Abstract
Herpesviruses are among the most complex and widespread viruses, infection and propagation of which depend on envelope proteins. These proteins serve as mediators of cell entry as well as modulators of the immune response and are attractive vaccine targets. Although envelope proteins are known to carry glycans, little is known about the distribution, nature, and functions of these modifications. This is particularly true for O-glycans; thus we have recently developed a “bottom up” mass spectrometry-based technique for mapping O-glycosylation sites on herpes simplex virus type 1. We found wide distribution of O-glycans on herpes simplex virus type 1 glycoproteins and demonstrated that elongated O-glycans were essential for the propagation of the virus. Here, we applied our proteome-wide discovery platform for mapping O-glycosites on representative and clinically significant members of the herpesvirus family: varicella zoster virus, human cytomegalovirus, and Epstein-Barr virus. We identified a large number of O-glycosites distributed on most envelope proteins in all viruses and further demonstrated conserved patterns of O-glycans on distinct homologous proteins. Because glycosylation is highly dependent on the host cell, we tested varicella zoster virus-infected cell lysates and clinically isolated virus and found evidence of consistent O-glycosites. These results present a comprehensive view of herpesvirus O-glycosylation and point to the widespread occurrence of O-glycans in regions of envelope proteins important for virus entry, formation, and recognition by the host immune system. This knowledge enables dissection of specific functional roles of individual glycosites and, moreover, provides a framework for design of glycoprotein vaccines with representative glycosylation.
Introduction
Herpesviridae is a family of enveloped viruses that infect a wide range of hosts (1), including humans, causing clinical manifestations of varying severity. Herpesvirus infection is characterized by a primary lytic infection followed by a lifelong latency established in the host (2). The Herpesviridae family is classified into three subfamilies: alphaherpesviruses (HSV-1, HSV-2, and VZV),2 betaherpesviruses (HCMV, HHV-6, and HHV-7), and gammaherpesviruses (EBV and Kaposi's sarcoma-associated herpesvirus) (3). Although most herpesvirus infections are self-limiting, they can lead to severe complications, particularly in immunocompromised patients (4–9). Furthermore, gammaherpesvirus infections have been associated with cancer development (10, 11).
Herpesviruses have large genomes, encoding from ∼70 to more than 230 distinct viral proteins depending on the virus type (12). The viral proteins include enzymes involved in viral DNA replication, proteins that form the viral particle, and viral envelope glycoproteins (3). The viral envelope glycoproteins play major roles in the early interactions, attachment, and penetration of the viral particle into the host cell and are involved in modulation of the immune system necessary to maintain the lifelong relationship between a herpesvirus and the infected host (13–15).
The envelope glycoproteins are processed and modified in the secretory compartment of the host cell where they are decorated with glycans that contribute to their biological properties (16). The most common classes of glycans found on viral envelope proteins are N-linked glycans, attached to asparagine residues of the polypeptide, and GalNAc-type O-linked glycans, attached to serine, threonine, or tyrosine residues (hereafter referred to as O-linked glycans) (17, 18). Whereas the functions and structures of N-linked glycans of viral glycoproteins have been elucidated in detail for most enveloped viruses, including herpesviruses, the distribution, structures, and functions of the O-glycans on viral glycoproteins have mostly remained elusive (19–28). This is largely due to lack of a reliable prediction algorithm and analytical difficulties in characterizing O-glycan sites on a proteome-wide basis. Moreover, the O-glycosylation capacity of cells varies, and thus analysis of O-glycosylation of a virus needs to take the host cell type into account. O-Glycosylation is initiated by up to 20 polypeptide GalNAc transferase isoforms with distinct substrate specificities, and cells express different subsets of these isoforms to regulate the O-glycosylation capacity (18, 29, 30). Furthermore, a large repertoire of elongating and branching enzymes creates a heterogeneous pool of O-glycan structures within a given cell and even at a given site, making it technically challenging to identify O-glycosylation sites in the right biological context. These characteristics of O-glycosylation have all contributed to the elusive nature of site localization. However, our recent introduction of a mass spectrometry-based proteome-wide discovery platform for mapping viral O-glycosylation sites has changed this view considerably (31, 32). Using HSV-1 and HSV-2 as models, we uncovered an unprecedented number of O-glycosites in functionally relevant regions of viral envelope glycoproteins (31, 33). In contrast to the general consensus, we also demonstrated that HSV-1 envelope glycoproteins carry more O-glycans than N-glycans (31).
O-Linked glycans may be found as single scattered glycans, or they may be concentrated in dense clusters of glycans in mucin-like domains of viral glycoproteins (31, 33). Important biological functions are associated with both types of glycans. For instance, clustered O-glycans of the distinct HSV-1 glycoprotein C have been found to be necessary not only for adjusting viral binding to its initial receptor, heparan sulfate, but also for preventing progeny virus from entrapment on the dying cell in which the virus replicated (21). An example of the functional role of single O-glycans emerged with the demonstration of a few O-glycosites on HSV-1 gB involved in the interaction with entry receptor paired immunoglobulin-like type 2 receptor α, possibly important for immune evasion (20, 31). Altogether, such data, including our recent demonstration of the importance of O-glycans for HSV-1 and HSV-2 propagation as well as HSV-2 immune sensing (31, 33, 34), emphasize important functions of O-glycans on herpesvirus envelope glycoproteins.
Based on these findings, we hypothesized that other herpesvirus family members may also be extensively O-glycosylated. To provide the first global map of O-glycosylation of a large virus family, we in the present study characterized the O-glycoproteomes of three clinically relevant Herpesviridae family members: VZV, HCMV, and EBV. Using our proteome-wide discovery platform, we here identified a large number of O-glycosylation sites in multiple glycoproteins from the three viruses. In addition, we for the first time report the O-glycoproteome obtained from a clinical VZV specimen. The presented data sets serve as a resource for exploring biological functions of specific O-glycan sites and structures, as well as a reference for design and testing of vaccines.
Experimental Procedures
Cells and Viruses
Diploid human embryonic lung fibroblasts (35) (HEL, obtained from the cell culture collection at the Sahlgrenska University Hospital, Department of Clinical Microbiology, Gothenburg, Sweden) at a low passage level were cultivated in Eagle's minimum essential medium (Gibco, Life Technologies) with 10% FCS (Sigma), 100 IU/ml penicillin, 100 μg/ml streptomycin (Gibco, Life Technologies), and 2 mm l-glutamine. P3HR1 (ATCC HTB-62) B lymphocytes isolated from Burkitt's lymphoma were maintained at a concentration between 4 × 105 and 8 × 105 cells/ml in RPMI 1640 (Gibco, Life Technologies), supplemented as above. All cells were maintained at 37 °C and 5% CO2. The HCMV laboratory strain Towne (ATCC-977) was used throughout the study. The virus titers were determined by plaque titration on HEL cells as previously described (36). A VZV patient isolate C821 that was typed by PCR (37) and subsequently passaged in HEL cells was used for the VZV cell culture experiments. The patient VZV isolate DE14 8565 from a zoster blister was obtained with a specimen collection swab (eSwab; Copan Diagnostics, Murrieta, CA) and stored in 1 ml of Amies medium until preparation. Written informed consent was obtained from the patient prior to sampling. Except for age, no clinical information about the patient was registered. Because patient and sample identity was anonymized, ethical approval was not required.
VZV Infection in Cell Culture
Cell-associated VZV (180,000 particles/cell as determined by quantitative PCR) was added to HEL fibroblasts in T175 cell culture flasks (6×). 5 ml of VZV-infected HEL fibroblasts was added to each T175 cell culture flask, generating a final ratio of 0.25 VZV-infected cells per uninfected HEL fibroblast. The virus was allowed to attach to the cells for 3 h at 37 °C and 5% CO2 before the inoculum was removed and fresh growth medium was added. The cells were incubated for 3–6 days until a strong cytopathic effect (+++ CPE) was detected. The cells were washed with ice-cold PBS and harvested by scraping with a rubber policeman in ice-cold PBS, followed by centrifugation (500 × g 10 min at 4 °C).
HCMV Infection in Cell Culture
HCMV Towne at a multiplicity of infection of 0.1 pfu/cell was added to confluent monolayers of HEL fibroblasts in T175 cell culture flasks (5×). The viral particles were allowed to attach to the cells for 1 h at 37 °C and 5% CO2 before the inoculum was removed, and new growth medium was added. The cells were harvested after 14 days by scraping with a rubber policeman in ice-cold PBS, followed by centrifugation (500 × g 10 min at 4 °C).
Stimulation of P3HR1 Cells for EBV Activation
4 × 107 to 8 × 107 pelleted cells (216 × g for y5 min at room temperature) were resuspended in 100 ml of growth medium supplemented with 20 ng/ml phorbol 12-myristate 13-acetate and 3 mm sodium butyrate (Sigma-Aldrich) in a 165-cm2 culture flask. The cells were incubated at 37 °C and 5% CO2 for 4 days and then harvested by centrifugation (320 × g for 10 min at 4 °C).
O-Glycoproteomic Analysis
O-Glycoproteomic analysis of infected cell lysates was performed as previously described (31) with several modifications. Briefly, cell pellet was resuspended in 0.1% RapiGest (Waters) in 50 mm ammonium bicarbonate and lysed using a sonic probe. Cleared cell lysates were reduced, alkylated, and treated with 5 units of peptide-N-glycosidase F (Roche) overnight at 37 °C, followed by digestion with trypsin (Roche) or chymotrypsin (Roche) for 12 h at 37 °C. Clinical VZV sample was only digested with trypsin because of a limited amount of material. The peptide-N-glycosidase F treatment was then repeated followed by a 2-h incubation with trypsin/chymotrypsin. 1:100–1:200 protease to protein ratio by weight was used with 75% of the protease amount added for the 12-h incubation and 25% added for the 2-h incubation. The samples were then treated with concentrated trifluoroacetic acid (up to 0.5% (v/v), 20 min at 37 °C) and cleared by centrifugation (10,000 × g 10 min). The cleared digests were purified on C18 Sep-Pak (Waters) and treated with 0.1 unit/ml of Clostridium perfringens neuraminidase (Sigma) in 50 mm sodium acetate, pH 5.0, at 37 °C for 2 h. T (Galβ1–3GalNAcα1-O-Ser/Thr) and Tn (GalNAcα1-O-Ser/Thr) glycopeptides were sequentially enriched using peanut agglutinin and Vicia villosa lectin weak affinity chromatography. C18 Stage-tip desalted lectin weak affinity chromatography fractions were screened by preliminary LC-MS for glycopeptide content, and those most enriched in glycopeptides were pooled together and further fractionated by isoelectric focusing.
nLC-MS2
EASY-nLC 1000 UHPLC (Thermo Scientific) interfaced via nanoSpray Flex ion source to an Orbitrap Fusion MS (Thermo) was used for analysis. The nLC was operated in a single analytical column set up using PicoFrit Emitters (New Objectives; 75-μm inner diameter) packed in-house with Reprosil-Pure-AQ C18 phase (Dr. Maisch, 1.9-μm particle size, 19–21-cm column length). Each sample dissolved in 0.1% formic acid was injected onto the column and eluted in a gradient from 2 to 20% B in 95 min, from 20% to 80% B in 10 min, and 80% B for 15 min at 200 nl/min (solvent A, 100% H2O; solvent B, 100% acetonitrile; both containing 0.1% (v/v) formic acid). A precursor MS1 scan (m/z 350–1,700) of intact peptides was acquired in the Orbitrap at a nominal resolution setting of 120,000, followed by Orbitrap HCD-MS2 and ETD-MS2 (m/z of 75–2,000) of the five most abundant multiply charged precursors in the MS1 spectrum; a minimum MS1 signal threshold of 50,000 was used for triggering data-dependent fragmentation events; MS2 spectra were acquired at a resolution of 60,000 for both HCD-MS2 and ETD-MS2. Maximum injection times were 75 and 150 ms for HCD and ETD fragmentation, respectively; isolation width was at 3 with quadrupole, and usually one microscan was collected for each spectrum. Automatic gain control targets were 5 × 104 for MS1 and 1 × 105 for MS2 scans. Supplemental activation (25%) of the charge-reduced species was used in the ETD analysis to improve fragmentation. Dynamic exclusion for 60 s was used to prevent repeated analysis of the same components. Polysiloxane ions at m/z 445.12003 were used as a lock mass in all runs. For clinical VZV sample, LTQ-Orbitrap Velos Pro spectrometer (Thermo Scientific) was used as previously described (31).
