Abstract
Keratins 8/18 (K8/18) are phosphoglycoproteins and form the major intermediate filament network of simple epithelia. The three O-GlcNAcylation (Ser29, Ser30, and Ser48) and two phosphorylation (Ser33 and Ser52) serine sites on K18 are well characterized. Both of these modifications have been reported to increase K18 solubility and regulate its filament organization. In this report, we investigated the site-specific interplay between these two modifications in regulating the functional properties of K18, like solubility, stability, and filament organization. An immortalized hepatocyte cell line (HHL-17) stably expressing site-specific single, double, and triple O-GlcNAc and phosphomutants of K18 were used to identify the site(s) critical for regulating these functions. Keratin 18 mutants where O-GlcNAcylation at Ser30 was abolished (K18-S30A) exhibited reduced phosphorylation induced solubility, increased stability, defective filament architecture, and slower migration. Interestingly, K18-S30A mutants also showed loss of phosphorylation at Ser33, a modification known to regulate the solubility of K18. Further to this, the K18 phosphomutant (K18-S33A) mimicked K18-S30A in its stability, filament organization, and cell migration. These results indicate that O-GlcNAcylation at Ser30 promotes phosphorylation at Ser33 to regulate the functional properties of K18 and also impact cellular processes like migration. O-GlcNAcylation and phosphorylation on the same or adjacent sites on most proteins antagonize each other in regulating protein functions. Here we report a novel, positive interplay between O-GlcNAcylation and phosphorylation at adjacent sites on K18 to regulate its fundamental properties.
Keywords: cytoskeleton, intermediate filament, keratin, migration, O-GlcNAcylation, phosphorylation, protein degradation
Introduction
Keratins are the major intermediate filament proteins of epithelia. They form a 10-nm filamentous cytoskeleton made up of a non-covalent obligate heteropolymer of type I and type II keratins and are expressed in a tissue-specific manner (1, 2). Keratin pair 8/18 forms the intermediate filament scaffold predominantly in simple epithelia such as the lining of the alimentary canal, liver, and pancreas (2, 3). The expression pattern of keratins is highly regulated among various epithelia, suggesting a cell type-specific role for various keratins (3). Ectopic expression of K8/18 in other epithelia is highly associated with a malignant phenotype (4). Keratins 8/18 provide mechanical support for cellular integrity and are central to various non-mechanical functions like protein biosynthesis, protection from apoptosis, regulation of cell cycle progression, motility, and organelle transport (5–10). Their biological roles are majorly dependent on their functional properties like solubility, filament organization, and dynamics (11, 12). Keratins 8/18 undergo several posttranslational modifications like phosphorylation, O-GlcNAcylation, acetylation, sumoylation, and transamidation (13). Of these, site-specific phosphorylation is very well characterized for its role in regulating the functional properties, like solubility and filament organization, of both keratin 8 and 18 (5, 9, 14–16). Recent evidence shows that these functional properties are also regulated by other modifications, like O-GlcNAcylation, sumoylation of keratins 8/18, and acetylation of keratin 8 (17–19). However, the site-specific roles of O-GlcNAcylation and its cross-talk with phosphorylation are yet to be uncovered.
The addition of a single GlcNAc to serine and threonine residues to nuclear and cytoplasmic proteins, termed O-GlcNAcylation, was first identified in 1984 by Torres and Hart (20). Two enzymes, O-GlcNAc transferase and O-GlcNAcase, regulate the cycling of O-GlcNAc on proteins (21, 22). Unlike complex classical glycosylation, which is static in nature, O-GlcNAcylation is a dynamic modification similar to phosphorylation (23). A wide range of nuclear and cytoplasmic proteins get abundantly modified with this single sugar modification, viz. transcription factors, nuclear pore proteins, enzymes, and cytoskeletal proteins (21). On most proteins, O-GlcNAcylation exhibits cross-talk with phosphorylation to regulate essential properties of proteins like protein-protein/DNA-protein interactions (24), subcellular localization (25), and proteasome-mediated protein degradation (26, 27), thereby controlling cellular processes like transcription, signal transduction, stress response, and cell cycle progression (28, 29). Aberrant O-GlcNAcylation is associated with several pathological conditions like cancer, diabetes, and neurodegenerative diseases (30–33).
There is no consensus sequence for O-GlcNAc modification. However, it is often observed on the same or proximal Ser/Thr residues that are used for phosphorylation, thereby negatively regulating phosphorylation (23). These two modifications can regulate each other by competitively blocking the site (34), sterically hindering the addition at an adjacent or proximal site (26), or by influencing their respective enzymes (35, 36). This type of reciprocal interplay between O-GlcNAcylation and phosphorylation to regulate protein functions is observed on several proteins, including intermediate filaments like neurofilaments (neurofilament–M) and cytoskeleton-associated proteins like Tau (37, 38). With recent advances in mass spectrometry, more complex and extensive cross-talk between O-GlcNAcylation and phosphorylation on many cellular proteins has been discovered (36).
Although O-GlcNAcylation (gSer29,5 gSer30, and gSer48) (39) and phosphorylation (Ser(P)33 and Ser(P)52) (5, 40) occur at proximal sites on keratin 18, their mutual interplay in regulating its functional properties is largely unexplored. In our previous report, we showed that total O-GlcNAcylation on keratins 8/18 can regulate solubility, filament organization, and stability (17). In this study, we aim to understand the site-specific role of K18 O-GlcNAcylation in regulating proximal phosphorylation and functional properties. Using a panel of site-specific O-GlcNAcylation and phosphorylation mutants of K18 along with antibodies that can detect site-specific phosphorylation, we demonstrate that these two modifications exhibit substantial cross-talk at proximal sites on K18 to regulate solubility, stability, and filament organization and also cellular processes such as migration. We also uncover a novel role for O-GlcNAcylation to promote phosphorylation at proximal sites. These findings represent the first detailed characterization of site-specific interplay between O-GlcNAcylation and phosphorylation of intermediate filament protein along with identification of a unique, phosphorylation-promoting role for O-GlcNAcylation.
Experimental Procedures
Reagents
HHL17 (human hepatocyte line 17) cells were a gift from Dr. A. H. Patel (Institute of Virology, Glasgow, UK) (41). The K18 phosphorylation-specific antibodies K18-Ser(P)33 (clone IB4) and anti K18-Ser(P)52 (clone 3055) were a gift from Prof. Bishr Omary (Michigan Medical School). Cell culture reagents were obtained from Invitrogen. The antibodies used in this study were mouse keratin 18 clone CY-90 (C8541), mouse keratin 8 clone M20 (C5301), and mouse HRP (A4416) (Sigma); mouse anti-O-GlcNAc clone RL-2 (MA1-072, Affinity Bioreagents); mouse Ser/Thr phosphorylation clone 22a (612549, BD Biosciences, Clontech); anti-GAPDH clone ABM22C5 (10-10011, Abgenex); rabbit HRP (sc-2004, Santa Cruz Biotechnology); and mouse Alexa Fluor 568 (A11004, Molecular Probes, Invitrogen). Cycloheximide, okadaic acid, PUGNAc, and fibronectin were purchased from Sigma-Aldrich (St. Louis, MO). WGA-Sepharose beads, the PVDF membrane, and the ECL kit were purchased from Amersham Biosciences. Cultureware was obtained from Nunclon. Phusion polymerase and restriction enzymes were from New England Biolabs. All other chemicals were purchased locally and were of analytical grade.
Cell Culture
The HHL17 cell line was routinely cultured and maintained in DMEM containing 0.03% glutamine, 10 units/ml penicillin G-sodium, 10 μg/ml streptomycin sulfate, 25 μg/ml amphotericin B, and 10% FBS at 37 °C and 5% CO2. For different drug/inhibitor treatments, cycloheximide, PUGNAc, and okadaic acid were used at concentrations of 200 μm/ml, 100 μm/ml, and 400 nm/ml, respectively, for various times.
Plasmids, Site-directed Mutagenesis, Cloning, and Stable Expression
Human K18 WT-YFP cDNA (a gift from Dr. Rudolf Leube and Dr. Reinhard Windoffer, Johannes Gutenberg University) and K18 O-GlcNAcylation triple mutant (TM) (S29A/S30A/S48A) cDNA (a gift from Prof. Bishr Omary, Michigan Medical School) were used to generate site-specific (Ser-to-Ala) single and double O-GlcNAc mutants of K18 using the QuikChange II site-directed mutagenesis kit according to the protocol of the manufacturer. K18-WT and all mutants were further cloned into EYFP-lentiviral vector (42). Using the QuikChange II site-directed mutagenesis kit, K18-S33A, K18-S52A, K18-S33D, and K18-S30A/S33D were generated from EYFP-K18-WT cloned in the lentiviral vector. The primers used for site-directed mutagenesis are listed in supplemental Table 1.
