Abstract
Thermophilic rhodopsin (TR) is a photoreceptor protein with an extremely high thermal stability and the first characterized light-driven electrogenic proton pump derived from the extreme thermophile Thermus thermophilus JL-18. In this study, we confirmed its high thermal stability compared with other microbial rhodopsins and also report the potential availability of TR for optogenetics as a light-induced neural silencer. The x-ray crystal structure of TR revealed that its overall structure is quite similar to that of xanthorhodopsin, including the presence of a putative binding site for a carotenoid antenna; but several distinct structural characteristics of TR, including a decreased surface charge and a larger number of hydrophobic residues and aromatic-aromatic interactions, were also clarified. Based on the crystal structure, the structural changes of TR upon thermal stimulation were investigated by molecular dynamics simulations. The simulations revealed the presence of a thermally induced structural substate in which an increase of hydrophobic interactions in the extracellular domain, the movement of extracellular domains, the formation of a hydrogen bond, and the tilting of transmembrane helices were observed. From the computational and mutational analysis, we propose that an extracellular LPGG motif between helices F and G plays an important role in the thermal stability, acting as a “thermal sensor.” These findings will be valuable for understanding retinal proteins with regard to high protein stability and high optogenetic performance.
Keywords: membrane protein, optogenetics, photoreceptor, proton pump, X-ray crystallography, MD simulation, thermal stability
Introduction
Light is one of the most essential factors for most organisms. On Earth, photoreceptor proteins have continuously evolved to use solar energy efficiently in organisms over the past 4.6 billion years. Rhodopsins, a type of photoreceptor protein, are known to exist in all domains of life (i.e. archaea, eubacteria, and eukarya) (1), indicating their biological importance. Indeed, rhodopsins use light energy to exert a variety of biological functions, such as light-induced ion transportation and photosensing (2, 3). Despite such functional differences, rhodopsins are commonly composed of seven transmembrane α-helices (called helices A to G) with the retinal chromophore bound to a specific Lys residue in the seventh or G-helix via a protonated Schiff base linkage. Light absorption by rhodopsins triggers trans-cis or cis-trans isomerization of the retinal, and the photoisomerization induces structural changes of the transmembrane helices and soluble domains to exhibit their cognate biological functions. Rhodopsins are categorized into two types, type 1 for microbes and type 2 for animals (1).
Recently, a significant number of genes encoding type 1 rhodopsins have been identified from microbes living in a wide range of environments. Many of such newly discovered rhodopsins act as proton pumps in the cell membrane to produce adenosine triphosphate (ATP) upon photo-accumulation in collaboration with ATP-synthase (4, 5). Among the newly discovered rhodopsins, the eubacterial ones form the largest phylogenetic family. In 2012, a gene encoding a eubacterial proton pumping rhodopsin was identified in the genome of an extreme thermophile, Thermus thermophilus JL-18 strain, which was isolated in the Great Boiling Spring in the United States Great Basin at ∼75 °C (6). Since that time, this protein, named thermophilic rhodopsin (TR),4 has been investigated to elucidate its molecular properties, including its light-driven proton pump activity, thermal stability, and photoreaction (7, 8). Those studies represent the first discovery and characterization of a rhodopsin from a thermophilic organism.
One of the physical characteristics of TR is its high thermal stability. TR maintains its visible color after incubation at 75 °C for more than 3 h even in the presence of a detergent (7). Because retinal is readily degraded above 50 °C (9), a robust protein structure is required to hold retinal inside the protein and retain its optimal function at the high temperature in the native habitat of T. thermophilus JL-18. Here, we confirmed the high thermal stability of TR compared with other microbial rhodopsins and characterized the robust neural silencing activity of TR as the first eubacterial rhodopsin available for optogenetics. Furthermore, we determined its x-ray crystal structure and elucidated possible mechanisms for its thermal stability together with the results of molecular dynamics (MD) simulations. These findings, which advance the understanding of ion transport mechanisms and the thermal stability of membrane proteins, should lead to the rational design of rhodopsin-based optogenetic tools.
Experimental Procedures
Plasmid DNA Preparation
The cDNA of the codon-optimized wild-type TR (7) was inserted into the pET21c expression vector (Novagen, Madison, WI). The cDNA of the codon-optimized TR mutant called TR-ΔLPGG, which was the deleted Leu211-Pro212-Gly213-Gly214 sequence in the extracellular loop between the F- and G-helices from the wild-type TR, was purchased from GenScript Inc., Tokyo, Japan. The cDNA of TR-ΔLPGG was then inserted into the pET22b expression vector (Novagen). NdeI and XhoI restriction enzymes were used. The correctness of the constructed plasmid DNA was confirmed by dideoxy sequencing (Applied Biosystems, Forster City, CA).
Preparation of TR for Crystallization
TR was prepared using essentially the same procedures as described previously (7). Codon-optimized TR was heterologously expressed in Escherichia coli BL21(DE3) cells. The protein expression was induced in the presence of 1 mm isopropyl β-d-1-thiogalactopyranoside (Wako Pure Chemical Industries, Ltd., Osaka, Japan) and 10 μm all-trans-retinal (Sigma-Aldrich). TR was then purified from the crude membrane fraction using a three-step column chromatographic method with the detergent n-dodecyl-β-d-maltoside (DDM): 1) Ni2+-nitrilotriacetic acid affinity chromatography using a HisTrap HP column (GE Healthcare), 2) anion-exchange chromatography using a HiTrap-Q HP column (GE Healthcare), and 3) size-exclusion chromatography using a Superdex-200 column (GE Healthcare).
The degree of purification was verified by SDS-PAGE and UV-visible absorption spectroscopy (data not shown). The purified TR showed an absorption maximum at 530 nm in DDM micelles without any other absorption band in the visible region. Based on data from SDS-PAGE and the absorption spectrum, the purity of TR was determined to be ∼95%.
The purified sample was concentrated, and the buffer was exchanged using Amicon Ultra centrifugal filter units (Merck Millipore Corp., Darmstadt, Germany). Finally, the sample contained ∼170 mg/ml protein suspended in 50 mm Tris-HCl buffer (pH 7.5) (Wako Pure Chemical Industries, Ltd.) including 30 mm NaCl (Wako Pure Chemical Industries) and 0.05% (w/v) DDM (Dojindo Laboratories, Kumamoto, Japan).
Measurements of Thermal Stability
TR, bacteriorhodopsin (BR), and archaerhodopsin-3 (AR3) were prepared in E. coli cells (7, 10), and xanthorhodopsin (XR) was prepared using an E. coli cell-free expression system (11). BR from Halobacterium salinarum was expressed in E. coli cells and purified by HisTrap HP column chromatography as described previously (10). For XR, we developed an E. coli cell-free expression system as described previously for Acetabularia rhodopsin II (ARII) (11) because XR without salinixanthin (SX), the secondary chromophore, is difficult to obtain not only from the native Salinibacter ruber membranes but also from the E. coli expression system as a recombinant protein. In this system, XR was obtained as the lipid-reconstituted form lacking SX.
To carry out the experiment in the solubilized form, the lipid-reconstituted XR was suspended in 50 mm Tris-HCl buffer (pH 8.0) in the presence of 1 m NaCl and was then solubilized by the addition of 5.0% n-octyl-β-d-glucoside (Dojindo Laboratories) to the suspension for 1 h. After ultracentrifugation (100,000 × g) at 4 °C for 30 min, the supernatant containing the n-octyl-β-d-glucoside-solubilized fraction was concentrated using an Amicon Ultra filter (30,000 molecular weight cut-off, Merck Millipore). Thereafter, the sample was applied to a HisTrap FF column (GE Healthcare) that had been equilibrated previously with the same buffer and washed with 50 mm Tris buffer (pH 8.0) containing 1 m NaCl, 20 mm imidazole, and 0.05% DDM. The sample was eluted with 50 mm Tris buffer (pH 7.0) containing 1 m NaCl, 1 m imidazole, and 0.05% DDM. As a result, all samples for measurements of thermal stability contained the same medium composition (50 mm Tris-Cl (pH ∼7.7), 1 m NaCl, ∼300 mm imidazole, and 0.05% (w/v) DDM).