Data Analysis
Data processing was performed using Proteome Discoverer 1.4 software (Thermo Scientific) as previously described with small changes (32). Sequest HT node was used instead of Sequest. All spectra were initially searched at the full cleavage specificity, filtered according to the confidence level (medium, low, and unassigned) and further searched with the semi-specific enzymatic cleavage. Up to two missed cleavages were allowed. For Orbitrap Fusion MS-derived data the precursor mass tolerance was set to 5 ppm, and the fragment ion mass tolerance was set to 20 mmu (milli mass units). For LTQ-Orbitrap Velos Pro MS-derived data, the precursor mass tolerance was set to 7 ppm. Carbamidomethylation on cysteine residues was used as a fixed modification. Methionine oxidation and HexNAc and HexHexNAc attachment to serine, threonine, and tyrosine were used as variable modifications for ETD-MS2. All HCD-MS2 were preprocessed as described (32) and searched under the same conditions mentioned above using only methionine oxidation as variable modification. All spectra were searched against a concatenated forward/reverse human-specific database (UniProt, January 2013, containing 20,232 canonical entries. In addition, another 251 common contaminants and 3187 entries of viruses known to infect humans were included in the search) using a target false discovery rate of 1%. An additional database of HCMV Towne protein entries was used because they were not present in the above-mentioned database. The false discovery rate was calculated using target decoy PSM validator node, a part of the Proteome Discoverer workflow. The resulting list was filtered to include only peptides with glycosylation as a modification. This resulted in a final glycoprotein list identified by at least one unique glycopeptide. ETD-MS2 data were used for unambiguous site assignment. HCD-MS2 data were used for unambiguous site assignment only if the number of HexNAc residues was equal to the number of potential sites on the peptide. Data analysis was assisted by manual validation.
Results
Mapping O-Glycosites in Human Herpesviruses
We applied our recently developed mass spectrometry-based approach (31) to map O-glycosites in VZV, HCMV, and EBV. VZV- or HCMV-infected HEL fibroblasts, as well as the EBV-transformed P3HR1 human Burkitt's lymphoma B cell line, were used for O-glycoproteomic analysis. In addition, we had a unique opportunity to analyze VZV-infected clinical material taken directly from a herpes zoster patient and compare it with the glycoproteome derived from a cell lysate. The major O-glycan structures produced in HEL fibroblasts are sialylated core-1 O-glycans (ST; Neu5Acα2–3Galβ1–3GalNAcα1-O-Ser/Thr), and during HSV-1 infection we have observed an increased amount of truncated O-glycan structure Tn (GalNAcα1-O-Ser/Thr) (31). Because it is essential to enrich glycopeptides in total protease digests of complex mixtures of proteins, we used our established two-step sequential lectin enrichment strategy by peanut agglutinin and V. villosa lectin to capture desialylated T (Galβ1–3GalNAcα1-O-Ser/Thr) and Tn (GalNAcα1-O-Ser/Thr) glycopeptides from virus-infected cell digests. To increase protein sequence coverage, we used trypsin and chymotrypsin digestion in parallel. We identified 53, 122, and 41 novel GalNAc-type O-glycosites on 6, 28, and 6 glycoproteins in VZV, HCMV, and EBV, respectively (Tables 1–3 and supplemental Data Sets S1–S5). Comparable numbers of T and Tn glycopeptides were identified in VZV-infected samples (Table 1 and supplemental Data Sets S1–S3), whereas markedly higher numbers of Tn glycopeptides were found in HCMV- and EBV-infected samples (Tables 2 and 3 and supplemental Data Sets S1, S4, and S5). Among the identified viral glycopeptides, by far the majority (>98.5%) belonged to proteins exposed to the lumenal side of the secretory pathway (supplemental Data Set S1). We did identify a few glycopeptides that were mapped to proteins described as nuclear or cytoplasmic proteins and hence not known to enter the secretory pathway (supplemental Data Set S1). These were not included in the analysis and most likely represent random contamination of O-GlcNAc glycopeptides found in the enriched fraction, where some peptides are also found (38). The individual glycoproteomes are discussed in detail in the following sections.
TABLE 1.
Summary of O-glycoproteins and O-glycosites identified in VZV
| Uniprot IDa | Protein name | Function | Total sites | Unambiguous sites | T (Hex-HexNAc) | Tn (HexNAc) | List of O-glycosylated amino acid positions |
|---|---|---|---|---|---|---|---|
| VZV total | 53 (68b) | 42 (57b) | 39 (54b) | 44 (59b) | |||
| P09257 | gB | Fusion (43, 44) | 7 | 3 | 4 | 7 | 73–88 (2x), 86, 92, 94, 126–129 (1x), 265–269 (1x) |
| P09256 | gC | Skin tropism (53) | 7 (22b) | 6 (21b) | 6 (21b) | 7 (22b) | 52–68 (1x), 78 (92,c 106,c 134,c 162c), 79 (93,c 107,c 135,c 163c), 82 (96,c 110,c 138,c 166c), 120 (148c), 121 (149c), 124 (152c) |
| P09259 | gE | Cell-cell spread, Fc receptor (47, 129) | 20 | 18 | 17 | 16 | 47–54 (1x), 55, 62, 70, 71, 79, 102, 118, 137, 154, 183, 224–225 (1x), 311, 325, 329, 333, 512, 519, 520, 526 |
| P09260 | gH | Entry, fusion (42) | 8 | 5 | 3 | 7 | 23, 25, 54–56 (1x), 123–131 (1x), 150–151 (1x), 177, 184, 598 |
| P09258 | gI | Cell-cell spread, Fc receptor, skin and T-cell tropism (46, 51, 129, 130) | 10 | 9 | 9 | 6 | 180–183 (1x), 205, 209, 212, 225, 238, 239, 241, 256, 259 |
| P09298 | gM | Cell-cell spread (39) | 1 | 1 | 0 | 1 | 312 |
a Uniprot IDs of reference VZV strain (Dumas) are provided.
b Given all gC tandem repeats are glycosylated.
c Alternative positions within gC tandem repeats.
TABLE 2.
Summary of O-glycoproteins and O-glycosites identified in HCMV
| Uniprot IDa | Protein name | Function | Total sites | Unambiguous sites | T (Hex-HexNAc) | Tn (HexNAc) | List of O-glycosylated amino acid positionsb |
|---|---|---|---|---|---|---|---|
| HCMV total | 122 | 97 | 44 | 119 | |||
| P13201 | gB | Fusion (131) | 2 | 2 | 1 | 2 | 58, 64 |
| P17176 | gH | Entry, fusion (59, 132) | 4 | 4 | 2 | 4 | 31, 172, 173, 546 |
| B9VXR5 | gL | Entry, fusion (59, 132) | 3 | 3 | 3 | 3 | 32, 34, 38 |
| B9VXN2 | gO | Entry (59) | 1 | 1 | 1 | 1 | 293 |
| B9VXN1 | gN | Assembly (133) | 4 | 1 | 2 | 3 | 23–27 (2x), 37–38 (1x), 39 |
| B9VXH1 | RL11 | Fcγ-binding (61) | 7 | 7 | 1 | 7 | 25, 26, 32, 44, 49, 51, 161 |
| B9VXH2 | RL12 | Fcγ-binding (62) | 18 | 14 | 5 | 18 | 41–47 (3x), 50, 51, 52, 126–128 (1x), 129, 131, 134, 136, 318, 319, 323, 325, 329, 331, 332 |
| P17175 | UL4 | Unknown | 3 | 3 | 2 | 3 | 27, 28, 29 |
| B9VXH7 | UL5 | Unknown | 7 | 4 | 2 | 7 | 42–48 (1x), 51, 55, 60, 69–71 (1x), 72, 74–76 (1x) |
| B9VXH9 | UL7 | Proangiogenic and anti-inflammatory factor (134, 135) | 1 | 1 | 1 | 1 | 172c |
| B9VXI0 | UL8 | Unknown | 3 | 1 | 1 | 3 | 172c, 246–249 (2x) |
| B9VXI2 | UL10 | Temperance for RPE cells (57) | 8 | 8 | 1 | 8 | 43, 145, 146, 147, 148, 155, 156, 196 |
| B9VXI3 | UL11 | Interacts with CD45 (110) | 5 | 5 | 0 | 5 | 189, 190, 194, 195, 199 |
| B9VXI4 | UL13 | Unknown | 4 | 4 | 3 | 4 | 156, 187, 337, 407 |
| B9VXI7 | UL16 | Interacts with NKG2D ligands to down-regulate expression at the cell surface (72, 136) | 4 | 3 | 2 | 3 | 47–53 (1x), 162, 168, 175 |
| B9VXJ3 | UL22A | Secreted chemokine receptor (69) | 14 | 13 | 6 | 13 | 30, 35, 37, 38, 40, 44, 45, 51, 53, 57, 58, 66–70 (1x), 75, 97 |
| B9VXN6 | UL78 | Chemokine receptor (7TM) (64) | 2 | 1 | 0 | 2 | 8–14 (1x), 24 |
| Q6SWQ0 | UL119 | Fcγ-binding (61) | 8 | 8 | 4 | 8 | 69, 70, 73, 74, 78, 79, 134, 140 |
| Q6SWP5 | UL124 | Unknown; latent protein (57) | 2 | 0 | 0 | 2 | 61–66 (2x) |
| Q6SWN8 | UL146 (vCXCL1) | Viral chemokine, attracts neutrophils for dissemination (70) | 1 | 1 | 0 | 1 | 111 |
| Q6SWP1 | UL148 | Regulates composition of alternative gH/gL complexes (137) | 1 | 1 | 0 | 1 | 86 |
| B9VXD7 | US3 | Down-regulates MHC-I (138) | 2 | 0 | 0 | 2 | 137–139 (2x) |
| B9VXE0 | US8 | Down-regulates MHC-I (73) | 2 | 2 | 2 | 2 | 45, 84 |
| B9VXE8 | US16 | Virus tropism factor; putative 7TM (139) | 4 | 2 | 2 | 4 | 284–289 (1x), 299, 300–305 (1x), 306 |
| Q03307 | US20 | NK cell evasion (promotes MICA degradation); replication in endothelial cells; putative 7TM (67, 136) | 3 | 1 | 1 | 3 | 334, 337–339 (2x) |
| B9VXF3 | US21 | Unknown; putative 7TM (66) | 2 | 1 | 0 | 2 | 225–230 (1x), 234 |
| B9VXF9 | US28 | GPCR; facilitates cell-cell spread, binds chemokines (140) | 1 | 0 | 0 | 1 | 2–14 (1x) |
| B9VXG0 | US29 | Unknown | 6 | 6 | 2 | 6 | 24, 31, 41, 66, 67, 69 |
a Uniprot IDs of HCMV strain Towne are provided.
b Sites mapping to secreted proteins/protein regions facing the lumenal part of the secretory pathway are listed (see all identified glycopeptides in supplemental Data Set S1).
c The same glycopeptide is mapped to both UL7 and UL8.
TABLE 3.
Summary of O-glycoproteins and O-glycosites identified in EBV
| Uniprot IDa | Protein name | Function | Total sites | Unambiguous sites | T (Hex-HexNAc) | Tn (HexNAc) | List of O-glycosylated amino acid positionsb |
|---|---|---|---|---|---|---|---|
| EBV total | 41 | 31 | 16 | 39 | |||
| P0C763 | gB | Fusion (76) | 5 | 5 | 3 | 5 | 24, 33, 34, 458, 621 |
| P0C6Z3 | gN | Egress, assembly (79) | 4 | 1 | 3 | 3 | 33–36 (2x), 44, 52–59 (1x) |
| Q1HVD8 | gp150 (BDLF3) | Deletion enhances infection of epithelial cells (141) | 6 | 2 | 1 | 6 | 62–71 (4x), 163, 164 |
| Q1HVC5 | gp78 (BILF2) | Unknown | 6 | 4 | 4 | 6 | 154–164 (2x), 165, 183, 187, 192 |
| P68343 | gp350 | Binding (80) | 19 | 18 | 4 | 19 | 249–253 (1x), 746, 750, 752, 753, 754, 774, 775, 776, 779, 780, 786, 787, 788, 791, 802, 803, 804, 805 |
| Q1HVG2 | gp42 | B-cell tropism factor (76, 106) | 1 | 1 | 1 | 0 | 43 |
a Uniprot IDs of reference EBV strain (AG876) are provided.
b Sites mapping to secreted proteins/protein regions facing the lumenal part of the secretory pathway are listed (see all identified glycopeptides in supplemental Data Set S1).
