoption of different quaternary states. The present work involving MtbAldR further suggests that adoption of open quaternary structures plays an important part in facilitating interactions with target DNA sites (Fig. 10A). As the modeling suggests, interactions with binding sites separated by ∼26–30 bases are conceptually possible with a “closed” oligomer with smaller adjustments/changes to the DNA-binding domains. On the other hand, binding to sites that are separated by ∼40 bases like in the ald promoter region (e.g. between sites O1 and O2), will require the protein to adopt the open quaternary association. It will be interesting to see whether binding of different effectors/ligands can elicit nuanced effects (e.g. can these proteins form open quaternary structures with different extents of “openness”?) or whether the protein can bind to the target site(s) in different quaternary structural states to elicit diverse functional consequences.
FIGURE 10.
A, superposition of open (Protein Data Bank code 2W29; MtbFFRP) and closed octamers (Protein Data Bank code 4PCQ; MtbAldR). This was modeled onto MtbAldR binding sites O1–O4, respectively, located upstream to the ald gene. The numbers between the two adjacent AldR-binding sites indicate the distances between the respective central T nucleotides in the base pairs. The model clearly suggests that the protein has to adopt the open quaternary structure in order to bind to the complete region. B, the sequence of the 400-bp region upstream to the M. tuberculosis ald gene is shown. This was divided into four 100-bp fragments, called S1–S4, as indicated, for the present EMSA studies. The MtbAldR binding sites that were identified later are also indicated and are marked as O1–O4, respectively, in the figure.
In line with the above, we tested the ability of 20 amino acids to bind to the protein as well as for the respective ability to disrupt the MtbAldR-DNA complex. With the exception of Cys and Val, the other amino acids showed binding to the protein. Of these, Ala, Tyr, Trp, and Asp, could completely inhibit the MtbAldR-DNA complex in our assays. In this context, our result involving Ala is in contrast with the earlier report involving M. smegmatis AldR (15), where binding of the protein to the upstream region of ald was reportedly enhanced in the presence of Ala. The differences in the observations may be due to the different DNA target sequences used in their assays (Fig. 10B), and consequently we hypothesize that different quaternary states of AldR might be binding to the respective tested sequences. This underscores the need for more complementary in vivo and in vitro experiments to probe the molecular mechanisms of AldR.
Subsequently, the oligomeric status of selected MtbAldR-amino acid complexes was probed using size exclusion chromatography. Binding of some of the amino acids increased the oligomeric status of MtbAldR, as assessed by the latter experiments (Table 2). A marginal increase in the oligomeric status (e.g. hexamer to heptamer) can be plausibly attributed to formation of an open structure. This would also explain the small retardation in the gel matrix, especially in light of the above discussion. The native PAGE analysis and DLS experiments involving Ala-bound MtbAldR and the apoprotein support this hypothesis. The increase in the hydrodynamic diameter of Ala-bound MtbAldR by ∼9.8% can be attributed to the formation of an open quaternary structure. However, larger oligomeric changes (e.g. hexamer to decamer, dodecamer, and tetradecamer) were also observed, and the latter changes cannot be attributed only to formation of an open structure. Of course, further structural evidence is needed to understand the exact changes that occur upon effector binding.
TABLE 2.
Effects of adding amino acids to MtbAldR and MtbAldR-DNA complex, respectively
| Amino acids | ANS intensity change upon adding respective amino acids to MtbAldR (7 μm) | Effect on the MtbAldR (650 nm)-DNA complex | Changes to the quaternary structure of MtbAldR (0.5 mg/ml)a |
|---|---|---|---|
| Tyrosine | Increase | Inhibition | —b |
| Phenylalanine | Increase | No effect | Hexamer to heptamer |
| Tryptophan | Increase | Inhibition | —b |
| Histidine | Increase | No effect | |
| Aspartate | Increase | Inhibition | Hexamer to heptamer |
| Leucine | Increase | No effect | No effect |
| Glutamate | Increase | Partial inhibition | Hexamer to decamer |
| Asparagine | Decrease | Partial inhibition | Hexamer to tetradecamer |
| Glutamine | Decrease | No effect | Hexamer to dodecamer |
| Alanine | Decrease | Inhibition | —a |
a Smaller changes to the oligomeric state of MtbAldR in the presence of amino acids like alanine can be interpreted as a transition to an “open” quaternary association, as hypothesized under “Discussion.”
b Inconclusive results; experiment was not performed.
