Abstract
Specific proteins are modified by ubiquitin at the endoplasmic reticulum (ER) and are degraded by the proteasome, a process referred to as ER-associated protein degradation. In Saccharomyces cerevisiae, two principal ER-associated protein degradation ubiquitin ligases (E3s) reside in the ER membrane, Doa10 and Hrd1. The membrane-embedded Doa10 functions in the degradation of substrates in the ER membrane, nuclear envelope, cytoplasm, and nucleoplasm. How most E3 ligases, including Doa10, recognize their protein substrates remains poorly understood. Here we describe a previously unappreciated but highly conserved C-terminal element (CTE) in Doa10; this cytosolically disposed 16-residue motif follows the final transmembrane helix. A conserved CTE asparagine residue is required for ubiquitylation and degradation of a subset of Doa10 substrates. Such selectivity suggests that the Doa10 CTE is involved in substrate discrimination and not general ligase function. Functional conservation of the CTE was investigated in the human ortholog of Doa10, MARCH6 (TEB4), by analyzing MARCH6 autoregulation of its own degradation. Mutation of the conserved Asn residue (N890A) in the MARCH6 CTE stabilized the normally short lived enzyme to the same degree as a catalytically inactivating mutation (C9A). We also report the localization of endogenous MARCH6 to the ER using epitope tagging of the genomic MARCH6 locus by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-mediated genome editing. These localization and CTE analyses support the inference that MARCH6 and Doa10 are functionally similar. Moreover, our results with the yeast enzyme suggest that the CTE is involved in the recognition and/or ubiquitylation of specific protein substrates.
Keywords: E3 ubiquitin ligase, endoplasmic-reticulum-associated protein degradation (ERAD), proteasome, proteolysis, ubiquitin, Doa10, MARCH6, TEB4
Introduction
Selective protein degradation in eukaryotic cells is largely carried out by the ubiquitin-proteasome system. Proteins identified for degradation by the ubiquitin-proteasome system are covalently modified with ubiquitin, a highly conserved 76-residue protein (1, 2). In most cases, ubiquitin is conjugated to the target protein via an amide linkage between its C-terminal glycine and ϵ-amino groups on substrate lysine residues. Ubiquitylation requires an enzyme cascade beginning with an E1 ubiquitin-activating enzyme, which adenylates the ubiquitin C-terminal carboxyl group and subsequently forms a high energy thioester bond with it. Ubiquitin is then transferred from the E1 active site cysteine to a cysteine in an E2 ubiquitin-conjugating enzyme. Finally, an E3 ubiquitin ligase mediates ubiquitin transfer from the E2 to the target protein. E3s come in several mechanistically distinct classes, but the most abundant are the RING E3s. The RING domain is characterized by a set of Cys and His residues that coordinate a pair of zinc ions; the RING directly contacts the E2-ubiquitin thioester-linked complex and promotes ubiquitin transfer to a substrate (3). Polyubiquitin chains are formed when the C terminus of one donor ubiquitin is conjugated to one of seven lysine residues, or the α-amino group, of an acceptor ubiquitin. Ubiquitin chains, typically linked through lysine 48 (Lys-48) of ubiquitin, are recognized by a large protease called the 26S proteasome, which will degrade the ubiquitylated substrate (2).
One branch of the ubiquitin-proteasome system operates at the ER4 and contiguous nuclear membranes. Removal of ubiquitylated proteins from the ER lumen and membrane bilayer is required for them to be destroyed by the 26S proteasome. The activity of the ubiquitin-proteasome system at the ER is commonly referred to as ER-associated protein degradation (ERAD) (4). Ubiquitylation of ERAD substrates in the yeast Saccharomyces cerevisiae is predominantly dependent on two membrane-embedded E3 ligases, Doa10 and Hrd1 (5). ERAD substrate specificity of these two E3s is dictated by localization of the ligase as well as the location and nature of the degradation signals within the substrate. Hrd1 localizes to the ER membrane and targets misfolded proteins with lesions in their luminal or transmembrane domains, termed ERAD-L and ERAD-M substrates, respectively. Doa10 is localized in the ER and inner nuclear membranes (6) and targets misfolded cytosolic/nucleoplasmic domains of soluble and membrane-embedded proteins, termed ERAD-C substrates (7). Recently, Doa10 was also found to target an ERAD-M substrate (8).
Both Hrd1 and Doa10 enzymes are part of distinct protein complexes with components required for substrate targeting, ubiquitylation, or retrotranslocation across the ER membrane (5, 9, 10). The 1319-residue Doa10 E3 ligase has 14 transmembrane segments, a topology that appears to be conserved in the mammalian ortholog MARCH6 (TEB4) (11). Both the N and C termini of Doa10/MARCH6 are exposed to the cytosol with ubiquitin ligase activity derived from an N-terminal RING-CH domain, a subclass of RING domains (12). Ubiquitylation of Doa10 substrates requires the activity of two E2 ubiquitin-conjugating enzymes, Ubc6 and Ubc7 (5).
Degradation of Doa10 substrates serves either regulatory or quality control purposes. One model substrate, a soluble nuclear protein, is the MATα2 transcriptional repressor. Within its N-terminal 67 residues, MATα2 bears a degradation signal (degron) for recognition by the Doa10 pathway (13, 14). This degron, named Deg1, is transferable, being sufficient to cause rapid degradation when fused to a variety of normally stable proteins (15). The molecular mechanisms underlying Doa10 recognition of Deg1 fusions, or other classes of substrates, are only partially understood. At least some Doa10 substrates require molecular chaperones for their ubiquitylation and degradation (see “Discussion”) (16–18). An unresolved question is whether Doa10 recognizes its substrates directly or requires a bridging chaperone(s) for binding.
As part of our attempts to determine mechanisms driving Doa10 substrate recognition, we uncovered a highly conserved but previously unrecognized C-terminal element (CTE) in Doa10 that plays a crucial role in the degradation of a specific subset of Doa10 substrates. An intact C-terminal domain in Doa10 was previously inferred to be required for the degradation of several substrates as they appear to be stabilized in a C-tail-deleted doa10 allele (11). Point mutagenesis of the tail has now revealed that the CTE is required for degradation of select Doa10 substrates, including ones carrying the Deg1 degron. The degradation defects in a doa10-N1314A mutant were traced to a loss of E3-mediated substrate ubiquitylation, suggesting a role for the CTE either before or at the ubiquitin ligation step. The CTE is not required for the turnover of two of the tested Doa10 substrates, Ubc6 and Ste6*, suggesting that the tail participates in specific substrate recognition rather than general ubiquitin ligase activity. We also initiated a parallel analysis of human MARCH6, the little studied ortholog of yeast Doa10. Besides demonstrating that endogenous MARCH6 is an ER-localized enzyme, we show that its CTE is essential for rapid autodegradation of MARCH6 mediated by its RING domain. These results highlight the evolutionarily conserved function of the CTE in these ERAD ubiquitin ligase orthologs and provide evidence for a ligase element that promotes recognition of specific substrates either directly or indirectly.
Experimental Procedures
Media and Methods for Yeast and Bacteria
Yeast rich (yeast extract-peptone-dextrose) and minimal (SD) media were prepared as described; yeast strains were genetically manipulated using standard techniques (19). For all experiments, yeast was grown at 30 °C. Standard methods were used for recombinant DNA manipulations (20).