Infectious lentivirus to express K18-WT and various mutants was produced as described previously (42). To generate K18-WT and various K18 mutants expressing stable cell lines, HHL-17 cells were infected with infectious lentivirus in the presence of Polybrene (8 μg/ml) at 80–90% confluence. After infection (24 h), stable cells expressing the transgene were selected and maintained in complete medium containing puromycin (0.8 μg/ml).
Protein Isolation and Immunoblotting
Total cell lysates were prepared by solubilizing HHL17 cells in 2% SDS cell lysis buffer (62.5 mm Tris and 2% SDS (pH 6.8)). Differential extraction of soluble and filamentous keratin was performed as described in Ref. 17. In brief, soluble keratins were extracted by resuspending cells in phosphate-buffered saline containing 1% Nonidet P40, 5 mm EDTA, protease inhibitor mixture (1 mm phenylmethylsulfonyl fluoride, 10 μm leupeptin, 10 μm pepstatin, and 25 μg/ml aprotinin), and phosphatase inhibitor mixture for 20 min at 4 °C, followed by centrifugation (16,000 × g, 1 h.). The supernatant was collected as the soluble fraction, and the resulting insoluble pellet was solubilized in 2% SDS cell lysis buffer (pellet). Proteins from total, soluble, and insoluble lysates in 1× Laemmli sample buffer were resolved on 12% SDS-PAGE, transferred to PVDF membranes, and probed with antibodies as indicated. Membranes were washed three times with Tris-buffered saline (20 mm Tris and 500 mm NaCl (pH 7.5)) containing 0.1% Tween 20 for 30 min, incubated with anti-mouse or anti-rabbit secondary antibodies coupled to horseradish peroxidase, and visualized using enhanced chemiluminescence. After Western blotting, the membrane was stained with Coomassie Brilliant Blue R-250, which served as a control for equal loading. For immunoprecipitation, cells were lysed in phosphate buffer (PBS (150 mm NaCl and 10 mm phosphate (pH 7.4)) containing 2% Empigen, phosphatase, and protease inhibitor mixture, followed by immunoprecipitation as described in Ref. 17.
Purification of the Glycosylated Form of K8 and 18
To differentially extract cellular K8 and K18 fractions, total cell lysates were resolved on 10% PAGE along with a prestained protein ladder. After electrophoresis, the gel was incubated in 4 m sodium acetate solution for 2 h at −20 °C. Proteins in the gel can be visualized as clear transparent bands against the white opaque gel because of precipitation of free SDS in the gel. The protein bands in the region of K8 (52 kDa) and keratin 18 (48 kDa) were separately collected in microcentrifuge tubes by excising the gel with reference to standard molecular weight markers. The gel pieces were crushed in 500 μl of extraction buffer (0.5% SDS, 150 mm NaCl, and 62.5 mm Tris (pH 6.8)), and the tubes were vortexed vigorously for 45 min at room temperature. Following this, the samples were centrifuged for 5 min at 10,000 × g, and the supernatant was collected into a fresh tube. SDS from the extracted samples was removed by adding 150 mg of activated SM-2 Biobeads, followed by incubation at room temperature for 2 h with gentle rocking. The sample was centrifuged at 2000 × g for 5 min at room temperature to pellet the SM-2 Biobeads, and the supernatant containing cellular fractions of either keratin 8 or 18 was collected and stored at −20 °C. O-GlcNAcylated proteins from these fractions were extracted using WGA-Sepharose beads as described previously (17). Glycosylated K8 and 18 from these samples were immunoprecipitated using K8- and K18-specific monoclonal antibodies as described previously (17).
Immunostaining and Fluorescence Imaging of Cells Expressing EYFP-K18
HHL17 cells stably expressing EYFP-K18-WT; single, double, or triple O-GlcNAcylation mutants; and phosphomutants were immunostained for K8 as described previously (17). Images were acquired on a Zeiss LSM510 META confocal microscope (Carl Zeiss Microimaging, Inc., Thornwood, NY) using an oil immersion ×63 Plan Apochromat phase-contrast objective (numerical aperture, 1.4) and processed using LSM510 imaging software, version 4.2.
Scratch Healing Assay
For wound healing assays, 6-well culture dishes were coated overnight with 10 μg/ml fibronectin in serum-free DMEM at 4 °C. HHL17 cells stably expressing O-GlcNAcylation and phosphorylation site-specific mutants were seeded at a density of 1 million cells/ml of complete medium and incubated at 37 °C for 24 h in a CO2 incubator. The cells were serum-starved for 24 h for inhibiting cell proliferation. A straight, uniform wound (∼400 μm in width) was made using a micropipette tip on the monolayer, and the cells were maintained in serum-free DMEM. The wound closure of cells in response to the immobilized fibronectin was measured for 25 h by time-lapse imaging of at least three different positions across the length of the wound using a Carl Zeiss inverted microscope at ×10 magnification.
Densitometry Quantitation and Statistical Analysis
Densitometric quantitation of scanned images was performed by ImageJ 1.43 software (National Institutes of Health). Band intensities were normalized to respective loading controls. Statistical analysis was performed using GraphPad Prism 5. Significance was analyzed by unpaired Student's t test for two samples and two-way analysis of variance for grouped data. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 were considered significant. Photoshop (CS2, Adobe) was used for preparing the figures.
Results
Phosphorylation-induced Solubility of Keratin 18 Is Dependent on Its O-GlcNAcylation at Serine 30
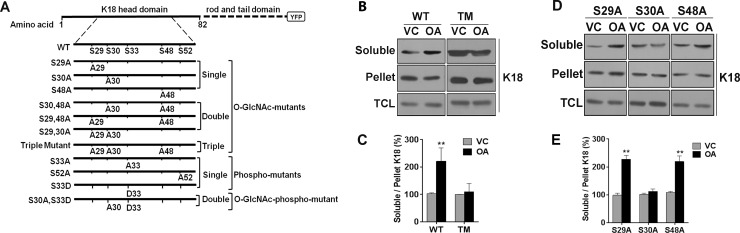
Because both O-GlcNAcylation and phosphorylation on keratin 18 are known to increase its solubility (17), we investigated whether perturbing keratin 18 O-GlcNAcylation alters its phosphorylation-dependent solubility. For this purpose, we generated stable lines of HHL17 cells expressing various O-GlcNAcylation and phosphorylation mutants of K18, as depicted in Fig. 1A. The expression of the YFP-tagged K18 transgene in these stable lines was confirmed by immunofluorescence and immunoblotting (data not shown). To assess phosphorylation-induced solubility, we treated stable cell lines with OA, a broad-spectrum phosphatase inhibitor that increases phosphorylation and thereby the solubility of keratin 8/18 filaments. OA treatment led to a notable increase in solubility of K18-WT, whereas the solubility of K18-TM (S29A/S30A/S48A) was unaltered (Fig. 1, B and C). Similarly, stable HHL17 cells expressing K18-S29A and K18-S48A mutants exhibited an increase in solubility, whereas mutation of Ser30 prevented OA-induced solubility of K18 (K18-S30A) (Fig. 1, D and E), similar to the triple O-GlcNAc mutant of K18 (Fig. 1, B and C). This suggests that phosphorylation-induced solubility of K18 is dependent on its O-GlcNAcylation at Ser30.
FIGURE 1.
Phosphorylation-dependent solubility is impaired in Ser30O-GlcNAc mutants of keratin 18. A, schematic outlining YFP-tagged keratin 18, the WT, and various mutants of O-GlcNAcylation sites (gSer29, gSer30, and gSer48) and phosphorylation sites (Ser(P)33 and Ser(P)52) used to generate stable HHL17 lines. B and D, similar numbers of cultured cells were treated for 2 h either with DMSO vehicle control (VC) or OA to extract the soluble and pellet fraction or total cell lysate (TCL) in identical volumes. Three parts of the soluble fraction (45 μl) and one part each of pellet and total cell lysate (15 μl) were immunoblotted for K18. B, Fractions of HHL17 cells expressing K18-WT and K18-TM and D, Fractions of HHL17 cells expressing single O-GlcNAc mutants of K18 (S29A, S30A, and S48A). C and E, densitometric quantification (average of three independent experiments, including B and D) indicating the percent change in keratin 18 solubility, which is calculated as the ratio of K18 in soluble and pellet fractions. The ratios in VC were taken as 100%. **, p < 0.01; unpaired Student's t test. Error bars represent mean ± S.E.