The VZV O-Glycoproteome
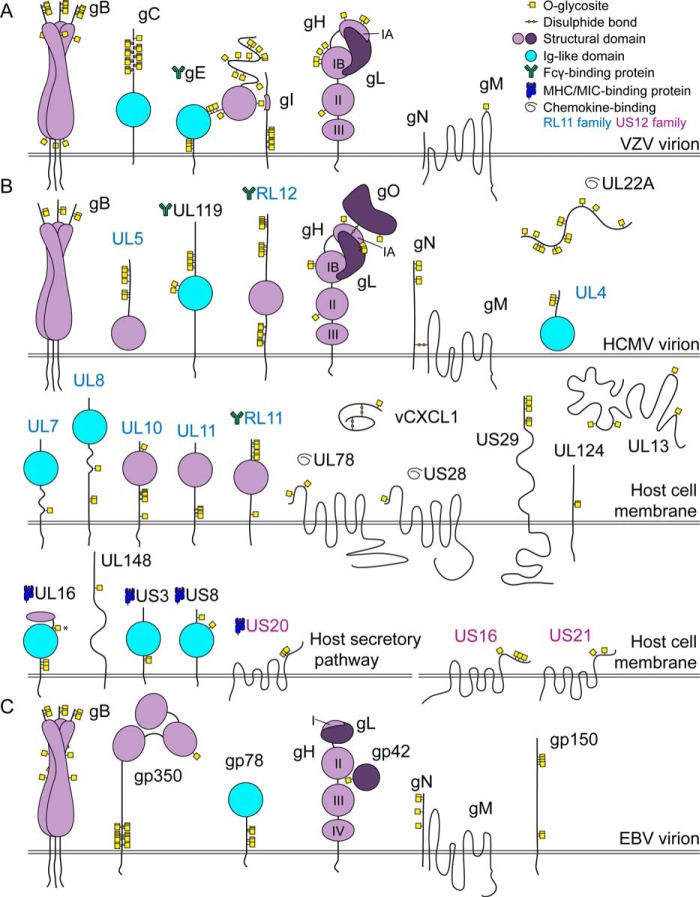
We identified 53 O-glycosylation sites on six of nine VZV envelope glycoproteins (Table 1 and supplemental Data Sets S1–S3), combining results obtained from infected cell lysate and clinical VZV sample (39, 40). Four envelope glycoproteins are essential for VZV replication in vitro: gB, gH, gL, and gE, of which gB and the gH-gL complex is thought to mediate both virion-cell and cell-cell fusion (41–44). Two of the fusion complex proteins were found to be O-glycosylated, with seven and eight sites identified in gB and gH, respectively (Table 1). Six gB O-glycosites localized to the N-terminal region of gB, whereas one O-glycosite was situated at the tip of the membrane-proximal domain potentially involved in cell fusion (Fig. 1A) (45). In gH, seven of the eight glycosites were found within the N-terminal domain I, which interacts with gL. Three of gH glycosites were located at the exposed N-terminal tip of domain I (subdomain IA) important for viral replication in human skin in vivo (42). The remaining four glycosites were found in the poorly structured region of subdomain IB. In addition, a single O-glycosite was situated in domain II, which is critical for gH maturation (Fig. 1A) (42).
FIGURE 1.
The O-glycoproteomes of VZV, HCMV, and EBV. A graphical depiction of identified O-linked glycosylation sites with respect to known structural features of viral glycoproteins (59, 60, 67, 72, 76, 96, 112, 120, 126–128). Domains of gH are labeled with Roman numerals. The O-glycosylation site marked with an asterisk can potentially have a slightly different location because of site ambiguity. Sites mapping to secreted proteins/regions facing the lumenal side of the secretory pathway (>98.5% of glycopeptides) are shown (see all identified glycopeptides in supplemental Data Set S1). A, VZV, all identical tandem repeats are shown occupied in VZV gC. B, HCMV. C, EBV.
VZV gE is the most abundant VZV glycoprotein and is essential for infectivity and cell-to-cell spread in cell culture (46, 47). Twenty O-glycosites were identified on gE (Table 1), 13 of which were dispersed throughout the N-terminal part of the ectodomain. The remaining glycosites were found in the unstructured linker region or juxtamembrane stem region (Fig. 1A). Eleven of the sites were situated within the unique and non-conserved (48) extreme N-terminal domain (amino acids 1–188), five of which were located within the region (amino acids 24–71) essential for binding the cellular entry receptor insulin-degrading enzyme (49, 50). Two O-glycosites, Thr183 and an ambiguous site spanning amino acids 224–225 (224–225 (1x)), mapped to two distinct regions important for gE-gI interaction, which determines gE trafficking and VZV virulence in skin (46, 47, 50). The binding partner gI, which is indispensable for infectivity of T cells and skin in vivo (51), possessed 10 O-glycosites, all of which were located in the stem region and away from the gE-binding domain (52). A single O-glycosite was identified in the C-terminal loop of the multi-span gM, which is also important for efficient cell-cell spread (Table 1 and Fig. 1A) (39).
Another important protein for VZV virulence in skin in vivo is gC (53). In accordance with the predicted dense glycosylation of the mucin-like tandem repeat domain in gC (54), we found seven O-glycosites in this region (Table 1 and Fig. 1A). Six of the sites were situated on two tryptic peptides, KPDPAVAPTSAASR and KPDPAVAPTSAATR, found five and two times within the protein sequence, respectively. Clearly it is not possible to discriminate by mass spectrometry how many of the identical repeats are glycosylated within the region. However, presuming that all tandem repeats are occupied with O-glycans, the total number of sites would increase to 22.
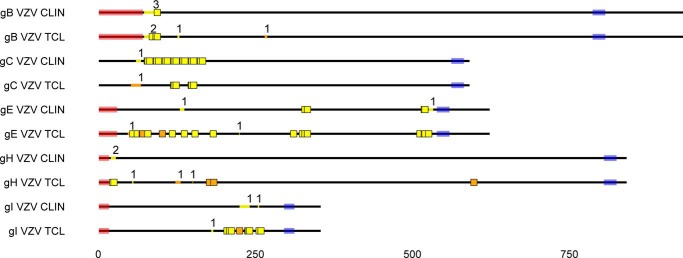
In VZV blister and the infected fibroblasts, 21 and 50 viral O-glycosites were identified, respectively (Table 4 and supplemental Data Sets S1–S3). The fewer sites obtained in the clinical material could in part be explained by the limited amount of the material, only allowing single digestion with trypsin. Five of the six viral proteins, gB, gC, gE, gH, and gI, were found to be glycosylated in both the clinical sample and the infected cell lysate, with the remaining viral protein (one glycosite on gM) only identified with chymotrypsin digestion of infected fibroblasts. Except for gC, a relatively smaller number of sites were identified in most of the viral proteins found in the clinical specimen (Table 4 and supplemental Data Sets S1 and S2). Despite differences in coverage, the O-glycosites found in the clinical sample correlated well with the ones found in the infected cells (Fig. 2). In both samples, tryptic digest-derived glycosites located to analogous parts of proteins, including the N terminus of gB and gH, the membrane-proximal region of gI, and the tandem repeat region of gC (Fig. 2). In contrast to the clinical sample, we only identified one of the two different tandem-repeated gC glycopeptides in the more abundant total cell lysate. For gE, similar clusters of sites were found in both samples, corresponding to the linker region between two structural domains and the juxtamembrane stem region. In contrast, only one O-glycosite was identified at the N-terminal unique region of gE in the clinical sample, compared with seven in infected cells (Fig. 2).
TABLE 4.
Comparison of O-glycosites identified in VZV derived from infected fibroblasts (TCL) and a clinical sample
| Uniprot IDa | Protein name | Total sites |
Unambiguous sites |
T (Hex-HexNAc) |
Tn (HexNAc) |
List of O-glycosylated amino acid positions (clinical) | List of O-glycosylated amino acid positions (TCL)d | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cb | Tc | All | Cb | Tc | All | Cb | Tc | All | Cb | Tc | All | ||||
| P09257 | gB | 4 | 7 | 7 | 1 | 3 | 3 | 4 | 3 | 4 | 1 | 7 | 7 | 73–92 (3x), 94 | 73–88 (2x), 86, 92, 94, 126–129 (1x), 265–269 (1x) |
| P09256 | gC | 7 | 4 | 7 | 6 | 3 | 6 | 6 | 0 | 6 | 7 | 4 | 7 | 60–68 (1x), 78 (92,e 106,e 134,e 162e), 79 (93,e 107,e 135,e 163e), 82 (96,e 110,e 138,e 166e), 120 (148e), 121 (149e), 124 (152e) | 52–63 (1x), 120 (148e), 121 (149e), 124 (152e) |
| P09259 | gE | 6 | 20 | 20 | 4 | 18 | 18 | 6 | 17 | 17 | 2 | 16 | 16 | 130–137 (1x), 329, 333, 519, 520, 526–533 (1x) | 47–54 (1x), 55, 62, 70, 71, 79, 102, 118, 137, 154, 183, 224–225 (1x), 311, 325, 329, 333, 512, 519, 520, 526 |
| P09260 | gH | 2 | 8 | 8 | 0 | 5 | 5 | 2 | 1 | 3 | 1 | 7 | 7 | 20–28 (2x) | 23, 25, 54–56 (1x), 123–131 (1x), 150–151 (1x), 177, 184, 598 |
| P09258 | gI | 2 | 10 | 10 | 0 | 9 | 9 | 2 | 9 | 9 | 2 | 6 | 6 | 225–241 (1x), 254–256 (1x) | 180–183 (1x), 205, 209, 212, 225, 238, 239, 241, 256, 259 |
| P09298 | gM | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | N/A | 312 |
a Uniprot IDs of reference VZV strain (Dumas) are provided.
b Clinical sample.
c Total infected cell lysate (TCL).
d Underlined glycosites are only found in the chymotryptic digests.
e Alternative positions within gC tandem repeats.
FIGURE 2.
O-Glycosites found in VZV derived from infected fibroblasts (TCL) and a clinical sample. VZV (strain Dumas) gB, gC, gE, gH, and gI protein sequences are shown as black lines, drawn to scale. Predicted signal peptides and transmembrane regions are shaded in pink and blue, respectively. Unambiguous O-glycosylation sites are shown as colored squares, whereas ambiguous sites are marked as colored lines within the protein backbone, where the number above indicates the number of glycosites. Trypsin and unique chymotrypsin digestion-derived glycosites are marked in yellow and orange, respectively. All identical potentially glycosylated VZV tandem repeats are shown occupied. Reference VZV sequences were used because of unavailable annotation of investigated isolate sequences. Glycoprotein M is not shown, because it was only found glycosylated in the infected cell lysate. CLIN, clinical sample; TCL, total infected cell lysate.
The HCMV O-Glycoproteome
HCMV has one of the largest genomes between human herpesviruses (more than 160 open reading frames) and encodes at least six well characterized virion-associated envelope glycoproteins with known functions in viral replication: gB, gH, gL, gM, gN, and gO (55, 56). However, at least 40 more protein sequences contain a predicted signal peptide, which would allow their potential modification with glycans in the secretory pathway. We identified 122 novel O-glycosylation sites on 28 HCMV proteins, including gB, gH, gL, gN, and gO (Table 2 and supplemental Data Sets S1 and S4). Most of the identified glycoproteins had a relatively small number of sites (1–8), with the exception of RL12 and UL22A, in which we found 18 and 14 sites, respectively. Five HCMV envelope glycoproteins are indispensable for replication: gB, gH, gL, gM, and gN (57). Glycoprotein B and gH-gL comprise the conserved fusion machinery; however, gH-gL can additionally be complexed with gO or UL128–131A to promote infectivity (58). In agreement with alphaherpesviruses, we found two O-glycosites at the N terminus of gB (Fig. 1B) (31, 33). Four O-glycosites were identified on gH, whereas three were found on gL. It is difficult to compare the site localization to alphaherpesviruses because of low sequence identity for both gH and gL. However, presuming similar protein architecture (59), three of four gH sites were located at the membrane-distal domain I, similarly to alphaherpesviruses (Fig. 1B). In addition, a single glycosite was found in domain II consistent with the findings for VZV gH. The gH-gL-associated protein gO was glycosylated at one position (Fig. 1B), whereas four O-glycosites were seen at the N terminus of gN (Fig. 1B).