Construction of Yeast Strains
All strains used in this study are derived from the MHY500 background (21) except YPH499 (22) and are listed in Table 1. MHY1685 is the MATa form of MHY1631 (12). Integrated missense mutations within the chromosomal DOA10 open reading frame (ORF) were introduced by delitto perfetto as described previously (23). In brief, mutations were generated by integrating the counterselectable reporter cassette KlURA3/kanMX4 (24) at nucleotide position 3928 (MHY3996) or 141 (MHY6669 and MHY6670) of the DOA10 ORF. The counterselectable reporter cassette was subsequently exchanged with an oligonucleotide duplex containing the missense mutation flanked on each end by 45–50 base pairs (bp) of DOA10-homologous sequence. MHY3996 was the strain used to generate MHY3998 and MHY4242, and MHY6669 and MHY6670 were used to generate MHY6758 and MHY6734, respectively. C-terminal epitope tagging of the WT allele encoding full-length Doa10 (MHY8655 and MHY2916), the C-terminally truncated protein encoded by doa10(1–1291) (MHY8663), and various CTE doa10 mutants (MHY6753, MHY6754, MHY6755, and MHY8688) was performed by in-frame integration of the 13myc-HIS3MX6 cassette from pFA6a-13myc-His3MX6 (25). All genomic DOA10 gene alterations were confirmed by DNA sequencing. MHY6605 was generated by replacing PDR5 with a kanMX cassette in the diploid MHY6498. After sporulation, a G418-resistant segregant was selected and confirmed by PCR. MHY6948 was derived from a cross between MHY1685 and MHY6605. MHY6948 was crossed with MHY3998, MHY6758, and MHY4242 to obtain MHY7038, MHY7040, and MHY7041, respectively. SKY307 is a derivative of SKY254 (23).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source/Ref. |
|---|---|---|
| MHY496 | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 ubc6-Δ1::HIS3 | 21 |
| MHY500 | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 | 13 |
| MHY551 | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 ubc7Δ::LEU2 | 13 |
| MHY1685 | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa10Δ::HIS3 | 12 |
| MHY2916 | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 DOA10-13myc-HISMX6 | This study |
| MHY3996 | MATa his3-Δ200 leu2-3,112::LEU2-Deg1-lacZ ura3-52 lys2-801 trp1-1 doa10::CORE cassette at position 3928 (KIURA3/kanMX4) | This study |
| MHY3998 | MATa his3-Δ200 leu2-3,112::LEU2-Deg1-lacZ ura3-52 lys2-801 trp1-1 doa10-G1309L | This study |
| MHY4242 | MATa his3-Δ200 leu2-3,112::LEU2-Deg1-lacZ ura3-52 lys2-801 trp1-1 doa10-N1314A | This study |
| MHY6605 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 pdr5Δ::kanMX6 | This study |
| MHY6669 | MATa his3-Δ200 leu2-3,112::LEU2-Deg1-lacZ ura3-52 lys2-801 trp1-1 doa10-G1309L::CORE cassette at position 141 (KIURA3/kanMX4) | This study |
| MHY6670 | MATa his3-Δ200 leu2-3,112::LEU2-Deg1-lacZ ura3-52 lys2-801 trp1-1 doa10-N1314A::CORE cassette at position 141 (KIURA3/kanMX4) | This study |
| MHY6734 | MATa his3-Δ200 leu2-3,112::LEU2-Deg1-lacZ ura3-52 lys2-801 trp1-1 doa10-N1314A-C39S | This study |
| MHY6753 | MATa his3-Δ200 leu2-3,112::LEU2-Deg1-lacZ ura3-52 lys2-801 trp1-1 doa10-G1309L-13myc-HISMX6 | This study |
| MHY6754 | MATa his3-Δ200 leu2-3,112::LEU2-Deg1-lacZ ura3-52 lys2-801 trp1-1 doa10-N1314A-13myc-HISMX6 | This study |
| MHY6755 | MATa his3-Δ200 leu2-3,112::LEU2-Deg1-lacZ ura3-52 lys2-801 trp1-1 doa10-G1309L-N1314A-13myc-HISMX6 | This study |
| MHY6758 | MATa his3-Δ200 leu2-3,112::LEU2-Deg1-lacZ ura3-52 lys2-801 trp1-1 doa10-C39S-G1309L | This study |
| MHY6948 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 pdr5Δ::kanMX6 doa10Δ::HIS3 | This study |
| MHY7038 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 pdr5Δ::kanMX6 doa10-G1309L | This study |
| MHY7040 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 pdr5Δ::kanMX6 doa10-G1309L-C39S | This study |
| MHY7041 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 pdr5Δ::kanMX6 doa10-N1314A | This study |
| MHY8655 | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 DOA10-13myc-HISMX6 | This study |
| MHY8663 | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa10 1-1291-13myc-HISMX6 | This study |
| MHY8688 | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa10-Y1306F-13myc-HISMX6 | This study |
| SKY307 | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa10-E633D | 23 |
| YPH499 | MATa his3-Δ200 leu2-Δ1 ura3-52 lys2-801 trp1-Δ63 ade2-101 | 22 |
Plasmids
All yeast plasmids used in this study were reported previously (14–16, 26–30). For expression of human MARCH6 (TEB4), the pcDNA3.1(−)-TEB4–3xFLAG-His6 vector was constructed as described (31). Oligonucleotides JMB409 (5′-GGTAAGCTTGACTATAAAGACCATGACGGTGATTATAAAGATCATGATATCGATTACAAG-3′) and JMB410 (5′-AATCTTAAGTTAATGATGGTGATGGTGATGAGAACCCTTGTCATCGTCATCCTTGTAATC-3′) were annealed and extended, generating a 3xFLAG-His6 DNA fragment with flanking HindIII and AflII sites. After digestion, the insert was ligated into HindIII-, AflII-cut pcDNA3.1(−)-TEB4-Myc-His and pcDNA3.1(−)-TEB4-C9A-Myc-His (obtained from Emmanuel Wiertz, Leiden University Medical Center, Leiden, Netherlands). Missense mutations G885L and N890A were introduced into pcDNA3.1(−)-TEB4–3xFLAG-His6 by site-directed mutagenesis (32). For each TEB4–3xFLAG-His6 plasmid, DNA for the entire ORF was sequenced. To generate pX459 gRNA-TEB4-HA, oligos JMB369 (5′-CACCGTGAGACAACTACTTTATTCT-3′) and JMB370 (5′-AAACAGAATAAAGTAGTTGTCTCAC-3′) were annealed, 5′-phosphorylated with T4 polynucleotide kinase, and ligated into BbsI-digested pX459 (33). The insert was verified by DNA sequencing.
Antibodies and Immunoblotting
The following mouse monoclonal antibodies were used for immunoblotting: anti-HA (16B12, Covance), anti-phosphoglycerate kinase (459250, ThermoFisher), anti-FLAG (F3165, Sigma), anti-β-gal (Z3781, Promega), and anti-β-actin (A5441, Sigma). The following rabbit polyclonal antibodies were used: anti-Ubc6 (gift from Thomas Sommer; validated in Ref. 12), anti-Doa10 (11), and the commercial anti-glucose-6-phosphate dehydrogenase (A9521, Sigma). Primary antibody incubations were followed by incubation with peroxidase-coupled anti-IgG antibodies (NA931V and NA934V, GE Healthcare) and visualized by enhanced chemiluminescence (34) with imaging on a G:Box system (Syngene) for quantification (Gene Tools, Syngene) or on film (Denville) as indicated in the figure legends.
Pulse-Chase Assays
Pulse-chase analysis was performed as described (13, 16, 20, 35). Briefly, log phase yeast cultures grown in SD medium (plus 200 μm CuSO4 for pOC9-URA3-HA-CL1-transformed strains) were labeled with ∼20 μCi of Tran35S-label (MP Biomedicals, GE Healthcare) per A600 unit of cells for 5–10 min in SD medium lacking methionine and cysteine. Labeled cultures were chased with excess unlabeled Met and Cys (10 mm each), and 2.2 A600 units were collected and processed for each time point. Following immunoprecipitation, proteins were eluted in Laemmli sample buffer and resolved by SDS-PAGE. Dried gels were analyzed on a Storm 860 phosphorimaging system and quantified with ImageQuant 5.2 software (GE Healthcare).
Cycloheximide-Chase Assays and Protein Extraction
Cycloheximide (CHX)-chase assays were performed as described previously (12, 27). In brief, yeast cultures were grown in yeast extract-peptone-dextrose or selective SD medium to log phase, 12.5 A600 units were collected, and CHX (10 mg/ml stock in EtOH; C7698, Sigma) was added to a final concentration of 0.25 mg/ml. At each time point, 2.5 A600 units were collected and later lysed with 300 mm NaOH, 100 mm 2-mercaptoethanol followed by 5% trichloroacetic acid (TCA) precipitation (36). Precipitated proteins were resuspended in TCA sample buffer (3.5% SDS, 0.5 m DTT, 80 mm Tris, 8 mm EDTA, 15% glycerol, 0.1 mg/ml bromophenol blue), and 0.5 A600 eq per lane were evaluated by immunoblotting.
In Vivo Ubiquitylation
Detection of the ubiquitin-modified Deg1-β-gal substrate was performed as described (37). Deg1-β-gal and HA-ubiquitin were co-expressed from pLR1 (YEplac195-Deg1-lacZ) and YEp112 plasmids, respectively. Substrate was immunoprecipitated using mouse anti-β-gal (1:500), resolved by SDS-PAGE, and assayed by immunoblotting.
Yeast Growth Analysis
Yeast cells were grown in selective SD medium overnight. Each culture was diluted to an A600 of 0.2, and 10-fold serial dilutions were made. Diluted cultures were spotted on solid SD medium, incubated at 30 °C for 2 days, and imaged.
Human Cell Culturing and Transfection
HeLa cells (ATCC) were cultured in standard conditions, namely in complete medium (DMEM supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin) at 37 °C in 5% CO2 in a humidified incubator. Cells were plated at 30–40% density 24 h prior to transfecting with a ratio of 150 μl Opti-MEM (Life Technologies), 6 μl of X-tremeGENE HP (Roche Applied Science), and 2 μg of DNA/2 ml of antibiotic-free complete medium according to the protocol from Roche Applied Science.
Human Cell Cycloheximide-Chase Analysis and Protein Extraction
HeLa cells in 6-well plates were treated with CHX at a final concentration of 0.1 mg/ml (0.1 g/ml stock in EtOH) 24 h after MARCH6–3xFLAG-His6 transfection. Cells were collected at each time point (0, 2, and 4 h) by tryptic detachment (0.05% trypsin and EDTA), neutralized with complete medium, transferred to a 1.6-ml microfuge tube, and placed on ice. Cells were pelleted at 500 × g for 4 min at 4 °C, washed once in ice-cold PBS, pelleted, and flash frozen in liquid nitrogen. Thawed cell pellets were lysed in 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1% (v/v) Triton X-100, 2 mm EDTA, 1 mm PMSF, and cOmplete EDTA-free protease inhibitor tablet (Roche Applied Science) (31) by rotating for 15 min at 4 °C. The insoluble fraction was pelleted at 11,200 × g for 10 min, supernatant was collected, and protein concentration was determined by the BCA assay (Thermo). Protein lysates were subjected to immunoblotting analysis. Cell extracts for measuring steady-state protein levels were generated as above, preceded in some cases by incubation for 4 h with 10 μm MG132 (10 mm stock in dimethyl sulfoxide; sc-201270, Santa Cruz Biotechnology) or dimethyl sulfoxide vehicle control.
Immunofluorescence Imaging of HeLa Cells
For transient transfections, HeLa cells were plated at 15–20% confluence on round, 18-mm diameter, Number 1 thickness Vista Vision Cover Glass (VWR), and the cells were transfected 24 h and processed 48 h after seeding. CRISPR/Cas9-generated MARCH6–2HA HeLa cells were seeded at 30–40% confluence 24 h prior to processing. Cells were fixed for 15 min in 3.7% (v/v) formaldehyde in Tris-buffered saline (TBS), permeabilized for 20 min with 0.2% (v/v) Triton X-100 in TBS, and blocked for 1 h in TBS containing 5% (w/v) BSA. Coverslips were incubated with the following primary antibodies diluted in 5% (w/v) BSA and 0.1% (v/v) Triton X-100 in TBS for 1 h or overnight at 4 °C: mouse anti-HA (1:500) or mouse anti-FLAG (1:1000) and rabbit anti-calnexin (1:500; ab75801, Abcam). Coverslips were incubated for 1 h with Alexa Fluor 488-conjugated anti-mouse and Alexa Fluor 568-conjugated anti-rabbit secondary antibodies (Life Technologies) (diluted 1:700 as above). DNA was stained with Hoechst 33342 (Invitrogen). Coverslips were mounted on glass slides using Fluoromount-G (Southern Biotech) and visualized with a 63×/1.4 numerical aperture lens on a Zeiss Axio Observer D1 microscope. Images were captured at room temperature using an AxioCam MRm charge-coupled device camera (Zeiss) paired with AxioVision (release 4.8.2.0) imaging software (Zeiss). Images were processed with Photoshop CS4 (Adobe).