O-GlcNAcylation at Serine 30 Regulates the Stability of K18
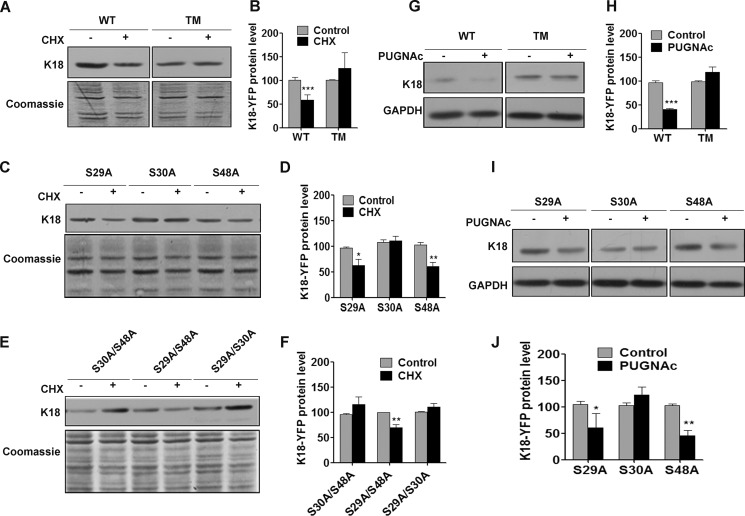
O-GlcNAcylation of keratin 18 reduces its stability by mediating ubiquitination-dependent proteasomal degradation (17). Because disassembly and sequestration of keratin 18 subunits into a soluble pool could be a prerequisite for its degradation (43), we predicted that K18 O-GlcNAcylation at serine 30 could target it for degradation and reduce its stability. The stability of the WT and various K18 O-GlcNAcylation mutants was assessed after inhibiting protein synthesis with cycloheximide. Compared with K18-WT, which shows a significant reduction upon CHX treatment, the level of K18-TM remained unchanged (Fig. 2, A and B). It was interesting to note that, among all K18 O-GlcNAc mutants, those that harbored S30A were highly stable with an unaltered protein level upon cycloheximide treatment (Fig. 2, C–F). To test whether increasing O-GlcNAcylation at Ser30 could reduce the stability of K18, K18-WT and various O-GlcNAc mutant-expressing cells were treated with PUGNAc, a potent O-GlcNAcase inhibitor. PUGNAc treatment is known to cause increased O-GlcNAcylation and subsequent degradation of K8/18 (17). However, K18-TM, which lacks all sites of glycosylation, exhibited resistance to PUGNAc-mediated degradation (data not shown) (17). Both K18 mutants with the S30A mutation (K18-TM and K18-S30A) exhibited no reduction upon PUGNAc treatment, whereas K18 with Ser30 (K18-WT, K18-S29A, and K18-S48A) showed a marked reduction in protein levels (Fig. 2, G–J). These results conclusively indicate that O-GlcNAcylation specifically at Ser30 determines the stability of K18.
FIGURE 2.
O-GlcNAcylation at Ser30 regulates the stability of K18. A, C, and E, K18 immunoblots of equal total proteins from CHX-treated (200 μm for 24 h) or control HHL17 cells stably expressing K18-WT or TM (A) or single O-GlcNAc mutants (C) of K18 and double O-GlcNAc mutants of K18 (E). Coomassie staining of blots serves as a loading control. B, D, and F, densitometric quantification (average of three independent experiments, including A, C, and E) indicating changes in total keratin 18 levels in control and CHX-treated cells (***, p < 0.001 (B); *, p < 0.05 (D); **, p < 0.01 (F); unpaired Student's t test). G and I, K18 immunoblot of equal total proteins from PUGNAc-treated (100 μm for 48 h) or control HHL17 cells expressing K18- WT or TM (G) and single O-GlcNAc mutants (I) of K18. The GAPDH immunoblot served as a loading control. H and J, densitometric quantification of G and I, indicating changes in total keratin 18 levels in control and PUGNAc-treated cells (average of three independent experiments, including G and I) (***, p < 0.001 (H); *, p < 0.05; **, p < 0.01 (J); unpaired Student's t test). Error bars represent mean ± S.E.
O-GlcNAcylation and Phosphorylation Exhibit Both a Cooperative and Antagonistic Relationship at Proximal Sites on K18
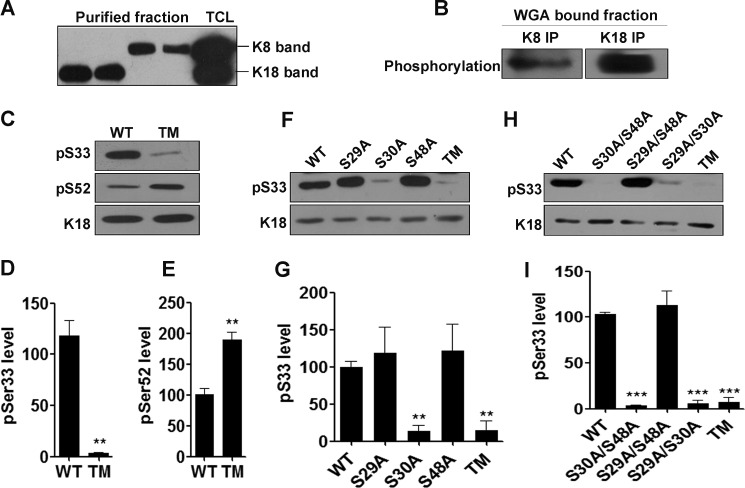
Both O-GlcNAcylation and phosphorylation on keratin 18 correlate positively with solubility (17). To address whether these two modifications on keratin 18 exhibit cross-talk or act independently to regulate K18 properties, we first investigated whether these two modifications coexist on K18. To achieve this, keratin 8 and keratin 18 were separated, purified, and renatured from preparative SDS-PAGE gels (as described under “Experimental Procedures”). The purified K8 and K18 fractions were clean, with no detectable cross-contamination of keratin 8 and 18 isoforms (Fig. 3A). O-GlcNAcylated proteins from these fractions were purified on WGA beads, followed by immunoprecipitation to enrich O-GlcNAcylated K8 and K18. Purified O-GlcNAcylated K8 and K18 showed notable levels of phosphorylation (Fig. 3B), suggesting co-existence of these two modifications on K8 and K18. We further aimed to investigate the relationship/cross-talk between these two modifications on K18 by assessing the levels of site-specific phosphorylation (Ser(P)33 and Ser(P)52) on various O-GlcNAc mutants of K18. The site-specific K18-Ser(P)33 and K18-Ser(P)52 phosphoantibodies have been reported previously (5, 40). The specificity of K18-Ser33 phosphoantibody was further confirmed using cells expressing phosphorylation mutants of K18 (K18-S33A and S52A) (data not shown). K18-TM showed a significant reduction in basal levels of Ser(P)33, whereas the levels of Ser(P)52 were notably higher compared with K18-WT (Fig. 3, C–E), suggesting that O-GlcNAcylation on K18 exhibits a cooperative relationship with Ser(P)33 and a reciprocal relationship with Ser(P)52. The existence of an inverse relationship between gSer48 and Ser(P)52 on K18 has been indicated earlier in in vitro studies (44). To investigate the O-GlcNAc site/s on K18 that aid in phosphorylation of Ser33, we assessed the basal levels of Ser(P)33 in various single and double O-GlcNAc mutants of K18. Interestingly, all O-GlcNAc mutants of K18 harboring S30A exhibited significantly lower levels of Ser(P)33 (Fig. 3, F–I), suggesting a dependence on O-GlcNAcylation of K18 at Ser30 for the occurrence of phosphorylation at Ser33.
FIGURE 3.
O-GlcNAcylation at serine 30 positively regulates phosphorylation of K18 at serine 33. Immunoblots of K8, K18, O-phosphorylation, Ser(P)33 (IB4), and Ser(P)52 (3055) as indicated. A, K8 and K18 immunoblot of purified keratin 8 and 18 fractions (as described under “Experimental Procedures”) to ensure the purity of the preparation. Total cell lysate (TCL) served as a marker to indicate the molecular weight of K8 and K18. B, K8 and K18 were immunoprecipitated from WGA-bound K8 or K18 fractions and immunoblotted with phospho-Ser/Thr. C, F, and H, total cell lysates of HHL17 cells stably expressing K18-WT and TM (C), single O-GlcNAc mutants of K18 (F), and double O-GlcNAc mutants (H) of K18 were immunoblotted with K18Ser(P)33 (IB4) or K18Ser(P)52 (3055) antibodies. D, E, G, and I, densitometric quantification (average of three independent experiments, including C, F, and H) showing levels of K18 Ser(P)33 and K18 Ser(P)52 (**, p < 0.01, unpaired Student's t test (D and E); **, p < 0.01; *** p < 0.001; one-way analysis of variance (G and I). Error bars represent mean ± S.E.