The purified TR-WT and TR-ΔLPGG were suspended in 50 mm Tris-HCl (pH 7.0) buffer containing 1 m NaCl and 0.05% DDM (Dojindo Laboratories). To adjust the DDM concentration, the samples were prepared by dialysis against the same buffer described above for 3 weeks.
For the measurement of thermal denaturation by UV-visible spectroscopy, the concentration of each protein was adjusted to ∼1 μm. The measurement was carried out by a method similar to that reported previously (7). The optical density (OD) of each sample (∼0.1 OD) at the wavelength of the absorption maximum (530, 550, 548, and 552 nm for TR-WT and TR-ΔLPGG, BR, XR, and AR3, respectively) was plotted against time for the incubation. The data were analyzed by a single exponential decay function to estimate the denaturation rate constant. Three independent experiments were averaged.
Optogenetic Control of Caenorhabditis elegans
Transgenic C. elegans worms expressing AR3 or TR were generated by microinjection of DNA into the distal arms of gonads of N2 hermaphrodites as described previously (12–14). The plasmid myo-3p::TR::gfp, aex-3p::AR3::gfp or aex-3p::TR::gfp was injected with pRF4 (rol-6d), a transgenic marker conferring the roller phenotype, to create these three lines. All-trans-retinal (Sigma-Aldrich) was taken up by feeding C. elegans warm on an agar plate in the presence of retinal as described previously (12, 13). Light-induced neural silencing was monitored by locomotion paralysis with green light (550 nm, 0.8 milliwatt/mm2 for muscle silencing or 4.4 milliwatts/mm2 for neural silencing) and was quantified by the change in body length as described previously (12, 13). In this system, the body was divided along its entire length into 11 segments by placing 12 dots at equal intervals. The sum of the length of each line connecting adjacent dots was defined as the body length of the worm. The average time for half of the body length change was calculated with standard error.
Crystallization, Data Collection, and Structure Determination
Purified TR was crystallized in lipidic cubic phase (LCP) at 23 °C as follows. The protein was diluted to ∼34 mg/ml and mixed with monoolein (Nu-Chek Prep, Elysian, MN) at room temperature using a syringe mixing apparatus. The final protein:lipid ratio was 2:3 (w/w). After formation of a transparent LCP by overnight incubation at 23 °C, the mixture was dispensed on 96-well glass sandwich plates in a 50-nl drop and was overlaid with 800 nl of precipitant solution by a crystallization robot, Mosquito LCP (TTP Labtech Ltd., Melbourn, Hertfordshire, UK). TR crystals were grown in 100 mm sodium citrate (pH 3.6), 34% PEG 400 (v/v). Typical crystals matured to full size in 1 week. The crystals were harvested directly from the LCP using a MicroMount loop (MiTeGen LLC, Ithaca, NY) and were flash-cooled in liquid nitrogen.
TR crystals were screened at the BL-1A beamline at the Photon Factory and the BL32XU beamline at SPring-8 (15). Diffraction data were collected at BL-1A using a Pilatus 2M detector. The crystal was exposed to a 25-μm-wide, 10-μm-high beam for 0.5 s at 0.5° oscillation/frame with a 2-fold attenuation at a wavelength of 1.1 Å. Diffraction images were processed by XDS (16); the statistics are summarized in Table 1. General data manipulation was performed using the CCP4 suite of programs (17).
TABLE 1.
Data collection and refinement statistics
Highest resolution shell is shown in parentheses.
| Data collection | |
| Beamline | PF-BL1A (Photon Factory, Tsukuba, Japan) |
| Wavelength (Å) | 1.1 |
| Oscillation angle (o) | 0.5 |
| Number of images | 360 (180o) |
| Space group | P1 |
| Cell dimensions | a = 63.186 Å, b = 67.693 Å, c = 73.045 Å |
| α = 116.48o, β = 114.14o, γ = 90.82o | |
| Resolution range (Å) | 46.52–2.8 (2.896–2.796) |
| Completeness (%) | 93.19 (89.08) |
| Rmerge | 0.1192 (0.4538) |
| Rmeas | 0.1683 |
| 〈I/σ(I)〉 | 6.26 (1.77) |
| CC½a | 0.989 (0.621) |
| CC*b | 0.997 (0.875) |
| Multiplicity | 1.8 (1.8) |
| Wilson B-factor | 37.59 |
| Refinement | |
| Rwork | 0.2284 (0.2872) |
| Rfree | 0.2777 (0.3065) |
| r.m.s.d. | |
| Bond (Å) | 0.003 |
| Angle (o) | 0.74 |
| Ramachandran statistics | |
| Favored regions (%) | 98.7 |
| Allowed regions (%) | 1.2 |
| Outliers (%) | 0.1 |
| Clash score | 6.19 |
| Average B-factor | 45.90 |
a The Pearson correlation coefficient of two half data sets that were derived by averaging half of the observations for a given reflection.
b CC* = √2CC1/2/(1 + CC1/2).
The structure was solved by molecular replacement using the structure of XR (18) (PDB ID: 3DDL) as a search model with the program Phaser (19). The correct molecular replacement solution contained four TR molecules packed in parallel in the asymmetric unit. Manual model building was performed in Coot (20) during iterative refinement. Early stage refinement was carried out with Refmac5 (21) using the jelly-body option. The later stages of refinement were performed using autoBUSTER (22) and Phenix (23) with TLS (translation-libration-screw), XYZ coordinates, and individual B-factor refinement. Structural analysis using Rampage (24) and the final refinement statistics are summarized in Table 1. Figures for the three-dimensional structures (Figs. 2, 4, a, c, d, and e, and 5) were prepared using PyMOL (Schrödinger LLC), and figures for the electrostatic potential surfaces (Fig. 4, e and f) were prepared using CueMol2 with PDB2PQR (25) and APBS (26). The intramolecular interactions of TR were analyzed using the PIC (Protein Interaction Calculator) Web server (27) using default parameters.
FIGURE 2.
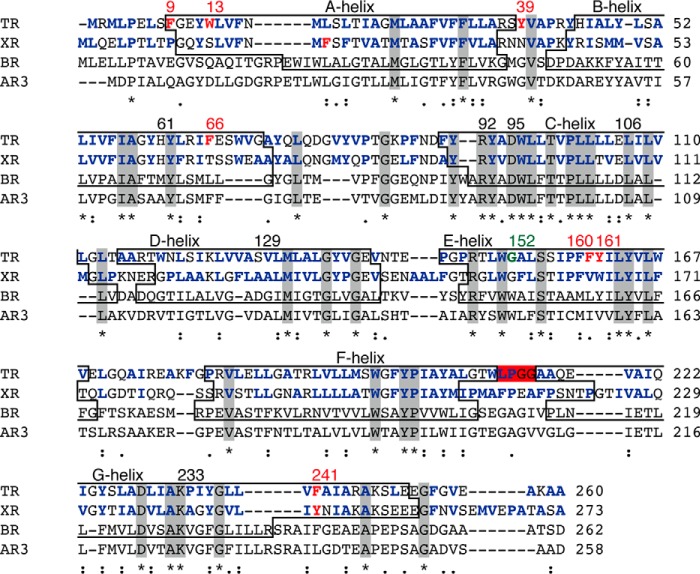
Amino acid sequence alignments of microbial proton-pumping rhodopsins generated by MUSCLE (multiple sequence comparison by log-expectation). Identical residues among these molecules are shown in gray boxes and by asterisks. The transmembrane helices (A-to-G helices), are shown in boxes. The helices belonging to AR3 are not shown because its crystal structure is lacking. The hydrophobic residues of TR and XR are shown in bold blue letters. TR and XR have non-identical aromatic residues, shown in bold red letters, and involve the aromatic-aromatic interactions. The LPGG sequence in the FG loop is shown in a red box. The conserved residue Gly152, which is assumed to contribute to secondary chromophore binding in TR, is highlighted in bold green letters. The residual numbers of TR are shown at the top of the sequence.