CRISPR/Cas9 Modification of Chromosomal MARCH6
HeLa cells were electroporated with pX459-gRNA3-TEB4-HA and a single-stranded donor oligonucleotide (ssODN) as template for homologous recombination as described previously (33, 38). For each electroporation, 2 × 106 pelleted HeLa cells were resuspended in 400 μl of ice-cold electroporation buffer (Bio-Rad) along with 5 μg of pX459-gRNA3-TEB4-HA and 1 μl of 100 μm stock ssODN in a 4-mm Gene Pulser cuvette (Bio-Rad) and electroporated at 200 V, 850 microfarads, and infinite resistance using a Gene Pulser Xcell (Bio-Rad). Cells were immediately transferred with an equal volume of complete medium and allowed to recover in 6-well dishes. Cells were expanded and verified by indirect immunofluorescence to contain modified MARCH6. The ssODN sequence is gRNA3–6xGly-2xHA (5′-AATCTGGCAAACAAGGCTCATCTCCACCACCTCCACAGTCATCACAAGAAGGGGGAGGCGGGGGTGGATACCCATACGATGTTCCAGATTACGCTTACCCATACGATGTTCCAGATTACGCTTAGAGTAGTTGTCTCAACAACTTGACCTTCCCCTTTACATGTCCTTTTTTGTG-3′). The ssODN contains a 2xHA epitope tag and a silent mutation at the gRNA protospacer adjacent motif site. Correct integration of the ssODN sequence was confirmed by PCR and subsequent sequencing of PCR products. Transformation occurred at <1% efficiency, and initial screens for clonal isolates were unsuccessful.
Use of Biological Replicates
Each experiment performed in the current study used unique and independent clonal transformants (biological replicates) except in the case of MARCH6–2HA cell lines (see above).
Results
Doa10 Contains a Highly Conserved Region at Its C Terminus
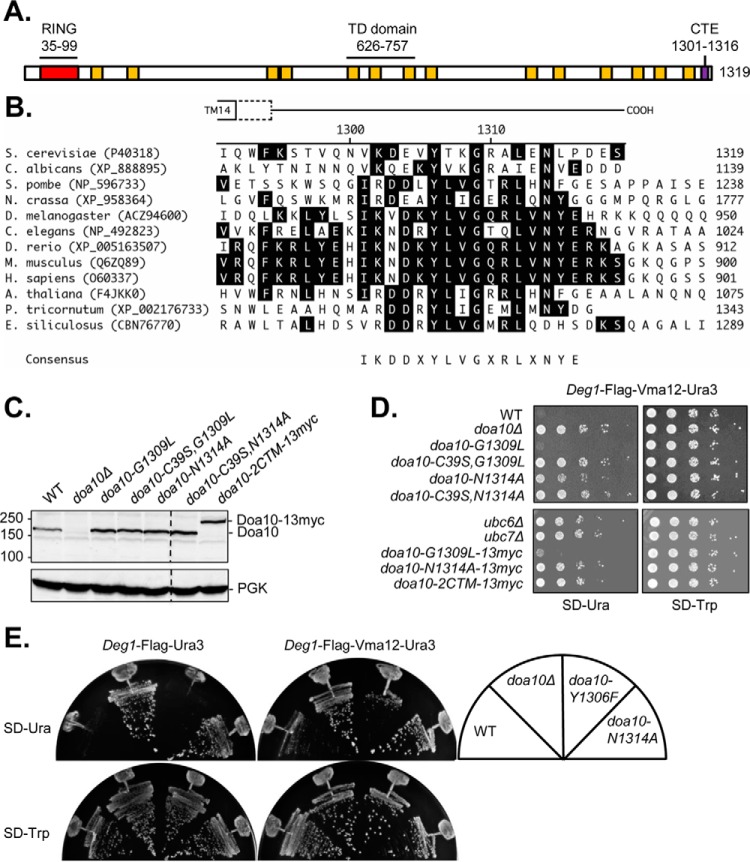
Doa10 and its orthologs are all large proteins with a dozen or more predicted transmembrane helices (TMs) (Fig. 1A) (12). Several doa10 mutant alleles obtained in the initial screen that identified Doa10 as an ERAD ligase encoded C-terminally truncated Doa10 proteins.5 The 1319-residue Doa10 protein bears a cytosolic C-terminal segment of ∼28 residues after the final TM (Fig. 1, A and B) (11). Sequence alignment of Doa10 against a broad set of orthologs revealed a highly conserved 16-residue stretch (residues 1301–1316), which we dubbed the CTE (Fig. 1B). The same element was identified with ScanProsite (39). Strong conservation was observed for an aliphatic residue at position 1301 (Val/Ile/Ala; S. cerevisiae Doa10 numbering), a basic residue (Lys/Arg) at 1302, a negatively charged residue (Asp/Glu) at position 1304, and an aliphatic residue (Ile/Leu) at 1312. Other residues, namely, Tyr-1306, Gly-1309, and Asn-1314, were invariant or nearly so in all investigated species.
FIGURE 1.
The cytosolic C-terminal tail of Doa10 contains a highly conserved region. A, diagram of full-length Doa10 highlighting the RING domain (red), TMs (yellow), CTE (purple), and TD domain (spanning TMs 5–7). B, sequence alignment of the yeast Doa10 C-terminal tail (residues 1291–1319) with Doa10 orthologs. The top schematic shows the predicted TM14 end point (11) and a potential extended end point at Lys-1295 (dashed box) followed by the cytosolic C terminus. Conserved residues among aligned sequences are highlighted in black. C, immunoblotting analysis of Doa10 CTE mutant protein levels. Doa10 is expressed from its endogenous locus in strains MHY500 (WT), MHY1685 (doa10Δ), MHY3998 (doa10-G1309L), MHY6758 (doa10-C39S,G1309L), MHY4242 (doa10-N1314A), MHY6734 (doa10-C39S,N1314A), and MHY6755 (doa10-G1309L,N1314A-13myc = doa10-2CTM-13myc). Doa10 was detected with an anti-Doa10 antibody (11). PGK, phosphoglycerate kinase loading control. Chemiluminescence was detected on film. The experiment was performed twice. D, independently of C-terminal epitope-tagging, N1314A CTE mutant exhibits Deg1-Vma12-Ura3 turnover defects based on degradation-dependent growth selection. Yeast strains were plated on SD-Trp (spotting control) or SD-Ura (growth selection). The substrate was expressed from the pRS414-MET25-Deg1-F-VMA12-URA3 plasmid. The strains used, from top, were: MHY500, MHY1685, MHY3998, MHY 6758, MHY4242, MHY6734, MHY496, MHY551, MHY6753, MHY6754, and MHY6755. The experiment with the above combination of strains was performed once. Phenotypes are reproducible and corroborated by substrate degradation assays in other figures. E, the Y1306F CTE mutant does not block Deg1 fusion protein turnover. Yeast strains transformed with pRS414-MET25-Deg1-F-URA3 or pRS414-MET25-Deg1-F-VMA12-URA3 were streaked on the indicated plates. Strains used, from left to right, were: MHY8655, MHY1685, MHY8688, and MHY6754. The experiment was performed with two biological replicates; one representative set is shown.
The high sequence conservation of the CTE, in combination with the evidence of functionally defective C-terminally truncated Doa10 proteins, prompted us to determine whether the CTE was required for Doa10 activity. Single point mutations were introduced at conserved residues Tyr-1306, Gly-1309, and Asn-1314 via in vivo site-directed mutagenesis (24) in the genomic locus of DOA10. Mutagenesis at each site introduced a nonpolar, hydrophobic residue. Given its identity across species, Tyr-1306 was conservatively changed to a Phe residue. The G1309L and N1314A mutations were potentially more severe. The G1309L and N1314A substitutions, alone, combined (“2CTM”), or in cis with an inactivating catalytic RING domain mutation (C39S), did not reduce Doa10 protein expression (Fig. 1C).
Mutant protein activity was first assessed in degradation-sensitive growth assays. Yeast strains expressing the artificial, membrane-embedded substrate Deg1-Vma12-Ura3 (26) as the sole source of Ura3 (required for uracil synthesis) were grown on SD minimal medium lacking uracil. Rapid degradation of the fusion protein in WT cells impairs growth on such SD-Ura plates. Strains with the doa10-G1309L or WT DOA10 allele grew equally poorly, suggesting that the G1309L mutation did not strongly impair Deg1-Vma12-Ura3 degradation (Fig. 1D). Based on growth assays of doa10-Y1306F cells expressing Deg1 fusion proteins (Fig. 1E), no degradation defect was observed even though this mutation is also in an absolutely conserved CTE residue. The C-terminal 13myc epitope tag did not obviously alter Doa10 activity either (Fig. 1D). By contrast, yeast with a doa10-N1314A mutation exhibited growth comparable with the doa10Δ, ubc6Δ, or ubc7Δ strains, suggesting a strong defect in substrate degradation (Fig. 1, D and E).
The Doa10 CTE Is Essential for Deg1 Substrate Degradation
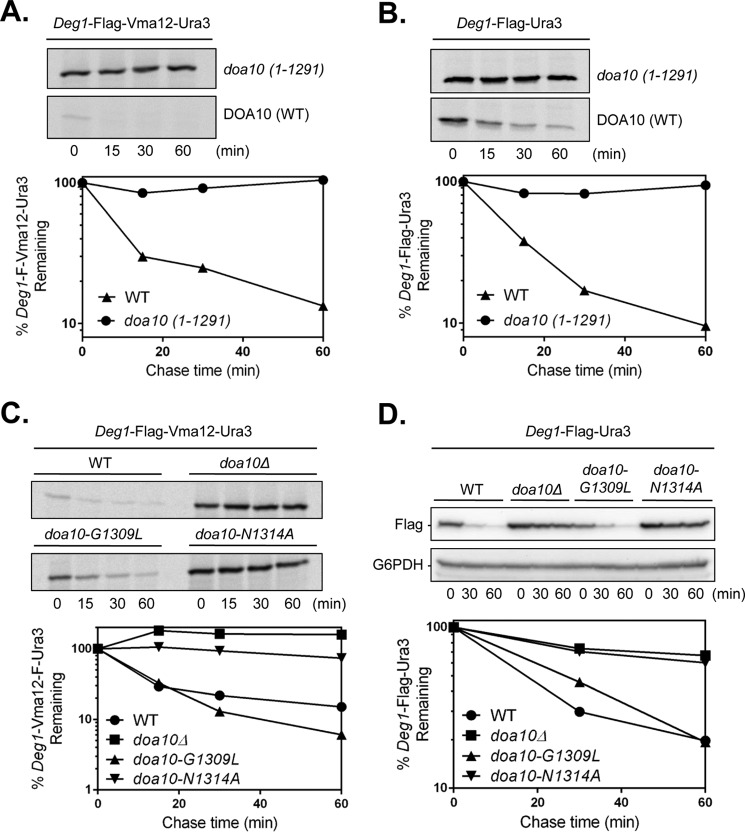
To determine directly whether the CTE was required for degradation of substrates bearing the Deg1 degron, we used in vivo degradation assays to test the turnover of Deg1-Vma12-Ura3 and the soluble nucleoplasmic substrate Deg1-Ura3 (6). We first tested a doa10 mutant allele lacking the entire C-terminal tail, doa10(1–1291)-13myc. Based on radioactive pulse-chase analysis, the Doa10 truncation led to complete stabilization of both Deg1-Vma12-Ura3 (Fig. 2A) and Deg1-Ura3 (Fig. 2B), whereas DOA10-13myc cells rapidly degraded both substrates with half-lives of ∼11–12 min. The doa10(1–1291)-13myc mutant protein is expressed at levels similar to the tagged WT protein, Doa10-13myc (11).