Most of the other identified glycoproteins with known functions were involved in counteracting the host defense mechanisms. Eight of the identified glycoproteins, RL11, RL12, UL4, UL5, UL7, UL8, UL10, and UL11, are members of the RL11 multigene family of HCMV proteins (60). Two of them, RL11 and RL12, are known to bind human IgG (61, 62). O-Glycosites mainly localized to either the very N terminus or the juxtamembrane stem region of the RL11 family members and not the characteristic RL11D domain (Fig. 1B) (60). UL119, which is an unrelated virion glycoprotein with eight identified O-glycosites, also has the capacity of Fcγ binding (61). Two of the sites were situated on the Ig-like domain of the protein (Fig. 1B). Two of the identified glycoproteins, US28 and UL78, belong to the GPCR family of seven-transmembrane domain receptors and have known functions in binding host chemokines (63) or chemokine receptors (64). The O-glycosites were located at the extracellular N termini of the proteins (Fig. 1B), which are often found O-glycosylated in human GPCRs (38). For US28, the O-glycosite (2–14 (1x)) was potentially located in the region essential for chemokine binding (amino acids 10–16) (65). US16, US20, and US21, members of the HCMV US12 family of putative seven-transmembrane domain proteins (66), were O-glycosylated at the C terminus (Fig. 1B), suggesting that their orientation in the membrane could be opposite to that of GPCRs (67). The secreted RANTES-specific chemokine receptor UL22A (also known as UL21.5) was found heavily glycosylated, as previously reported (68, 69). One of the HCMV-encoded chemokines, vCXCL1, which is thought to attract neutrophils for dissemination, was also found to be O-glycosylated, as recently shown (70, 71). Four of HCMV immunoevasins, UL16, US3, US8, and US20, were identified to carry O-glycans (Fig. 1B). UL16 interacts with NKG2D ligands and reduces their expression on the cell surface. Three of the four O-glycosites were found at the membrane-proximal stem region of the molecule, one of which (Tyr162) mapped to a distinct protein-protein contact area with MICB (Fig. 1B) (72). US3 and US8, which have the capacity of binding MHC class I, were glycosylated at the stem region and the N terminus, respectively (Fig. 1B) (73).
The EBV O-Glycoproteome
At least nine EBV virion-associated envelope glycoproteins are known (74). Six of those—gB, gN, gp42, gp78, gp150 and gp350—were found to be O-glycosylated, with a total of 41 glycosites identified (Table 3 and supplemental Data Sets S1 and S5). EBV depends on gB and gH-gL for entry into host cells. Fusion effector gB was O-glycosylated at five positions, three of which were located at the extreme N terminus of the protein, in accordance with VZV and HCMV gB glycosylation (Fig. 1, A–C). In contrast to other herpesviruses, we did not identify any O-glycosites on gH or gL (Fig. 1C). Infection of B cells requires an additional viral protein gp42 (75). gp42 is proteolytically cleaved releasing a secreted form that links gH-gL on the virion to HLA class II on B cells, thereby bringing the two membranes to close proximity (76). A single O-glycosite was identified on gp42 localized in one of the regions essential for high affinity binding to gH-gL and just C-terminally to the proteolytic cleavage site required for release of gp42 from the membrane (Fig. 1C) (77, 78). Similar to betaherpesviruses, gM-gN protein complex is particularly important for EBV viral particle formation (79). In agreement with the results obtained for HCMV, four O-glycosites were found at the N terminus of EBV gN (Fig. 1C). Although dispensable for infectivity, gp350 is important for the initial attachment to B cells (80) and has been shown to be highly O-glycosylated (81). gp350 also represents a very potent immunogen (82, 83). On gp350, 19 O-glycosites were identified, 18 of which were located in the Pro/Ser/Thr-rich mucin-like stem region, and 1 glycosite was found at the tip of one of the N-terminal domains (Fig. 1C). The remaining glycosites observed in EBV were situated on late proteins gp150 (BDLF3) and gp78 (BILF2), with most O-glycosites found in the stem region or at the N terminus, respectively (Fig. 1C).
Discussion
Until recently, most evidence for viral O-glycosylation has originated from the interrogation of densely glycosylated Pro/Ser/Thr-rich mucin-like sequences such as those found in HSV-1 gC and Ebola virus glycoprotein (84, 85). With our recent introduction of a mass spectrometric strategy for global mapping of viral O-glycosylation sites, we substantially expanded the number of identified O-glycosylation sites in alphaherpesviruses HSV-1 and -2 and demonstrated the importance of elongated viral O-glycans for virus propagation (31), as well as early immune recognition (33). The aim of the present study was to provide knowledge on the global O-glycosylation of three additional clinically important members of the Herpesviridae family, VZV, HCMV, and EBV, representing the three distinct herpesvirus subfamilies. The identified O-glycosites were widely distributed on most of the viral envelope glycoproteins, and importantly we identified conserved glycosylation patterns in distinct regions of homologous viral proteins, suggesting that O-glycosylation at certain regions is of importance for herpesviruses. In addition, we generated an O-glycoproteome from a clinical VZV sample obtained from a zoster blister representing the first O-glycoproteome from a primary source of virus unaffected by artificial propagation in cell culture.
Identification of O-glycosylation sites is hampered not only by the lack of reliable prediction algorithms but also by the unique differential biosynthetic regulation of O-glycosylation, underlining the importance of direct experimental analysis (18). The glycoproteomic strategy employed here is highly sensitive and combines enrichment of the most prevalent glycoforms (simple core-1 O-glycans) produced in the host cells used for viral propagation (31). Limitations, however, include the failure to enrich for peptides exclusively expressing core-2 or other more complex glycoforms and lack of stoichiometry for the glycosites identified (86). We have previously estimated that simple core-2 structures constitute ∼10–15% of total O-glycans in HEL fibroblasts (31). It is noteworthy to mention that in the present study we have identified a small fraction (∼6%) of the virus-derived glycosites potentially carrying both core-2 and core-1 O-glycans on the same sites in the peanut agglutinin-enriched peptides (87). The finding suggests that despite the presence of more complex O-glycans at certain sites, we might still detect them as biosynthetic intermediates. We are not, however, in a position to predict the exact proportion of glycosites missed by our method. Another limitation is that available protease cleavage sites will determine protein coverage. To increase the coverage we therefore utilized both trypsin and chymotrypsin digestion in parallel, which expanded the number of identified sites for certain proteins. Despite these limitations, we were able to capture the majority of glycoforms expressed in the infected cells and achieve high coverage for abundant glycoproteins.
The majority of identified glycopeptides (>98.5%) were mapped to proteins exposed to the lumenal side of the secretory pathway. A few glycopeptides, however, were mapped to nuclear or cytoplasmic proteins that are not known to enter the secretory pathway. Such glycopeptides most likely represent a minor contamination with cytoplasmic O-GlcNAc glycopeptides. An argument against this interpretation is that several of the identified HexNAc residues were elongated with hexose, suggesting that they represent genuine T-structures. It cannot, however, be completely excluded that an initial GlcNAc residue could be elongated by a galactose residue by the highly efficient galactosyltransferases present in cell lysates alongside released donor substrates (88), and more detailed analyses are required to establish the exact nature of the identified glycans.
The characterization of the O-glycoproteomic landscape of herpesviruses provides a first step in being able to appreciate and probe the biological functions of this prevalent modification of herpesvirus envelope glycoproteins. Information on site-specific O-glycosylation of virus and viral glycoproteins produced in different cellular systems could prove to be important because we predict that O-glycosylation may not only vary with respect to structures, but more importantly also vary considerably with respect to sites of O-glycan attachment. This is because the repertoire of polypeptide GalNAc transferases that controls the O-glycosylation capacity is cell-specific and may also be influenced by the viral infection itself, as evidenced by the induction of GalNAc-T3 by influenza A virus (18, 89). We have recently developed a quantitative differential O-glycoproteomic strategy to address non-redundant contributions of individual GalNAc transferase isoforms to the O-glycosylation capacity of a cell (90), and this could be applied to address changes in viral O-glycosylation between clinical isolates or samples propagated in different cell types. Because O-glycans may affect immunity by shielding protein epitopes or introducing glycopeptide epitopes (91–93), it is important to consider O-glycans in the context of vaccine design. It is also important to consider O-glycans for innate immune targeting ligands to augment immunity (33, 94).
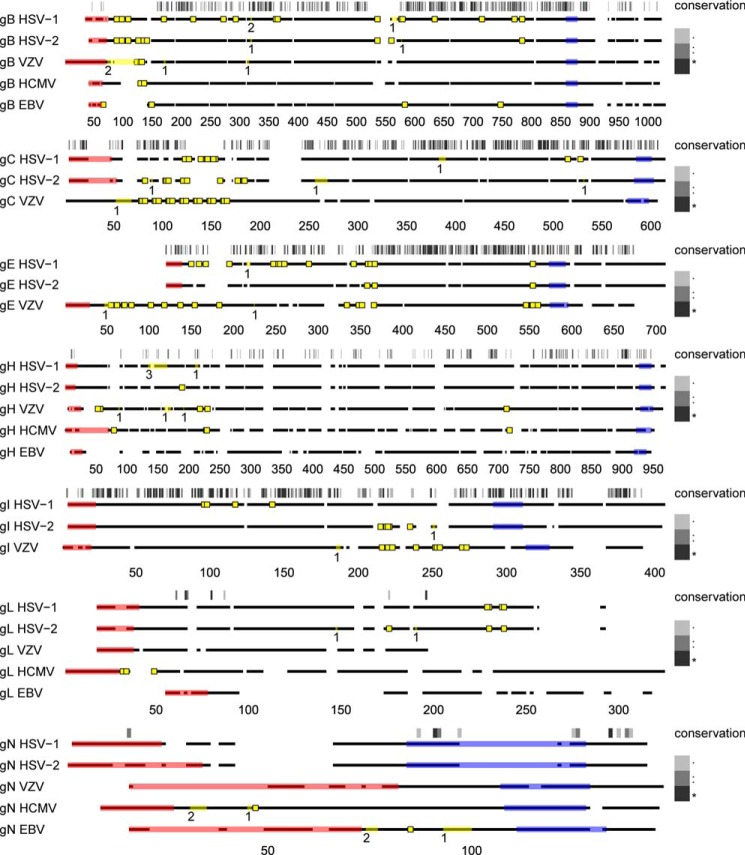
The three Herpesviridae subfamilies, alphaherpesviruses, betaherpesviruses, and gammaherpesviruses, diverged from a common ancestor more than 200 million years ago (1). More than 40 genes are conserved between all herpesviruses and are referred to as core genes. Of those, gB, gH, gL, gM, and gN are the conserved envelope glycoproteins (3). In addition, four more envelope glycoproteins, gC, gE, gI, and gK, are conserved between alphaherpesviruses (95). Using the O-glycoproteomes from five different human herpesviruses, we sought to investigate the extent to which sites and patterns of O-glycosylation were conserved on homologous envelope glycoproteins between HSV-1, HSV-2, VZV, HCMV, and EBV (Fig. 3 and supplemental Figs. S1–S7). One of the most conserved patterns of O-glycosites found in all herpesviruses was localized at the extreme N terminus of the fusogenic protein gB, despite high variability in the protein sequence. In addition, we found several more glycosites in common between VZV and the other two alphaherpesviruses, HSV-1 and -2, that were located on more highly conserved regions, including the O-glycans on the membrane-proximal domain that contains putative fusion loops of gB (96) (Fig. 3 and supplemental Fig. S1). Concordant glycosylation was also found in the N-terminal mucin-like regions of gC, as well as in two clusters of O-glycosites in VZV gE, homologous to sites in HSV-1 and HSV-2 gE (Fig. 3 and supplemental Figs. S1 and S3). It should be mentioned, however, that we also found O-glycosites that were not shared between the different family members. In conclusion, certain regions of homologous Herpesviridae envelope glycoproteins share similar patterns of O-glycosylation that potentially could be linked to specific functions, although virus-specific differences are also observed.