O-GlcNAcylation at Ser30 Determines the Occurrence of Phosphorylation at Ser33 on K18
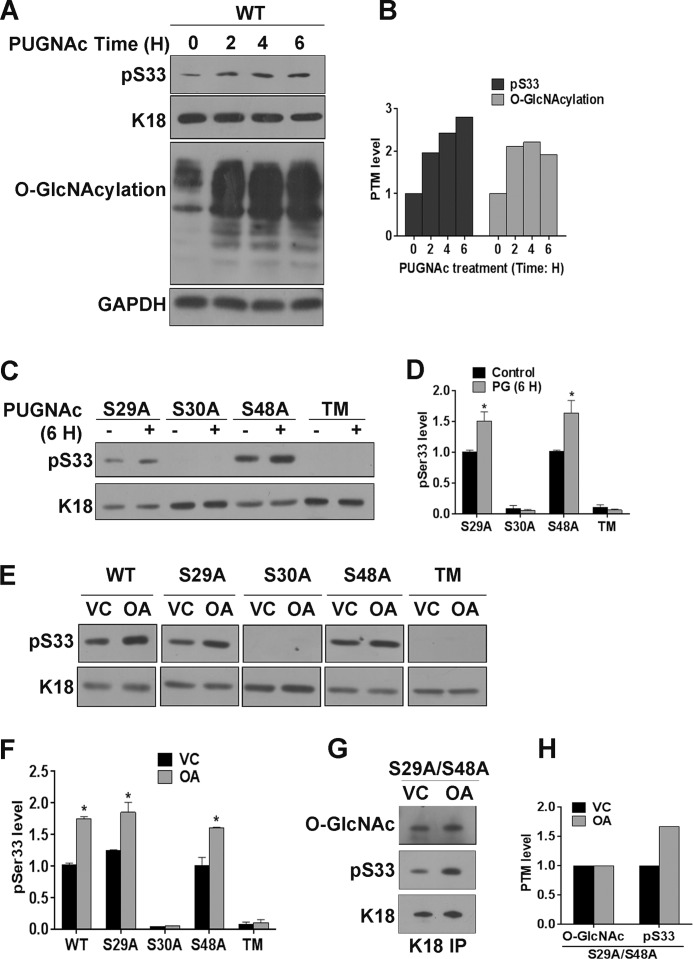
Because abrogation of O-GlcNAcylation of K18 at Ser30 prevents its phosphorylation at Ser33, we set to investigate whether this cooperative relationship can be observed when O-GlcNAc levels on K18 are altered dynamically. PUGNAc treatment of K18-WT-expressing cells led to a time-dependent increase in total O-GlcNAcylation with a concomitant increase in phosphorylation of K18 at Ser33 (Fig. 4, A and B). This PUGNAc-mediated increase in Ser(P)33 levels was seen only in K18 mutants where Ser30 O-GlcNAcylation site was available, viz. K18-S29A and K18-S48A (Fig. 4, C and D). These results confirm that O-GlcNAcylation of K18 at Ser30 can positively influence phosphorylation at Ser33. Because the dynamics of phosphorylation are regulated by the activities of specific kinases and phosphatases acting on that site, we predicted that inhibition of phosphatases by OA could restore Ser(P)33 levels in K18-S30A mutants. OA treatment led to a notable increase in Ser(P)33 levels on K18-WT and K18 O-GlcNAc mutants where Ser30 O-GlcNAcylation can occur, viz. K18-S29A and K18-S48A (Fig. 4, E and F). This suggests that O-GlcNAcylation of K18 at Ser30 is essential for the occurrence of phosphorylation at serine 33. We aimed to further investigate whether this cooperativity is mutual, with Ser(P)33 regulating O-GlcNAcylation at Ser30. To address this, we used stable cells where the K18 mutant can be O-GlcNAcylated only at serine 30 (K18-S29A/S48A) because antibodies recognizing site-specific O-GlcNAcylation on keratin 18 are not available. It was interesting to note that treating these cells with OA led to an increase in Ser(P)33 with no notable change in O-GlcNAcylation at serine 30 (Fig. 4, G and H). This suggests that the cooperative interplay between modifications at these two sites is one-sided, with gSer30 acting as an upstream switch for phosphorylation at Ser33.
FIGURE 4.
O-GlcNAcylation of keratin 18 at serine 30 supports dynamic phosphorylation at serine 33. A, C, and E, immunoblot of equal total proteins from HHL-17 cells expressing WT or O-GlcNAc mutants. A, cells expressing K18-WT were treated with PUGNAc for different times (0, 2, 4 and 6 h). VC, vehicle control. Shown are cells expressing single and triple O-GlcNAc mutants treated with PUGNAc for 6 h (C) and OA for 2 h (E). B, densitometric analysis of total O-GlcNAcylation and K18 phosphorylation at Ser33 in A. D and F, densitometric analysis of Ser(P)33 levels on K18 (average of three independent experiments, including C and E; *, p < 0.05; Student's t test). Error bars represent mean ± S.E. G, immunoprecipitate (IP) of the double O-GlcNAc mutant K18-S29A/S48A immunoblotted with O-GlcNAc, Ser(P)33, and K18. H, densitometric analysis of O-GlcNAcylation and Ser(P)33 on K18. The K18 immunoblot served as a control for loading.
O-GlcNAcylation-mediated Phosphorylation of Keratin 18 at Serine 33 Regulates Its Stability
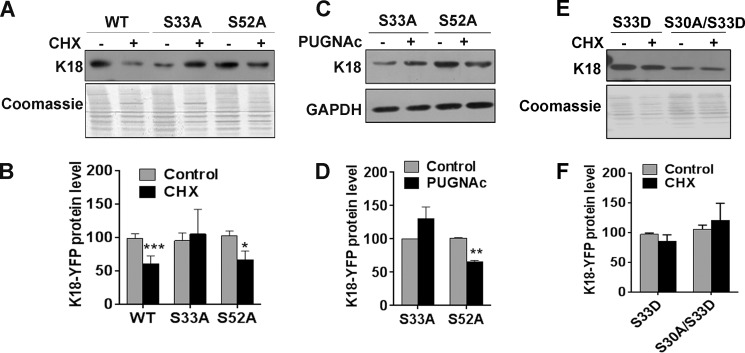
Because O-GlcNAcylation at Ser30 induces solubility and subsequent degradation of K18 along with increased phosphorylation at Ser33 (Figs. 1–4), it is possible that Ser(P)33 mediates the effect of gSer30 on the solubility and stability of K18. Although phosphorylation at Ser33 has been shown previously to regulate solubility (5, 45) and shear stress-induced reorganization of keratin 8/18 filaments (14), its role in regulating the stability of keratin 18 is still unclear (46). To investigate the role of site-specific phosphorylation on K18 in regulating its stability, cells expressing K18-WT and phosphorylation mutants (K18-S33A and K18-S52A) were treated with cycloheximide. Although K18-WT and K18-S52A exhibited reduced K18 levels upon cycloheximide treatment, the protein levels of K18-S33A remained unchanged, suggesting a role for Ser(P)33 in regulating the stability of K18 (Fig. 5, A and B). It is possible that O-GlcNAcylation at serine 30 can influence the stability of K18 either directly by aiding ubiquitination or indirectly by stabilizing Ser(P)33, which would lead to increased solubility and subsequent degradation. To understand this, stable cells expressing K18 phosphorylation mutants (K18-S33A and K18-S52A) were treated with PUGNAc to increase O-GlcNAcylation on K18. PUGNAc treatment led to reduced levels of K18-S52A but not K18-S33A (Fig. 5, C and D). These results indicate that O-GlcNAcylation at Ser30 regulates the solubility and stability of keratin 18 mainly by stabilizing Ser(P)33. To confirm this, we assessed the stability of K18 after rescuing the phosphorylation at Ser33 by substituting with a phosphomimetic mutation (S33D). However, both K18-S33D and K18-S30A/S33D showed higher stability, as assessed by cycloheximide treatment, suggesting that S33D does not substitute for phosphorylation at Ser33 and therefore could not rescue the enhanced stability of K18 upon loss of phosphorylation at Ser33 (Fig. 5, E and F).