The CD experiments involving MtbAldR in the presence of various amino acids also suggest that larger structural change takes place upon amino acid binding. It is known that binding of effectors to this protein family can lead to conformational changes (2, 8, 9). One such observed change is to the relative spatial disposition of the N-terminal domains. Another large change observed by us (8, 10) in a homologous mycobacterial protein was a displacement of the ligand binding loop in the C-terminal domain by nearly 4 Å upon effector binding. The conformational changes reflect the structural fine tuning that these transcriptional regulators adopt to modulate their DNA binding activity in the presence/absence of suitable effector molecules.
So far, a clear functional role for binding of only Ala to M. smegmatis AldR and consequent regulation of the ald gene has been reported. Our results have identified three other amino acids (viz. Tyr, Trp, and Asp) that bind to the protein and disrupt the DNA complex, and the functional significance of this is yet to be elucidated. Interestingly, aromatic amino acids are important for the pathogen's viability (35), and the components of the Shikimate pathway have been identified as important M. tuberculosis targets due to their involvement in aromatic amino acid metabolism (36). Asp has been identified as an effector for the Lrp from Halobacterium salinarum for regulating aspartate transaminase and glutamine synthetase gene expression (37). Asp has also been suggested to bind to archaeal FL-11 and regulate aspartate oxidase expression (38). Indeed, a sequence homology search of the M. tuberculosis H37Rv genome sequence identified potential AldR/FFRP binding sites upstream to the aspartate transaminase, aspartate oxidase, and chorismate synthase genes (data not shown). It will be interesting to examine whether MtbAldR is also involved in the regulation of some of these other genes.
The binding experiments with the G131T mutant of MtbAldR gave further insights into the interactions with target DNA. It may be recalled that the mutation of the same conserved glycine residue (Gly102) in MtbFFRP (10), and also binding of arginine to FL11 (12), results in the open quaternary association observed in Rv3291c and FL11. Gly102 in MtbFFRP is an essential residue for effector binding that is common to two separate binding sites in the protein. The type II binding site occurs in the intradimer interface binding domain, and the type I binding site is located at the interdimer interface. Ligands that bind to the type II site can disrupt DNA interactions by modulating the relative spatial disposition of the DNA-binding domains in the dimer, although the site itself is located relatively distal to the DNA binding residues. On the other hand, ligands that bind to the type I sites can modulate DNA binding by changing the oligomeric association. Interestingly, the G131T mutant of MtbAldR does not bind to amino acids/ligands but retains the ability to form a complex with DNA. Because the amino acids are unable to bind to the effector sites, they cannot interact with the protein and disrupt the DNA complex. This supports our earlier hypothesis that binding of the amino acids can cause changes to the relative spatial disposition of the DNA binding motifs, among other things. It also highlights that it is possible to identify small molecule inhibitors that can inhibit the MtbAldR-DNA complex whose mode of action is as hypothesized above.
We initially used virtual screening strategies to identify potential inhibitors that can bind to either the type I or type II site of MtbAldR. We tested about 25 compounds in the EMSAs, and this resulted in the identification of a tetrahydroquinoline carbonitrile derivative (S010-0261), levothyroxine, and liothyronine as small molecule inhibitors that can disrupt the MtbAldR-DNA complex. The binding of the latter two compounds to MtbAldR and its G131T mutant was also checked by fluorescence displacement assays and CD experiments. These compounds cannot bind to the type I site due to the predicted steric hindrance but can bind well to the type II site. Importantly, these inhibitors do not bind to the G131T mutant, and the latter result, taken together with the docking analysis, clearly indicates that the inhibitors bind to the type II site. To the best of our knowledge, these compounds represent the very first inhibitors of the Lrp/AsnC/FFRP family of proteins and demonstrate that the FFRP-DNA complex can be disrupted by the binding of compounds to the site located relatively distal to the DNA-interacting helix-turn-helix motif.