FIGURE 4.
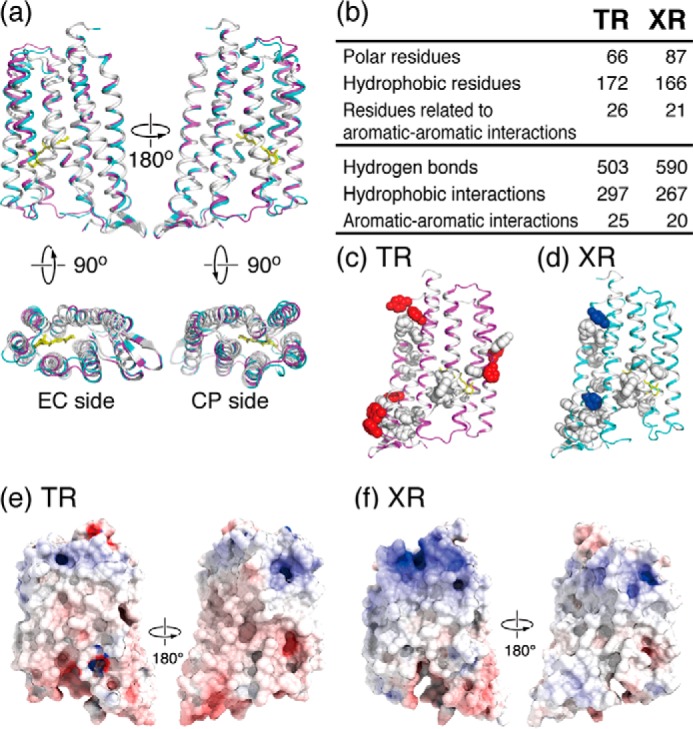
Structural comparison of TR and XR. a, superimposed structures of TR (pink) and XR (blue) in which the identical and non-identical residues in their amino acid sequence alignments (also shown in Fig. 2) are shown in white and color, respectively. The retinal is shown as a yellow stick. b, table showing the difference in the number of residues and intramolecular interactions of TR and XR. Polar residues: Asn, Gln, Ser, Thr, Asp, Glu, His, Lys, and Arg. Hydrophobic residues: Ala, Phe, Ile, Leu, Met, Val, Trp, Pro, and Tyr. Aromatic residues: Phe, Tyr, and Trp. c and d, aromatic-aromatic interactions of TR and XR. Side chains of the identical and non-identical aromatic residues forming aromatic-aromatic interactions are shown in white and colored spherical representations, respectively. e and f, surface potential distributions of TR and XR. The electrostatic potential surfaces are mapped at contouring levels from −10 kT/e (blue) to 10 kT/e (red).
FIGURE 5.
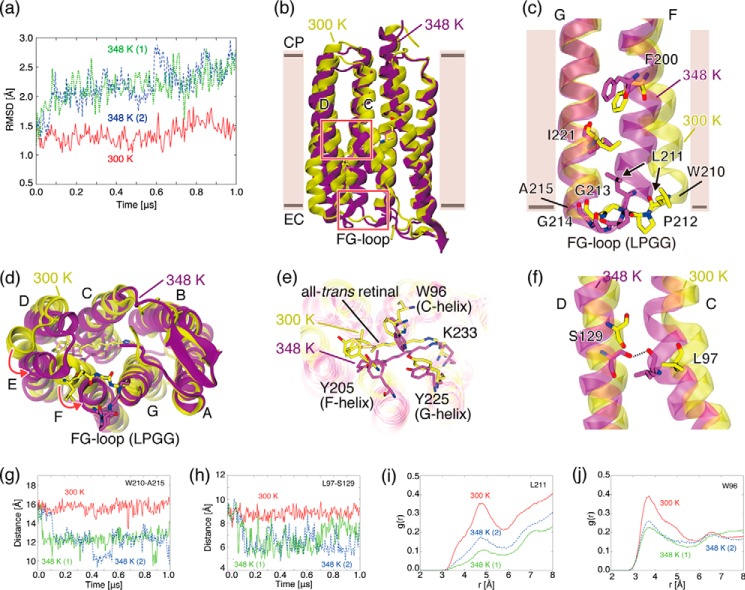
Temperature-induced conformational changes of TR observed in MD simulations. a, r.m.s.d. values of Cα atoms of TR in MD simulations with respect to the x-ray crystallographic structure at 300 K (solid red line) and at 348 K (dotted green line and dashed blue line). b, overall structures of TR simulated at 300 K (yellow) and 348 K (purple). The upper and lower boxed areas indicate regions where a hydrogen bond between the C- and D-helices is newly formed at 348 K and the extracellular region of the F- and G-helices, respectively. c, extracellular regions of the F- and G-helices simulated at 300 and 348 K. View is from the back side of the protein depicted in a. d, view of the extracellular side of the protein depicted in a. e, the region near the retinal chromophore simulated at 300 and 348 K. f, the region around Leu97 in the C-helix simulated at 300 and 348 K. A hydrogen bond between Leu97 and Ser129 is newly formed at 348 K. g, time evolution of distance between Cα atoms of Trp210 and Ala215, which are located at both ends of the FG loop. h, time evolution of the distance between the Cα atoms of Leu97 and Ser. i, RDF of water molecules from the Cγ atom of Leu211. j, RDF of water molecules from the Cη2 atom of Trp96.
Molecular Dynamics Simulation
All MD simulations were performed with NAMD (versions 2.8 and 2.9) (28) under isothermal isobaric conditions using CHARMM22/CMAP protein force field (29, 30), CHARMM36 lipid force field (31), and a TIP3P water model. The previously determined force field of the retinal protonated Schiff base was employed for the chromophore (32, 33). Long-range electrostatic interactions were treated using the particle mesh Ewald method. The short-range interactions were cut off at 9 Å with a smoothing function ranging between 8 and 9 Å. The initial TR structure was taken from the x-ray crystallographic structure reported in the present study. Water molecules were placed by the DOWSER program (34) to fill internal cavities in TR. Glu106 was protonated according to the pKa value predicted by the PROPKA program (35). A monomer protein of TR was embedded into a lipid bilayer solvated in a water solution containing 150 mm NaCl added with an Autoionize plugin for VMD. The simulation system consisted of 206 POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine) lipids, 20,202 waters, 58 Cl− ions, and 58 Na+ ions. The simulation box size was ∼94 × 77 × 121 Å so that the closest distance between the proteins in the periodic images was sufficiently large (>50 Å), indicating that the numbers of lipid and water molecules in the simulation box was sufficient. The entire system was equilibrated at 300 K under 1 bar for 50 ns followed by the production runs. MD simulations were performed at two different temperatures, i.e. at low temperature (300 K) and at high temperature (348 K), under 1 bar for 1 μs each. For confirmation of reproducibility, we performed an additional MD simulation at a high temperature (348 K) starting with different initial coordinates and velocities taken from the trajectory at 60 ns in the production simulation at 300 K. The conformational changes characterized by the Cα distance matrices observed in the two independent MD simulations at 348 K (data not shown) were essentially the same, confirming the reproducibility and consistency of the thermally induced conformational changes.