FIGURE 5.
Phosphorylation at Ser33 regulates the stability of keratin 18. A and E, equal total proteins from untreated or CHX-treated (200 μm for 24 h) cells expressing WT, S33A, and S52A (A) and S33D and S30A/S33D (E) mutants of K18 immunoblotted with K18. C, untreated or PUGNAc-treated (100 μm for 48 h) cells expressing phosphomutants (S33A and S52A) of K18 were immunoblotted with K18 and GAPDH as indicated. The Coomassie-stained blot in A and E and GAPDH in C served as a loading control. B, D, and F, densitometric analysis indicating relative levels of K18 (average of three independent experiments, including A, C, and E; *, p < 0.05; **, p < 0.01; ***, p < 0.001; Student's t test). Error bars represents mean ± S.E.
O-GlcNAcylation at Serine 30 Is Essential for Maintaining Normal Keratin 18 Filament Organization and Cell Migration
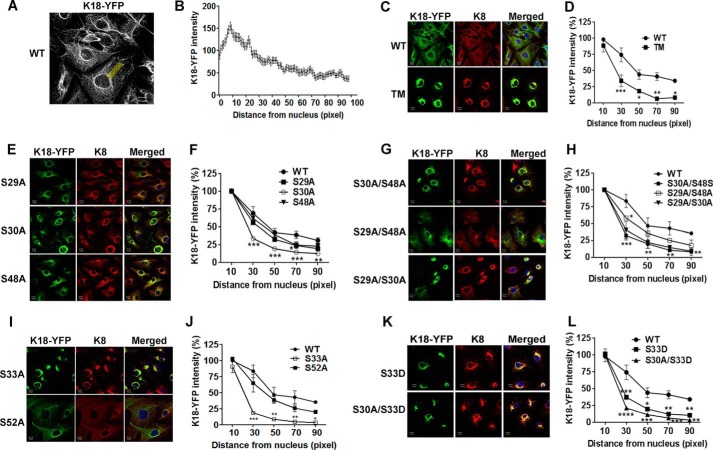
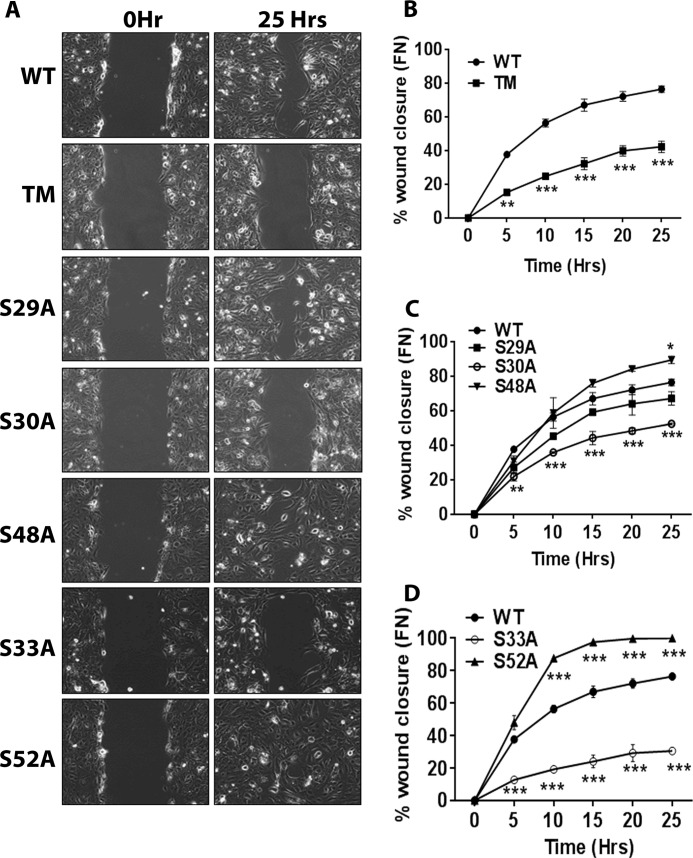
Because gSer30 and Ser(P)33 cooperate to regulate the solubility and stability of K18, we hypothesized that modifications at these two sites could also regulate filament organization of K18. Intensity quantification of K18-WT filaments across the cell reveal that filament density is highest around the nucleus and gradually decreases toward the cell periphery (Fig. 6, A and B) (19). This organization was severely affected in both K18-TM and K18-S30A, which displayed increased filament accumulation around the nucleus with collapse of peripheral filaments, whereas the filament organization of the K18-S29A and K18-S48A mutants was similar to K18-WT (Fig. 6, C–F). Moreover, rescue of O-GlcNAcylation at Ser30 in K18-S29A/S48A could restore the abnormal filament organization of K18-TM (Fig. 6, G and H), implying a key role for gSer30 in supporting the filament organization of K18. Because gSer30 on K18 leads to phosphorylation at Ser33, we predicted that loss of Ser(P)33 could phenocopy the filament organization of K18-S30A and K18-TM. As expected, the K18-S33A mutant exhibited perinuclear aggregation and loss of peripheral filaments, whereas the filament organization of the K18-S52A mutant was similar to K18-WT (Fig. 6, I and J). This is also supported by previous reports that show that K18-Ser(P)33 plays a key role in regulating filament organization and that loss of phosphorylation at this site (K18-S33A) causes the collapse of filaments around the nucleus (5). To confirm these results, we assessed the filament organization in the K18-S33D and K18-S30A/S33D phosphomimetic mutants. The collapsed filament organization was not rescued in either of these stable lines, suggesting that S33D is unable to compensate for phosphorylation at Ser33 (Fig. 6, K and L). These results imply that gSer30-dependent phosphorylation of Ser33 is essential for maintaining the normal filament organization of keratin 18. To further investigate the cellular impact of the cooperativity between gSer30 and Ser(P)33, we assessed the migration of site-specific O-GlcNAcylation and phosphorylation mutants on fibronectin substrate in a scratch wound assay. It is interesting to note that only S30A and S33A O-GlcNAc and phosphomutants of K18, respectively, exhibited reduced migration and wound closure compared with the WT (Fig. 7, A–D). The other O-GlcNAcylation and phosphorylation mutants of K18 (S29A, S48A, and S52A) showed migration similar to K18-WT (Fig. 7, A–D). These results further highlight the importance of Ser30 O-GlcNAcylation-mediated Ser33 phosphorylation on keratin 18 in modulating cellular processes like migration.
FIGURE 6.
O-GlcNAcylation of K18 at Ser30 is essential for maintaining normal filament organization. A, immunofluorescence image of cells expressing WT-K18, demonstrating the method adopted to quantitate K18-YFP intensity from the perinuclear region to the cell periphery. Fluorescence intensity across a straight line of uniform length and width (100 and 30 pixels, respectively) was measured at three different regions per cell. B, fluorescence intensity of the keratin network across a cell (perinuclear region to cell periphery). Each point represents mean ± S.E. of three independent experiments with 20 cells/experiment. C, E, G, I, and K, immunofluorescence images of cells expressing K18-WT and K18-TM (C), single O-GlcNAc mutants (E), double mutants (G), phosphomutants (I), and phosphomimetic mutant (K) of K18. Images show K18-YFP (green), K8 (red), and merged images with DAPI (blue). Scale bars = 10 μm. D, F, H, J, and L, quantification of fluorescence intensity of the K18-YFP filament network of cells in C, E, G, I, and K (average of three independent experiments; 20 cells/experiment; *, p < 0.05; **, p < 0.01; ***, p < 0.001; two-way analysis of variance followed by Bonferroni post-tests). Error bars represent mean ± S.E.
FIGURE 7.
O-GlcNAcylation and phosphorylation of K18 regulate the cellular migration on fibronectin. A, comparison of migration of HHL17 cells stably expressing K18-WT and the indicated O-GlcNAcylation and phosphorylation mutants of K18 by scratch wound assay on fibronectin (FN). B, C, and D, quantitation of percent wound closure of two independent experiments, each experiment at three different points. *, p < 0.05; **, p < 0.01; ***, p < 0.001; two-way analysis of variance followed by Bonferroni post-tests). Error bars represent mean ± S.E.
