Summary
Structures of biomolecular systems are increasingly computed by integrative modeling that relies on varied types of experimental data and theoretical information. We describe here the proceedings and conclusions from the first wwPDB Hybrid/Integrative Methods Task Force Workshop held at the European Bioinformatics Institute in Hinxton, UK, October 6 and 7, 2014. At the workshop, experts in various experimental fields of structural biology, experts in integrative modeling and visualization, and experts in data archiving addressed a series of questions central to the future of structural biology. How should integrative models be represented? How should the data and integrative models be validated? What data should be archived? How should the data and models be archived? What information should accompany the publication of integrative models?
Keywords: integrative modeling, hybrid modeling, integrative structural biology, Protein Data Bank
1 Background
1.1 Historical rationale for the Workshop
The Protein Data Bank (PDB; http://wwpdb.org) was founded in 1971 with seven protein structures as its first holdings (Protein Data Bank, 1971). The global PDB archive now holds more than 100,000 atomic structures of biological macromolecules and their complexes, all of which are freely accessible. Most structures in the PDB archive (~90%) have been determined by X-ray crystallography, with the remainder contributed by two newer 3D structure determination methods, nuclear magnetic resonance (NMR) spectroscopy and electron microscopy (3DEM).
Considerable effort has gone into understanding how to best curate the structural models and experimental data produced with these methods. Over the past several years, the Worldwide Protein Data Bank (wwPDB; the global organization responsible for maintaining the PDB archive) (Berman et al., 2003) has established expert, method-specific Task Forces to advise on which experimental data and metadata from each method should be archived and how these data and the resulting structure models should be validated. The wwPDB X-ray Validation Task Force (VTF) made detailed recommendations on how to best validate structures determined by X-ray crystallography (Read et al., 2011). These recommendations have been implemented as a software pipeline used within the wwPDB Deposition and Annotation (D&A) system. Initial recommendations of the wwPDB NMR (Montelione et al., 2013) and Electron Microscopy (Henderson et al., 2012) VTFs have also been implemented. In addition, the wwPDB and, in later years, the Structural Biology Knowledgebase (SBKB), spearheaded three workshops focused on validation, archiving, and dissemination of comparative protein structure models (Berman et al., 2006; Schwede et al., 2009). It is anticipated that as new validation methods are developed and as more experience is gained with existing ones, additional validation procedures will be implemented in the wwPDB D&A system.
Increasingly, structures of very large macromolecular machines are being determined by combining observations from complementary experimental methods, including X-ray crystallography, NMR spectroscopy, 3DEM, small-angle scattering (SAS), crosslinking, and many others (Figure 1, Table 1). Data from these complementary methods are used to compute integrative or hybrid models (Ward et al., 2013). Atomic models produced in this fashion have been deposited into the PDB, but there is currently no mechanism available within the PDB framework for archiving the experimental data generated by methods other than X-ray crystallography, NMR spectroscopy, and 3DEM. The most recently established task force, the wwPDB SAS Task Force (Trewhella et al., 2013b), recommended creation of a SAS data and model repository that would interoperate with the PDB. The SAS Task Force also recommended that an international meeting be held to consider how best to deal with the archiving of data and models coming from integrative structure determination approaches.
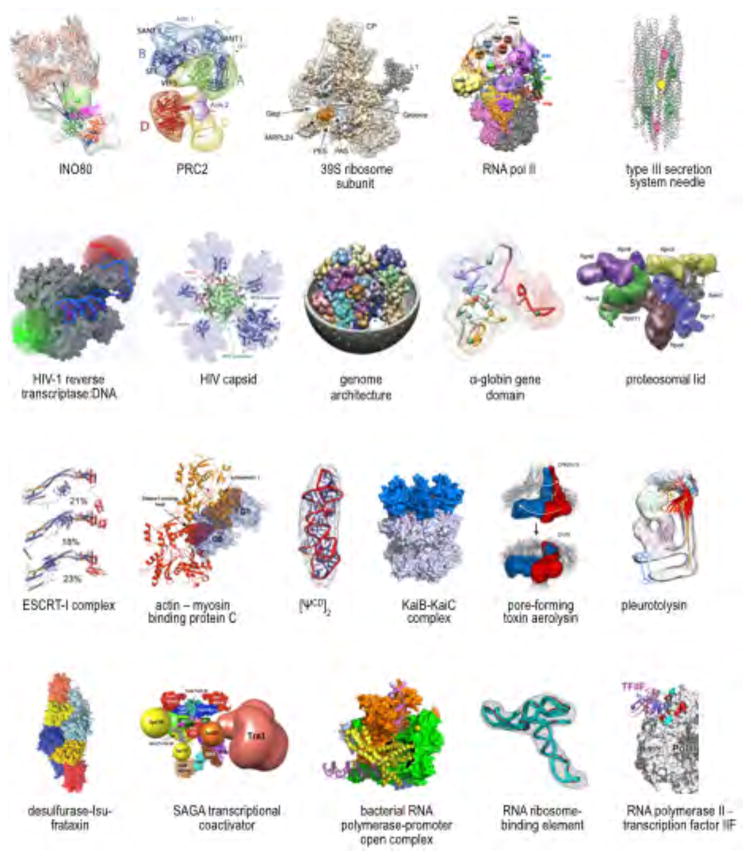
Figure 1. Examples of recently determined integrative structures.