FIGURE 3.
Conservation of O-linked glycosylation sites on homologous envelope glycoproteins of human herpesviruses. Clustal Omega server was used to align amino acid sequences of gB, gH, gL, and gN between HSV-1 (31), HSV-2 (33), VZV, HCMV, and EBV, as well as gC, gE, and gI between the alphaherpesviruses. Conserved glycoprotein M was not included, because it was only found glycosylated in one of the investigated viruses. Protein backbones are depicted as broken black lines, where spaces represent gaps in the alignment. Individual alignments were drawn to scale (indicated below each graph). Sequence conservation is indicated above the aligned sequences for each set and is represented by a grayscale barcode that maps to the Clustal alignment score, as shown in the legend. In brief, for the Clustal alignment score, an asterisk indicates a position with a fully conserved residue, and a colon indicates conservation of an amino acid with strongly similar properties, whereas a period indicates conservation of an amino acid with weakly similar properties. Predicted signal peptides and transmembrane regions are shaded in pink and blue, respectively. Unambiguous O-glycosylation sites are shown as yellow squares, whereas ambiguous sites are marked as yellow lines within the protein backbone, where the number below indicates the number of glycosites. It should be noted that O-glycosylation sites on VZV are derived both from the clinical sample and the infected total cell lysate. All identical potentially glycosylated VZV tandem repeats are shown occupied. Two ambiguous O-glycosylation sites from our previous publication (31) (HSV-1 gB 109–123 (HexHexNAc) and gE 135–143 (HexHexNAc)) were omitted from the graph, because we cannot exclude the possibility they could be part of an elongated structure on an adjacent site. Reference strain sequences were used for HSV-2, VZV, and EBV because of incomplete or unavailable annotation of investigated strains.
O-Glycosylation occurs in two principally distinct patterns on proteins that may be related to their biosynthesis and function. Isolated O-glycosites are often important for regulated proteolytic processing and exert co-regulatory functions in basic cellular processes (97). Densely clustered sites, on the other hand, are often present in vulnerable protein regions and confer protection from non-regulated proteolysis. Similarly to human proteins, viral proteins accommodate a substantial number of isolated O-glycosites outside the mucin-like regions (31, 33, 98, 99). In mammalian proteins, such sites have been demonstrated to play important regulatory roles in basic cellular processes such as secretion, pro-protein processing, and ectodomain shedding (100–104). It could thus be speculated that single-site O-glycosylation on viral proteins affects the cleavage of viral proteins with importance for infection. As an example, we identified a single O-glycosite adjacent to a proteolytic cleavage site of EBV gp42 essential for infection of B cells (78, 105). Given that O-glycosylation often protects from cleavage, the different extent of glycosylation could potentially be a co-regulatory mechanism for cell tropism, because gp42 is not required for infection of epithelial cells (75, 106). The same site might also play a role in interaction with gH-gL, because it mapped to one of the regions required for high affinity binding (107). The immunoevasin UL16 in HCMV represents another example where a glycosylation site is mapped to an interaction surface, which down-regulates NKG2D ligand MICB expression at the cell surface and subsequent detection by NK cells (72). We also identified a number of single or clustered O-glycans at the extracellular termini of five different HCMV seven-transmembrane domain receptors similar to what has been observed in human multi-span receptors (38). Viral chemokine receptor UL78 has been demonstrated to heteromerize with human chemokine receptors, modulating their function (64). Based on the relatively short N-terminal regions of these receptors, it is quite unlikely for them to be involved in dimerization, as seen for GPCRs bearing large extracellular domains (108). However, glycosylation could potentially modulate ligand binding or limited proteolysis-associated receptor turnover, as hypothesized for the N terminus of the β1-adrenergic receptor (109). Among other specific functions, O-linked glycans may contribute to transport and stable cell surface expression of viral proteins. This has been suggested for HCMV UL11 and for VZV gB (28, 110). Interestingly, we identified an ambiguous site in VZV gB spanning a region (amino acids 126–129) containing the potential O-glycosylation site at Thr129, investigated for Ala substitution, suggesting that the O-glycan in question may indeed be important for surface expression of gB (28). In addition, we found glycosylation in the EBV gB linker region (T621), which has been suggested to be relevant for gB oligomerization and surface expression (111, 112).
In agreement with our previous findings in HSV-1 and HSV-2, we found dense glycosylation at the N-terminal tandem repeat region of gC in VZV. The function of these densely glycosylated areas are not clear, but it is proposed that O-glycans in the distally located mucin-like region in gC has a direct role in modulating the interaction with cell surface proteoglycans (21). In a similar way it can be speculated that the abundant O-glycosylation found on the unique N-terminal domain of VZV gE affects the multiple functions specified by the region, including interactions with cell entry receptor insulin-degrading enzyme and binding partner gI (50). O-Glycosylation was also enriched in multiple members of the RL11 family and at the N-terminal domain of gN in HCMV. N-terminal HCMV gN glycosylation has been suggested to protect from neutralization by antibodies (91). Moreover, dense glycosylation was found in the stem region of several glycoproteins in VZV, HCMV, and EBV, suspected to protect the otherwise vulnerable region from unspecific proteolytic cleavage (113). Another potential function could be to provide structural support for keeping the ectodomain away from the membrane (114).
Viruses such as HIV-1, HCV, and Hendra virus exploit N-glycans to shield themselves from the host immune response (23, 25, 115, 116). In a similar way, O-glycans have also been suggested to shield immunodominant epitopes in herpesviruses (91, 92). Nevertheless, there is limited information regarding the impact of individual O-glycans on shielding underlying peptide epitopes or the capacity of host immunity to present and recognize glycosylated antigens (93, 94). Here we show that herpesviruses are broadly O-glycosylated including protein regions subject to immune recognition. In this context we found O-glycans (Ser62, Ser70, and Ser71; Ser71 and Ser79; Tyr102; Tyr154) localized to four distinct immunodominant human B cell epitopes, previously mapped to the N-terminal region of VZV gE using non-glycosylated synthetic peptides (117). Two of these epitopes represent neutralizing antibody epitopes (Ser71 and Ser79; Tyr154) (118). In addition, three T cell epitopes in mice were also mapped to VZV gE regions with identified O-glycans (major epitope: Ser71 and Ser79; minor epitopes: Tyr102; Thr118) (118). Despite immunogens being produced in yeast and thereby lacking mucin-type O-glycosylation (18), mouse immune sera were able to detect and neutralize virus produced in mammalian cells, suggesting either that the epitope recognition is not affected by adjacent glycosylation or that putative O-linked glycans only partly occupy and protect the epitopes. We also found O-glycans located within a neutralizing peptide epitope at the N terminus of HCMV gH (S31) (119) and a discontinuous neutralizing antibody epitope on VZV gH (Ser177 and Thr184) (120). These findings suggest that O-linked glycans could have a role in the masking of immunodominant epitopes from antibodies and cytotoxic T cells. However, detailed studies are required to investigate the contribution of distinct O-glycans to epitope masking or recognition by immune cells. It should be mentioned that we could only confirm a subset of the gE glycosylation sites in the clinical VZV sample. This could be due to the low coverage caused by the scarce material available for analysis, or it could simply signify incomplete protection of these epitopes. Finally, specific O-glycans have not only been shown to protect but also to evoke immune responses (121, 122) and hence could serve as potential diagnostic markers, as well as vaccine targets.
As discussed, one of the potential caveats of O-glycoproteomic analysis in vitro is that viral glycosylation is placed in the context of host cell glycosylation capacity. To compare O-glycosylation in artificially infected cells to O-glycosylation in vivo, we analyzed VZV-infected fibroblasts and virus obtained from a primary clinical isolate in parallel. A relatively high degree of glycosylation overlap was identified between the clinical VZV sample and VZV derived from infected cells, although a significant number of O-glycosites were not found in the clinical sample. There could be several explanations for the lower number of glycosylation sites, including substantially scarcer clinical material compared with infected cell lysate, which only allowed us to perform a single digestion with trypsin. Another factor that could influence the identification of sites is genetic differences between the clinical and laboratory VZV isolates. For example, variable numbers of mucin-like tandem repeats can be found in viral glycoproteins, such as HSV-1 gI (123) and VZV gC, derived from different clinical isolates and laboratory strains. Even though VZV is one of the most conserved human herpesviruses (124), it cannot be excluded that minor deviations of the isolate-specific VZV peptide sequences from the sequences available in the search database could prevent the identification of all peptides. The same issues are valid for the investigated EBV P3HR1 and HCMV Towne strains, which are not completely annotated in the available protein databases. Despite the lower overall coverage in the clinical sample, O-glycosites identified in the laboratory VZV strain represent the in vivo glycosylation well, with the only sites exclusively identified in the clinical isolate residing within specific tandem repeats of VZV gC. This would support the use of laboratory strain-derived glycoproteins for addressing relevant biological questions. This represents the first attempt to characterize the O-glycoproteome of a clinical virus specimen. In the future, efforts should be made to evaluate the occupancy of individual glycosites and the individual structures on viruses. Comprehensive glycomic characterization of clinical isolates may lead to identification of sites and structures important for protein-protein interaction or raising potent immune responses. Using our expanding library of glycoengineered cell lines would enable production of designer viruses presenting defined glycostructures on envelope glycoproteins for antiviral vaccine development (125).
In conclusion, we generated the most comprehensive O-glycoproteomes of VZV, HCMV, and EBV to date and showed that certain regions of conserved proteins are consistently glycosylated in herpesviruses. O-Glycans on viral envelope glycoproteins can play multiple roles from inducing extended molecular conformations and protection from unspecific cleavage to more regulated events, such as protein-protein interaction, modulation of limited proteolysis, and immune recognition. The results should enable more focused studies of O-glycosylation at individual sites, which may confer new knowledge in specific areas of herpesvirus biology. Moreover, the results provide a reference base for design and development of vaccines taking both N- and O-glycosylation into account.
Author Contributions
I. B., R. N., S. O., and H. H. W. conceived and designed the experiments. I. B., R. N., and S. Y. V. performed the experiments. I. B., H. J. J., S. L. K., S. Y. V., and H. H. W. analyzed the data. I. B. and H. H. W. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript
Supplementary Material
This work was supported in part by Danish Research Council Grant 1331-00133B; Program of Excellence 2016 Grant 2016CDO04210 (Copenhagen as the next leader in precise genetic engineering, Program CDO2016) from the University of Copenhagen; Danish National Research Foundation Grant DNRF107; and by funds from the Carl Emil Friis og hustru Olga Doris Friis Foundation, the Lundbeck Foundation, the A. P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til Almene Formaal, the Kirsten og Freddy Johansen Fonden, the Mizutani Foundation, and the Novo Nordisk Foundation. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Data Sets S1–S5 and Supplemental Figures S1–S7.
- HSV
- herpes simplex virus
- VZV
- varicella zoster virus
- HCMV
- human cytomegalovirus
- EBV
- Epstein-Barr virus
- HEL
- human embryonic lung
- GPCR
- G protein-coupled receptor
- HCD
- higher energy collisional dissociation
- ETD
- electron-transfer dissociation.