Discussion
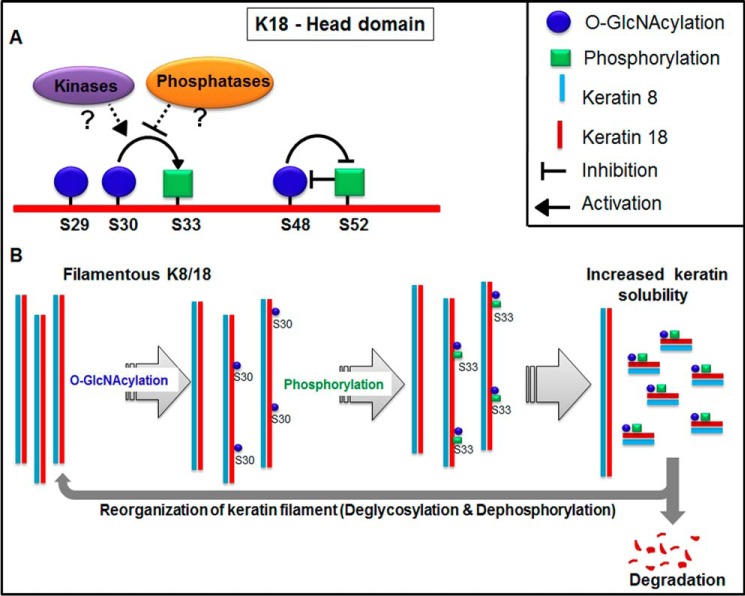
On most phosphoglycoproteins discovered so far, O-GlcNAcylation and phosphorylation antagonize each other in site occupancy and regulation of protein functions (47). However, this seems unclear for K18 because both of these modifications exhibit similar changes under certain conditions, like mitotic arrest (48) and heat stress (49), whereas, during induced hepatotoxicity, they exhibit an inverse relationship (50). Of the two phosphorylation sites mapped on keratin 18, Ser(P)33 causes increased solubility during mitosis and filament reorganization in response to shear stress (5, 14), whereas Ser(P)52 is known to regulate filament reorganization under stress conditions (51). Mutating the three known O-GlcNAcylation sites of K18 (Ser29, Ser30, and Ser48 to Ala) led to altered filament organization and decreased both solubility and degradation of keratin 18 (17). However, it is still unclear whether this regulatory role of O-GlcNAcylation is site-specific and whether it is dependent on its cross-talk with phosphorylation. Here we demonstrate a hitherto unreported, positive cross-talk between Ser30 O-GlcNAcylation and Ser33 phosphorylation on K18, and this cross-talk is responsible for regulating K18 solubility, filament organization, and stability (Fig. 8).
FIGURE 8.
Model depicting cross-talk between O-GlcNAcylation and phosphorylation at proximal sites on keratin 18 to regulate the solubility, degradation, and reorganization of the keratin filament network. A, O-GlcNAcylation at serine 30 facilitates proximal phosphorylation at serine 33 either by aiding the interaction with specific kinases or hindering the interaction with phosphatases. O-GlcNAcylation at serine 48 exhibits a mutually exclusive relationship with phosphorylation at serine 52. B, serine 30 O-GlcNAcylation of keratin 18 filaments promotes stabilization of serine 33 phosphorylation, which leads to filament disassembly and a concomitant increase in soluble keratin 18. The soluble keratin 18 subunits are either targeted for proteasomal degradation or incorporated into the filament network upon loss of O-GlcNAcylation and phosphorylation at serines 30 and 33, respectively.
Solubility of the K8/18 network is known to play an essential role in degradation because filament or aggregated K8/18, although ubiquitinated, cannot be degraded by proteasomes unless they are disassembled into soluble form (43). Phosphorylation at serine 33 plays an important role in the solubility and reorganization of keratin 8/18 filament network mainly during mitosis and shear stress (5, 14). Moreover, it is also a binding site for the 14-3-3 class of proteins, which further aids in the solubility of K8/18 filaments (45). In addition to this, OA-mediated inhibition of phosphatases led to an increase in Ser(P)33 levels along with enhanced binding to 14-3-3 in HT-29 epithelial cells (5, 45), whereas abrogation of phosphorylation at Ser33 had no effect on OA-induced solubility of the K18-S33A mutant when expressed in NIH3T3 fibroblasts (5). Although these reports strongly indicate a role for Ser(P)33 in mediating K18 solubility, they also point to varied effects of modifications on the solubility of keratin 18 in different cell types (5, 45). Of the three sites of O-GlcNAcylation, it was interesting to observe that loss of gSer30 was enough to stall the phosphorylation-induced solubility during OA treatment (Fig. 1). Further to this, the loss of gSer30 also led to an increased stability of K18 (Fig. 2), suggesting that O-GlcNAcylation at Ser30 could be central to not only phosphorylation-dependent solubility but also subsequent degradation. On most proteins, O-GlcNAcylation protects against proteasomal degradation either directly or by antagonizing phosphorylation (52). O-GlcNAcylation could also target the protein for degradation, as in the case of the kinase CK2, where O-GlcNAcylation at Ser347 leads to degradation by antagonizing phosphorylation at Thr344 (53). In either of the cases, O-GlcNAcylation regulates protein stability by antagonizing phosphorylation. Interestingly, this seemed contrary on K18 because phosphorylation-induced solubility and degradation were totally dependent on O-GlcNAcylation at serine 30 (Figs. 1 and 2).
The co-existence of both these modifications on K18 seems plausible because they exhibit similar changes during conditions associated with increased solubility and filament reorganization. Indeed, the presence of notable phosphorylation on O-GlcNAcylated K8 and K18 species confirms the co-occurrence of these modifications (Fig. 3B). However, because of the presence of multiple sites of both these modifications, they could exhibit a mutually exclusive relationship at proximal sites but can still co-exist at distal sites. Further analysis using site-specific K18 O-GlcNAc mutants along with site-specific K18 phosphorylation antibodies indicate both a cooperative and antagonistic interplay between these modifications at proximal sites (Fig. 3, C–E). Although O-GlcNAcylation at Ser30 promotes phosphorylation at Ser33 (Fig. 3, F–I), O-GlcNAcylation at Ser48 antagonizes phosphorylation at Ser52 (data not shown). Although a previous study supports the presence of a reciprocal relationship between gSer48 and Ser(P)52 (44), the existence of a promotive/cooperative relationship between gSer30 and Ser(P)33 is both interesting and novel. Furthermore, global high-throughput phosphoproteomics upon elevated O-GlcNAc levels does support the prevalence of such a non-reciprocal, co-operative interplay on many proteins (36). Such a type of synergistic relationship between O-GlcNAcylation and phosphorylation exists in the catalytic cleft of the calcium/calmodulin-dependent kinase IV (CAMKIV) kinase, albeit at distal sites, where mutation of the glycosylation sites Thr57/Ser58 to alanine on calcium/calmodulin-dependent kinase IV causes a drastic reduction in basal activating phosphorylation at Thr200 (35).
This cooperative interplay between gSer30 and Ser(P)33 not only responds to an acute pharmacological increase in O-GlcNAcylation but also seems to be unidirectional, with gSer30 acting as a switch for phosphorylation at Ser33 (Fig. 4). Thus, O-GlcNAcylation at Ser30 can facilitate phosphorylation at Ser33 either by aiding or hindering interactions with kinases and phosphatases, respectively. The possibility of gSer30 facilitating interaction with kinase(s) seems more probable because the loss of Ser(P)33 in K18-TM and S30A mutants could not be restored upon inhibiting phosphatases (Fig. 4, E and F). It is also important to note that OA-induced enhanced association of 14-3-3 with K18 is marginally reduced but not completely diminished in the triple glycosylation mutant when expressed in baby hamster kidney fibroblasts (5). Although this suggests that Ser33 could still be basally phosphorylated in the absence of glycosylation at Ser30, it also hints at the possibility of different kinases involved in phosphorylating Ser33 in different cell systems (5, 14, 54). Overall, these results suggest that gSer30 controls the solubility and degradation of K18 by stabilizing phosphorylation at Ser33. This seems plausible because phosphorylation on many proteins acts as a signal for ubiquitination-mediated proteasomal degradation (55). Even on K8, shear stress-induced phosphorylation at Ser73 enables E2 ligases to mediate proteasomal degradation (56). Indeed, loss of phosphorylation at Ser33 increased the stability of K18, which could not be rescued by increasing O-GlcNAcylation on K18 (Fig. 5, A–D) or by phosphomimetic substitution at Ser33 (Fig. 5, E and F). Although substitution with negatively charged residues like aspartate rescues loss of Ser/Thr phosphorylation on many proteins, on keratin 18, S33D does not seem to substitute for phosphorylation (Fig. 5, E and F) (5). These findings imply that O-GlcNAcylation at Ser30 predominantly aids in the phosphorylation at Ser33, which in turn causes increased solubility and subsequent degradation of K18.