The molecular architecture of INO80 was determined with a 17 Å resolution cryo-EM map and 212 intra-protein and 116 inter-protein crosslinks (Russel et al., 2009). The molecular architecture of Polycomb Repressive Complex 2 (PRC2) was determined with a 21 Å resolution negative-stain EM map and ~60 intra-protein and inter-protein crosslinks (Shi et al., 2014). The molecular architecture of the large subunit of the mammalian mitochondrial ribosome (39S) was determined with a 4.9 Å resolution cryo-EM map and ~70 inter-protein cross-links (Ward et al., 2013). The molecular architecture of the RNA polymerase II transcription pre-initiation complex was determined with a 16 Å resolution cryo-EM map plus 157 intra-protein and 109 inter-protein crosslinks (Alber et al., 2008). The atomic model of Type III secretion system needle was determined with a 19.5 Å resolution cryo-EM map and solid-state NMR data (Loquet et al., 2012). Molecular architecture of the productive HIV-1 reverse transcriptase:DNA primer-template complex in the open educt state was determined by FRET positioning and screening (FPS) using a known HIV-1 reverse transcriptase structure (Kalinin et al., 2012). The structure of HIV-1 capsid protein was determined using residual dipolar couplings (RDC) and SAXS data (Deshmukh et al., 2013). The human genome architecture was determined based on tethered chromosome conformation capture (TCC) and population-based modeling (Kalhor et al., 2012). The structural model of α-globin gene domain was determined based on Chromosome Conformation Capture Carbon Copy (5C) experiments (Bau et al., 2011). The molecular architecture of the proteosomal lid was determined using native MS and 28 cross-links (Politis et al., 2014). Atomic resolution conformations of ESCRT-I complex were determined with SAXS, double electron-electron transfer (DEER), and FRET (Boura et al., 2011). Integrative model of actin and the cardiac myosin binding protein C was developed from a combination of crystallographic and NMR structures of subunits and domains, with positions and orientations optimized against SAXS and SANS data to reveal information about the quaternary interactions (Whitten et al., 2008). The ensemble of [ψCD]2 NMR structures were fitted into the averaged cryo-electron tomography map (Miyazaki et al., 2010). Integrative model of the cyanobacterial circadian timing KaiB-KaiC complex was obtained based on H/D exchange and collision cross section data from MS (Snijder et al., 2014). The prepore and pore conformations of the pore-forming toxin aerolysin were obtained combining cryo-EM data and molecular dynamics simulations (Degiacomi and Dal Peraro, 2013; Degiacomi et al., 2013). Segment of a pleurotolysin pore map (~11 Å resolution) with an ensemble of conformations showing the trajectory of β-sheet opening during pore formation (Lukoyanova et al., 2015). A SAXS-based rigid body model of a ternary complex of the iron-sulphur cluster assembly proteins desulfurase (orange) and scaffold protein Isu (blue) with bacterial orthologue of frataxin (yellow) was validated by NMR chemical shifts and mutagenesis (Prischi et al., 2010). The molecular architecture of the SAGA transcription coactivator complex was determined with 199 inter- and 240 intra-subunit crosslinks, several comparative models based on X-ray crystal structures, and a TFIID core EM map at 31 Å resolution (Han et al., 2014). Structural Organization of the bacterial (T. aquaticus) RNA polymerase-promoter open complex obtained by FRET (Mekler et al., 2002), subsequently validated by a crystal structure (Zhang et al., 2012). The RNA ribosome-binding element from turnip crinkle virus genome, determined using NMR, SAXS, and EM data (Gong et al., 2015). The molecular architecture of the complex between RNA polymerase II and transcription factor IIF was determined using a deposited crystal structure of RNA polymerase II, homology models of some domains in transcription factor IIF, as well as 95 intra-protein and 129 inter-protein cross links (Chen et al., 2010).
Table 1. Types of structural data used in integrative modeling.
Example methods that are informative about a variety of structural aspects of biomolecular systems are listed.
| Structural information | Method |
|---|---|
| Atomic structures of parts of the studied system | X-ray and neutron crystallography, NMR spectroscopy, 3DEM, comparative modeling, and molecular docking |
| 3D maps and 2D images | Electron microscopy and tomography |
| Atomic and protein distances | NMR, FRET and other fluorescence techniques, DEER, EPR, and other spectroscopic techniques; chemical crosslinks detected by mass spectrometry and disulfide bonds detected by gel electrophoresis |
| Binding site mapping | NMR spectroscopy, mutagenesis, FRET |
| Size, shape, and pairwise atomic distance distributions | SAS |
| Shape and size | Atomic force microscopy, ion mobility mass spectrometry, fluorescence correlation spectroscopy and fluorescence anisotropy |
| Component positions | Super-resolution optical microscopy, FRET imaging |
| Physical proximity | Co-purification, native mass spectrometry, genetic methods, and gene/protein sequence covariance |
| Solvent accessibility | Footprinting methods, including H/D exchange assessed by mass spectrometry or NMR, and even functional consequences of point mutations |
| Proximity between different genome segments | Chromosome Conformation Capture and other data |
| Propensities for different interaction modes | Molecular mechanics force fields, potentials of mean force, statistical potentials, and sequence co-variation |
In response, a Hybrid/Integrative Methods Task Force was assembled by the wwPDB organization. Its inaugural meeting was held at the EMBL European Bioinformatics Institute (EBI) October 6th and 7th 2014 (http://wwpdb.org/task/hybrid.php). In all, 38 participants from 35 academic and government institutions worldwide attended the workshop, which was co-chaired by Andrej Sali (University of California, San Francisco, USA), Torsten Schwede (SIB and University of Basel, Switzerland), and Jill Trewhella (University of Sydney, Australia). Attendees included experts in relevant experimental techniques, integrative modeling, visualization, and data and model archiving.
The workshop began with plenary talks followed by focused discussions. Gerard Kleywegt introduced the workshop objectives. Andrej Sali outlined the current state of integrative modeling. Helen Berman gave an overview of the history and status of the wwPDB organization. Jill Trewhella described the increasing role of SAS in integrative structural modeling, the need for the development of community standards and validation tools for biomolecular modeling using SAS data, and how SAS data and modeling resources could interoperate with the PDB. Claus Seidel outlined state-of-the-art single molecule and ensemble Förster resonance energy transfer (FRET) spectroscopy (Kalinin et al., 2012), live cell imaging, as well as related label-based spectroscopic methods for measuring select interatomic distances in macromolecular systems. Torsten Schwede presented the Protein Model Portal (Haas et al., 2013), including its linking of large databases of comparative models with experimental structure information in the PDB, and the Model Archive repository for all categories of in silico structural models.
1.2 Current archives for models and/or supporting data
In this section, we review the PDB and management of data derived from crystallography, NMR spectroscopy, 3DEM, and SAS, plus archives for models derived exclusively based on theoretical information.
1.2.1 Protein Data Bank
For more than four decades, the PDB has served as the single global archive for atomic models of biological macromolecules; first for those derived from crystallography, and subsequently for models from NMR spectroscopy and 3DEM. The PDB also archives experimental data necessary to validate the structural models determined using these three methods. In addition, descriptions of the chemistry of polymers and ligands are collected, as are metadata describing sample preparation, experimental methods, model building, refinement statistics, literature references, etc. For all structural models in the PDB, geometric features are assessed with respect to standard valence geometry and intermolecular interactions, as recommended by the three wwPDB VTFs mentioned above.
1.2.2 Crystallography: Models and Data
For structures derived using X-ray, Neutron, and combined X-ray/Neutron crystallography, it has been mandatory to deposit structure factor amplitudes into the PDB since 2008 (http://www.wwpdb.org/news/news?year=2007#29-November-2007); until then, the submission of these primary data was optional. Additional validation against deposited structure factor amplitudes is carried out using procedures recommended by the X-ray VTF (Read et al., 2011). The resulting validation report includes graphical summaries of the quality of the overall model plus residue-specific features. Detailed assessments of various aspects of the model and its agreement with experimental and stereochemical data are also provided. In the near future, unmerged intensities will also be collected, enabling further validation activities.