Measurement of Proton Pump Activity
The proton pump activity was measured by the light-induced pH changes of E. coli suspension as described previously (7). Briefly, TR-expressed E. coli BL21(DE3) cells were washed three times in 100 mm NaCl and then suspended in the same solution. The pH change was monitored by using a LAQUA F-72 pH meter equipped with a micro ToupH pH electrode (HORIBA, Ltd., Kyoto, Japan). The cell suspension was kept in the dark and then illuminated for 3 min with a Xe light source, MAX-302 (Asahi Spectra Co., Ltd., Tokyo, Japan), equipped with a 520 ± 10-nm band-pass filter for TR-WT and the TR-ΔLPGG mutant. When necessary, a proton-selective ionophore, carbonyl cyanide 3-chlorophenylhydrazone (CCCP, Sigma-Aldrich), was used.
Results and Discussion
Thermal Stability of TR
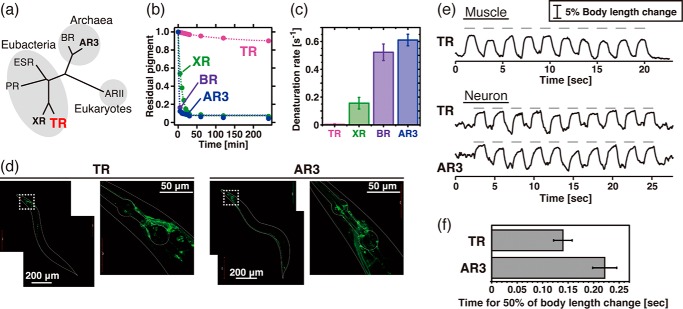
The proton-pumping rhodopsins are divided into distinct archaeal, eubacterial, and eukaryotic groups (Fig. 1a). Fig. 1b shows the time-dependent decrease in visible absorption, which is considered thermal denaturation and therefore was used to measure the thermal stability of the rhodopsins. As reported previously, TR maintains its visible color even when it is incubated at physiological temperature for T. thermophilus (75 °C) for several hours (Fig. 1b) (7). To compare its thermal stability with other proton-pumping rhodopsins, we performed similar experiments with BR from the archaeon H. salinarum, archaerhodopsin-3 from the archaeon Halorubrum sodomense, and xanthorhodopsin from the eubacterium S. ruber (Fig. 1, a and b). Of note, we succeeded in expressing carotenoid-free XR using a cell-free protein expression system (11). As shown in Fig. 1b, BR, AR3, and XR lost their visible color within 20 min at 75 °C. The denaturation rates were estimated by fitting to a single exponential function. These experiments reveal that the thermal stability of TR (denaturation rate constant, 8.0 × 10−3 [min−1]) is much higher (20∼100-fold) than those of the others (denaturation rate constants, 1.6 × 10−1 [min−1] for XR, 5.2 × 10−1 [min−1] for BR, and 6.1 × 10−1 [min−1] for AR3) (Fig. 1c), although the functionally important residues located in the proton transport pathway are conserved among them (Fig. 2).
FIGURE 1.
Thermal stability and optogenetic availability of TR. a, phylogenetic tree of microbial proton-pumping rhodopsins of known structure. PR, ESR, and ARII stand for proteorhodopsin, Exiguobacterium sibiricum rhodopsin, and Acetabularia rhodopsin II, respectively. b, denaturation kinetics of TR, XR, BR, and AR3 at 75 °C. The broken lines represent fitting curves of a single exponential function. c, denaturation rates at 75 °C estimated from the kinetics shown in b (n = 3). d, expression of TR::GFP and AR3::GFP in transgenic C. elegans by the pan-neuronal promoter. e, paralyzing activity of TR and AR3. Upper panel, elongation of the body length of C. elegans worms expressing TR::GFP in body wall muscle cells (n = 7). Ten pulses of green light (0.8 milliwatt/mm2, 550 nm) of 1.0 s in duration with 1.0-s interstimulus intervals were applied as indicated by the gray bars. Lower panel, elongation of the body length of worms expressing either TR::GFP (n = 4) or AR3::GFP (n = 4) in neurons with 10 green pulses (4.4 milliwatts/mm2, 550 nm) of 1.0 s with 1.0-s interstimulus intervals. f, the average time for the half of the body length change in worms expressing TR and AR3 calculated from the data shown in e, lower panel.
Applicability of TR as an Optogenetic Tool
In 2010, Chow et al. reported that proton-pumping rhodopsins can be applied to optogenetics technology as light-dependent neural silencers (36). They found the archaeal AR3 to be the most powerful among the seven proton pumps tested (36). Here, to investigate the availability of TR as a silencer of excitable cells, we first expressed TR as a fusion construct with green fluorescent protein (GFP) in body wall muscle cells of C. elegans, which drive locomotion of worms, under the control of a promoter of the myosin heavy chain gene myo-3 (12–14). Then the locomotion of the worms was measured with and without green light (550 nm), which overlaps the absorption spectra of both TR (530 nm) and AR3 (552 nm). Green light illumination of worms expressing TR-GFP led to locomotion paralysis, and the body length was increased significantly during 1 s of illumination, as shown in Fig. 1e. Upon switching off the light, the body length immediately returned to normal. The increased body length indicates that the paralysis was elicited by the relaxation of the body wall muscles, consistent with the presumed silencing ability of TR.
We next expressed GFP-tagged TR and AR3 in neurons by using a pan-neuronal promoter. As shown by the green-colored cells in Fig. 1d, GFP signals were clearly observed in the soma and the axon of many neurons in the head, mid body, and tail for both TR and AR3, indicating their successful expression. Similar to the worms carrying the transgene myo-3p::TR, green light illumination led to locomotion paralysis with ∼5% increased body length in worms carrying either TR or AR3 (Fig. 1e). These changes are likely to be mediated by the silencing of motor neurons, which govern muscular contraction, as the paralysis was associated with an increase in the body length. The body length immediately returned to normal upon switching off the light. The time for half of the body length change, indicating the apparent neural silencing activities of the rhodopsins, was then calculated, and their averages are shown in Fig. 1f with error bars. Because the wavelength of light used here (550 nm) corresponds well to the absorption maximum of AR3 (552 nm) rather than TR (530 nm), we concluded that TR receives the light with a lower efficiency than AR3. These results indicate that TR is an efficient and powerful neural silencer, comparable to AR3. Although a light-driven chloride-pumping rhodopsin, halorhodopsin, is also used as a neural silencer in the optogenetics technique (37), the silencing activity of halorhodopsin is comparably smaller than that of AR3.
Some eubacterial proton pumps can bind and use carotenoids as a secondary chromophore for their photoreactive functions (38, 39). The amino acid residues necessary for carotenoid binding are also conserved in TR, as described in the following section. Therefore, in addition to the availability of TR itself, the TR-carotenoid complex will be available for optogenetics studies in the near future. A recent report has demonstrated the availability of animal rhodopsin and melanopsin for the optogenetics of C. elegans worm (40). The technique is advantageous for direct monitoring of the cellular behavior caused by the coupling reaction between these proteins and their cognate G proteins. On the other hand, microbial rhodopsins including TR can directly depolarize or hyperpolarize the host neurons without involving other cellular responses. From the point of basic molecular properties, microbial rhodopsins are advantageous for duplicate measurements because these molecules can continue to hold the retinal chromophore after one photoreaction. Although the use of microbial and animal rhodopsins in optogenetics has both advantages and disadvantages, the usefulness of TR shown here would give the researchers valuable information. To understand and utilize those properties, we determined the three-dimensional structure of TR at an atomic resolution as described below.