FIGURE 2.
The Doa10 CTE is required for Deg1 substrate degradation. Deg1-FLAG-Vma12-Ura3 (A) and Deg1-FLAG-Ura3 (B) degradation was examined by pulse-chase analysis. Extracts from yeast strains MHY8655 (DOA10-13myc) and MHY8663 (doa10(1–1291)-13myc) were immunoprecipitated with mouse anti-FLAG-conjugated resin. Substrates were expressed from pRS414-MET25-Deg1-F-VMA12-URA3 and pRS414-MET25-Deg1-F-URA3 plasmids, respectively. A cycloheximide-chase (not shown) confirmed the result in A. The experiment in B was performed twice. Quantifications are from the single assays presented for A and B. C, pulse-chase analysis of Deg1-FLAG-Vma12-Ura3 turnover was performed by immunoprecipitation with anti-MATα2 antibody (15) from MHY500 (WT), MHY1685 (doa10Δ), MHY3998 (doa10-G1309L), and MHY4242 (doa10-N1314A). The pulse-chase experiment was repeated with WT, doa10Δ, and N1314A strains and confirmed the results shown. Quantifications are from the assay shown at top. D, CHX-chase analysis of Deg1-FLAG-Ura3 degradation. Substrate was expressed in the same strains as in C from pRS416-GPD-Deg1-F-URA3 and detected using anti-FLAG antibody. The experiment was performed twice. Quantification is from the blot shown. G6PDH, glucose-6-phosphate dehydrogenase loading control.
To specifically test the requirement for the CTE in Deg1 substrate turnover, we analyzed the G1309L and N1314A point mutants of Doa10. Pulse-chase analysis of Deg1-Vma12-Ura3 degradation revealed that the doa10-N1314A allele led to a stabilization of the substrate (t½ ∼133 min) comparable with that seen in doa10Δ (Fig. 2C), whereas doa10-G1309L cells degraded the substrate at rates comparable with those in WT cells (t½ ∼10–11 min). Similarly, blocking protein synthesis with CHX and following the resulting rates of substrate disappearance from cells by immunoblotting analysis showed that the doa10-N1314A mutation impaired Deg1-Ura3 degradation (t½ ∼77 min) to a degree comparable with that seen with doa10Δ (Fig. 2D). Deg1-Ura3 degradation was similar in DOA10 and doa10-G1309L mutant cells (t½ ∼20 min). Consistent with these observations, the endogenous nucleoplasmic MATα2 protein, which bears the Deg1 degradation signal, was also stabilized only in the doa10-N1314A strain (data not shown). These data indicate that the CTE of Doa10 is required for the degradation of both soluble and membrane-embedded Deg1 substrates with the conserved residue Asn-1314 being critical.
Doa10 Substrates with Different Degrons Require the Doa10 CTE
We next asked whether substrates other than Deg1-bearing proteins were dependent on the Doa10 CTE for their degradation. A subset of Doa10-dependent substrates are fully stabilized by loss of Ubc6 or Ubc7, such as Deg1-protein fusions (13), whereas others, such as Ste6*, are only mildly impaired upon loss of the Ubc6 E2 (16, 18). Ubc6 itself is relatively short lived and targeted by Doa10 in a way that depends on the conserved TEB4-Doa10 (TD) domain (23). These observations suggest that substrate ubiquitylation by the Doa10 ligase can occur by distinct mechanisms. It could be, for instance, that the Doa10 CTE functions specifically with Ubc6 and would only be required for the ubiquitylation of substrates strongly dependent on this E2.
First we tested Vma12-Ndc10C′. Ndc10C′ comprises the last 100 residues of the Ndc10 kinetochore protein, which, when expressed as a separate fragment, acts as a degron (28). The degron is a bipartite signal composed of two amphipathic helices (“DegA”) followed by an unstructured region at the C terminus (“DegB”) (28, 40). Ndc10C′-based substrates are exclusively targeted for degradation by Doa10. As is true of Deg1 fusions, we found that degradation of Vma12-Ndc10C′ was fully dependent on Ubc6 (Fig. 3A). In WT cells, Vma12-Ndc10C′ was degraded with a half-life of ∼53 min. Vma12-Ndc10C′ was stabilized in the doa10-N1314A CTE mutant to a degree similar to that observed in ubc6Δ or ubc7Δ mutant cells (Fig. 3A).
FIGURE 3.
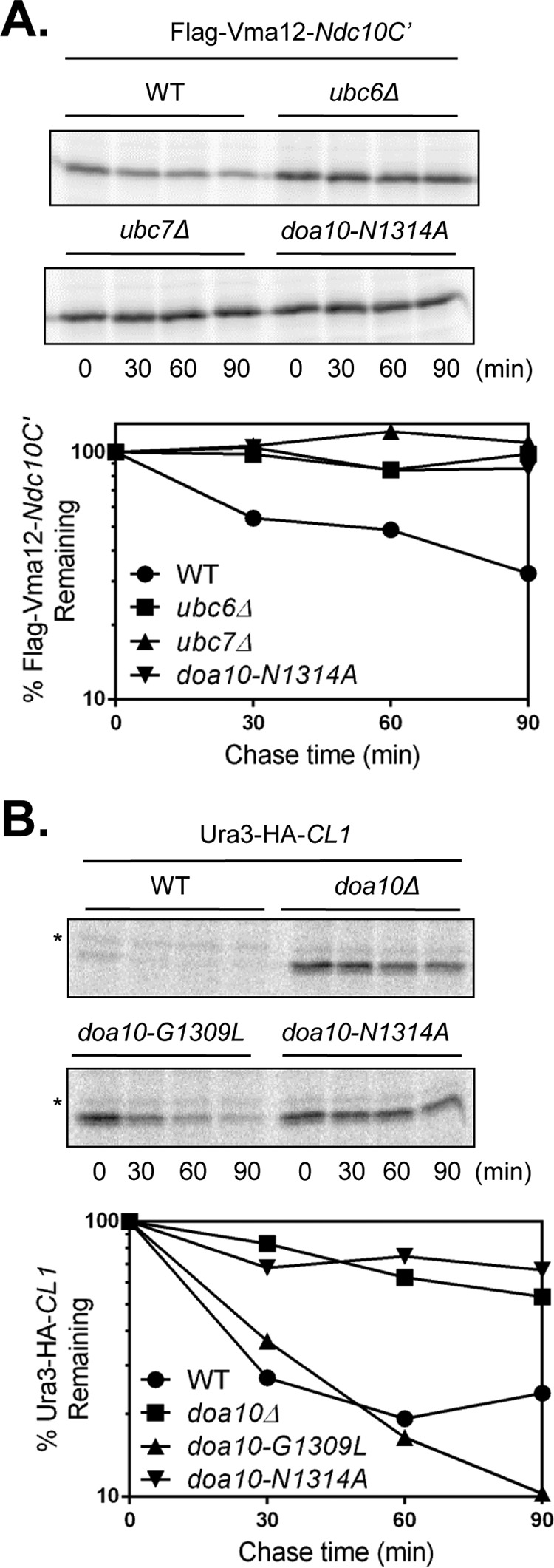
The CTE is required for degradation of multiple Doa10 substrates. A, pulse-chase analysis of FLAG-Vma12-Ndc10C′ degradation was performed in strains MHY500 (WT), MHY496 (ubc6Δ), MHY551 (ubc7Δ), and MHY4242 (doa10-N1314A). Substrate was expressed from p414GPD FLAG-Vma12-pHis-Ndc10C′ (28) and immunoprecipitated with anti-FLAG-agarose. One experiment was completed. B, pulse-chase analysis was performed on Ura3-HA-CL1. Yeast strains corresponding to those used in Fig. 2C were transformed with pOC9-Ura3-HA-CL1 (29). Immunoprecipitation was performed with a mouse anti-HA antibody. An independent experiment by CHX-chase analysis supported the result. Asterisks, nonspecific bands.
We then tested the artificial degron CL1 (29). CL1-fused substrates are short lived cytoplasmic proteins that require Doa10 for their turnover (26) but appear to be only partially stabilized in ubc6Δ or ubc7Δ single mutants, although the E2 double mutant is strongly defective (18, 29). By pulse-chase analysis, Ura3-HA-CL1 was stabilized to a similar extent by either doa10-N1314A or doa10Δ (Fig. 3B). In doa10-G1309L and WT (DOA10) cells, Ura3-HA-CL1 was rapidly degraded (t½ ∼23 min). Together with the foregoing examination of nuclear and nuclear membrane Deg1-based substrates of Doa10, the analyses of the ER-membrane embedded Vma12-Ndc10C′ and the cytosolic Ura3-HA-CL1 proteins indicate that Doa10 substrates with diverse subcellular localizations and degrons require the CTE.