1.2.3 Nuclear Magnetic Resonance Spectroscopy: Models and Data
The Biological Magnetic Resonance Data Bank (BioMagResBank or BMRB; http://www.bmrb.wisc.edu) is a repository for experimental and derived data gathered from NMR spectroscopic studies of biological molecules. The BMRB archive contains quantitative NMR spectral parameters, including assigned chemical shifts, coupling constants, and peak lists together with derived data, including relaxation parameters, residual dipolar couplings, hydrogen exchange rates, pKa values, etc. Other data contained in the BMRB include: NMR restraints processed from original author depositions available from the PDB; time-domain spectral data from NMR experiments used to assign spectral resonances and determine structures of biological macromolecules; chemical shift and structure validation reports; and a database of one- and two-dimensional 1H and 13C NMR spectra for over 1200 metabolites. The BMRB website also provides tools for querying and retrieving data.
Since 2006, BMRB has been a member of the wwPDB organization (Markley et al., 2008). Chemical shift and restraint data that accompany model data are housed in both the BMRB and PDB archives. Deposited NMR data without model coordinates reside exclusively in the BMRB archive. The wwPDB D&A system provides for deposition, annotation, and validation of NMR models and related experimental data. Depositors of chemical shift and other data sets without accompanying models are automatically redirected to BMRB to deposit their data. Data exchange between the BRMB and PDB archives is facilitated by software tools utilizing correspondences maintained between the PDB Exchange Dictionary (PDBx) and the BMRB NMRSTAR Dictionary. Validation methods for NMR-derived models, measured chemical shifts, and restraint data are currently under development, in response to recommendations of the NMR VTF (Montelione et al., 2013). A working group composed of the major biomolecular NMR software developers has created a common NMR exchange format (NEF) for structural restraints, similar to NMR-STAR. The adoption of this NEF by NMR software developers will simplify data exchange and the archiving of NMR structural restraints by the wwPDB.
1.2.4 Electron Microscopy: Models and Maps
Atomistic structural models determined using 3DEM methods were first archived in the PDB in the 1990s. In 2002, the EM Data Bank (EMDB) was created by the Macromolecular Structure Database (now PDBe) at the EBI. In 2006, the EMDataBank (http://www.EMDataBank.org) was established as the unified global portal for one-stop deposition and retrieval of 3DEM density maps, atomic models, and associated metadata (Lawson et al., 2011). EMDataBank is a joint effort among PDBe, the Research Collaboratory for Structural Bioinformatics (RCSB) at Rutgers, and the National Center for Macromolecular Imaging (NCMI) at Baylor College of Medicine. EMDataBank also serves as a resource for news, events, software tools, data standards, raw data, and validation methods for the 3DEM community. 3DEM model and map data are now stored in separate branches of the wwPDB ftp archive site.
As for NMR-based models, the wwPDB D&A system supports processing of atomistic models and map data from 3DEM structure determinations. 3DEM map data deposited without atomistic models are stored exclusively in EMDB. Again, as for NMR, a mapping is maintained between the PDBx data dictionary and the EMDB XML-based data model. Validation methods for 3DEM maps and atomistic models are currently under development in response to recommendations from the EM VTF (Henderson et al., 2012).
1.2.5 Small-Angle Scattering: Data and Model Archiving
The report from the first meeting of the wwPDB SAS Task Force (Trewhella et al., 2013a) made the case for establishing “a global repository that holds standard format X-ray and neutron SAS data that is searchable and freely accessible for download” and that “options should be provided for including in the repository SAS-derived shape and atomistic models based on rigid-body refinement against SAS data along with specific information regarding the uniqueness and uncertainty of the model, and the protocol used to obtain it.”
At present, there are two databases available for storing SAS data and models with associated metadata and analyses, both of which are freely accessible without limitations on data utilization via the Internet. As of March 2015, BIOISIS (http://www.bioisis.net/) contained 99 structures and is supported by teams at the Advanced Light Source and Diamond, while SASBDB (http://www.sasbdb.org/) (Valentini et al., 2015) contained 195 models and 114 experimental datasets and is supported by a team at EMBL-Hamburg.
Having evolved separately, these databases are distinctive in character. There was in principle agreement within the wwPDB SAS Task Force that BIOISIS and SASBDB will exchange datasets. Such exchange would be a step toward developing a federated approach to SAS data and model archiving, which in turn could ultimately be federated with the PDB, BMRB, and EMDB.
Further development of the sasCIF dictionary is required to permit full data exchange between the two SAS data repositories. sasCIF is a core Crystallographic Information File (CIF) developed to facilitate the SAS data exchange (Malfois and Svergun, 2000). As its name implies, sasCIF was implemented as an extension of the core CIF dictionary and has recently been extended to include new elements related to models, model fitting, validation tools, sample preparation, and experimental conditions (M. Kachala, J. Westbrook and D.I. Svergun, in preparation). sasCIFtools were developed as a documented set of publicly available programs for sasCIF data processing and format conversion; currently, SASBDB supports both import and export of sasCIF files.
1.2.6 Protein Model Portal
Comparative or homology modeling is routinely used to generate structural models of proteins for which experimentally determined structural models are not yet available (Marti-Renom et al., 2000; Schwede et al., 2009). Until 2006, such in silico models could be archived in the PDB, albeit in the absence of clear policies and procedures for their validation. Following recommendations from a stakeholder workshop convened in November 2005 (Berman et al., 2006), depositions to the PDB archive are limited to structural models substantially determined by experimental measurements from a defined physical sample (effective date October 15, 2006). The workshop also recommended that a central, publicly available archive or portal should be established for exclusively in silico models, and that methodology for estimating the accuracy of such computational models should be developed.
The Protein Model Portal (PMP) (Arnold et al., 2009; Haas et al., 2013) was developed at the Swiss Institute of Bioinformatics (SIB) at the University of Basel as a component of the SBKB (Berman et al., 2009; Gabanyi et al., 2011). Today, the SBKB integrates experimental information provided by the PDB with in silico models computed by automated modeling resources. In addition, the PMP provides access to several state-of-the-art model quality assessment services (Schwede et al., 2009). Since 2013, the Model Archive (http://modelarchive.org) resource has also served as a repository for individually generated in silico models of macromolecular structures, primarily those described in peer-reviewed publications. Finally, the Model Archive hosts all legacy models that were available from the PDB archive prior to 2006.
Each model in the PMP is assigned a stable, unique accession code (and digital object identifier or DOI) to ensure accurate cross-referencing in publications and other data repositories. Unlike experimentally determined structural models, in silico models are not the product of experimental measurements of a physical sample. They are generated computationally using various molecular modeling methods and underlying assumptions. Examples include comparative modeling, virtual docking of ligand molecules to protein targets, virtual docking of one protein to another, simulations of molecular dynamics and motions, and de novo (ab initio) protein modeling.
Effective archival storage of such models depends critically on capturing sufficient detail regarding underlying assumptions, parameters, methodology, and modeling constraints, to allow for assessment and faithful re-computation of the model. It is also essential that these models be accompanied by reliable estimates of uncertainty. In October 2013, a workshop on “Theoretical Model Archiving, Validation and PDBx/mmCIF Data Exchange Format” (http://www.proteinmodelportal.org/workshop-2013/) was hosted at Rutgers University to launch development of community standards for theoretical model archiving.