References
- 1. Davison A. J. (2002) Evolution of the herpesviruses. Vet. Microbiol. 86, 69–88 [DOI] [PubMed] [Google Scholar]
- 2. Grinde B. (2013) Herpesviruses: latency and reactivation—viral strategies and host response. J. Oral Microbiol. 5, 22766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McGeoch D. J., Rixon F. J., and Davison A. J. (2006) Topics in herpesvirus genomics and evolution. Virus Res. 117, 90–104 [DOI] [PubMed] [Google Scholar]
- 4. Fishman J. A. (2013) Overview: cytomegalovirus and the herpesviruses in transplantation. Am. J. Transplant. 3, (Suppl. 3) 1–8 [DOI] [PubMed] [Google Scholar]
- 5. Shiley K., and Blumberg E. (2010) Herpes viruses in transplant recipients: HSV, VZV, human herpes viruses, and EBV. Infect. Dis. Clin. North Am. 24, 373–393 [DOI] [PubMed] [Google Scholar]
- 6. Astuto M., Palermo C. I., Costanzo C. M., Ettorre G. C., Palmucci S., Franchina C., Russo R., Valastro P., Timpanaro V., and Scalia G. (2014) Fatal pulmonary disease and encephalic complication in a man with HSV-1 infection: A case report. J. Clin. Virol. 59, 59–62 [DOI] [PubMed] [Google Scholar]
- 7. Rowe A. M., St Leger A. J., Jeon S., Dhaliwal D. K., Knickelbein J. E., and Hendricks R. L. (2013) Herpes keratitis. Prog. Retin. Eye Res. 32, 88–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sabugo F., Espinoza-Araya R., Meneses M. F., and Cuchacovich M. (2014) Acute herpes simplex virus 1 pneumonitis in a patient with systemic lupus erythematosus. J. Clin. Rheumatol. 20, 42–44 [DOI] [PubMed] [Google Scholar]
- 9. Sili U., Kaya A., Mert A., and HSV Encephalitis Study Group (2014) Herpes simplex virus encephalitis: clinical manifestations, diagnosis and outcome in 106 adult patients. J. Clin. Virol. 60, 112–118 [DOI] [PubMed] [Google Scholar]
- 10. Tsao S. W., Tsang C. M., To K. F., and Lo K. W. (2015) The role of Epstein-Barr virus in epithelial malignancies. J. Pathol. 235, 323–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gramolelli S., and Schulz T. F. (2015) The role of Kaposi sarcoma-associated herpesvirus in the pathogenesis of Kaposi sarcoma. J. Pathol. 235, 368–380 [DOI] [PubMed] [Google Scholar]
- 12. Davison A. J. (2007) Overview of classification. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis (Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R., and Yamanishi K., eds) pp. 3–9, Cambridge University Press, Cambridge, UK: [PubMed] [Google Scholar]
- 13. Krummenacher C., Carfí A., Eisenberg R. J., and Cohen G. H. (2013) Entry of herpesviruses into cells: the enigma variations. Adv. Exp. Med. Biol. 790, 178–195 [DOI] [PubMed] [Google Scholar]
- 14. Corrales-Aguilar E., Hoffmann K., and Hengel H. (2014) CMV-encoded Fcγ receptors: modulators at the interface of innate and adaptive immunity. Semin. Immunopathol. 36, 627–640 [DOI] [PubMed] [Google Scholar]
- 15. Favoreel H. W., Van Minnebruggen G., Van de Walle G. R., Ficinska J., and Nauwynck H. J. (2006) Herpesvirus interference with virus-specific antibodies: bridging antibodies, internalizing antibodies, and hiding from antibodies. Vet. Microbiol. 113, 257–263 [DOI] [PubMed] [Google Scholar]
- 16. Hobman T. C. (1993) Targeting of viral glycoproteins to the Golgi complex. Trends Microbiol. 1, 124–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Olofsson S., and Hansen J. E. (1998) Host cell glycosylation of viral glycoproteins: a battlefield for host defence and viral resistance. Scand. J. Infect. Dis. 30, 435–440 [DOI] [PubMed] [Google Scholar]
- 18. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., and Tabak L. A. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Serafini-Cessi F., Dall'Olio F., Scannavini M., and Campadelli-Fiume G. (1983) Processing of herpes simplex virus-1 glycans in cells defective in glycosyl transferases of the Golgi system: relationship to cell fusion and virion egress. Virology 131, 59–70 [DOI] [PubMed] [Google Scholar]
- 20. Wang J., Fan Q., Satoh T., Arii J., Lanier L. L., Spear P. G., Kawaguchi Y., and Arase H. (2009) Binding of herpes simplex virus glycoprotein B (gB) to paired immunoglobulin-like type 2 receptor α depends on specific sialylated O-linked glycans on gB. J. Virol. 83, 13042–13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Altgärde N., Eriksson C., Peerboom N., Phan-Xuan T., Moeller S., Schnabelrauch M., Svedhem S., Trybala E., Bergström T., and Bally M. (2015) Mucin-like region of herpes simplex virus type 1 attachment protein gC modulates the virus-glycosaminoglycan interaction. J. Biol. Chem. 290, 21473–21485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davis C. W., Mattei L. M., Nguyen H. Y., Ansarah-Sobrinho C., Doms R. W., and Pierson T. C. (2006) The location of asparagine-linked glycans on West Nile virions controls their interactions with CD209 (dendritic cell-specific ICAM-3 grabbing nonintegrin). J. Biol. Chem. 281, 37183–37194 [DOI] [PubMed] [Google Scholar]
- 23. Helle F., Vieyres G., Elkrief L., Popescu C. I., Wychowski C., Descamps V., Castelain S., Roingeard P., Duverlie G., and Dubuisson J. (2010) Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J. Virol. 84, 11905–11915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lennemann N. J., Rhein B. A., Ndungo E., Chandran K., Qiu X., and Maury W. (2014) Comprehensive functional analysis of N-linked glycans on Ebola virus GP1. mBio 5, e00862–e00813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang W., Nie J., Prochnow C., Truong C., Jia Z., Wang S., Chen X. S., and Wang Y. (2013) A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology 10, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chu F. L., Wen H. L., Hou G. H., Lin B., Zhang W. Q., Song Y. Y., Ren G. J., Sun C. X., Li Z. M., and Wang Z. (2013) Role of N-linked glycosylation of the human parainfluenza virus type 3 hemagglutinin-neuraminidase protein. Virus Res. 174, 137–147 [DOI] [PubMed] [Google Scholar]
- 27. Luo S., Hu K., He S., Wang P., Zhang M., Huang X., Du T., Zheng C., Liu Y., and Hu Q. (2015) Contribution of N-linked glycans on HSV-2 gB to cell-cell fusion and viral entry. Virology 483, 72–82 [DOI] [PubMed] [Google Scholar]
- 28. Suenaga T., Matsumoto M., Arisawa F., Kohyama M., Hirayasu K., Mori Y., and Arase H. (2015) Sialic acids on varicella-zoster virus glycoprotein B required for cell-cell fusion. J. Biol. Chem. 290, 19833–19843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pedersen J. W., Bennett E. P., Schjoldager K. T., Meldal M., Holmér A. P., Blixt O., Cló E., Levery S. B., Clausen H., and Wandall H. H. (2011) Lectin domains of polypeptide GalNAc transferases exhibit glycopeptide binding specificity. J. Biol. Chem. 286, 32684–32696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wandall H. H., Hassan H., Mirgorodskaya E., Kristensen A. K., Roepstorff P., Bennett E. P., Nielsen P. A., Hollingsworth M. A., Burchell J., Taylor-Papadimitriou J., and Clausen H. (1997) Substrate specificities of three members of the human UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J. Biol. Chem. 272, 23503–23514 [DOI] [PubMed] [Google Scholar]
- 31. Bagdonaite I., Nordén R., Joshi H. J., Dabelsteen S., Nyström K., Vakhrushev S. Y., Olofsson S., and Wandall H. H. (2015) A strategy for O-glycoproteomics of enveloped viruses: the O-glycoproteome of herpes simplex virus type 1. PLoS Pathog. 11, e1004784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Steentoft C., Vakhrushev S. Y., Vester-Christensen M. B., Schjoldager K. T., Kong Y., Bennett E. P., Mandel U., Wandall H., Levery S. B., and Clausen H. (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 [DOI] [PubMed] [Google Scholar]
- 33. Iversen M. B., Reinert L. S., Thomsen M. K., Bagdonaite I., Nandakumar R., Cheshenko N., Prabakaran T., Vakhrushev S. Y., Krzyzowska M., Kratholm S. K., Ruiz-Perez F., Petersen S. V., Goriely S., Bibby B. M., Eriksson K., et al. (2016) An innate antiviral pathway acting before interferons at epithelial surfaces. Nat. Immunol. 17, 150–158 [DOI] [PubMed] [Google Scholar]
- 34. Nordén R., Halim A., Nyström K., Bennett E. P., Mandel U., Olofsson S., Nilsson J., and Larson G. (2015) O-linked glycosylation of the mucin domain of the herpes simplex virus type 1-specific glycoprotein gC-1 is temporally regulated in a seed-and-spread manner. J. Biol. Chem. 290, 5078–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lundström M., Jeansson S., and Olofsson S. (1987) Host cell-induced differences in the O-glycosylation of herpes simplex virus gC-1: II. demonstration of cell-specific galactosyltransferase essential for formation of O-linked oligosaccharides. Virology 161, 395–402 [DOI] [PubMed] [Google Scholar]
- 36. Chiba S., Striker R. L. Jr., and Benyesh-Melnick M. (1972) Microculture plaque assay for human and simian cytomegaloviruses. Appl. Microbiol. 23, 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bergström T. (1996) Polymerase chain reaction for diagnosis of varicella zoster virus central nervous system infections without skin manifestations. Scand. J. Infect. Dis. Suppl. 100, 41–45 [PubMed] [Google Scholar]
- 38. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., et al. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sadaoka T., Yanagi T., Yamanishi K., and Mori Y. (2010) Characterization of the varicella-zoster virus ORF50 gene, which encodes glycoprotein M. J. Virol. 84, 3488–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zerboni L., Sen N., Oliver S. L., and Arvin A. M. (2014) Molecular mechanisms of varicella zoster virus pathogenesis. Nat. Rev. Microbiol. 12, 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suenaga T., Satoh T., Somboonthum P., Kawaguchi Y., Mori Y., and Arase H. (2010) Myelin-associated glycoprotein mediates membrane fusion and entry of neurotropic herpesviruses. Proc. Natl. Acad. Sci. U.S.A. 107, 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vleck S. E., Oliver S. L., Brady J. J., Blau H. M., Rajamani J., Sommer M. H., and Arvin A. M. (2011) Structure-function analysis of varicella-zoster virus glycoprotein H identifies domain-specific roles for fusion and skin tropism. Proc. Natl. Acad. Sci. U.S.A. 108, 18412–18417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oliver S. L., Sommer M., Zerboni L., Rajamani J., Grose C., and Arvin A. M. (2009) Mutagenesis of varicella-zoster virus glycoprotein B: putative fusion loop residues are essential for viral replication, and the furin cleavage motif contributes to pathogenesis in skin tissue in vivo. J. Virol. 83, 7495–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oliver S. L., Brady J. J., Sommer M. H., Reichelt M., Sung P., Blau H. M., and Arvin A. M. (2013) An immunoreceptor tyrosine-based inhibition motif in varicella-zoster virus glycoprotein B regulates cell fusion and skin pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, 1911–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hannah B. P., Cairns T. M., Bender F. C., Whitbeck J. C., Lou H., Eisenberg R. J., and Cohen G. H. (2009) Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J. Virol. 83, 6825–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mallory S., Sommer M., and Arvin A. M. (1997) Mutational analysis of the role of glycoprotein I in varicella-zoster virus replication and its effects on glycoprotein E conformation and trafficking. J. Virol. 71, 8279–8288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berarducci B., Rajamani J., Reichelt M., Sommer M., Zerboni L., and Arvin A. M. (2009) Deletion of the first cysteine-rich region of the varicella-zoster virus glycoprotein E ectodomain abolishes the gE and gI interaction and differentially affects cell-cell spread and viral entry. J. Virol. 83, 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Berarducci B., Rajamani J., Zerboni L., Che X., Sommer M., and Arvin A. M. (2010) Functions of the unique N-terminal region of glycoprotein E in the pathogenesis of varicella-zoster virus infection. Proc. Natl. Acad. Sci. U.S.A. 107, 282–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li Q., Ali M. A., and Cohen J. I. (2006) Insulin degrading enzyme is a cellular receptor mediating varicella-zoster virus infection and cell-to-cell spread. Cell 127, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Q., Krogmann T., Ali M. A., Tang W. J., and Cohen J. I. (2007) The amino terminus of varicella-zoster virus (VZV) glycoprotein E is required for binding to insulin-degrading enzyme, a VZV receptor. J. Virol. 81, 8525–8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moffat J., Ito H., Sommer M., Taylor S., and Arvin A. M. (2002) Glycoprotein I of varicella-zoster virus is required for viral replication in skin and T cells. J. Virol. 76, 8468–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oliver S. L., Sommer M. H., Reichelt M., Rajamani J., Vlaycheva-Beisheim L., Stamatis S., Cheng J., Jones C., Zehnder J., and Arvin A. M. (2011) Mutagenesis of varicella-zoster virus glycoprotein I (gI) identifies a cysteine residue critical for gE/gI heterodimer formation, gI structure, and virulence in skin cells. J. Virol. 85, 4095–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moffat J. F., Zerboni L., Kinchington P. R., Grose C., Kaneshima H., and Arvin A. M. (1998) Attenuation of the vaccine Oka strain of varicella-zoster virus and role of glycoprotein C in alphaherpesvirus virulence demonstrated in the SCID-hu mouse. J. Virol. 72, 965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grose C., Carpenter J. E., Jackson W., and Duus K. M. (2010) Overview of varicella-zoster virus glycoproteins gC, gH and gL. Curr. Top. Microbiol. Immunol. 342, 113–128 [DOI] [PubMed] [Google Scholar]