Ubc6 and Ste6* Turnover Does Not Require the Doa10 CTE
The data presented to this point would seem to imply that Doa10 CTE function is linked to the basic catalytic activity of the ligase. However, only a limited set of substrates were tested. Besides functioning as a Doa10 cofactor, the E2 Ubc6 is also a Doa10 substrate and requires a functional RING and intact TD domain for its ubiquitin-dependent degradation (23). Mutating Glu-633 to an Asp residue (E633D) within TM5 of the TD domain strongly stabilizes Ubc6 but has little to no effect on Deg1 fusion protein degradation. We performed CHX-chase analyses of endogenous Ubc6 in DOA10-13myc (WT), doa10-2CTM, and doa10(1–1291)-13myc strains (Fig. 4A). Neither point mutation of the CTE nor its full deletion impaired Ubc6 degradation. This suggests that the CTE is not central to the catalytic activity of Doa10 and instead has a more specialized, substrate-restricted role.
FIGURE 4.
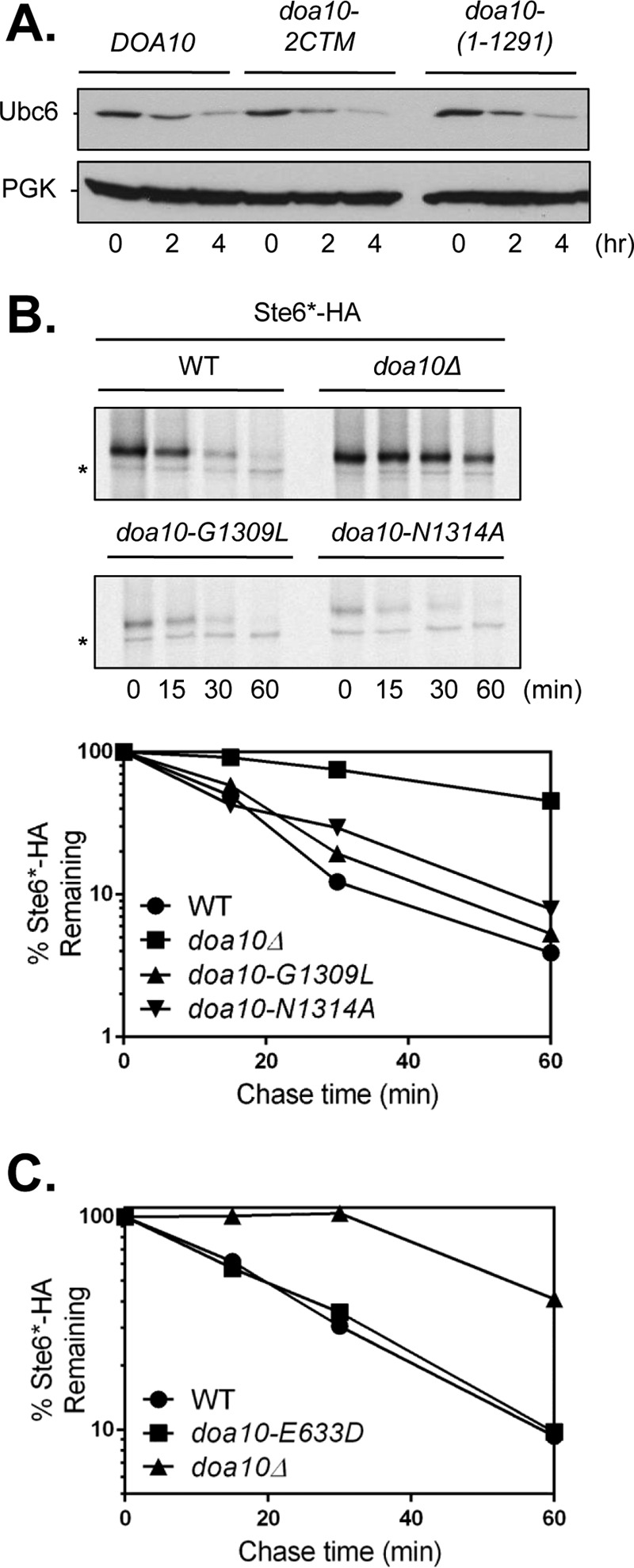
The C-terminal tail of Doa10 is not required for Ubc6 or Ste6* degradation. A, CHX-chase assay of endogenous Ubc6 turnover using anti-Ubc6 immunoblotting. PGK, phosphoglycerate kinase loading control. The chemiluminescence signal was detected with film. Strains used were: MHY8655 (DOA10-13myc), MHY6755 (doa10-2CTM-13myc), and MHY8663 (doa10(1–1291)-13myc). The experiment was performed once. B, pulse-chase analysis of Ste6*-HA degradation. Cell lysis was performed as described (36). Immunoprecipitations were done with a mouse anti-HA antibody. Substrate was expressed from pSM1082 plasmid (16). Asterisks, cross-reactive bands. Strains used were the same as in Fig. 2C. Effects on Ste6*-HA degradation among WT, doa10Δ, and doa10-N1314A strains were reproduced by a CHX-chase experiment. Quantification is from the pulse-chase assay above. C, quantification of Ste6*-HA turnover via pulse-chase. Strains used were: MHY500 (WT), MHY1685 (doa10Δ), and SKY305 (doa10-E633D). The experiment was executed twice. Quantification represents one experiment.
Support for this last conclusion came from analysis of an additional substrate, Ste6*. Ste6 is a polytopic membrane protein normally localized at the cell surface. Ste6* (Ste6-166) is a truncated derivative that remains in the ER and is targeted for rapid degradation by ERAD (36). Ste6* degradation is mostly Doa10-dependent but differs from most other Doa10 substrates in that Ste6* can also be targeted by Hrd1 and is only weakly stabilized (∼2-fold or less) by loss of Ubc6 (16). The Ubc7 E2, which functions with both Doa10 and Hrd1, has much greater impact on Ste6* degradation than does Ubc6 (16, 41). By pulse-chase analysis, neither CTE point mutation caused a significant change in Ste6* degradation rates (t½ ∼15 min) in contrast to a full deletion of Doa10 (t½ ∼55 min) (Fig. 4B).
Given that Ubc6 and Ste6* were the only tested substrates not affected by CTE mutation, we wondered whether Ste6* degradation requires an intact TD domain as is true for Ubc6 proteolysis (23). Pulse-chase analysis, however, revealed normal degradation kinetics for Ste6* in doa10-E633D (Fig. 4C). These results indicate that different Doa10 substrates show distinct requirements for the CTE and TD domain of Doa10.
The Doa10 CTE Promotes Efficient Deg1 Substrate Ubiquitylation
To determine whether the CTE is required for substrate ubiquitylation, we evaluated the in vivo ubiquitylation of a model Deg1 substrate, Deg1-β-gal, which has been used previously for this type of analysis (12, 37). Concordant with the results with Deg1-Ura3 (Fig. 2D), the soluble nucleoplasmic Deg1-β-gal protein was also stabilized in the doa10-N1314A strain, whereas degradation still occurred in doa10-G1309L cells (Fig. 5A). To examine its ubiquitylation, Deg1-β-gal was immunoprecipitated from denatured extracts of yeast expressing HA-tagged ubiquitin followed by anti-HA immunoblotting (Fig. 5B). As expected, high molecular mass, ubiquitylated forms of Deg1-β-gal were seen for WT yeast and were sharply reduced in cells lacking Doa10 (doa10Δ). Ubiquitylated species were still detected but at lower levels in the doa10-G1309L CTE mutant. By contrast, these species were barely detectable in the doa10-N1314A strain, which mirrored doa10Δ (Fig. 5B). The residual Deg1-β-gal ubiquitylation in doa10-G1309L cells was abrogated if the RING domain was also inactivated (doa10-C39S,G1309L), confirming that this ubiquitin modification is Doa10-dependent. These results place Doa10 CTE function at or before the ubiquitylation step in the degradation pathway. Potential roles include substrate recruitment or stimulation of the ubiquitin ligation reaction (or both).
FIGURE 5.
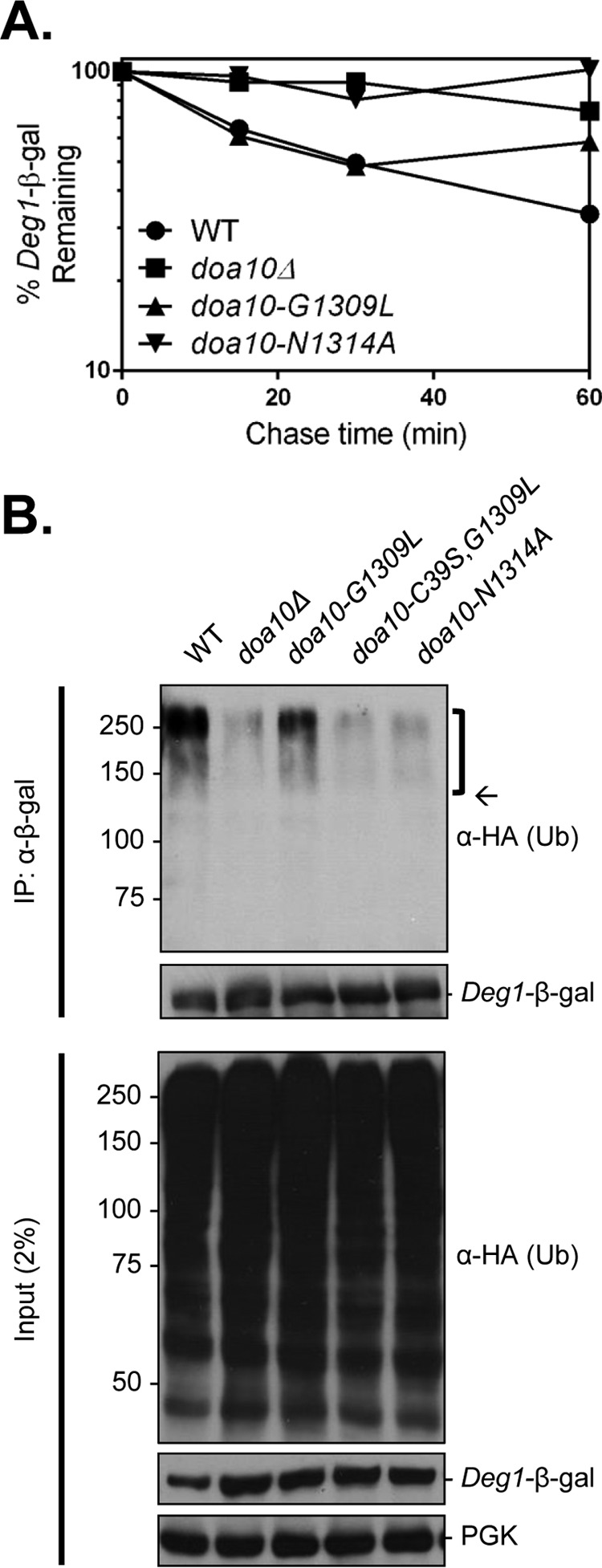
A functional Doa10 CTE is required for Deg1 substrate ubiquitylation. A, pulse-chase quantification of Deg1-β-gal degradation kinetics. Immunoprecipitation (IP) was performed using an anti-β-gal antibody. Substrate was expressed from the pLR1 plasmid in the following yeast strains: MHY6605 (DOA10), MHY6948 (doa10Δ), MHY7038 (doa10-G1309L), and MHY7041 (doa10-N1314A). Quantification is from one pulse-chase assay. B, in vivo ubiquitylation assay of Deg1-β-gal. Substrate was precipitated with an anti-β-gal antibody. For immunoblotting and film-based detection, anti-β-gal was used to detect Deg1-β-gal, and anti-HA was used to detect ubiquitylated proteins. Bracket, ubiquitylated Deg1-β-gal species. Arrow, expected molecular size of unmodified Deg1-β-gal. The pLR1 and YEp112 (HA-ubiquitin) plasmids were transformed into MHY7040 (doa10-C39S,G1309L) and the strains from A. The experiment was performed twice. Ub, ubiquitin.