2 Integrative/Hybrid Structure Modeling
2.1 Motivation
Samples of many biological macromolecules prove recalcitrant to mainstream structural biology methods (i.e., crystallography, NMR, or 3DEM), because they are not crystallizable, are insoluble, are not of adequate purity, are conformationally heterogeneous, are too large or small, or do not remain intact during the course of the experiment. In such cases, integrative modeling is increasingly being used to compute structural models based on complementary experimental data and theoretical information (Figures 1 and 2; Table 1) (Alber et al., 2007; Alber et al., 2008; Robinson et al., 2007; Russel et al., 2012; Sali et al., 2003; Sali et al., 1990; Schneidman-Duhovny et al., 2014; Ward et al., 2013). Structural biology is no stranger to integrative models. Insights into the molecular details of the B-DNA double helix (Watson and Crick, 1953), the α-helix, and the β-sheet (Pauling et al., 1951) all depended on constructing structural models that encompassed data derived from multiple sources (albeit without the benefit of digital computation). Integrative structure modeling of today has its origins in attempts to fit X-ray derived substructures into an EM density map of a larger assembly (Rayment et al., 1993). Other early examples include the model of the Gla-EGF domains from coagulation Factor X based on NMR and SAS data (Sunnerhagen et al., 1996), and the superhelical assembly of the bacteriophage fd gene 5 protein with single stranded DNA based on neutron and X-ray SAS data, EM data, and the crystal structure of G5P (Olah et al., 1995); the latter study was inspired in part by molecular dynamics simulations guided by contacts from an NMR structure of the G5P dimer and EM data (Folmer et al., 1994).
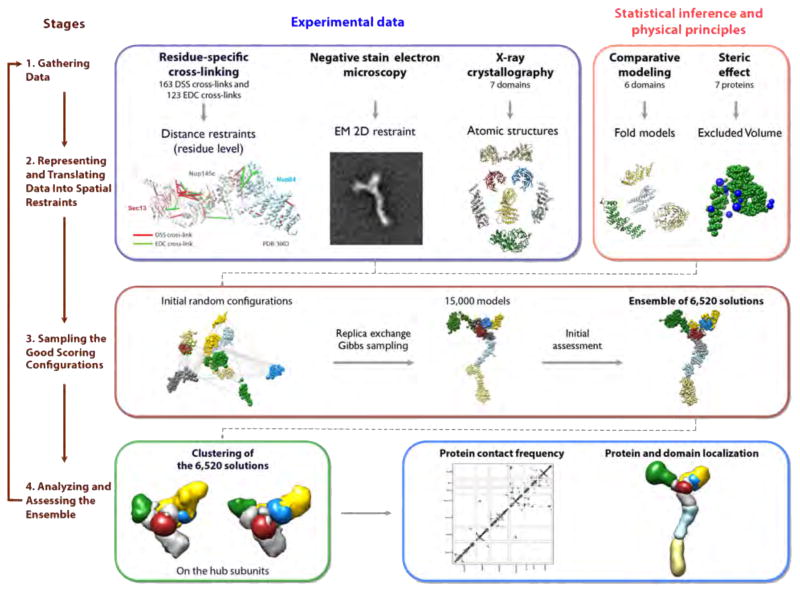
Figure 2. The four stages of integrative structure determination.
The approach is illustrated by its application to the heptameric Nup84 subcomplex of the Nuclear Pore Complex (Shi et al., 2014).
Beyond overcoming sample limitations, the integrative approach has several additional advantages (Alber et al., 2007). First, synergy among the input data minimizes the drawbacks of sparse, noisy, and ambiguous data obtained from compositionally and structurally heterogeneous samples. Each individual piece of data may contain relatively little structural information, but by simultaneously fitting a model to all data derived from independent experiments, the uncertainty of the structures that fit the data can be markedly reduced. Second, the integrative approach can be used to produce all structural models consistent with available data, instead of myopically focusing on just one model. Third, comparison of an ensemble of structural models permits estimation of precision and, sometimes, the accuracy of both the experimental data and the model. Fourth, the integrative approach can make structural biologists more efficient by identifying which additional measurements are likely to have the greatest impact on integrative model precision and accuracy. Finally, integrative modeling provides a framework for considering perturbations of the system that are often required to collect the data; for example, spin labels are required for EPR experiments, membrane proteins are often reconstituted in micelles for NMR spectroscopy, and point mutations or even entire domains are introduced to stabilize preferred conformations for crystallization. While such perturbations complicate structural analysis, integrative modeling may allow us to distinguish biologically relevant states from artifacts of any individual approach. In summary, integrative structure determination maximizes the accuracy, precision, completeness, and efficiency of the structural coverage of biomolecular systems.
2.2 Experimental and computational methods for generating structural information
Input information for integrative modeling can come from various experimental methods, physical theories, and statistical analyses of databases of known structures, biopolymer sequences, and interactions. These methods probe different structural aspects of the system (Table 1). In addition to information about average structures, a number of methods provide dynamical insights, which can also be incorporated into integrative modeling procedures (Russel et al., 2009). For example, both NMR spectroscopy and X-ray crystallography provide access to various measures of conformational dynamics; FRET, time-dependent Double Electron-Electron Resonance (DEER) spectroscopies, and even quantitative cross-linking/mass spectrometry (qCLMS) (Fischer et al., 2013) can map distance changes in time; SAXS can provide time-resolved information on the structures and processes with the temporal resolution of a millisecond; molecular dynamics simulations can map the dynamics of an atomic structure up to the millisecond time scale; and high-speed AFM imaging can provide the dynamic live images of single molecules (Ando, 2014).
2.3 Approach
All structural characterization approaches correspond to finding models that best fit input information, as judged by use of a scoring function quantifying the difference between the observed data and the data computed from the model. Thus, any information about a structure determination target must always be converted to an explicit structural model through computation. Integrative approaches explicitly combine diverse experimental and theoretical information, with the goal of increasing accuracy, precision, coverage, and efficiency of structure determination. Input information can vary greatly in terms of resolution (i.e., precision, noise, uncertainty), accuracy, and quantity. All structure determination methods are integrative, albeit with differences in degree. At one end of the spectrum, even structure determination using predominantly crystallographic, NMR, or high-resolution single particle EM data also generally requires a molecular mechanics force field description of atomic structure. At the other end of the spectrum, integrative methods rely more evenly on different types of information, often resulting in coarser models with higher uncertainty (Figure 1). Examples of such integrative methods include docking of comparative models of subunits into a 3DEM density map of the macromolecular assembly (Lasker et al., 2009); rigid-body fitting of multi-domain structures and complexes determined by crystallography or NMR to SAS data (Petoukhov and Svergun, 2005); and use of conformational sampling methods with low-density NMR data (Lange et al., 2012; Mueller et al., 2000), chemical cross links (Young et al., 2000), or even chemical shift data alone (Shen et al., 2008). It is not difficult to appreciate how integrative methods blur distinctions between models based primarily on theoretical considerations and those based primarily on experimental measurements from a physical sample.