- 55. Varnum S. M., Streblow D. N., Monroe M. E., Smith P., Auberry K. J., Pasa-Tolic L., Wang D., Camp D. G. 2nd, Rodland K., Wiley S., Britt W., Shenk T., Smith R. D., and Nelson J. A. (2004) Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78, 10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mocarski E., Jr. (2007) Betaherpes viral genes and their functions. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis (Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R., and Yamanishi K., eds) pp. 204–230, Cambridge University Press, Cambridge, UK: [PubMed] [Google Scholar]
- 57. Dunn W., Chou C., Li H., Hai R., Patterson D., Stolc V., Zhu H., and Liu F. (2003) Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U.S.A. 100, 14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou M., Lanchy J. M., and Ryckman B. J. (2015) Human cytomegalovirus gH/gL/gO promotes the fusion step of entry into all cell types, whereas gH/gL/UL128–131 broadens virus tropism through a distinct mechanism. J. Virol. 89, 8999–9009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ciferri C., Chandramouli S., Donnarumma D., Nikitin P. A., Cianfrocco M. A., Gerrein R., Feire A. L., Barnett S. W., Lilja A. E., Rappuoli R., Norais N., Settembre E. C., and Carfi A. (2015) Structural and biochemical studies of HCMV gH/gL/gO and pentamer reveal mutually exclusive cell entry complexes. Proc. Natl. Acad. Sci. U.S.A. 112, 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sekulin K., Görzer I., Heiss-Czedik D., and Puchhammer-Stöckl E. (2007) Analysis of the variability of CMV strains in the RL11D domain of the RL11 multigene family. Virus Genes 35, 577–583 [DOI] [PubMed] [Google Scholar]
- 61. Corrales-Aguilar E., Trilling M., Hunold K., Fiedler M., Le V. T., Reinhard H., Ehrhardt K., Mercé-Maldonado E., Aliyev E., Zimmermann A., Johnson D. C., and Hengel H. (2014) Human cytomegalovirus Fcγ binding proteins gp34 and gp68 antagonize Fcγ receptors I, II and III. PLoS Pathog. 10, e1004131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cortese M., Calò S., D'Aurizio R., Lilja A., Pacchiani N., and Merola M. (2012) Recombinant human cytomegalovirus (HCMV) RL13 binds human immunoglobulin G Fc. PLoS One 7, e50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vomaske J., Nelson J. A., and Streblow D. N. (2009) Human Cytomegalovirus US28: a functionally selective chemokine binding receptor. Infectious Disorders Drug Targets 9, 548–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tadagaki K., Tudor D., Gbahou F., Tschische P., Waldhoer M., Bomsel M., Jockers R., and Kamal M. (2012) Human cytomegalovirus-encoded UL33 and UL78 heteromerize with host CCR5 and CXCR4 impairing their HIV coreceptor activity. Blood 119, 4908–4918 [DOI] [PubMed] [Google Scholar]
- 65. Casarosa P., Waldhoer M., LiWang P. J., Vischer H. F., Kledal T., Timmerman H., Schwartz T. W., Smit M. J., and Leurs R. (2005) CC and CX3C chemokines differentially interact with the N terminus of the human cytomegalovirus-encoded US28 receptor. J. Biol. Chem. 280, 3275–3285 [DOI] [PubMed] [Google Scholar]
- 66. Lesniewski M., Das S., Skomorovska-Prokvolit Y., Wang F. Z., and Pellett P. E. (2006) Primate cytomegalovirus US12 gene family: a distinct and diverse clade of seven-transmembrane proteins. Virology 354, 286–298 [DOI] [PubMed] [Google Scholar]
- 67. Cavaletto N., Luganini A., and Gribaudo G. (2015) Inactivation of the human cytomegalovirus US20 gene hampers productive viral replication in endothelial cells. J. Virol. 89, 11092–11106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Müllberg J., Hsu M. L., Rauch C. T., Gerhart M. J., Kaykas A., and Cosman D. (1999) The R27080 glycoprotein is abundantly secreted from human cytomegalovirus-infected fibroblasts. J. Gen. Virol. 80, 437–440 [DOI] [PubMed] [Google Scholar]
- 69. Wang D., Bresnahan W., and Shenk T. (2004) Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc. Natl. Acad. Sci. U.S.A. 101, 16642–16647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lüttichau H. R. (2010) The cytomegalovirus UL146 gene product vCXCL1 targets both CXCR1 and CXCR2 as an agonist. J. Biol. Chem. 285, 9137–9146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Geyer H., Hartung E., Mages H. W., Weise C., Belužić R., Vugrek O., Jonjic S., Kroczek R. A., and Voigt S. (2014) Cytomegalovirus expresses the chemokine homologue vXCL1 capable of attracting XCR1+ CD4− dendritic cells. J. Virol. 88, 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Müller S., Zocher G., Steinle A., and Stehle T. (2010) Structure of the HCMV UL16-MICB complex elucidates select binding of a viral immunoevasin to diverse NKG2D ligands. PLoS Pathog. 6, e1000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tirabassi R. S., and Ploegh H. L. (2002) The human cytomegalovirus US8 glycoprotein binds to major histocompatibility complex class I products. J. Virol. 76, 6832–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Johannsen E., Luftig M., Chase M. R., Weicksel S., Cahir-McFarland E., Illanes D., Sarracino D., and Kieff E. (2004) Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U.S.A. 101, 16286–16291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang X., Kenyon W. J., Li Q., Müllberg J., and Hutt-Fletcher L. M. (1998) Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72, 5552–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sathiyamoorthy K., Jiang J., Hu Y. X., Rowe C. L., Möhl B. S., Chen J., Jiang W., Mellins E. D., Longnecker R., Zhou Z. H., and Jardetzky T. S. (2014) Assembly and architecture of the EBV B cell entry triggering complex. PLoS Pathog. 10, e1004309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liu F., Marquardt G., Kirschner A. N., Longnecker R., and Jardetzky T. S. (2010) Mapping the N-terminal residues of Epstein-Barr virus gp42 that bind gH/gL by using fluorescence polarization and cell-based fusion assays. J. Virol. 84, 10375–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sorem J., Jardetzky T. S., and Longnecker R. (2009) Cleavage and secretion of Epstein-Barr virus glycoprotein 42 promote membrane fusion with B lymphocytes. J. Virol. 83, 6664–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lake C. M., and Hutt-Fletcher L. M. (2000) Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 74, 11162–11172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nemerow G. R., Houghten R. A., Moore M. D., and Cooper N. R. (1989) Identification of an epitope in the major envelope protein of Epstein-Barr virus that mediates viral binding to the B lymphocyte EBV receptor (CR2). Cell 56, 369–377 [DOI] [PubMed] [Google Scholar]
- 81. Serafini-Cessi F., Malagolini N., Nanni M., Dall'Olio F., Campadelli-Fiume G., Tanner J., and Kieff E. (1989) Characterization of N- and O-linked oligosaccharides of glycoprotein 350 from Epstein-Barr virus. Virology 170, 1–10 [DOI] [PubMed] [Google Scholar]
- 82. Janz A., Oezel M., Kurzeder C., Mautner J., Pich D., Kost M., Hammerschmidt W., and Delecluse H. J. (2000) Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 74, 10142–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Thorley-Lawson D. A., and Poodry C. A. (1982) Identification and isolation of the main component (gp350-gp220) of Epstein-Barr virus responsible for generating neutralizing antibodies in vivo. J. Virol. 43, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Olofsson S., Sjöblom I., Lundström M., Jeansson S., and Lycke E. (1983) Glycoprotein C of herpes simplex virus type 1: characterization of O-linked oligosaccharides. J. Gen. Virol. 64, 2735–2747 [DOI] [PubMed] [Google Scholar]
- 85. Yang Z. Y., Duckers H. J., Sullivan N. J., Sanchez A., Nabel E. G., and Nabel G. J. (2000) Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 6, 886–889 [DOI] [PubMed] [Google Scholar]
- 86. Levery S. B., Steentoft C., Halim A., Narimatsu Y., Clausen H., and Vakhrushev S. Y. (2015) Advances in mass spectrometry driven O-glycoproteomics. Biochim. Biophys. Acta 1850, 33–42 [DOI] [PubMed] [Google Scholar]
- 87. Halim A., Westerlind U., Pett C., Schorlemer M., Rüetschi U., Brinkmalm G., Sihlbom C., Lengqvist J., Larson G., and Nilsson J. (2014) Assignment of saccharide identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides. J. Proteome Res. 13, 6024–6032 [DOI] [PubMed] [Google Scholar]
- 88. Wandall H. H., Rumjantseva V., Sørensen A. L., Patel-Hett S., Josefsson E. C., Bennett E. P., Italiano J. E. Jr., Clausen H., Hartwig J. H., and Hoffmeister K. M. (2012) The origin and function of platelet glycosyltransferases. Blood 120, 626–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nakamura S., Horie M., Daidoji T., Honda T., Yasugi M., Kuno A., Komori T., Okuzaki D., Narimatsu H., Nakaya T., and Tomonaga K. (2015) Influenza A virus-induced expression of a GalNAc transferase, GALNT3, via miRNAs is required for enhanced viral replication. J. Virol. 90, 1788–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Schjoldager K. T., Joshi H. J., Kong Y., Goth C. K., King S. L., Wandall H. H., Bennett E. P., Vakhrushev S. Y., and Clausen H. (2015) Deconstruction of O-glycosylation-GalNAc-T isoforms direct distinct subsets of the O-glycoproteome. EMBO Reports 16, 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kropff B., Burkhardt C., Schott J., Nentwich J., Fisch T., Britt W., and Mach M. (2012) Glycoprotein N of human cytomegalovirus protects the virus from neutralizing antibodies. PLoS Pathog. 8, e1002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Machiels B., Lété C., Guillaume A., Mast J., Stevenson P. G., Vanderplasschen A., and Gillet L. (2011) Antibody evasion by a gammaherpesvirus O-glycan shield. PLoS Pathog. 7, e1002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Olofsson S., Blixt O., Bergstrom T., Frank M., and Wandall H. H. (2015) Viral O-GalNAc peptide epitopes: a novel potential target in viral envelope glycoproteins. Rev. Med. Virol. 26, 34–48 [DOI] [PubMed] [Google Scholar]
- 94. Madsen C. B., Petersen C., Lavrsen K., Harndahl M., Buus S., Clausen H., Pedersen A. E., and Wandall H. H. (2012) Cancer associated aberrant protein O-glycosylation can modify antigen processing and immune response. PLoS One 7, e50139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Roizman B., and Campadelli-Fiume G. (2007) Alphaherpes viral genes and their functions. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis (Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R., and Yamanishi K., eds) pp. 70–92, Cambridge University Press, Cambridge, UK: [PubMed] [Google Scholar]