Structure Determination of TR and Comparison with XR and BR
TR was crystalized into LCP. The structure was solved by molecular replacement using XR structure (18) as a searching model and was determined at 2.8 Å resolution (PDB ID: 5AZD). In the crystal, the asymmetric unit contains four TR molecules (denoted as chains A–D) that are almost identical in structure (data not shown). Therefore, we used the data from chain A in all figures (Figs. 3–5) and in the following discussion; the structural statistics of TR are summarized in Table 1. Similar to the known structures of other microbial rhodopsins, the overall structure of TR is composed of seven transmembrane α-helices, with the all-trans-retinal chromophore covalently bound to Lys233 at the center of the G-helix via a protonated Schiff base linkage (Fig. 3a) (1). Note that about 50% of Asp95, one of the putative counterions for the protonated Schiff base, is protonated at pH 3.6 for the crystallization, because the pKa for Asp95 was determined to be 3.4 in our previous study (7).
FIGURE 3.
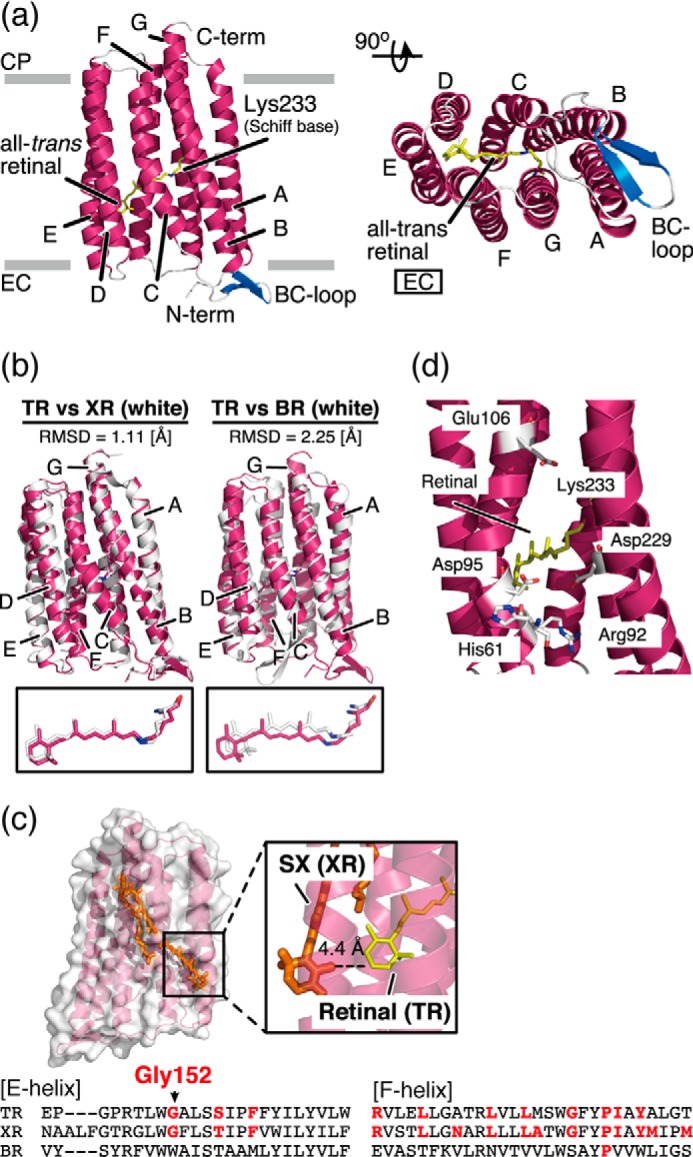
Structural details of TR. a, overall structure of TR shown in ribbon representation. Transmembrane α-helices A–G are shown in pink, interhelical loops in the cytoplasmic (CP) and extracellular (EC) sides of the membrane are shown in gray, and β-strands in the BC loop are shown in blue. All-trans-retinal connected to Lys233 via a protonated Schiff base linkage is shown as a yellow stick. b, superimposed structures of TR (pink) and XR (18) (white; PDB ID: 3DDL) and TR and BR (40) (white; PDB ID: 1C3W). r.m.s.d. values were calculated using carbonyl carbon. Magnified views of the retinal chromophore are shown below. c, expected carotenoid-binding cavity in TR. The carotenoid SX in the XR structure (18) is superimposed on the E- and F-helices in the TR structure. The magnified view shows the geometry between SX in XR and the all-trans-retinal in TR. The amino acid sequence alignments of the E- and F-helices are shown at the bottom of the panel. The residues involved in carotenoid binding in XR are conserved in the TR sequence (bold red letters). The most important Gly residue is conserved in TR as Gly152. d, putative proton transport pathway in TR. The retinal and residues involved in proton transport are shown in stick representation.
As shown in Fig. 3b, the structure of TR was compared with the archaeal proton pump BR (41) and the eubacterial proton pump XR (18) by superimposition of those structures. The structure of TR was significantly different from that of BR (root mean square deviation (r.m.s.d.) = 2.25 [Å]), which corresponds to a low sequence identity and similarity between TR and BR (∼25 and ∼59%, respectively). Several structural differences between TR and BR were found; (i) an extension of the B- and G-helices; (ii) a slight tilting of the A-, D-, F-, and G-helices; and (iii) a planar structure of the all-trans-retinal (Fig. 3b). In contrast, the structure of TR was similar to that of XR (r.m.s.d. = 1.11 [Å]), as expected from the high sequence identity and similarity between TR and XR (∼57% and ∼85%, respectively). It has been reported that XR binds a carotenoid molecule called SX as a secondary chromophore in the crystal structure because XR was purified from the native habitat having a carotenoid synthesis system (38). In the XR-SX complex, light energy transfer from SX to the retinal in XR occurs (38). The residues responsible for SX binding, including a critical Gly residue (Gly152 in TR) (18, 39), are completely conserved in TR, and SX can be fitted into the structure of TR with an appropriate energy-transferable distance between SX and retinal (Fig. 3c) (18). Thus, in addition to the characteristics described above, TR is assumed to have a putative carotenoid-binding cavity in the E-helix (Fig. 3c). Because SX has a broad and blue-shifted absorption spectrum with a large molecular extinction coefficient, the TR-SX complex would be useful for optogenetics as a molecule that absorbs a wide range of the visible region with high efficiency. Notably, it has been reported that the native T. thermophilus has a carotenoid synthesis system and a kind of carotenoid named thermozeaxanthin, which shares a similar structure with SX, as a major component (42). Thus, it is assumed that TR has a potential carotenoid binding ability. Further studies to confirm the carotenoid binding are now under way.
Regarding the proton pumping function of TR, Fig. 3d shows an expanded view of the putative proton transport pathway in TR. As with the other proton-pumping rhodopsins, in TR there are two putative counterions, Asp95 and Asp229 (7), for the protonated Schiff base nitrogen of Lys233. When aspartate works as a primary proton acceptor, a point mutation of that residue leads to a large spectral red-shift (∼20 nm), because of the delocalization of the positive charge of Lys233, and to a loss of its proton pumping activity. Consistent with this, the D95N mutant showed a 25-nm spectral red-shift and no significant proton pumping activity, indicating that Asp95 is the primary proton acceptor (data not shown). It is assumed that a proton of the protonated Schiff base moves to Asp95 upon illumination, after which a proton is transferred from the putative proton-donating residue, Glu106 (8), to the deprotonated Schiff base. During the reaction, His61 and Arg92 are thought to be involved in the maintenance of both the pKa values of the charged residues and the proper structural changes of the peptide backbone analogous to the other eubacterial proton pumps including XR (18, 43–46). Unfortunately, we cannot discuss the structural details of TR at the atomic level by precisely comparing them with XR or other microbial rhodopsins because of the present modest resolution (2.8 Å). However, the proton-transporting mechanism of TR is basically the same as those of conventional proton-pumping rhodopsins (8). The improvement in the crystallization of TR to achieve a much better resolution is currently in progress.