The practice of integrative structure determination is iterative, consisting of four stages (Figure 2): gathering of data; choosing the representation and encoding of all data within a numerical scoring function consisting of spatial restraints; configurational sampling to identify structural models with good scores; and analyzing the models, including quantifying agreement with input spatial restraints and estimating model uncertainty. Input information about the system can be used (i) to select the set of variables that best represent the system (system representation), (ii) to rank the different configurations (scoring function), (iii) to search for good scoring solutions (sampling); and (iv) to further filter good-scoring solutions produced by sampling.
2.4 Types of integrative models
A structural model of a macromolecular assembly is defined by the relative positions and orientations of its components (e.g., atoms, pseudo-atoms, residues, secondary structure elements, domains, subunits, and subcomplexes). While traditional structural biology methods usually produce a single atomistic model, integrative models tend to be more complex in at least four respects. First, a model can be multi-scale (Grime and Voth, 2014), representing different levels of structural detail by a collection of geometrical primitives (e.g., points, spheres, tubes, 3D Gaussians, or probability densities). Thus, the same part of a system can be described with multiple representations and different parts of a system can be represented differently. An optimal representation facilitates accurate formulation of spatial restraints together with efficient and complete sampling of good-scoring solutions, while retaining sufficient detail (without over fitting) such that the resulting models are maximally useful for subsequent biological analysis (Schneidman-Duhovny et al., 2014). Second, a model can be multi-state, specifying multiple discrete states of the system required to explain the input information (each state may differ in structure and/or composition) (Molnar et al., 2014; Pelikan et al., 2009). Third, a model can also specify the order of states in time and/or transitions between the states. This feature allows representation of a multi-step biological process, a functional cycle (Diez et al., 2004), a kinetic network (Pirchi et al., 2011), time evolution of a system (e.g., a molecular dynamics trajectory) (Bock et al., 2013), or FRET trajectories; for a comprehensive description of biomolecular function, it is essential to register state lifetimes, characteristic relaxation times, and direct rate constants. Finally, an ensemble of models may be provided to underscore the uncertainty in the input information, with each individual model satisfying the input information within an acceptable threshold (e.g., NMR-derived ensembles currently available in the PDB (Clore and Gronenborn, 1991; Snyder et al., 2005; Snyder et al., 2014) and the ensembles generated from SAXS (Tria et al., 2015)). This aspect of the representation allows us to describe model uncertainty and to assess the completeness of input information; such ensembles are distinct from multiple states that represent actual variations in the structure, as implied by experimental information that cannot be accounted for by a single representative structure (Schneidman-Duhovny et al., 2014; Schroder, 2015).
3 Task Force Deliberations and Recommendations
3.1 Charge to the Task Force
A healthy debate is underway about how to classify structural models. A major motivation for this discussion is the lack of accurate general methods to assess the precision and accuracy of any model. As a result, models are often classified based on the predominant type of information used to compute them, which in turn tends to reflect the data-to-parameter ratio and thus model accuracy. However, as previously discussed, all structures are in fact integrative models that have been derived both from experimental measurements involving a physical sample of a biological macromolecule and prior knowledge of the underlying stereochemistry. It is, therefore, difficult if not impossible to draw definitive lines on the spectrum ranging from very well-determined ultra-high resolution crystallographic structures (>40 experimental observations per non-hydrogen atom in the crystallographic asymmetric unit) and structural models based on a single or even no experimental observation.
Reflecting this debate about model classification, there are in principle several possibilities for archiving the models and associated data among distinct, publicly accessible model/data repositories, including (i) a single mega archive that serves as the repository for every type of structural model and data; (ii) independent, free-standing repositories that house distinct types of models and data; and (iii) a federated system of inter-operating repositories that archive models and data, with “spheres of influence” based on community consensus.
To address some of the challenges ahead and make recommendations about how best to proceed, the community stakeholders who assembled at the October 2014 meeting of the wwPDB Hybrid/Integrative Methods Task Force were divided into three discussion groups, each tasked with considering a series of related questions. What experimental data (beyond crystallography, NMR, and 3DEM) should be archived? Where and how should it be validated? What kinds of non-atomistic models can we expect and how should they be validated? What are the criteria for deciding where models should be archived? How should non-atomistic and mixed atomistic/non-atomistic models be archived? Should there be a separate archive for integrative (mixed) models (and data)? Should we establish a federated system of data and model archives to support integrative structural biology? The three breakout groups were asked to address these questions, report back with their findings, and make recommendations for the future. Each group independently approached the same set of questions. At the close of the meeting, the teams converged to compare notes, identify areas of commonality and diversity, and determine how best to move forward. The resulting consensus is reflected in this document.
3.2 Recommendations
Recommendation 1: In addition to archiving the models themselves, all relevant experimental data and metadata as well as experimental and computational protocols should be archived; inclusivity is key
Ideally, structural models of any kind, derived by any method, should be archived.
Models are of greatest value when they are independently tested, potentially improved, and serve to further our understanding of how the function of a biological system is determined by its 3D structure(s). Therefore, models and necessary annotations must be freely available to the research community. The modeling process should be reproducible. Information concerning all aspects of a model should be deposited, including input data, corresponding spatial restraints, output models, and protocols used to convert input data into models. In addition to the input experimental data, the archival deposition should specify or include theoretically derived restraints used to compute the model (e.g., a statistical potential and a molecular mechanics force field). In practice, frequently used data types (e.g., distance information) should be prioritized for early complete implementation. Uncertainty in the input data needs to be well documented; some data uncertainty estimates may require modeling (e.g., Bayesian error estimates (Rieping et al., 2005)). Consistency between input data and the structural model should be documented as part of model validation.
Each expert community should drive decisions as to how much raw data, processed data, and metadata to deposit, subject to the minimal requirement that the spatial restraints used for modeling must be derivable from the deposited information. Attention needs to be paid to annotating measurement conditions, such as temperature (Fenwick et al., 2014), sample concentration, environmental conditions (e.g., buffer), construct definition, and identification of all assembly components, all of which can significantly influence the experimental outcome. Cost benefit analyses should be used to help guide which data should be archived. As much data as practical should be deposited, to facilitate model validation, future improvements of the model, and methods development (e.g., benchmarking sets). Of particular importance will be availability of some raw data to help drive improvement of data processing methods and for use by methods developers, who are often not generating the experimental data themselves.
Recommendation 2: A flexible model representation needs to be developed, allowing for multi-scale models, multi-state models, ensembles of models, and models related by time or other order
Model representation should allow for as many types of “structural” models as possible, thereby encouraging collaboration among developers of integrative modeling software (Russel et al., 2012). At a minimum, the model representation should allow encoding of an ensemble of multi-scale multi-state time-ordered models (Section 2.4). Uncertainty of the model coordinates should be tightly associated with the model coordinates in the model representation. Any model resident within an archive should be “self-contained” to facilitate utilization (e.g., for visualization). A common representation and format for models are useful for reasons of software interoperability. Particle-based representations/primitives need to be prioritized; non-particle-based model representations (e.g., continuum representations) merit further consideration by appropriate community stakeholders.