Phosphorylation at Ser33 is also a key regulator of K18 solubility and filament organization both in cultured cells and hepatocyte in vivo (5, 45, 57). In general, keratins 8/18 exhibit a gradient of filament network with highly bundled filament near the nucleus and fine, thin filaments at the cell periphery (58). As reported previously, expression of K18-S33A mutant led to disruption of filament organization, visualized as highly concentrated perinuclear filaments and a collapsed peripheral filament network, whereas the architecture of K8/18 is unaltered in K18-S52A mutants or K18-WT-expressing cells (Fig. 6, C, D, I, and J) (5). Among the O-GlcNAcylation mutants, only K18-S30A mimicked the perinuclear collapsed network of K18, as seen in K18-S33A phosphomutant (Fig. 6, C–H). The defects in filament organization in K18-S33A and K18-S30A could not be rescued by S33D mutations, suggesting that, similar to K18 stability, S33D cannot substitute for Ser(P)33 to rescue filament organization. These observations further strengthen the regulatory role of gSer30 in promoting Ser(P)33, which in turn dictates the solubility, filament organization, and turnover of keratin 18.
The dynamic reorganization of keratin 8/18 filaments is essential for the plasticity of the keratin filament network during many cellular processes, including cell migration. Rapid changes in keratin network dynamics are initiated especially in the lamellipodia of migrating cells (58). It is also important to note that phosphorylation of keratin 8 alone can play a critical role in mediating the migration of epithelial tumor cells and hepatocytes (9, 59). Interestingly, loss of O-GlcNAcylation at Ser30 or phosphorylation at Ser33 on K18 was sufficient to reduce the migrating potential of hepatocytes, indicating that both gSer30 and gSer33 act cooperatively to impact filament dynamics during cell migration (Fig. 7).
Taken together these results demonstrate a synergism between S30 O-GlcNAcylation and Ser33 phosphorylation on keratin 18 in regulating its functional properties like solubility, filament organization, and stability (Fig. 7). The cycling of O-GlcNAc on cellular proteins is tightly linked to glucose metabolism (60, 61) and is often deregulated in multiple pathological conditions like diabetes and cancer (32). These findings raise the possibility of factors like glucose metabolism to regulate keratin 8/18, mainly in metabolically active cells like hepatocytes. The positive regulation of phosphorylation by proximal O-GlcNAcylation opens up new avenue to understand the functional regulation of phosphoglycoproteins. Understanding how O-GlcNAcylation can facilitate phosphorylation on K18 will be important because this type of interplay might be as abundant as reciprocal cross-talk.
Author Contributions
P. S. K. performed the experiments, generated mutants, designed the experiments, analyzed the data, interpreted the results, and wrote the manuscript. S. B. conceived and supervised the work, generated the mutants, performed the purification and immunoprecipitation of WGA-bound (glycosylated) keratins, designed the experiments, supervised data analysis, interpreted the results, and wrote the manuscript. R. D. K. conceived and supervised the work, designed the experiments, supervised data analysis, and wrote the manuscript. M. M. V. supervised the work and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Prof. M. Bishr Omary (University of Michigan Medical School) for the keratin 18 O-GlcNAc mutant (S29A/S30A/S48A) cDNA construct and the K18 phosphorylation site-specific antibodies Ser(P)52 (clone 3055) and Ser(P)33 (clone IB4). We also thank Dr. Rudolf E. Leube and Dr. Reinhard Windoffer (Johannes Gutenberg University) for the YFP-tagged keratin 18 wild-type construct; Dr. A. H. Patel (Institute of Virology, Glasgow, UK) for HHL17 cells; V. Kailaje and T. Dighe for help with confocal imaging; Priyanka Parekh, Geeta Iyer, Laxmi Varma, and Sonam Hatkar for assistance; and D. Chavan and A. Pawar for technical help.
This work was supported by Department of Biotechnology Grant BT/PR3201/MED/30/643/2011 and the Advanced Centre for Treatment, Research and Education in Cancer. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table 1.
- gSer29
- O-GlcNAcylated Ser29
- PUGNAc
- O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate
- WGA
- wheat germ agglutinin
- TM
- triple mutant
- EYFP
- enhanced YFP
- OA
- okadaic acid
- CHX
- cycloheximide.
References
- 1. Schweizer J., Bowden P. E., Coulombe P. A., Langbein L., Lane E. B., Magin T. M., Maltais L., Omary M. B., Parry D. A., Rogers M. A., and Wright M. W. (2006) New consensus nomenclature for mammalian keratins. J. Cell Biol. 174, 169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coulombe P. A., and Omary M. B. (2002) “Hard” and “soft” principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 14, 110–122 [DOI] [PubMed] [Google Scholar]
- 3. Moll R., Divo M., and Langbein L. (2008) The human keratins: biology and pathology. Histochem. Cell Biol. 129, 705–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Raul U., Sawant S., Dange P., Kalraiya R., Ingle A., and Vaidya M. (2004) Implications of cytokeratin 8/18 filament formation in stratified epithelial cells: induction of transformed phenotype. Int. J. Cancer 111, 662–668 [DOI] [PubMed] [Google Scholar]
- 5. Ku N. O., Liao J., and Omary M. B. (1998) Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins. EMBO J. 17, 1892–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vijayaraj P., Kröger C., Reuter U., Windoffer R., Leube R. E., and Magin T. M. (2009) Keratins regulate protein biosynthesis through localization of GLUT1 and -3 upstream of AMP kinase and Raptor. J. Cell Biol. 187, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tao G. Z., Looi K. S., Toivola D. M., Strnad P., Zhou Q., Liao J., Wei Y., Habtezion A., and Omary M. B. (2009) Keratins modulate the shape and function of hepatocyte mitochondria: a mechanism for protection from apoptosis. J. Cell Sci. 122, 3851–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ku N. O., Toivola D. M., Strnad P., and Omary M. B. (2010) Cytoskeletal keratin glycosylation protects epithelial tissue from injury. Nat. Cell Biol. 12, 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Busch T., Armacki M., Eiseler T., Joodi G., Temme C., Jansen J., von Wichert G., Omary M. B., Spatz J., and Seufferlein T. (2012) Keratin 8 phosphorylation regulates keratin reorganization and migration of epithelial tumor cells. J. Cell Sci. 125, 2148–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pan X., Hobbs R. P., and Coulombe P. A. (2013) The expanding significance of keratin intermediate filaments in normal and diseased epithelia. Curr. Opin. Cell Biol. 25, 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loschke F., Seltmann K., Bouameur J. E., and Magin T. M. (2015) Regulation of keratin network organization. Curr. Opin. Cell Biol. 32, 56–64 [DOI] [PubMed] [Google Scholar]
- 12. Omary M. B., Ku N. O., Liao J., and Price D. (1998) Keratin modifications and solubility properties in epithelial cells and in vitro. Subcell. Biochem. 31, 105–140 [PubMed] [Google Scholar]
- 13. Snider N. T., and Omary M. B. (2014) Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 15, 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sivaramakrishnan S., Schneider J. L., Sitikov A., Goldman R. D., and Ridge K. M. (2009) Shear stress induced reorganization of the keratin intermediate filament network requires phosphorylation by protein kinase C ζ. Mol. Biol. Cell 20, 2755–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ridge K. M., Linz L., Flitney F. W., Kuczmarski E. R., Chou Y. H., Omary M. B., Sznajder J. I., and Goldman R. D. (2005) Keratin 8 phosphorylation by protein kinase C delta regulates shear stress-mediated disassembly of keratin intermediate filaments in alveolar epithelial cells. J. Biol. Chem. 280, 30400–30405 [DOI] [PubMed] [Google Scholar]
- 16. Snider N. T., Park H., and Omary M. B. (2013) A conserved rod domain phosphotyrosine that is targeted by the phosphatase PTP1B promotes keratin 8 protein insolubility and filament organization. J. Biol. Chem. 288, 31329–31337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Srikanth B., Vaidya M. M., and Kalraiya R. D. (2010) O-GlcNAcylation determines the solubility, filament organization, and stability of keratins 8 and 18. J. Biol. Chem. 285, 34062–34071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Snider N. T., Weerasinghe S. V., Iñiguez-Lluhí J. A., Herrmann H., and Omary M. B. (2011) Keratin hypersumoylation alters filament dynamics and is a marker for human liver disease and keratin mutation. J. Biol. Chem. 286, 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Snider N. T., Leonard J. M., Kwan R., Griggs N. W., Rui L., and Omary M. B. (2013) Glucose and SIRT2 reciprocally mediate the regulation of keratin 8 by lysine acetylation. J. Cell Biol. 200, 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Torres C. R., and Hart G. W. (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes: evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 [PubMed] [Google Scholar]