Thermal Stabilization Mechanism Suggested by the Structural Comparison
As shown in Fig. 1, b and c, TR showed a 20-fold higher thermal stability than XR. There are several potential reasons why TR is more stable than XR, which are discussed here in the context of their structural comparison. Fig. 4a shows the superimposed structures of TR and XR. The identical residues, shown in white, are especially localized to the inside of the helices, which are composed of the proton transport pathway. In contrast, non-identical residues, shown by color, are found mainly on the exposed surfaces of the proteins. From the comparison, it was revealed that TR has 172 hydrophobic residues involving in 297 hydrophobic interactions, the number of which is greater than that of XR, as shown in Fig. 4b. It is noteworthy that seven non-identical aromatic residues (Phe9, Trp13, Tyr39, Phe66, Phe160, Tyr161, and Phe241) of TR form aromatic-aromatic interactions, whereas only two non-identical residues (Phe20 and Tyr248) of XR contribute to such interactions (Fig. 4, c and d). On the other hand, TR has a lesser number of polar amino acids than XR (66 and 87, respectively), indicating that the number of polar interactions in TR is also less than in XR (Fig. 4b). These findings suggest that the high thermal stability of TR is achieved by the hydrophobic and aromatic-aromatic interactions within the protein and with the membrane lipid molecules surrounding TR under the native environment.
Atomistic Mechanisms of the Thermal Stability
To examine the structural and dynamic behaviors of TR at high temperature, we performed MD simulations by using TR structure in the technically well established lipid bilayer system at 348 K (75 °C) and compared them with those at an ambient temperature (300 K) (27 °C) for 1-μs duration. An increase in the temperature from 300 to 348 K induced a distinct conformational transition to a characteristic conformational substate where the r.m.s.d. values of the Cα atoms of the protein in the trajectories with respect to the x-ray structure were significantly altered (Fig. 5a). Although the r.m.s.d. at 300 K does not vary greatly from ∼1.4 Å over the entire simulation time, the r.m.s.d. values of the two independent MD trajectories at 348 K exhibit drastic increases from ∼1.4 to 2.2 Å within 200 ns after heating from 300 K (data not shown), indicating that TR undergoes a thermally induced conformational transition upon heating to 348 K. The thermally induced conformational changes are spatially extensive and involve three concerted local structural changes, which could contribute to stabilizing the conformational substate at that high temperature (Fig. 5b) as follows.
LPGG Sequence in the FG Loop
The structural changes were found in the extracellular side of the F-helix and in the extracellular loop between the F- and G-helices (called the FG loop (Fig. 5, c and d)). The loop includes a Leu211-Pro212-Gly213-Gly214 (LPGG) sequence, which is absent in BR and XR (Fig. 2). At 300 K, the F- and G-helices are nearly parallel and distant from each other. Accordingly, the FG loop forms in an extended conformation to the bulk water environment (Fig. 5, c and d), which can be monitored by the distance between the Cα atoms of Trp210 and Ala215 located at both ends of the FG loop connecting the F- and G-helices (Fig. 5, c and g) and by the radial distribution function (RDF) of water molecules from the Cγ atom of Leu211 located at the beginning of the FG loop (Fig. 5i). Upon heating to 348 K, on the other hand, the extracellular side of the F-helix moves toward the G-helix, and the FG loop intrudes into the hydrophobic space between the F- and G-helices (Fig. 5, c and d) establishing hydrophobic packing among them, accompanied by the exclusion of water molecules (Fig. 5i). The hydrophobic interaction in the extracellular region is considered to be formed by a gain of translational and rotational entropy and thus is more enhanced at a higher temperature.
Hydrophobic Core around Retinal
The movement of the F-helix toward the G-helix leads to the formation of a hydrophobic core around the retinal (Fig. 5e), accompanying the movement of the C-helix (Fig. 5d) and the exclusion of water molecules from this region, which can be measured by the RDF of water molecules from the Cη2 atom of Trp96 in the C-helix, located proximal to the retinal (Fig. 5j). The formation of the hydrophobic core is considered to stabilize the conformational substate at the higher temperature. To clarify the structural changes of TR upon thermal stimulation experimentally, spin-label EPR experiments and fluorescence measurements of Trp residues around the retinal chromophore are useful.
Hydrogen Bond
In addition, a hydrogen bond formation between the C- and D-helices at high temperature was observed. As a result of the displacement of the C-helix, the main-chain carbonyl group of Leu97, located around the center of the C-helix, which lacks a hydrogen bond partner at low temperature because of a kink in the C-helix introduced by Pro101, approaches the hydroxyl group of Ser129 in the D-helix (Fig. 5h) and forms an inter-helix hydrogen-bond with it (Fig. 5f).
As described above, TR has a large number of hydrophobic residues and characteristic aromatic residues involved in the aromatic-aromatic interaction (Fig. 4, b and c). Such unique structural properties, revealed by the x-ray structure and the structural transitions that enhance the hydrophobic interaction upon the thermal stimulation suggested by MD simulations, are a possible mechanism for the thermal stabilization of TR at high temperature. Note that no sequence similarity of the FG loops is found, and the hydroxyl group at position 129 in the D-helix of TR does not exist in the corresponding regions of BR and XR (Fig. 2). The thermally induced interactions in TR are therefore expected to contribute to retaining the structural folding at an increased temperature that would otherwise cause denaturation as observed in BR and XR (Fig. 1, b and c).
Thermal Stability of TR Lacking the FG Loop
To examine the effect of the formation of the tight packing structure in the FG loop indicated by the MD simulation as one of the possible mechanisms of high structural stability, we then prepared the mutant TR, which was the deleted Leu211-Pro212-Gly213-Gly214 sequence in the FG loop from the wild-type TR (named TR-ΔLPGG (Figs. 5c and 6a)). The TR-ΔLPGG mutant was functionally expressed in E. coli cells, showed outward proton pumping activity, and exhibited an absorption maximum at 530 nm in the presence of DDM, almost the same as the wild-type TR (Fig. 6, b and c). We then measured the thermal stability of the ΔLPGG and wild-type TR according to the same procedure reported previously (7). Fig. 6, d and e, shows the time-dependent decrease in absorption at 530 nm at 75 °C. The TR-ΔLPGG mutant was almost denatured in 210 min, whereas more than 60% of the wild-type TR remained. From the denaturation kinetics (Fig. 6f), the data of which were analyzed by the single exponential function, we determined the denaturation rate constants as (2.54 ± 0.10) × 10−3 min−1 for the wild-type TR and (1.06 ± 0.03) × 10−2 min−1 for the TR-ΔLPGG mutant, respectively (Fig. 6g).
FIGURE 6.