Endogenous MARCH6 and Exogenous MARCH6 CTE Mutants Localize to the ER
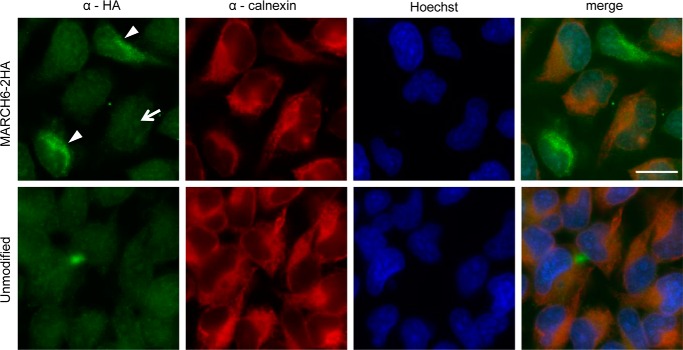
The human ortholog of Doa10, MARCH6 (TEB4), is also believed to be an ERAD ubiquitin ligase, although localization of the endogenously expressed protein to the ER has not been directly demonstrated (42). Like Doa10, MARCH6 appears to have 14 transmembrane domains with the N-terminal RING domain and C-terminal tail protruding into the cytoplasm (11). Several MARCH6 substrates have been identified, including squalene monooxygenase; correspondingly, the yeast squalene monooxygenase ortholog, Erg1, is a Doa10 substrate (43). Although the E2 enzymes that operate with MARCH6 are unknown, orthologs of Ubc6 (Ube2j1/Ube2j2) and Ubc7 (Ube2g1/Ube2g2) have been identified and are known to function with other ERAD E3 ligases (44–48). To study the in vivo properties of MARCH6, we utilized CRISPR/Cas9-mediated genome editing to modify genomic MARCH6 in HeLa cells with a sequence encoding a C-terminal 2xHA epitope tag (see “Experimental Procedures”). This led to the modification of at least one MARCH6 allele based on PCR and indirect immunofluorescence (Fig. 6). A small percentage (<1%) of treated cells gave an anti-HA-specific signal (Fig. 6, arrowheads); no signal was detected in the unmodified HeLa control cells. The signal for MARCH6–2HA using a mouse anti-HA antibody showed a strong overlap with the ER marker calnexin based on rabbit anti-calnexin staining. These data clearly indicate ER localization of HA-tagged MARCH6 expressed from its chromosomal locus even though we were not able to isolate clones in which all cells expressed the HA-modified allele.
FIGURE 6.
Endogenous MARCH6 localization at the ER based on chromosomally tagged MARCH6. Indirect immunofluorescence analysis of HeLa cells transformed with CRISPR/Cas9, gRNA targeting the 3′-end of MARCH6, and ssODNs was performed. MARCH6–2HA expressed from the genomically tagged locus was visualized by staining with mouse anti-HA antibody. The ER marker protein calnexin was visualized by staining with rabbit anti-calnexin antibody. DNA was stained with Hoechst 33342. Arrowheads denote HA-tag positive cells. Arrow, example of faint Hoechst 33342 bleed-through in Alexa Fluor 488 channel after long exposure. Scale bar, 20 μm. MARCH6–2HA was stained in two separate experiments with only one including calnexin staining.
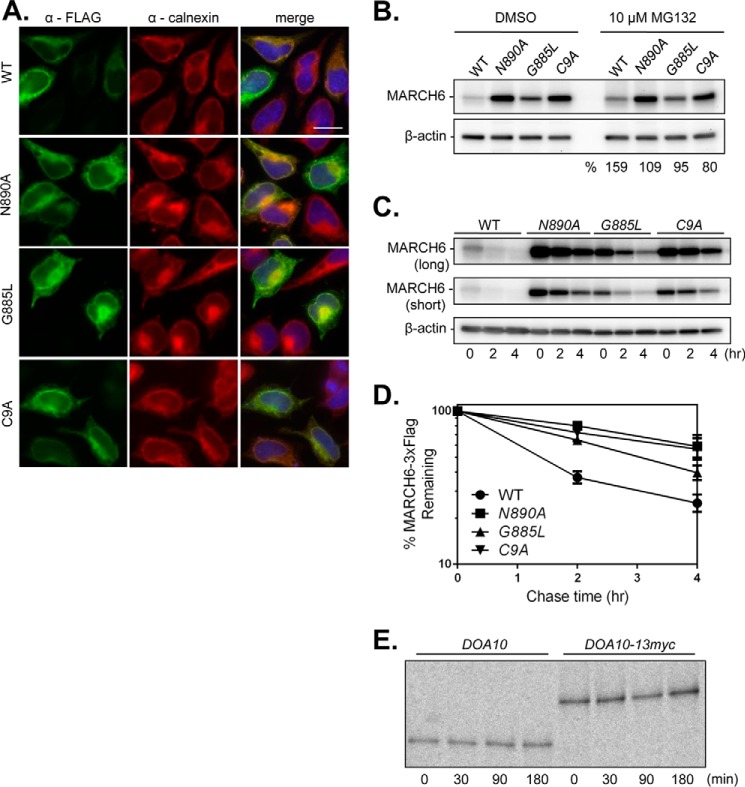
We also examined the subcellular localization of a number of MARCH6 variants that were transiently expressed in HeLa cells. Indirect immunofluorescence analysis was performed on cells expressing WT MARCH6–3xFLAG, catalytically inactive MARCH6-C9A-3xFLAG, MARCH6-G885L-3xFLAG, or MARCH6-N890A-3xFLAG; the latter two proteins bear mutations in the conserved CTE residues corresponding to Doa10 Gly-1309 and Asn-1314, respectively (Figs. 1B and 7A). As reported previously (42), WT and C9A co-localized with calnexin, and overexpression resulted in localization throughout the ER network. The G885L and N890A mutants both co-localized with calnexin and had patterns indistinguishable from WT MARCH6–3xFLAG. The exogenous MARCH6 variants were readily detectable in contrast to the low levels of endogenous MARCH6.
FIGURE 7.
MARCH6 autoregulation requires the conserved CTE asparagine residue. A, indirect immunofluorescence of exogenous MARCH6. HeLa cells transiently expressing MARCH6–3xFLAG-His6 WT or N890A, G885L, or C9A variants were fixed, and MARCH6 was visualized by mouse anti-FLAG staining (green). Calnexin (ER marker) was stained with rabbit anti-calnexin antibody (red). DNA was stained with Hoechst 33342 (blue in merge). Scale bar, 20 μm. MARCH6–3xFLAG-His6 localization was confirmed by staining a panel of HA-tagged MARCH6 variants. B, proteasome inhibition does not strongly affect mutant MARCH6 levels. Equivalent protein amounts (20 μg) from transiently transfected HeLa cells treated with vehicle alone (dimethyl sulfoxide (DMSO)) or 10 μm MG132 for 4 h were analyzed by anti-FLAG and anti-β-actin immunoblotting. Quantification (“%”) represents the relative MARCH6 steady-state levels in MG132- versus vehicle-treated cells. Two independent trials were completed. C, a representative CHX-chase experiment used to analyze MARCH6–3xFLAG turnover in transfected HeLa cells. Whole cell lysates (40 μg for WT and 20 μg for mutants) were processed and detected as in B. D, quantification of MARCH6–3xFLAG turnover kinetics from four independent CHX-chase experiments; error bars represent S.E. MARCH6-G885L-3xFLAG turnover was significantly reduced compared with WT at 2 and 4 h into the chase (p value <0.05, unpaired t test). E, pulse-chase analysis of endogenously expressed WT Doa10 and the WT Doa10-13myc-tagged variant. Cells were lysed as described (36), and proteins were precipitated with an anti-Doa10 antibody. Strains used were: YPH499 (DOA10) and MHY2916 (DOA10-13myc). The experiment was performed once.
The CTE Is Required for Autoregulation of MARCH6 Degradation
A common mechanism for E3 ligase regulation is through autoubiquitylation and degradation by the proteasome (49). MARCH6 is among the E3 ligases known to trigger its own turnover in a manner that requires its ubiquitin ligase activity (42). To investigate the conservation of MARCH6 CTE function in protein degradation, we compared the steady-state levels and degradation kinetics of MARCH6-G885L and MARCH6-N890A with the WT and catalytically inactive C9A RING mutant versions of the ligase.
Expression of all three mutants was much higher than that of WT MARCH6 with N890A mutant levels being comparable with the C9A RING-dead mutant (Fig. 7B, dimethyl sulfoxide-treated). Furthermore, proteasome inhibition by MG132 increased WT steady-state levels by 59% but had minimal effect on any of the mutant E3s, suggesting that proteasome-dependent degradation of the CTE mutants was already nearly completely blocked by the mutations (Fig. 7B). Differences in exogenous expression levels of the WT and RING-dead MARCH6 proteins were reported previously (31, 50) and are known to be due to differences in MARCH6 degradation (42). Our data suggest that the CTE is required for MARCH6 self-regulation through proteasome-dependent degradation, and both residues Gly-885 and Asn-890 are important for this.