- 96. Heldwein E. E., Lou H., Bender F. C., Cohen G. H., Eisenberg R. J., and Harrison S. C. (2006) Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313, 217–220 [DOI] [PubMed] [Google Scholar]
- 97. Goth C. K., Halim A., Khetarpal S. A., Rader D. J., Clausen H., and Schjoldager K. T. (2015) A systematic study of modulation of ADAM-mediated ectodomain shedding by site-specific O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 112, 14623–14628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bräutigam J., Scheidig A. J., and Egge-Jacobsen W. (2013) Mass spectrometric analysis of hepatitis C viral envelope protein E2 reveals extended microheterogeneity of mucin-type O-linked glycosylation. Glycobiology 23, 453–474 [DOI] [PubMed] [Google Scholar]
- 99. Go E. P., Hua D., and Desaire H. (2014) Glycosylation and disulfide bond analysis of transiently and stably expressed clade C HIV-1 gp140 trimers in 293T cells identifies disulfide heterogeneity present in both proteins and differences in O-linked glycosylation. J. Proteome Res. 13, 4012–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Breuza L., Garcia M., Delgrossi M. H., and Le Bivic A. (2002) Role of the membrane-proximal O-glycosylation site in sorting of the human receptor for neurotrophins to the apical membrane of MDCK cells. Exp. Cell Res. 273, 178–186 [DOI] [PubMed] [Google Scholar]
- 101. Leuenberger B., Hahn D., Pischitzis A., Hansen M. K., and Sterchi E. E. (2003) Human meprin β: O-linked glycans in the intervening region of the type I membrane protein protect the C-terminal region from proteolytic cleavage and diminish its secretion. Biochem. J. 369, 659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Maryon E. B., Zhang J., Jellison J. W., and Kaplan J. H. (2009) Human copper transporter 1 lacking O-linked glycosylation is proteolytically cleaved in a Rab9-positive endosomal compartment. J. Biol. Chem. 284, 28104–28114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Remacle A. G., Chekanov A. V., Golubkov V. S., Savinov A. Y., Rozanov D. V., and Strongin A. Y. (2006) O-glycosylation regulates autolysis of cellular membrane type-1 matrix metalloproteinase (MT1-MMP). J. Biol. Chem. 281, 16897–16905 [DOI] [PubMed] [Google Scholar]
- 104. Schjoldager K. T., and Clausen H. (2012) Site-specific protein O-glycosylation modulates proprotein processing: deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim. Biophys. Acta 1820, 2079–2094 [DOI] [PubMed] [Google Scholar]
- 105. Rowe C. L., Chen J., Jardetzky T. S., and Longnecker R. (2015) Membrane anchoring of Epstein-Barr virus gp42 inhibits fusion with B cells even with increased flexibility allowed by engineered spacers. mBio 6, e02285–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Borza C. M., and Hutt-Fletcher L. M. (2002) Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8, 594–599 [DOI] [PubMed] [Google Scholar]
- 107. Abaitua F., Zia F. R., Hollinshead M., and O'Hare P. (2013) Polarized cell migration during cell-to-cell transmission of herpes simplex virus in human skin keratinocytes. J. Virol. 87, 7921–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gurevich V. V., and Gurevich E. V. (2008) How and why do GPCRs dimerize? Trends Pharmacol. Sci. 29, 234–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hakalahti A. E., Vierimaa M. M., Lilja M. K., Kumpula E. P., Tuusa J. T., and Petäjä-Repo U. E. (2010) Human β1-adrenergic receptor is subject to constitutive and regulated N-terminal cleavage. J. Biol. Chem. 285, 28850–28861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gabaev I., Elbasani E., Ameres S., Steinbrück L., Stanton R., Döring M., Lenac Rovis T., Kalinke U., Jonjic S., Moosmann A., and Messerle M. (2014) Expression of the human cytomegalovirus UL11 glycoprotein in viral infection and evaluation of its effect on virus-specific CD8 T cells. J. Virol. 88, 14326–14339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Reimer J. J., Backovic M., Deshpande C. G., Jardetzky T., and Longnecker R. (2009) Analysis of Epstein-Barr virus glycoprotein B functional domains via linker insertion mutagenesis. J. Virol. 83, 734–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Backovic M., Longnecker R., and Jardetzky T. S. (2009) Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc. Natl. Acad. Sci. U.S.A. 106, 2880–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Magrané J., Casaroli-Marano R. P., Reina M., Gåfvels M., and Vilaró S. (1999) The role of O-linked sugars in determining the very low density lipoprotein receptor stability or release from the cell. FEBS Lett. 451, 56–62 [DOI] [PubMed] [Google Scholar]
- 114. Schuman J., Qiu D., Koganty R. R., Longenecker B. M., and Campbell A. P. (2000) Glycosylations versus conformational preferences of cancer associated mucin core. Glycoconj. J. 17, 835–848 [DOI] [PubMed] [Google Scholar]
- 115. Fournillier A., Wychowski C., Boucreux D., Baumert T. F., Meunier J. C., Jacobs D., Muguet S., Depla E., and Inchauspé G. (2001) Induction of hepatitis C virus E1 envelope protein-specific immune response can be enhanced by mutation of N-glycosylation sites. J. Virol. 75, 12088–12097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bradel-Tretheway B. G., Liu Q., Stone J. A., McInally S., and Aguilar H. C. (2015) Novel functions of Hendra virus G N-Glycans and comparisons to Nipah virus. J. Virol. 89, 7235–7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Fowler W. J., Garcia-Valcarcel M., Hill-Perkins M. S., Murphy G., Harper D. R., Jeffries D. J., Burns N. R., Adams S. E., Kingsman A. J., and Layton G. T. (1995) Identification of immunodominant regions and linear B cell epitopes of the gE envelope protein of varicella-zoster virus. Virology 214, 531–540 [DOI] [PubMed] [Google Scholar]
- 118. Garcia-Valcarcel M., Fowler W. J., Harper D. R., Jeffries D. J., and Layton G. T. (1997) Induction of neutralizing antibody and T-cell responses to varicella-zoster virus (VZV) using Ty-virus-like particles carrying fragments of glycoprotein E (gE). Vaccine 15, 709–719 [DOI] [PubMed] [Google Scholar]
- 119. Nejatollahi F., Hodgetts S. J., Vallely P. J., and Burnie J. P. (2002) Neutralising human recombinant antibodies to human cytomegalovirus glycoproteins gB and gH. FEMS Immunol. Med. Microbiol. 34, 237–244 [DOI] [PubMed] [Google Scholar]
- 120. Xing Y., Oliver S. L., Nguyen T., Ciferri C., Nandi A., Hickman J., Giovani C., Yang E., Palladino G., Grose C., Uematsu Y., Lilja A. E., Arvin A. M., and Carfí A. (2015) A site of varicella-zoster virus vulnerability identified by structural studies of neutralizing antibodies bound to the glycoprotein complex gHgL. Proc. Natl. Acad. Sci. U.S.A. 112, 6056–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Cló E., Kracun S. K., Nudelman A. S., Jensen K. J., Liljeqvist J. Å., Olofsson S., Bergström T., and Blixt O. (2012) Characterization of the viral O-glycopeptidome: a novel tool of relevance for vaccine design and serodiagnosis. J. Virol. 86, 6268–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. D'Arrigo I., Cló E., Bergström T., Olofsson S., and Blixt O. (2013) Diverse IgG serum response to novel glycopeptide epitopes detected within immunodominant stretches of Epstein-Barr virus glycoprotein 350/220: diagnostic potential of O-glycopeptide microarrays. Glycoconj. J. 30, 633–640 [DOI] [PubMed] [Google Scholar]
- 123. Norberg P., Olofsson S., Tarp M. A., Clausen H., Bergström T., and Liljeqvist J. A. (2007) Glycoprotein I of herpes simplex virus type 1 contains a unique polymorphic tandem-repeated mucin region. J. Gen. Virol. 88, 1683–1688 [DOI] [PubMed] [Google Scholar]
- 124. Norberg P. (2010) Divergence and genotyping of human alpha-herpesviruses: an overview. Infect. Genet. Evol. 10, 14–25 [DOI] [PubMed] [Google Scholar]
- 125. Yang Z., Wang S., Halim A., Schulz M. A., Frodin M., Rahman S. H., Vester-Christensen M. B., Behrens C., Kristensen C., Vakhrushev S. Y., Bennett E. P., Wandall H. H., and Clausen H. (2015) Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat. Biotechnol. 33, 842–844 [DOI] [PubMed] [Google Scholar]
- 126. Sharma S., Wisner T. W., Johnson D. C., and Heldwein E. E. (2013) HCMV gB shares structural and functional properties with gB proteins from other herpesviruses. Virology 435, 239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Matsuura H., Kirschner A. N., Longnecker R., and Jardetzky T. S. (2010) Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc. Natl. Acad. Sci. U.S.A. 107, 22641–22646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Szakonyi G., Klein M. G., Hannan J. P., Young K. A., Ma R. Z., Asokan R., Holers V. M., and Chen X. S. (2006) Structure of the Epstein-Barr virus major envelope glycoprotein. Nat. Struct. Mol. Biol. 13, 996–1001 [DOI] [PubMed] [Google Scholar]
- 129. Litwin V., Jackson W., and Grose C. (1992) Receptor properties of two varicella-zoster virus glycoproteins, gpI and gpIV, homologous to herpes simplex virus gE and gI. J. Virol. 66, 3643–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Christensen J., Steain M., Slobedman B., and Abendroth A. (2013) Varicella-zoster virus glycoprotein I is essential for spread in dorsal root ganglia and facilitates axonal localization of structural virion components in neuronal cultures. J. Virol. 87, 13719–13728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wille P. T., Wisner T. W., Ryckman B., and Johnson D. C. (2013) Human cytomegalovirus (HCMV) glycoprotein gB promotes virus entry in trans acting as the viral fusion protein rather than as a receptor-binding protein. mBio 4, e00332–00313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Vanarsdall A. L., Ryckman B. J., Chase M. C., and Johnson D. C. (2008) Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans. J. Virol. 82, 11837–11850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Mach M., Kropff B., Kryzaniak M., and Britt W. (2005) Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J. Virol. 79, 2160–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Engel P., Pérez-Carmona N., Albà M. M., Robertson K., Ghazal P., and Angulo A. (2011) Human cytomegalovirus UL7, a homologue of the SLAM-family receptor CD229, impairs cytokine production. Immunol. Cell Biol. 89, 753–766 [DOI] [PubMed] [Google Scholar]
- 135. MacManiman J. D., Meuser A., Botto S., Smith P. P., Liu F., Jarvis M. A., Nelson J. A., and Caposio P. (2014) Human cytomegalovirus-encoded pUL7 is a novel CEACAM1-like molecule responsible for promotion of angiogenesis. mBio 5, e02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Fielding C. A., Aicheler R., Stanton R. J., Wang E. C., Han S., Seirafian S., Davies J., McSharry B. P., Weekes M. P., Antrobus P. R., Prod'homme V., Blanchet F. P., Sugrue D., Cuff S., Roberts D., et al. (2014) Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation. PLoS Pathog. 10, e1004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Li G., Nguyen C. C., Ryckman B. J., Britt W. J., and Kamil J. P. (2015) A viral regulator of glycoprotein complexes contributes to human cytomegalovirus cell tropism. Proc. Natl. Acad. Sci. U.S.A. 112, 4471–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Noriega V. M., Hesse J., Gardner T. J., Besold K., Plachter B., and Tortorella D. (2012) Human cytomegalovirus US3 modulates destruction of MHC class I molecules. Mol. Immunol. 51, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Bronzini M., Luganini A., Dell'Oste V., De Andrea M., Landolfo S., and Gribaudo G. (2012) The US16 gene of human cytomegalovirus is required for efficient viral infection of endothelial and epithelial cells. J. Virol. 86, 6875–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Noriega V. M., Gardner T. J., Redmann V., Bongers G., Lira S. A., and Tortorella D. (2014) Human cytomegalovirus US28 facilitates cell-to-cell viral dissemination. Viruses 6, 1202–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Borza C. M., and Hutt-Fletcher L. M. (1998) Epstein-Barr virus recombinant lacking expression of glycoprotein gp150 infects B cells normally but is enhanced for infection of epithelial cells. J. Virol. 72, 7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.