Recommendation 3: Procedures for estimating the uncertainty of integrative models should be developed, validated, and adopted
Assessment of both an integrative model and the information on which it is based is of critical importance for guiding subsequent use of the model. For atomistic models, extant standard validation criteria from X-ray crystallography should be used. Beyond this test, validation of integrative models and data is a major research challenge that must be addressed and overcome. The following represent promising considerations (Alber et al., 2007; Schneidman-Duhovny et al., 2014): convergence of conformational sampling, fit of the model to the input information, test for clashes between geometrical primitives comprising the model, precision of the ensemble of solutions (visualized with, for example, ribbon plots), cross-validation and statistical bootstrapping based on available data, tests based on data determined after the model was computed, and sensitivity analysis of the model to input data. Bayesian approaches may be particularly well suited to describe model uncertainty by computing posterior model densities from a forward model, noise model, and priors (Muschielok et al., 2008; Rieping et al., 2005). Tools for visualizing model validation should be developed.
Communities generating data used in integrative modeling should agree on the standard set of descriptors for data quality, as has been done for crystallography, NMR, and 3DEM.
Recommendation 4: A federated system of model and data archives should be created
Integrative models can be based on a broad array of different experimental and computational techniques. While the specific spatial restraints implied by the data and used to construct an integrative model should be deposited with the model itself, the underlying experimental data often contain much richer information. This information should be captured in a federated system of domain-specific model and data archives. These individual member archives should be developed by community experts, based on method-specific standards for data archiving and validation. A federated system of model and data archives implies the need for a seamless exchange of information between independent archives. This seamless exchange requires a common dictionary of terms, agreed data formats, persistent and stable data object identifiers, and close synchronization of policies and procedures. Federated model and data archives need to develop efficient methods for data exchange to allow for transparent data access across the enterprise.
A single interface for the deposition of all data and models into the federated system is highly desirable. Such an interface would greatly facilitate the task of the depositor, and, thereby, maximize compliance with deposition standards and requirements. In addition, reliance on a single entry point will help to ensure consistency across the federation at the time of deposition. Following successful deposition, individual data sets can be transferred to member databases for data curation and archiving if domain-specific databases exist. There should also be provision for collecting unstructured information in a “data commons”, as proposed by the data science initiative at the NIH (Margolis et al., 2014).
Access to the contents of the federated database through a single portal is also most desirable, to facilitate dissemination of data, models, and experimental/computational protocols.
Of particular importance for integrative modeling will be the option to modify or update any aspect of the modeling procedure, for example, by adding new data. The federated archive should allow versioning for each deposited model. Such capabilities will facilitate the cycle of experiment and modeling, and accelerate production of more accurate, precise, and complete models (Russel et al., 2012).
Recommendation 5: Publication standards for integrative models should be established
Over the past decade, the wwPDB organization has worked with relevant scientific journals to help establish publication standards for structural models coming from crystallography, NMR spectroscopy, and 3DEM. Community standards now include requiring authors to make their validation reports available to reviewers and editors. Through the International Union of Crystallography (IUCr) Small Angle Scattering and Journals Commissions, the SAS community developed and agreed upon publication guidelines for structural modeling of biomolecules therefrom (Jacques et al., 2012). A set of standards for publishing integrative models should be developed along similar lines.
3.3 Implementation
Implementation of Recommendation 1 poses a host of cultural and technical challenges. Experimentalists and modelers need to provide the data, models, and protocols, thus at least partly addressing increasing concerns regarding reproducibility of scientific results. From a technical perspective, interoperating data dictionaries for all methods need to be created. In addition, potential storage bottlenecks need to be addressed.
Implementation of Recommendations 2 and 3 will require significant research as to how best to represent and validate the many different kinds of integrative models. In addition, the community will need to agree on a common set of standards that are sufficiently mutable to allow for future innovation. Efforts such as the “Cryo-EM Modeling Challenge” may facilitate this process (http://www.emdatabank.org/modeling_chllnge).
Implementation of Recommendation 4 will require agreement on a common data exchange system among member repositories. Based on past accomplishments, the wwPDB is well positioned to play a leadership role in establishing the proposed federated system, including provision of common deposition and access interfaces. The wwPDB should begin this process by providing training and advice on data archiving and curation to contributing domin-specific member repositories.
Implementation of Recommendation 5 will require continued work with the journals that publish structural models of biological macromolecules.
Significant resources will be required to implement these recommendations, including grants for research, infrastructure, and workshops. These efforts are international by their very nature and will require funding from multiple public and private sources, including in North America, Europe, and Asia.
Acknowledgments
The workshop was supported by funding to PDBe by Wellcome Trust 088944; RCSB PDB by NSF DBI 1338415; PDBj by JST-NBDC; BMRB by NLM P41 LM05799; EMDataBank by NIH GM079429, and tax-deductible donations made to the wwPDB Foundation in support of wwPDB outreach activities.
Footnotes
All attendees of the Workshop are listed as authors.