To verify that the CTE is important for proteolytic autoregulation, we performed CHX-chase/immunoblotting assays on transiently transfected HeLa cells (Fig. 7C). Compared with WT, the RING-dead and two CTE mutants were present at levels higher than WT at the initial time points (Fig. 7C) as expected. The degradation rates were also substantially slowed (Fig. 7, C and D). The C9A (t½ ∼4.7 h) and N890A (t½ ∼5.4 h) mutants were degraded about 3-fold more slowly than the WT enzyme (t½ ∼1.7 h). The G885L mutant (t½ ∼3.1 h) also exhibited a statistically significant stabilization compared with WT. In contrast to the instability of the human MARCH6 protein, yeast Doa10 is long lived based on pulse-chase analysis (Fig. 7E). Introducing CTE mutations in Doa10 does not alter the steady-state levels of the protein (Fig. 1C), whereas we see different degrees of stabilization upon introduction of the corresponding CTE mutations in MARCH6 (Fig. 7, B and C). We conclude that the evolutionarily conserved CTE is important for proteasome-mediated MARCH6 autoregulation in human cells.
Discussion
Despite Doa10 being one of the two major ERAD E3 ligases in yeast, little is known about its substrate recognition mechanisms. Multiple degrons and substrates for Doa10 have been identified and, in some cases, extensively characterized. However, no binding site(s) on Doa10 for specific substrates has been identified, and it is possible that binding is mediated by additional factors such as molecular chaperones. Here we have identified a previously unrecognized but highly conserved 16-residue region in the cytosolic C-terminal segment of Doa10 that has a crucial role in the degradation of select Doa10 substrates. Analysis of the CTE revealed a point mutant, doa10-N1314A, that is expressed at WT levels but is nonfunctional. The functional conservation of the CTE was confirmed by demonstrating that CTE mutations in the human Doa10 ortholog, MARCH6, impair the RING-dependent autodegradation of MARCH6. Other than the RING-CH domain, the MARCH6 CTE is the only region in this mammalian ubiquitin ligase shown to be important for its function. We also provide the first evidence that endogenous MARCH6 is localized to the ER, similarly to its yeast ortholog.
A Unique Conserved Region among Doa10 Orthologs
The Doa10 CTE is a sequence motif unique to Doa10 orthologs. The only significant hits from protein BLAST searches with this region against available protein databases (51) or ScanProsite motif searches (39) were large membrane proteins with N-terminal RING-CH and internal TD domains. These latter features have previously been used to define the Doa10/MARCH6 ubiquitin ligase family (12, 23). Other proteins that contain the RING-CH motif, such as the 10 additional membrane-associated RING-CH (MARCH) proteins in humans, do not have the CTE (or TD domain). The CTE motif appears to be important for a specific E3 functionality in ERAD. It is present even in very distant orthologs from plants, stramenopiles, and Rhizaria (Fig. 1B), suggesting a central and conserved role for this region in Doa10/MARCH6 enzymes throughout the Eukarya.
Multiple Substrate Classes Dependent on the Doa10 CTE
What might be the mechanistic contribution of the Doa10 CTE to ligase function? The major classes of substrates tested here that are completely dependent on the CTE for their degradation are Deg1, DegAB, and CL1 degron fusion proteins. These substrates differ in localization when fused to soluble proteins with CL1 fusions predominantly found in the cytoplasm and Deg1 fusions concentrated in the nucleus. This suggests that the CTE is crucial to Doa10 ligase function in both the ER and inner nuclear membrane (INM) (6). All three of these degrons appear to be recognized by the Doa10 machinery through exposure of a hydrophobic face of an amphipathic helix (14, 28, 52). It is possible that the hydrophobic residues in the Doa10 CTE create a hydrophobic interface for degron binding; however, we have not yet been able to detect Doa10 interactions with these substrates, probably due to the low affinity or transience of these associations.
Although no substrate interaction motifs in Doa10 have yet been identified, all three of these degron classes also require chaperone proteins for Doa10-mediated ubiquitylation and degradation, suggesting that certain substrate-Doa10 interactions could be chaperone-mediated (17, 18). The Ssa Hsp70 chaperone family is required for the degradation of all tested Doa10 substrates (CL1 and DegAB fusions, Deg1-Vma12, and Ste6*) (17, 18, 28, 53), so they could be universally required. However, more substrates need to be tested to verify this conclusion.
Hsp70-dependent Doa10 substrates require various Hsp40 co-chaperones as well. The Hsp40 Sis1 is required for Vma12-DegAB and partially required for Deg1-Vma12 degradation (17), whereas Ydj1 is required for Ura3-HA-CL1 turnover (18). A non-degraded variant of Vma12-DegAB was shown to interact with Doa10 only in the presence of Sis1 (17, 28). Given that the CTE is required for both Sis1- and Ydj1-dependent Doa10 substrates, it is unlikely to be a docking site for a single specific co-chaperone. Ydj1 is also required for Ste6* turnover but only when a second Hsp40, Hlj1, is also deleted (16). Redundancy for Hsp40 co-chaperones is common, making it challenging to determine all the co-chaperones required for turnover of a particular substrate (54, 55).
Endogenous MARCH6 Localization
Using CRISPR/Cas9 genome editing, we visualized the endogenous localization and expression level of MARCH6 at the single cell level by epitope tagging the MARCH6 locus. MARCH6–2HA co-localized with the ER marker calnexin. Based on immunofluorescence analysis, expression levels of endogenous MARCH6 are much lower than the ectopically expressed protein (compare Figs. 6 and 7A). Localization based solely on the ectopically produced protein is problematic because failure of the overexpressed protein to fold or assemble properly could cause it to accumulate in the ER as a result of ER quality control mechanisms (36). Low expression levels and/or poor reactivity likely explain why endogenous MARCH6 localization had not been reported previously using MARCH6-specific antibodies.
In yeast, two E3 ubiquitin ligase complexes are known to localize to the INM and target nucleoplasmic and INM substrates: the Doa10 complex (6) and the Asi RING finger protein complex (56, 57). No mammalian INM-embedded ubiquitin ligases have been identified to date. The Asi complex is not conserved in mammals, but we predict that MARCH6 will also reach the INM in mammalian cells to target soluble nuclear and INM substrates.
Implications of a Functionally Conserved MARCH6 CTE
The present characterization of the MARCH6 CTE provides the first functional data on a MARCH6 domain outside of the N-terminal RING-CH domain. Analysis of MARCH6 autodegradation revealed that mutation of residue Asn-890 within the CTE has as severe an impact as mutating the RING-CH domain. In HeLa cells, the CTE-G885L mutation also reduced MARCH6 degradation albeit more mildly. The G1309L mutation in the Doa10 CTE has even more subtle effects on substrate degradation. The mutation reduced the level of ubiquitylated Deg1-β-gal (Fig. 5), and it appeared to have a mild effect on the steady-state levels of substrates (Figs. 2C and 3B) but had little to no detectable effect on their degradation kinetics. Less conservative mutations might be needed to clarify the importance of individual residues in the CTE.
During our CHX-chase analyses, we found that MARCH6 degradation was only partially impaired by mutating a zinc-coordinating residue of the RING-CH domain. A similar degree of inhibition was seen with the CTE N890A mutation (Fig. 7, C and D). These partial effects could reflect the activity of a second E3 ligase capable of targeting MARCH6 at least when the latter is overexpressed. Supporting this hypothesis, both exogenously expressed MARCH6-C9A and MARCH6-N890A mutants were still ubiquitylated in vivo (data not shown). The mechanism for MARCH6 self-ubiquitylation and degradation remains unknown but appears to be CTE-mediated. The CTE is likely to contribute in some way to substrate discrimination; MARCH6 might recognize itself as a substrate by such a CTE-dependent mechanism.
Doa10/MARCH6 Conserved Domains
The conservation between Doa10 and MARCH6 is highest in the catalytic RING-CH domain, the newly characterized CTE, and the TD domain. The TD domain is the largest conserved region between the two orthologs, spanning roughly 130 residues and including TMs 5–7. Mutational analysis of the Doa10 TD domain, mainly of TM5, revealed a region required specifically for Ubc6 degradation (23). The conserved TD domain was proposed to contribute to Ubc6 recognition, possibly functioning in both the positioning of the E2 during exogenous substrate ubiquitylation and the binding of the E2 at a distinct site that promotes its degradation (23). A specific mutation in TM5, E633D, strongly stabilized Ubc6 but had no effect on Deg1 fusion substrates. The substrate selectivity of the CTE is opposite that of the TD domain as it targets Deg1 fusions and not Ubc6. Ste6* degradation requires neither the TD domain nor the CTE, suggesting additional mechanisms for substrate recognition. The TD domain could be required for Doa10-mediated ERAD-M as the transmembrane domain of Ubc6 is partially involved in its degradation (23, 58). The high conservation of the TD domain TMs makes this hypothesis appealing.
Future experiments should determine whether the MARCH6 TD domain, like its CTE, is required for its self-regulated turnover. Other MARCH6 substrates that require the CTE or TD domain for their degradation also need to be characterized. Our present analysis of Doa10 and MARCH6 has allowed direct functional comparison between these two otherwise fairly divergent ligase orthologs. The two proteins both localize to the ER and both require the newly described CTE for their function. Future studies will aim at determining the mechanisms of substrate recognition by these ERAD ubiquitin ligases, particularly the mechanistic contributions of the CTE.
Author Contributions
M. H. and S. G. K. conceived the study. J. M. B., D. Z., and M. H. designed the experiments and wrote the paper. D. Z. designed, performed, and analyzed the experiments in Figs. 1, C and D; 2, A–C; 4, A and B; and 5. J. M. B. designed, performed, and analyzed the experiments in Figs. 1E, 2D, 3, 4C, 6, and 7. S. G. K. generated multiple yeast strains, performed and analyzed the experiment in Fig. 7E, and gave critical input for the manuscript. All authors reviewed the results and approved a final version of the manuscript.
Acknowledgments
We thank Christian Schlieker for reagents and equipment, Bebiana Sá-Moura for generating the MHY6605 yeast strain, Tommer Ravid for generating the MHY2916 strain, and Emmanuel Wiertz for a clone of the MARCH6 gene. We also thank Christopher Hickey and Judith Ronau for helpful comments on the manuscript.