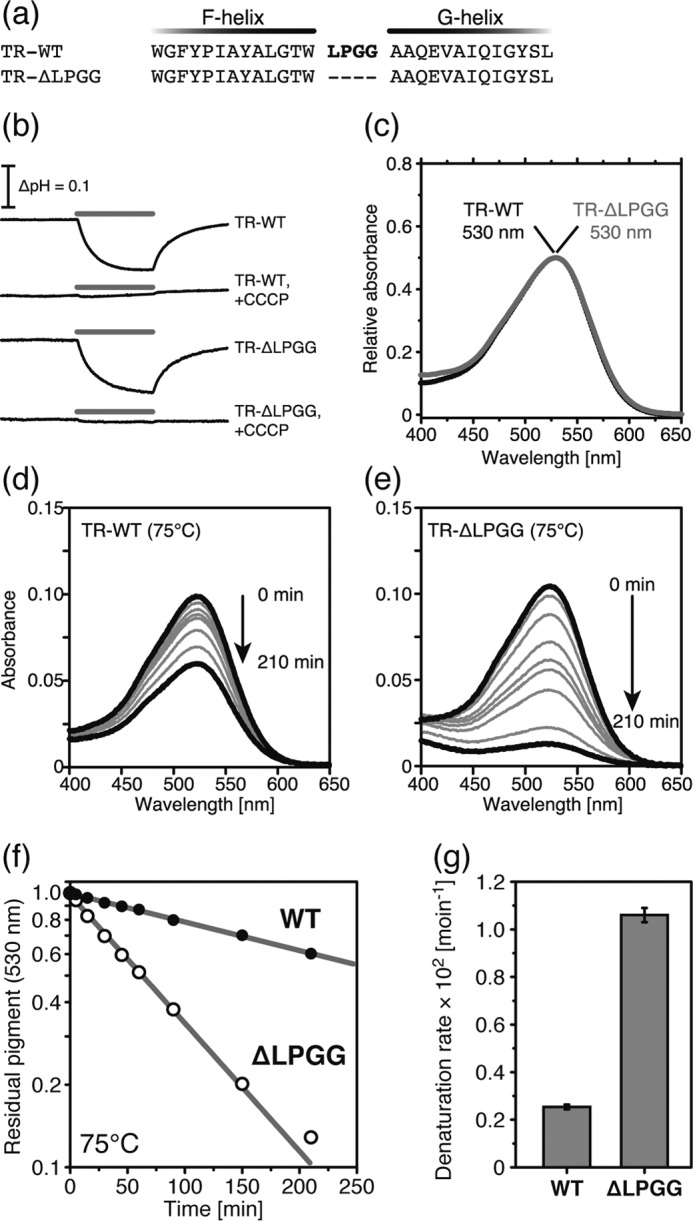
Comparison of TR-WT and TR-ΔLPGG. a, partial amino acid sequence from F- to G-helices. Bold red letters represent the LPGG sequence (Leu211-Pro212-Gly213-Gly21 4) in the extracellular loop between the F- and G-helices. b, light-induced pH changes of TR-WT and TR-ΔLPGG in E. coli suspension in the presence of 100 mm NaCl. The pH was decreased during the 520 ± 10-nm light irradiation for 3 min, shown by the gray bars, in the absence of CCCP. In the presence of 10 μm CCCP, the pH change disappeared. c, visible absorption spectra of TR-WT (black) and TR-ΔLPGG (gray). The absorption maxima were 530 nm. d and e, time-dependent thermal denaturation of TR-WT (d) and TR-ΔLPGG (e) at 75 °C. f, the denaturation kinetics of TR-WT (black) and TR-ΔLPGG (white) at 75 °C. The data were analyzed by a single exponential decay function shown by the gray lines. g, the denaturation rate constants are shown as a bar graph.
These data indicate that the deletion of the LPGG sequence in the FG loop affected the thermal stability of TR. We presume that the deletion of LPGG suppressed the conformational transition of the FG loop and the consequent formation of the tight packing caused by hydrophobic interactions at high temperature, which was simulated by the MD described above. However, at present it is difficult to conclude that the LPGG sequence in the FG loop is an essential factor in maintaining the high thermal stability of TR. Further experiments to prove the importance of the LPGG sequence, such as the addition of the LPGG sequence to AR3 and XR, are needed.
Conclusion
In this study, we have reported the high thermal stability and optogenetic availability of TR. To verify their molecular origins, we determined the crystal structure of TR at 2.8 Å resolution. This gave us information about the structure with a putative secondary chromophore-binding cavity at atomic resolution. Combined with the MD simulations, we assume the importance of the hydrophobic interactions in TR as contributing to its high thermal stability. Thus these findings will be valuable for understanding and utilizing retinal proteins in respect to high protein stability and high optogenetic performance.
Author Contributions
T. T. and K. M. contributed equally to this work. T. T., K. M., T. H., M. T., N. Honda, N. Hashimoto, K. S., S. H., T. M., and Y. S. performed the experiments. T. T., K. M., T. H., M. T., N. Hashimoto, N. Honda, K. Y., M. Y., S. M., S. H., T. M., and Y. S. analyzed the data. S. T., S. H., T. M., and Y. S. designed the research. T. T., S. H., T. M., and Y. S. wrote the paper.
Acknowledgments
We thank the staffs of BL-1A at the Photon Factory (Proposals 2012G132, 2013R22, and 2014G171) and of BL32XU at SPring-8 for their assistance in the synchrotron radiation experiments. We are grateful to Dr. Ayako Okazaki (Nagoya University, Japan) for kind advice regarding the optogenetic experiments on C. elegans and to Dr. Hidehito Kuroyanagi (Tokyo Medical and Dental University, Japan) for supplying plasmids. Some of the computations were performed at the Research Center for Computational Science, Okazaki, Japan.
This work was supported by Grants 15K18519 (to T. T.), 25104004 and 25291034 (to S. H.), and 15H04363 and 15H00878 (to Y. S.) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT). This work also was supported by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from MEXT and the Japan Agency for Medical Research and Development (AMED) (to T. M.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5AZD) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- TR
- thermophilic rhodopsin
- MD
- molecular dynamics
- DDM
- n-dodecyl-β-d-maltoside
- LCP
- lipidic cubic phase
- BR
- bacteriorhodopsin
- AR3
- archaerhodopsin-3
- XR
- xanthorhodopsin
- SX
- salinixanthin
- CCCP
- carbonyl cyanide 3-chlorophenylhydrazone
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- RDF
- radial distribution function.