References
- Alber F, Dokudovskaya S, Veenhoff L, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait B, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Alber F, Forster F, Korkin D, Topf M, Sali A. Integrating diverse data for structure determination of macromolecular assemblies. Annu Rev Biochem. 2008;77:443–477. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]
- Ando T. High-speed AFM imaging. Curr Opin Struct Biol. 2014;28:63–68. doi: 10.1016/j.sbi.2014.07.011. [DOI] [PubMed] [Google Scholar]
- Arnold K, Kiefer F, Kopp J, Battey JN, Podvinec M, Westbrook JD, Berman HM, Bordoli L, Schwede T. The Protein Model Portal. J Struct Funct Genomics. 2009;10:1–8. doi: 10.1007/s10969-008-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bau D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA. The three-dimensional folding of the alpha-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol. 2011;18:107–114. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Burley SK, Chiu W, Sali A, Adzhubei A, Bourne PE, Bryant SH, Dunbrack RL, Jr, Fidelis K, Frank J, et al. Outcome of a workshop on archiving structural models of biological macromolecules. Structure. 2006;14:1211–1217. doi: 10.1016/j.str.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook JD, Gabanyi MJ, Tao W, Shah R, Kouranov A, Schwede T, Arnold K, Kiefer F, Bordoli L, et al. The protein structure initiative structural genomics knowledgebase. Nucleic Acids Res. 2009;37:D365–368. doi: 10.1093/nar/gkn790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock LV, Blau C, Schroder GF, Davydov II, Fischer N, Stark H, Rodnina MV, Vaiana AC, Grubmuller H. Energy barriers and driving forces in tRNA translocation through the ribosome. Nat Struct Mol Biol. 2013;20:1390–1396. doi: 10.1038/nsmb.2690. [DOI] [PubMed] [Google Scholar]
- Boura E, Rozycki B, Herrick DZ, Chung HS, Vecer J, Eaton WA, Cafiso DS, Hummer G, Hurley JH. Solution structure of the ESCRT-I complex by small-angle X-ray scattering, EPR, and FRET spectroscopy. Proc Natl Acad Sci USA. 2011;108:9437–9442. doi: 10.1073/pnas.1101763108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM. Structures of larger proteins in solution: three- and four-dimensional heteronuclear NMR spectroscopy. Science. 1991;252:1390–1399. doi: 10.1126/science.2047852. [DOI] [PubMed] [Google Scholar]
- Degiacomi MT, Dal Peraro M. Macromolecular symmetric assembly prediction using swarm intelligence dynamic modeling. Structure. 2013;21:1097–1106. doi: 10.1016/j.str.2013.05.014. [DOI] [PubMed] [Google Scholar]
- Degiacomi MT, Iacovache I, Pernot L, Chami M, Kudryashev M, Stahlberg H, van der Goot FG, Dal Peraro M. Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism. Nat Chem Biol. 2013;9:623–629. doi: 10.1038/nchembio.1312. [DOI] [PubMed] [Google Scholar]
- Deshmukh L, Schwieters CD, Grishaev A, Ghirlando R, Baber JL, Clore GM. Structure and dynamics of full-length HIV-1 capsid protein in solution. J Am Chem Soc. 2013;135:16133–16147. doi: 10.1021/ja406246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez M, Zimmermann B, Börsch M, König M, Schweinberger E, Steigmiller S, Reuter R, Felekyan S, Kudryavtsev V, Seidel CAM, Gräber P. Proton-powered subunit rotation in single membrane-bound F0 F1 -ATP synthase. Nature Structural & Molecular Biology. 2004;11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- Fenwick RB, van den Bedem H, Fraser JS, Wright PE. Integrated description of protein dynamics from room-temperature X-ray crystallography and NMR. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E445–E454. doi: 10.1073/pnas.1323440111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer L, Chen ZA, Rappsilber J. Quantitative cross-linking/mass spectrometry using isotope-labelled cross-linkers. J Proteomics. 2013;88:120–128. doi: 10.1016/j.jprot.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer RH, Nilges M, Folkers PJ, Konings RN, Hilbers CW. A model of the complex between single-stranded DNA and the single-stranded DNA binding protein encoded by gene V of filamentous bacteriophage M13. J Mol Biol. 1994;240:341–357. doi: 10.1006/jmbi.1994.1449. [DOI] [PubMed] [Google Scholar]
- Gabanyi MJ, Adams PD, Arnold K, Bordoli L, Carter LG, Flippen-Andersen J, Gifford L, Haas J, Kouranov A, McLaughlin WA, et al. The Structural Biology Knowledgebase: a portal to protein structures, sequences, functions, and methods. J Struct Funct Genomics. 2011;12:45–54. doi: 10.1007/s10969-011-9106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Schwieters CD, Tang C. Conjoined Use of EM and NMR in RNA Structure Refinement. Plos One. 2015;10:e0120445. doi: 10.1371/journal.pone.0120445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime JMA, Voth GA. Highly Scalable and Memory Efficient Ultra-coarse-grained Molecular Dynamics Simulations. J Chem Theor Comp. 2014;10:423–431. doi: 10.1021/ct400727q. [DOI] [PubMed] [Google Scholar]
- Haas J, Roth S, Arnold K, Kiefer F, Schmidt T, Bordoli L, Schwede T. The Protein Model Portal--a comprehensive resource for protein structure and model information. Database: the journal of biological databases and curation. 2013;2013:bat031. doi: 10.1093/database/bat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Luo J, Ranish J, Hahn S. Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J. 2014;33:2534–2546. doi: 10.15252/embj.201488638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques DA, Guss JM, Svergun DI, Trewhella J. Publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution. Acta Crystallogr D: Biol Crystallogr. 2012;68:620–626. doi: 10.1107/S0907444912012073. [DOI] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinin S, Peulen T, Sindbert S, Rothwell PJ, Berger S, Restle T, Goody RS, Gohlke H, Seidel CAM. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nature Methods. 2012;9:1218–1225. doi: 10.1038/nmeth.2222. [DOI] [PubMed] [Google Scholar]
- Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, Baker D. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci U S A. 2012;109:10873–10878. doi: 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K, Topf M, Sali A, Wolfson HJ. Inferential optimization for simultaneous fitting of multiple components into a CryoEM map of their assembly. J Mol Biol. 2009;388:180–194. doi: 10.1016/j.jmb.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Baker ML, Best C, Bi C, Dougherty M, Feng P, van Ginkel G, Devkota B, Lagerstedt I, Ludtke SJ, et al. EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res. 2011;39:D456–464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Griesinger C, Kolbe M, Baker D, Becker S, Lange A. Atomic model of the type III secretion system needle. Nature. 2012;486:276–279. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukoyanova N, Kondos SC, Farabella I, Law RH, Reboul CF, Caradoc-Davies TT, Spicer BA, Kleifeld O, Traore DA, Ekkel SM, et al. Conformational changes during pore formation by the perforin-related protein pleurotolysin. PLoS Biol. 2015;13:e1002049. doi: 10.1371/journal.pbio.1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfois M, Svergun D. sasCIF: an extension of core Crystallographic Information File for SAS. J App Cryst. 2000;33:812–816. [Google Scholar]