- 21. Hart G. W., Housley M. P., and Slawson C. (2007) Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 22. Hurtado-Guerrero R., Dorfmueller H. C., and van Aalten D. M. (2008) Molecular mechanisms of O-GlcNAcylation. Curr. Opin. Struct. Biol. 18, 551–557 [DOI] [PubMed] [Google Scholar]
- 23. Slawson C., Housley M. P., and Hart G. W. (2006) O-GlcNAc cycling: how a single sugar post-translational modification is changing the way we think about signaling networks. J. Cell Biochem. 97, 71–83 [DOI] [PubMed] [Google Scholar]
- 24. Lewis B. A., and Hanover J. A. (2014) O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem. 289, 34440–34448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sayat R., Leber B., Grubac V., Wiltshire L., and Persad S. (2008) O-GlcNAc-glycosylation of β-catenin regulates its nuclear localization and transcriptional activity. Exp. Cell Res. 314, 2774–2787 [DOI] [PubMed] [Google Scholar]
- 26. Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., and Cho J. W. (2006) Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8, 1074–1083 [DOI] [PubMed] [Google Scholar]
- 27. Zhang F., Su K., Yang X., Bowe D. B., Paterson A. J., and Kudlow J. E. (2003) O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115, 715–725 [DOI] [PubMed] [Google Scholar]
- 28. Sakabe K., and Hart G. W. (2010) O-GlcNAc transferase regulates mitotic chromatin dynamics. J. Biol. Chem. 285, 34460–34468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bond M. R., and Hanover J. A. (2015) A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 208, 869–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma Z., and Vosseller K. (2014) Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J. Biol. Chem. 289, 34457–34465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuzwa S. A., Shan X., Macauley M. S., Clark T., Skorobogatko Y., Vosseller K., and Vocadlo D. J. (2012) Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol. 8, 393–399 [DOI] [PubMed] [Google Scholar]
- 32. Slawson C., Copeland R. J., and Hart G. W. (2010) O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci. 35, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Slawson C., and Hart G. W. (2011) O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer 11, 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng X., Cole R. N., Zaia J., and Hart G. W. (2000) Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor β. Biochemistry 39, 11609–11620 [DOI] [PubMed] [Google Scholar]
- 35. Dias W. B., Cheung W. D., Wang Z., and Hart G. W. (2009) Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification. J. Biol. Chem. 284, 21327–21337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Z., Gucek M., and Hart G. W. (2008) Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. U.S.A. 105, 13793–13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dong D. L., Xu Z. S., Chevrier M. R., Cotter R. J., Cleveland D. W., and Hart G. W. (1993) Glycosylation of mammalian neurofilaments: localization of multiple O-linked N-acetylglucosamine moieties on neurofilament polypeptides L and M. J. Biol. Chem. 268, 16679–16687 [PubMed] [Google Scholar]
- 38. Arnold C. S., Johnson G. V., Cole R. N., Dong D. L., Lee M., and Hart G. W. (1996) The microtubule-associated protein tau is extensively modified with O-linked N-acetylglucosamine. J. Biol. Chem. 271, 28741–28744 [DOI] [PubMed] [Google Scholar]
- 39. Ku N. O., and Omary M. B. (1995) Identification and mutational analysis of the glycosylation sites of human keratin 18. J. Biol. Chem. 270, 11820–11827 [DOI] [PubMed] [Google Scholar]
- 40. Liao J., Lowthert L. A., Ku N. O., Fernandez R., and Omary M. B. (1995) Dynamics of human keratin 18 phosphorylation: polarized distribution of phosphorylated keratins in simple epithelial tissues. J. Cell Biol. 131, 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clayton R. F., Rinaldi A., Kandyba E. E., Edward M., Willberg C., Klenerman P., and Patel A. H. (2005) Liver cell lines for the study of hepatocyte functions and immunological response. Liver Int. 25, 389–402 [DOI] [PubMed] [Google Scholar]
- 42. Sehgal L., Budnar S., Bhatt K., Sansare S., Mukhopadhaya A., Kalraiya R. D., and Dalal S. N. (2012) Generation of HIV-1 based bi-cistronic lentiviral vectors for stable gene expression and live cell imaging. Indian J. Exp. Biol. 50, 669–676 [PubMed] [Google Scholar]
- 43. Rogel M. R., Jaitovich A., and Ridge K. M. (2010) The role of the ubiquitin proteasome pathway in keratin intermediate filament protein degradation. Proc. Am. Thorac. Soc. 7, 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tao G. Z., Kirby C., Whelan S. A., Rossi F., Bi X., MacLaren M., Gentalen E., O'Neill R. A., Hart G. W., and Omary M. B. (2006) Reciprocal keratin 18 Ser48 O-GlcNAcylation and Ser52 phosphorylation using peptide analysis. Biochem. Biophys. Res. Commun. 351, 708–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liao J., and Omary M. B. (1996) 14-3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility cofactor. J. Cell Biol. 133, 345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ku N. O., and Omary M. B. (2000) Keratins turn over by ubiquitination in a phosphorylation-modulated fashion. J. Cell Biol. 149, 547–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hart G. W., Slawson C., Ramirez-Correa G., and Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chou C. F., and Omary M. B. (1993) Mitotic arrest-associated enhancement of O-linked glycosylation and phosphorylation of human keratins 8 and 18. J. Biol. Chem. 268, 4465–4472 [PubMed] [Google Scholar]
- 49. Liao J., Lowthert L. A., and Omary M. B. (1995) Heat stress or rotavirus infection of human epithelial cells generates a distinct hyperphosphorylated form of keratin 8. Exp. Cell Res. 219, 348–357 [DOI] [PubMed] [Google Scholar]
- 50. Ku N. O., Michie S. A., Soetikno R. M., Resurreccion E. Z., Broome R. L., Oshima R. G., and Omary M. B. (1996) Susceptibility to hepatotoxicity in transgenic mice that express a dominant-negative human keratin 18 mutant. J. Clin. Invest. 98, 1034–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ku N. O., Michie S. A., Soetikno R. M., Resurreccion E. Z., Broome R. L., and Omary M. B. (1998) Mutation of a major keratin phosphorylation site predisposes to hepatotoxic injury in transgenic mice. J. Cell Biol. 143, 2023–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ruan H. B., Nie Y., and Yang X. (2013) Regulation of protein degradation by O-GlcNAcylation: crosstalk with ubiquitination. Mol. Cell. Proteomics 12, 3489–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tarrant M. K., Rho H. S., Xie Z., Jiang Y. L., Gross C., Culhane J. C., Yan G., Qian J., Ichikawa Y., Matsuoka T., Zachara N., Etzkorn F. A., Hart G. W., Jeong J. S., Blackshaw S., Zhu H., and Cole P. A. (2012) Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat. Chem. Biol. 8, 262–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ku N. O., Fu H., and Omary M. B. (2004) Raf-1 activation disrupts its binding to keratins during cell stress. J. Cell Biol. 166, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 56. Jaitovich A., Mehta S., Na N., Ciechanover A., Goldman R. D., and Ridge K. M. (2008) Ubiquitin-proteasome-mediated degradation of keratin intermediate filaments in mechanically stimulated A549 cells. J. Biol. Chem. 283, 25348–25355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ku N. O., Michie S., Resurreccion E. Z., Broome R. L., and Omary M. B. (2002) Keratin binding to 14-3-3 proteins modulates keratin filaments and hepatocyte mitotic progression. Proc. Natl. Acad. Sci. U.S.A. 99, 4373–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kölsch A., Windoffer R., Würflinger T., Aach T., and Leube R. E. (2010) The keratin-filament cycle of assembly and disassembly. J. Cell Sci. 123, 2266–2272 [DOI] [PubMed] [Google Scholar]
- 59. Bordeleau F., Galarneau L., Gilbert S., Loranger A., and Marceau N. (2010) Keratin 8/18 modulation of protein kinase C-mediated integrin-dependent adhesion and migration of liver epithelial cells. Mol. Biol. Cell 21, 1698–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bond M. R., and Hanover J. A. (2013) O-GlcNAc cycling: a link between metabolism and chronic disease. Annu. Rev. Nutr. 33, 205–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zachara N. E., and Hart G. W. (2004) O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 1673, 13–28 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.