Author Contributions
A. D. performed experiments, analyzed data, and wrote the paper. S. S. performed the DLS, size exclusion, and native gel experiments. S. K. P. synthesized a compound, and R. P. T. synthesized a compound and analyzed its data. R. R. designed the experiments, analyzed data, and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
This work was funded by Council of Scientific and Industrial Research (CSIR), India, vide Grants BSC0104 (SPLenDID) and BSC0121 (GENESIS). This paper bears the CDRI communication number 9203. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 4PCQ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- FFRP
- feast/famine regulatory protein
- FAM
- 6-carboxyfluorescein
- ANS
- 1-anilino-8-naphthalenesulfonate
- DLS
- dynamic light scattering.
References
- 1. Beloin C., Jeusset J., Revet B., Mirambeau G., Le Hégarat F., and Le Cam E. (2003) Contribution of DNA conformation and topology in right-handed DNA wrapping by the Bacillus subtilis LrpC protein. J. Biol. Chem. 278, 5333–5342 [DOI] [PubMed] [Google Scholar]
- 2. Yokoyama K., Ishijima S. A., Clowney L., Koike H., Aramaki H., Tanaka C., Makino K., and Suzuki M. (2006) Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors. FEMS Microbiol. Rev. 30, 89–108 [DOI] [PubMed] [Google Scholar]
- 3. Kumarevel T., Nakano N., Ponnuraj K., Gopinath S. C., Sakamoto K., Shinkai A., Kumar P. K., and Yokoyama S. (2008) Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector l-glutamine: implications of effector binding in molecular association and DNA binding. Nucleic Acids Res. 36, 4808–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thaw P., Sedelnikova S. E., Muranova T., Wiese S., Ayora S., Alonso J. C., Brinkman A. B., Akerboom J., van der Oost J., and Rafferty J. B. (2006) Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 34, 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calvo J. M., and Matthews R. G. (1994) The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58, 466–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Newman E. B., D'Ari R., and Lin R. T. (1992) The leucine-Lrp regulon in E. coli: a global response in search of a raison d'etre. Cell 68, 617–619 [DOI] [PubMed] [Google Scholar]
- 7. Tani T. H., Khodursky A., Blumenthal R. M., Brown P. O., and Matthews R. G. (2002) Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. U.S.A. 99, 13471–13476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shrivastava T., and Ramachandran R. (2007) Mechanistic insights from the crystal structures of a feast/famine regulatory protein from Mycobacterium tuberculosis H37Rv. Nucleic Acids Res. 35, 7324–7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yokoyama K., Nogami H., Kabasawa M., Ebihara S., Shimowasa A., Hashimoto K., Kawashima T., Ishijima S. A., and Suzuki M. (2009) The DNA-recognition mode shared by archaeal feast/famine-regulatory proteins revealed by the DNA-binding specificities of TvFL3, FL10, FL11 and Ss-LrpB. Nucleic Acids Res. 37, 4407–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shrivastava T., Dey A., and Ramachandran R. (2009) Ligand-induced structural transitions, mutational analysis, and “open” quaternary structure of the M. tuberculosis feast/famine regulatory protein (Rv3291c). J. Mol. Biol. 392, 1007–1019 [DOI] [PubMed] [Google Scholar]
- 11. de los Rios S., and Perona J. J. (2007) Structure of the Escherichia coli leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J. Mol. Biol. 366, 1589–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamada M., Ishijima S. A., and Suzuki M. (2009) Interactions between the archaeal transcription repressor FL11 and its coregulators lysine and arginine. Proteins 74, 520–525 [DOI] [PubMed] [Google Scholar]
- 13. Singh P., Yadav G. P., Gupta S., Tripathi A. K., Ramachandran R., and Tripathi R. K. (2011) A novel dimer-tetramer transition captured by the crystal structure of the HIV-1 Nef. PLoS One 6, e26629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deng W., Wang H., and Xie J. (2011) Regulatory and pathogenesis roles of Mycobacterium Lrp/AsnC family transcriptional factors. J. Cell. Biochem. 112, 2655–2662 [DOI] [PubMed] [Google Scholar]