This work was supported by National Institutes of Health Grant R01 GM046904 (to M. H.). Additional training support was provided by National Institutes of Health Grants F32 GM113456 (to J. M. B.) and T32 GM7223 (to D. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
S. G. Kreft and M. Hochstrasser, unpublished data.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated protein degradation
- CTE
- C-terminal element
- CRISPR
- clustered regularly interspaced short palindromic repeats
- SD
- synthetic defined
- CHX
- cycloheximide
- ssODN
- single-stranded donor oligonucleotide
- TM
- transmembrane helix
- TD
- TEB4-Doa10
- MARCH
- membrane-associated RING-CH
- INM
- inner nuclear membrane
- gRNA
- guide RNA.
References
- 1. Pickart C. M. (2000) Ubiquitin biology: an old dog learns an old trick. Nat. Cell Biol. 2, E139–E141 [DOI] [PubMed] [Google Scholar]
- 2. Ravid T., and Hochstrasser M. (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Metzger M. B., Pruneda J. N., Klevit R. E., and Weissman A. M. (2014) RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McCracken A. A., and Brodsky J. L. (1996) Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 132, 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zattas D., and Hochstrasser M. (2015) Ubiquitin-dependent protein degradation at the yeast endoplasmic reticulum and nuclear envelope. Crit. Rev. Biochem. Mol. Biol. 50, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deng M., and Hochstrasser M. (2006) Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature 443, 827–831 [DOI] [PubMed] [Google Scholar]
- 7. Nakatsukasa K., and Brodsky J. L. (2008) The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Habeck G., Ebner F. A., Shimada-Kreft H., and Kreft S. G. (2015) The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. J. Cell Biol. 209, 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carvalho P., Goder V., and Rapoport T. A. (2006) Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 10. Neuber O., Jarosch E., Volkwein C., Walter J., and Sommer T. (2005) Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat. Cell Biol. 7, 993–998 [DOI] [PubMed] [Google Scholar]
- 11. Kreft S. G., Wang L., and Hochstrasser M. (2006) Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI). J. Biol. Chem. 281, 4646–4653 [DOI] [PubMed] [Google Scholar]
- 12. Swanson R., Locher M., and Hochstrasser M. (2001) A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 15, 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen P., Johnson P., Sommer T., Jentsch S., and Hochstrasser M. (1993) Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell 74, 357–369 [DOI] [PubMed] [Google Scholar]
- 14. Johnson P. R., Swanson R., Rakhilina L., and Hochstrasser M. (1998) Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 94, 217–227 [DOI] [PubMed] [Google Scholar]
- 15. Hochstrasser M., and Varshavsky A. (1990) In vivo degradation of a transcriptional regulator: the yeast α2 repressor. Cell 61, 697–708 [DOI] [PubMed] [Google Scholar]
- 16. Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., and Michaelis S. (2004) Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 279, 38369–38378 [DOI] [PubMed] [Google Scholar]
- 17. Shiber A., Breuer W., Brandeis M., and Ravid T. (2013) Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Mol. Biol. Cell 24, 2076–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Metzger M. B., Maurer M. J., Dancy B. M., and Michaelis S. (2008) Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J. Biol. Chem. 283, 32302–32316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guthrie C., and Fink G. R. (1991) Guide to Yeast Genetics and Molecular Biology: Methods in Enzymology, Academic Press, San Diego, CA: [PubMed] [Google Scholar]
- 20. Rubenstein E. M., Kreft S. G., Greenblatt W., Swanson R., and Hochstrasser M. (2012) Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase. J. Cell Biol. 197, 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sommer T., and Jentsch S. (1993) A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature 365, 176–179 [DOI] [PubMed] [Google Scholar]
- 22. Sikorski R. S., and Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kreft S. G., and Hochstrasser M. (2011) An unusual transmembrane helix in the endoplasmic reticulum ubiquitin ligase Doa10 modulates degradation of its cognate E2 enzyme. J. Biol. Chem. 286, 20163–20174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Storici F., Lewis L. K., and Resnick M. A. (2001) In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19, 773–776 [DOI] [PubMed] [Google Scholar]
- 25. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 26. Ravid T., Kreft S. G., and Hochstrasser M. (2006) Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 25, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zattas D., Adle D. J., Rubenstein E. M., and Hochstrasser M. (2013) N-terminal acetylation of the yeast Derlin Der1 is essential for Hrd1 ubiquitin-ligase activity toward luminal ER substrates. Mol. Biol. Cell 24, 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Furth N., Gertman O., Shiber A., Alfassy O. S., Cohen I., Rosenberg M. M., Doron N. K., Friedler A., and Ravid T. (2011) Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol. Biol. Cell 22, 4726–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gilon T., Chomsky O., and Kulka R. G. (1998) Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 17, 2759–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hochstrasser M., Ellison M. J., Chau V., and Varshavsky A. (1991) The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl. Acad. Sci. U.S.A. 88, 4606–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park S. E., Kim J. M., Seok O. H., Cho H., Wadas B., Kim S. Y., Varshavsky A., and Hwang C. S. (2015) Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science 347, 1249–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng L., Baumann U., and Reymond J. L. (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mruk D. D., and Cheng C. Y. (2011) Enhanced chemiluminescence (ECL) for routine immunoblotting: an inexpensive alternative to commercially available kits. Spermatogenesis 1, 121–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hickey C. M., and Hochstrasser M. (2015) STUbL-mediated degradation of the transcription factor MATα2 requires degradation elements that coincide with corepressor binding sites. Mol. Biol. Cell 26, 3401–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Loayza D., Tam A., Schmidt W. K., and Michaelis S. (1998) Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 9, 2767–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laney J. D., and Hochstrasser M. (2002) Analysis of protein ubiquitination. Curr. Protoc. Protein Sci. Chapter 14, Unit 14.5 [DOI] [PubMed] [Google Scholar]
- 38. Chen F., Pruett-Miller S. M., Huang Y., Gjoka M., Duda K., Taunton J., Collingwood T. N., Frodin M., and Davis G. D. (2011) High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8, 753–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de Castro E., Sigrist C. J., Gattiker A., Bulliard V., Langendijk-Genevaux P. S., Gasteiger E., Bairoch A., and Hulo N. (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alfassy O. S., Cohen I., Reiss Y., Tirosh B., and Ravid T. (2013) Placing a disrupted degradation motif at the C terminus of proteasome substrates attenuates degradation without impairing ubiquitylation. J. Biol. Chem. 288, 12645–12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vashist S., and Ng D. T. (2004) Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hassink G., Kikkert M., van Voorden S., Lee S. J., Spaapen R., van Laar T., Coleman C. S., Bartee E., Früh K., Chau V., and Wiertz E. (2005) TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem. J. 388, 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Foresti O., Ruggiano A., Hannibal-Bach H. K., Ejsing C. S., and Carvalho P. (2013) Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. Elife 2, e00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van den Boomen D. J., Timms R. T., Grice G. L., Stagg H. R., Skødt K., Dougan G., Nathan J. A., and Lehner P. J. (2014) TMEM129 is a Derlin-1 associated ERAD E3 ligase essential for virus-induced degradation of MHC-I. Proc. Natl. Acad. Sci. U.S.A. 111, 11425–11430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van de Weijer M. L., Bassik M. C., Luteijn R. D., Voorburg C. M., Lohuis M. A., Kremmer E., Hoeben R. C., LeProust E. M., Chen S., Hoelen H., Ressing M. E., Patena W., Weissman J. S., McManus M. T., Wiertz E. J., and Lebbink R. J. (2014) A high-coverage shRNA screen identifies TMEM129 as an E3 ligase involved in ER-associated protein degradation. Nat. Commun. 5, 3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sato T., Sako Y., Sho M., Momohara M., Suico M. A., Shuto T., Nishitoh H., Okiyoneda T., Kokame K., Kaneko M., Taura M., Miyata M., Chosa K., Koga T., Morino-Koga S., Wada I., and Kai H. (2012) STT3B-dependent posttranslational N-glycosylation as a surveillance system for secretory protein. Mol. Cell 47, 99–110 [DOI] [PubMed] [Google Scholar]
- 47. Burr M. L., Cano F., Svobodova S., Boyle L. H., Boname J. M., and Lehner P. J. (2011) HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. U.S.A. 108, 2034–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen B., Mariano J., Tsai Y. C., Chan A. H., Cohen M., and Weissman A. M. (2006) The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl. Acad. Sci. U.S.A. 103, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Bie P., and Ciechanover A. (2011) Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 18, 1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zelcer N., Sharpe L. J., Loregger A., Kristiana I., Cook E. C., Phan L., Stevenson J., and Brown A. J. (2014) The E3 ubiquitin ligase MARCH6 degrades squalene monooxygenase and affects 3-hydroxy-3-methyl-glutaryl coenzyme A reductase and the cholesterol synthesis pathway. Mol. Cell. Biol. 34, 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gilon T., Chomsky O., and Kulka R. G. (2000) Degradation signals recognized by the Ubc6p-Ubc7p ubiquitin-conjugating enzyme pair. Mol. Cell. Biol. 20, 7214–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Han S., Liu Y., and Chang A. (2007) Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J. Biol. Chem. 282, 26140–26149 [DOI] [PubMed] [Google Scholar]
- 54. Sahi C., Kominek J., Ziegelhoffer T., Yu H. Y., Baranowski M., Marszalek J., and Craig E. A. (2013) Sequential duplications of an ancient member of the DnaJ-family expanded the functional chaperone network in the eukaryotic cytosol. Mol. Biol. Evol. 30, 985–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sahi C., and Craig E. A. (2007) Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. U.S.A. 104, 7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Khmelinskii A., Blaszczak E., Pantazopoulou M., Fischer B., Omnus D. J., Le Dez G., Brossard A., Gunnarsson A., Barry J. D., Meurer M., Kirrmaier D., Boone C., Huber W., Rabut G., Ljungdahl P. O., and Knop M. (2014) Protein quality control at the inner nuclear membrane. Nature 516, 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Foresti O., Rodriguez-Vaello V., Funaya C., and Carvalho P. (2014) Quality control of inner nuclear membrane proteins by the Asi complex. Science 346, 751–755 [DOI] [PubMed] [Google Scholar]
- 58. Walter J., Urban J., Volkwein C., and Sommer T. (2001) Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 20, 3124–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]