References
- 1. Ernst O. P., Lodowski D. T., Elstner M., Hegemann P., Brown L. S., and Kandori H. (2014) Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grote M., Engelhard M., and Hegemann P. (2014) Of ion pumps, sensors and channels: perspectives on microbial rhodopsins between science and history. Biochim. Biophys. Acta 1837, 533–545 [DOI] [PubMed] [Google Scholar]
- 3. Inoue K., Tsukamoto T., and Sudo Y. (2014) Molecular and evolutionary aspects of microbial sensory rhodopsins. Biochim. Biophys. Acta 1837, 562–577 [DOI] [PubMed] [Google Scholar]
- 4. Martinez A., Bradley A. S., Waldbauer J. R., Summons R. E., and DeLong E. F. (2007) Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc. Natl. Acad. Sci. U.S.A. 104, 5590–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoshizawa S., Kawanabe A., Ito H., Kandori H., and Kogure K. (2012) Diversity and functional analysis of proteorhodopsin in marine Flavobacteria. Environ. Microbiol. 14, 1240–1248 [DOI] [PubMed] [Google Scholar]
- 6. Murugapiran S. K., Huntemann M., Wei C. L., Han J., Detter J. C., Han C. S., Erkkila T. H., Teshima H., Chen A., Kyrpides N., Mavrommatis K., Markowitz V., et al. (2013) Whole genome sequencing of Thermus oshimai JL-2 and Thermus thermophilus JL-18, incomplete denitrifiers from the United States Great Basin. Genome Announc. 1, e00106–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsukamoto T., Inoue K., Kandori H., and Sudo Y. (2013) Thermal and spectroscopic characterization of a proton pumping rhodopsin from an extreme thermophile. J. Biol. Chem. 288, 21581–21592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsukamoto T., Demura M., and Sudo Y. (2014) Irreversible trimer to monomer transition of thermophilic rhodopsin upon thermal stimulation. J. Phys. Chem. B. 118, 12383–12394 [DOI] [PubMed] [Google Scholar]
- 9. Sudo Y., Yamabi M., Iwamoto M., Shimono K., and Kamo N. (2003) Interaction of Natronobacterium pharaonis phoborhodopsin (sensory rhodopsin II) with its cognate transducer probed by increase in the thermal stability. Photochem. Photobiol. 78, 511–516 [DOI] [PubMed] [Google Scholar]
- 10. Hohenfeld I. P., Wegener A. A., and Engelhard M. (1999) Purification of histidine-tagged bacteriorhodopsin, pharaonis halorhodopsin and pharaonis sensory rhodopsin II functionally expressed in Escherichia coli. FEBS Lett. 442, 198–202 [DOI] [PubMed] [Google Scholar]
- 11. Wada T., Shimono K., Kikukawa T., Hato M., Shinya N., Kim S. Y., Kimura-Someya T., Shirouzu M., Tamogami J., Miyauchi S., Jung K. H., Kamo N., and Yokoyama S. (2011) Crystal structure of the eukaryotic light-driven proton-pumping rhodopsin, Acetabularia rhodopsin II, from marine alga. J. Mol. Biol. 411, 986–998 [DOI] [PubMed] [Google Scholar]
- 12. Okazaki A., Sudo Y., and Takagi S. (2012) Optical silencing of C. elegans cells with arch proton pump. PLoS One 7, e35370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sudo Y., Okazaki A., Ono H., Yagasaki J., Sugo S., Kamiya M., Reissig L., Inoue K., Ihara K., Kandori H., Takagi S., and Hayashi S. (2013) A blue-shifted light-driven proton pump for neural silencing. J. Biol. Chem. 288, 20624–20632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Okazaki A., Takahashi M., Toyoda N., and Takagi S. (2014) Optical silencing of C. elegans cells with light-driven proton pumps. Methods 68, 425–430 [DOI] [PubMed] [Google Scholar]
- 15. Hirata K., Kawano Y., Ueno G., Hashimoto K., Murakami H., Hasegawa K., Hikima T., Kumasaka T., and Yamamoto M. (2013) Achievement of protein micro-crystallography at SPring-8 beamline BL32XU. J. Phys. Conf. Ser. 425, 012002 [Google Scholar]
- 16. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 18. Luecke H., Schobert B., Stagno J., Imasheva E. S., Wang J. M., Balashov S. P., and Lanyi J. K. (2008) Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore. Proc. Natl. Acad. Sci. U.S.A. 105, 16561–16565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bricogne G., Blanc E., Brandl M., Flensburg C., and Keller P. (2011) BUSTER Version 2.10. Global Phasing Ltd., Cambridge, United Kingdom [Google Scholar]
- 23. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lovell S. C., Davis I. W., Arendall W. B. 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., and Richardson D. C. (2003) Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 25. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., and Baker N. A. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tina K. G., Bhadra R., and Srinivasan N. (2007) PIC: Protein Interactions Calculator. Nucleic Acids Res. 35, W473–W476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mackerell A. D. Jr., Feig M., and Brooks C. L. 3rd (2004) Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25, 1400–1415 [DOI] [PubMed] [Google Scholar]
- 30. Best R. B., Zhu X., Shim J., Lopes P. E., Mittal J., Feig M., and Mackerell A. D. Jr (2012) Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klauda J. B., Venable R. M., Freites J. A., O'Connor J.W., Tobias D. J., Mondragon-Ramirez C., Vorobyov I., MacKerell A. D. Jr., and Pastor R. W. (2010) Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tajkhorshid E., Baudry J., Schulten K., and Suhai S. (2000) Molecular dynamics study of the nature and origin of retinal's twisted structure in bacteriorhodopsin. Biophys. J. 78, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hayashi S., Tajkhorshid E., and Schulten K. (2002) Structural changes during the formation of early intermediates in the bacteriorhodopsin photocycle. Biophys. J. 83, 1281–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang L., and Hermans J. (1996) Hydrophilicity of cavities in proteins. Proteins 24, 433–438 [DOI] [PubMed] [Google Scholar]
- 35. Li H., Robertson A. D., and Jensen J. H. (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins 61, 704–721 [DOI] [PubMed] [Google Scholar]
- 36. Chow B.Y., Han X., Dobry A. S., Qian X., Chuong A. S., Li M., Henninger M. A., Belfort G. M., Lin Y., Monahan P. E., and Boyden E. S. (2010) High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 463, 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang F., Wang L. P., Brauner M., Liewald J. F., Kay K., Watzke N., Wood P. G., Bamberg E., Nagel G., Gottschalk A., and Deisseroth K. (2007) Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 [DOI] [PubMed] [Google Scholar]
- 38. Balashov S. P., Imasheva E. S., Boichenko V. A., Antón J., Wang J. M., and Lanyi J. K. (2005) Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science 309, 2061–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Imasheva E. S., Balashov S. P., Choi A. R., Jung K. H., and Lanyi J. K. (2009) Reconstitution of Gloeobacter violaceus rhodopsin with a light-harvesting carotenoid antenna. Biochemistry 48, 10948–10955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cao P., Sun W., Kramp K., Zheng M., Salom D., Jastrzebska B., Jin H., Palczewski K., and Feng Z. (2012) Light-sensitive coupling of rhodopsin and melanopsin to Gi/o and Gq signal transduction in Caenorhabditis elegans. FASEB J. 26, 480–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luecke H., Schobert B., Richter H. T., Cartailler J. P., and Lanyi J. K. (1999) Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 291, 899–911 [DOI] [PubMed] [Google Scholar]
- 42. Tian B., and Hua Y. (2010) Carotenoid biosynthesis in extremophilic Deinococcus-Thermus bacteria. Trends Microbiol. 18, 512–520 [DOI] [PubMed] [Google Scholar]
- 43. Bergo V. B., Sineshchekov O. A., Kralj J. M., Partha R., Spudich E. N., Rothschild K. J., and Spudich J. L. (2009) His-75 in proteorhodopsin, a novel component in light-driven proton translocation by primary pumps. J. Biol. Chem. 284, 2836–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hempelmann F., Hölper S., Verhoefen M. K., Woerner A. C., Köhler T., Fiedler S. A., Pfleger N., Wachtveitl J., and Glaubitz C. (2011) His75-Asp97 cluster in green proteorhodopsin. J. Am. Chem. Soc. 133, 4645–4654 [DOI] [PubMed] [Google Scholar]
- 45. Tsukamoto T., Kikukawa T., Kurata T., Jung K. H., Kamo N., and Demura M. (2013) Salt bridge in the conserved His-Asp cluster in Gloeobacter rhodopsin contributes to trimer formation. FEBS Lett. 587, 322–327 [DOI] [PubMed] [Google Scholar]
- 46. Balashov S. P., Petrovskaya L. E., Lukashev E. P., Imasheva E. S., Dioumaev A. .K, Wang J. M., Sychev S. V., Dolgikh D. A., Rubin A. B., Kirpichnikov M. P., and Lanyi J. K. (2012) Aspartate-histidine interaction in the retinal Schiff base counterion of the light-driven proton pump of Exiguobacterium sibiricum. Biochemistry 51, 5748–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]