- 15. Jeong J. A., Baek E. Y., Kim S. W., Choi J. S., and Oh J. I. (2013) Regulation of the ald gene encoding alanine dehydrogenase by AldR in Mycobacterium smegmatis. J. Bacteriol. 195, 3610–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mani Tripathi S., and Ramachandran R. (2006) Direct evidence for a glutamate switch necessary for substrate recognition: crystal structures of lysine ϵ-aminotransferase (Rv3290c) from Mycobacterium tuberculosis H37Rv. J. Mol. Biol. 362, 877–886 [DOI] [PubMed] [Google Scholar]
- 17. Tripathi S. M., and Ramachandran R. (2008) Crystal structures of the Mycobacterium tuberculosis secretory antigen alanine dehydrogenase (Rv2780) in apo and ternary complex forms captures “open” and “closed” enzyme conformations. Proteins 72, 1089–1095 [DOI] [PubMed] [Google Scholar]
- 18. Hasan S., Daugelat S., Rao P. S., and Schreiber M. (2006) Prioritizing genomic drug targets in pathogens: application to Mycobacterium tuberculosis. PLoS Comput. Biol. 2, e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giffin M. M., Modesti L., Raab R. W., Wayne L. G., and Sohaskey C. D. (2012) ald of Mycobacterium tuberculosis encodes both the alanine dehydrogenase and the putative glycine dehydrogenase. J. Bacteriol. 194, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dey A., and Ramachandran R. (2014) Cloning, overexpression, purification and preliminary x-ray analysis of a feast/famine regulatory protein (Rv2779c) from Mycobacterium tuberculosis H37Rv. Acta Crystallogr. F Struct. Biol. Commun. 70, 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 22. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brünger A. T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 26. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27. Davis I. W., Murray L. W., Richardson J. S., and Richardson D. C. (2004) MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lovell S. C., Davis I. W., Arendall W. B. 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., and Richardson D. C. (2003) Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 29. Tina K. G., Bhadra R., and Srinivasan N. (2007) PIC: protein interactions calculator. Nucleic Acids Res. 35, W473–W476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 31. Stryer L. (1965) The interaction of a naphthalene dye with apomyoglobin and apohemoglobin: a fluorescent probe of non-polar binding sites. J. Mol. Biol. 13, 482–495 [DOI] [PubMed] [Google Scholar]
- 32. Goodsell D. S., Morris G. M., and Olson A. J. (1996) Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recognit. 9, 1–5 [DOI] [PubMed] [Google Scholar]
- 33. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., and Liang J. (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jeong J. A., Hyun J., and Oh J. I. (2015) Regulation mechanism of the ald gene encoding alanine dehydrogenase by AldR in Mycobacterium smegmatis and Mycobacteirum tuberculosis by Lrp/AsnC family regulator AldR. J. Bacteriol. 197, 3142–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parish T., and Stoker N. G. (2002) The common aromatic aminoacid biosynthesis pathway is essential in Mycobacterium tuberculosis. Microbiology 148, 3069–3077 [DOI] [PubMed] [Google Scholar]
- 36. Ducati R. G., Basso L. A., and Santos D. S. (2007) Mycobacterial shikimate pathway enzymes as targets for drug design. Curr. Drug Targets 8, 423–435 [DOI] [PubMed] [Google Scholar]
- 37. Schwaiger R., Schwarz C., Furtwängler K., Tarasov V., Wende A., and Oesterhelt D. (2010) Transcriptional control by two leucine-responsive regulatory proteins in Halobacterium salinarum R1. BMC Mol. Biol. 11, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yokoyama K., Ishijima S. A., Koike H., Kurihara C., Shimowasa A., Kabasawa M., Kawashima T., and Suzuki M. (2007) Feast/famine regulation by transcription factor FL11 for the survival of the hyperthermophilic archeaon Pyrococcus OT3. Structure 15, 1542–1554 [DOI] [PubMed] [Google Scholar]