- Margolis R, Derr L, Dunn M, Huerta M, Larkin J, Sheehan J, Guyer M, Green ED. The National Institutes of Health’s Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. J Am Med Inform Assoc. 2014;21:957–958. doi: 10.1136/amiajnl-2014-002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley JL, Ulrich EL, Berman HM, Henrick K, Nakamura H, Akutsu H. BioMagResBank (BMRB) as a partner in the Worldwide Protein Data Bank (wwPDB): new policies affecting biomolecular NMR depositions. J Biomol NMR. 2008;40:153–155. doi: 10.1007/s10858-008-9221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, Niu W, Ebright YW, Levy R, Ebright RH. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y, Irobalieva RN, Tolbert BS, Smalls-Mantey A, Iyalla K, Loeliger K, D’Souza V, Khant H, Schmid MF, Garcia EL, et al. Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J Mol Biol. 2010;404:751–772. doi: 10.1016/j.jmb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar KS, Bonomi M, Pellarin R, Clinthorne GD, Gonzalez G, Goldberg SD, Goulian M, Sali A, DeGrado WF. Cys-scanning disulfide crosslinking and bayesian modeling probe the transmembrane signaling mechanism of the histidine kinase, PhoQ. Structure. 2014;22:1239–1251. doi: 10.1016/j.str.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelione GT, Nilges M, Bax A, Güntert P, Herrmann T, Markley JL, Richardson J, Schwieters C, Vuister GW, Vranken W, Wishart D. Recommendations of the wwPDB NMR Structure Validation Task Force. Structure. 2013;21:1563–1570. doi: 10.1016/j.str.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller GA, Choy WY, Yang D, Forman-Kay JD, Venters RA, Kay LE. Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein. J Mol Biol. 2000;300:197–212. doi: 10.1006/jmbi.2000.3842. [DOI] [PubMed] [Google Scholar]
- Muschielok A, Andrecka J, Jawhari A, Bruckner F, Cramer P, Michaelis J. A nano-positioning system for macromolecular structural analysis. Nature Methods. 2008;5:965–971. doi: 10.1038/nmeth.1259. [DOI] [PubMed] [Google Scholar]
- Olah GA, Gray DM, Gray CW, Kergil DL, Sosnick TR, Mark BL, Vaughan MR, Trewhella J. Structures of fd gene 5 protein.nucleic acid complexes: a combined solution scattering and electron microscopy study. J Mol Biol. 1995;249:576–594. doi: 10.1006/jmbi.1995.0320. [DOI] [PubMed] [Google Scholar]
- Pauling L, Corey RB, Branson HR. The structures of proteins. Proc Natl Acad Sci USA. 1951;37:205. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirchi M, Ziv G, Riven I, Cohen SS, Zohar N, Barak Y, Haran G. Single-molecule fluorescence spectroscopy maps the folding landscape of a large protein. Nature Communications. 2011;2 doi: 10.1038/ncomms1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis A, Stengel F, Hall Z, Hernandez H, Leitner A, Walzthoeni T, Robinson CV, Aebersold R. A mass spectrometry-based hybrid method for structural modeling of protein complexes. Nat Methods. 2014;11:403–406. doi: 10.1038/nmeth.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prischi F, Konarev PV, Iannuzzi C, Pastore C, Adinolfi S, Martin SR, Svergun DI, Pastore A. Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly. Nat Commun. 2010;1:95. doi: 10.1038/ncomms1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protein Data Bank. Protein Data Bank. Nature New Biol. 1971;233:223. [Google Scholar]
- Rayment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC, Milligan RA. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- Read RJ, Adams PD, Arendall WB, III, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lutteke T, Otwinowski Z, et al. A new generation of crystallographic validation tools for the Protein Data Bank. Structure. 2011;19:1395–1412. doi: 10.1016/j.str.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieping W, Habeck M, Nilges M. Inferential structure determination. Science. 2005;309:303–306. doi: 10.1126/science.1110428. [DOI] [PubMed] [Google Scholar]
- Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- Russel D, Lasker K, Phillips J, Schneidman-Duhovny D, Velazquez-Muriel J, Sali A. The structural dynamics of macromolecular processes. Curr Opin Cell Biol. 2009;21:97–108. doi: 10.1016/j.ceb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel D, Lasker K, Webb B, Velazquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, Sali A. Putting the pieces together: integrative structure determination of macromolecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Glaeser R, Earnest T, Baumeister W. From words to literature in structural proteomics. Nature. 2003;422:216–225. doi: 10.1038/nature01513. [DOI] [PubMed] [Google Scholar]
- Sali A, Overington JP, Johnson MS, Blundell TL. From comparisons of protein sequences and structures to protein modelling and design. Trends Biochem Sci. 1990;15:235–240. doi: 10.1016/0968-0004(90)90036-b. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Pellarin R, Sali A. Uncertainty in integrative structural modeling. Curr Opin Struct Biol. 2014;28:96–104. doi: 10.1016/j.sbi.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder GF. Hybrid methods for macromolecular structure determination: experiment with expectations. Curr Opin Struct Biol. 2015;31:20–27. doi: 10.1016/j.sbi.2015.02.016. [DOI] [PubMed] [Google Scholar]
- Schwede T, Sali A, Honig B, Levitt M, Berman HM, Jones D, Brenner SE, Burley SK, Das R, Dokholyan NV, et al. Outcome of a workshop on applications of protein models in biomedical research. Structure. 2009;17:151–159. doi: 10.1016/j.str.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Fernandez-Martinez J, Tjioe E, Pellarin R, Kim SJ, Williams R, Schneidman D, Sali A, Rout MP, Chait BT. Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Mol Cell Proteomics. 2014;13:2927–2943. doi: 10.1074/mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder J, Burnley RJ, Wiegard A, Melquiond ASJ, Bonvin AMJJ, Axmann IM, Heck AJR. Insight into cyanobacterial circadian timing from structural details of the KaiB-KaiC interaction. Proc Natl Acad Sci USA. 2014;111:1379–1383. doi: 10.1073/pnas.1314326111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder DA, Bhattacharya A, Huang YJ, Montelione GT. Assessing precision and accuracy of protein structures derived from NMR data. Proteins. 2005;59:655–661. doi: 10.1002/prot.20499. [DOI] [PubMed] [Google Scholar]
- Snyder DA, Grullon J, Huang YJ, Tejero R, Montelione GT. The expanded FindCore method for identification of a core atom set for assessment of protein structure prediction. Proteins. 2014;82(Suppl 2):219–230. doi: 10.1002/prot.24490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnerhagen M, Olah GA, Stenflo J, Forsen S, Drakenberg T, Trewhella J. The relative orientation of Gla and EGF domains in coagulation factor X is altered by Ca2+ binding to the first EGF domain. A combined NMR-small angle X-ray scattering study. Biochemistry. 1996;35:11547–11559. doi: 10.1021/bi960633j. [DOI] [PubMed] [Google Scholar]
- Trewhella J, Hendrickson WA, Kleywegt GJ, Sali A, Sato M, Schwede T, Svergun DI, Tainer JA, Westbrook J, Berman HM. Report of the wwPDB Small-Angle Scattering Task Force: data requirements for biomolecular modeling and the PDB. Structure. 2013a;21:875–881. doi: 10.1016/j.str.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Trewhella J, Hendrickson WA, Sato M, Schwede T, Svergun D, Tainer JA, Westbrook J, Kleywegt GJ, Berman HM. Meeting Report of the wwPDB Small-Angle Scattering Task Force: Data Requirements for Biomolecular Modeling and the PDB Structure. 2013b;21:875–881. doi: 10.1016/j.str.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Tria G, Mertens HDT, Kachala M, Svergun DI. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2015;2:207–217. doi: 10.1107/S205225251500202X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini E, Kikhney AG, Previtali G, Jeffries CM, Svergun DI. SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res. 2015;43:D357–363. doi: 10.1093/nar/gku1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A, Sali A, Wilson I. Integrative structural biology. Science. 2013;339:913–915. doi: 10.1126/science.1228565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Whitten AE, Jeffries CM, Harris SP, Trewhella J. Cardiac myosin-binding protein C decorates F-actin: implications for cardiac function. Proc Natl Acad Sci USA. 2008;105:18360–18365. doi: 10.1073/pnas.0808903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, Dollinger G. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc Natl Acad Sci U S A. 2000;97:5802–5806. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural Basis of Transcription Initiation. Science. 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]