Abstract
The development of three-dimensional (3D) cultures is increasing, as they are able to provide the utility of in vitro models and the strength of testing in physiologically relevant systems. When cultured in a scaffold-free agarose hydrogel system, MCF-7 human breast carcinoma cells organize and develop into microtissues that contain a luminal space, in stark contrast to the flat morphology of MCF-7 two-dimensional (2D) monolayer cultures. Following exposure to 1nM E2, expression of typical estrogen-responsive genes, including progesterone receptor (PGR), PDZ containing domain 1 (PDZK1) and amphiregulin (AREG) is increased in both 2D and 3D cultures. When examining expression of other genes, particularly those involved in cell adhesion, there were large changes in 3D MCF-7 microtissues, with little to no change observed in the MCF-7 monolayer cultures. Together, these results indicate that while the initial estrogen-regulated transcriptional targets respond similarly in 2D and 3D cultures, there are large differences in activation of other pathways related to cell-cell interactions.
Introduction
As a result of the National Research Council report “Toxicity Testing in the 21st Century: A Vision and a Strategy,” [1] there is a large need to develop more efficient and physiologically relevant models for evaluation of compound safety and toxicity. Due to the cost- and time-intensive nature of animal testing, in vitro based screening models are growing in favor. Additionally, in vitro cell culture models allow for the use of human cell lines, enabling researchers to better evaluate human health and disease states when compared to in vivo animal models.
Traditionally, human primary cells and cell lines have been cultured in 2-dimensional (2D) monolayer on plastic substrates. While 2D cultures are technically easy to conduct and inexpensive, they do not recapitulate the biology of cells in vivo, where tissues are highly cell dense and cells are in contact with other cell types and extracellular matrix (ECM). Previous work has demonstrated that this contact with ECM and cells can alter gene expression profiles and cellular morphology [2–5], potentially leading to differences in experimental results between 2D in vitro and intact in vivo systems.
As an alternative strategy to address several of the shortcomings of both in vivo and 2D in vitro experimental models, 3-dimensional (3D) culture models have been developed. A variety of models exist, with two of the most popular models being scaffold-based and scaffold-free systems. In scaffold-based 3D culture, cells of interest are seeded at low density within natural or synthetic matrices. When cultured in Matrigel, a laminin-rich matrix, both normal and malignant human breast cells form 3D cultures with morphologies dependent on their gene expression profiles [3]. Nonmalignant MCF-10A cells cultured in Matrigel have been demonstrated to form lumen-containing mammary acini through apoptosis, a structure reminiscent of the in vivo morphology of the adult mammary gland [6]. When transformed into malignant cells, MCF-10A cells become invasive, distorting the structure and morphology of the in vitro mammary acini in a process that promotes oncogenesis [6–8]. This and similar systems have provided important insight into the process of both morphogenesis and oncogenesis; however, these models are not without limitations. Typically, cells are seeded embedded within matrix at low densities that do not mimic the highly cellular nature of epithelial tissues in vivo. Scaffold-based models are highly dependent on the nature of the scaffold material, as previous work has shown that different matrices including collagen I and laminin can have distinct effects on cells, including altering their invasive potential and gene expression profiles [9–12]. Many scaffold materials do not accurately reconstruct the in vivo ECM content, and often rely on only one component such as collagen, while ignoring the heterogeneous nature of native ECM.
Scaffold-free culture models including hanging drop and non-adhesive agarose hydrogels allow for cells to assemble into 3D microtissues free from the influence of outside matrix. While the hanging drop method requires no specialized equipment to form 3D cultures, it is technically difficult to change media and the low media to cell ratio increases the rate of waste accumulation, making long-term cultures difficult to assess. Additionally, hanging drop cultures are susceptible to being physically disturbed, and this can lead to a loss of samples through handling. The use of nonadhesive agarose hydrogels to culture cells in a scaffold-free environment circumvents these problems, as the system allows for easy media changes and handling of cultures. In this system, cells are trypsinized and seeded into non-adhesive hydrogels, where they settle into recesses and come into contact with other cells [13]. Within 24 hours after seeding, cells self-assemble into microtissues free of the influence of outside matrix and substrates [14]. Previous work using MCF-7 cells has demonstrated that when cultured in this model, MCF-7 cells form 3D microtissues that are more differentiated than 2D cultures, have apical-basal polarity and contain a luminal space, reminiscent of the in vivo morphology of the adult mammary gland [15].
In the present study, we describe differences in gene expression between 2D monolayer and 3D MCF-7 microtissue cultures following exposure to estradiol (E2). These results indicate that E2 treatment alters expression of classical estrogen responsive genes in a similar matter in both 2D and 3D cultures. However, with longer exposure to E2, the similarities of the response in both culture systems subside and the responses diverge.
Materials and Methods
Chemicals and Reagents
Cell culture media and supplements were purchased from Life Technologies, Inc (Grand Island, NY). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Flowery Branch, GA) and dextran-coated-charcoal (DCC) stripped was purchased from Gemini Bioscience (Sacramento, CA). Estradiol (E2), tamoxifen (TAM), dimethylsulfoxide (DMSO), Trizol and insulin were purchased from Sigma Aldrich (St. Louis, MO). Agarose was purchased from Fisher Scientific (Agawam, MA).
Cell Culture
MCF-7 cells (HTB-22) [16] were purchased from ATCC (Manassas, VA) and maintained in DMEM-F12 complete media supplemented with 10% fetal bovine serum (FBS), MEM nonessential amino acids, gentamicin and 10μg/mL insulin in a 5%CO2 incubator at 37°C as previously described [15]. Media was changed every 2–3 days and cells were passaged when 65–80% confluent. MCF-7 cells were limited to use within the first 10 passages from the original purchased vial from ATCC, to control for genomic drift due to instability.
3D Scaffold-Free Cell Culture
MCF-7 cells were seeded into agarose hydrogels prepared as previously described [15, 17] at a density of 600,000 cells/mL. Cells were allowed to settle into wells for 30 minutes and then placed in 2mL complete growth media, with media changes every 2–3 days until estrogen treatment.
Estrogen Treatment of MCF-7 Cultures
For monolayer cultures, MCF-7 cells were seeded at a density of 300,000 cells/mL in 6-well plates, and allowed to grow for 3 days in complete media. Cells were then washed twice with PBS and placed in phenol red-free DMEMF12 media supplemented with 5% dextran-charcoal stripped fetal bovine serum, MEM nonessential amino acids, gentamicin and 6ng/mL bovine insulin for 48 hours. For 3D cultures, cells were seeded into agarose hydrogels as above and grown for 7 days in 10% complete media, with media changed every 2–3 days. After 7 days of growth in complete media, 3D MCF-7 cultures were washed twice with PBS and transferred to fresh treatment media for 48 hours. Following 48 hours, 2D and 3D cultures were exposed to estradiol or vehicle control (dimethylsulfoxide) for 8 hours and collected in Trizol for gene expression analysis. All experiments were performed in triplicate.
RNA Isolation, RT-PCR and PCR Array Analysis
Monolayer cultures were collected by scraping into Trizol. Microtissues were collected from hydrogels by centrifugation, pelleted and lysed in Trizol. Samples were then isolated using the RNEasy Mini Kit (Qiagen) per manufacturer’s instructions. RNA quantity was determined using a Nanodrop ND1000, and quality was determined using an Agilent 2100 Bioanalyzer. A RIN value of greater than 0.7 was used as a cutoff for further analysis using PCR arrays.
A previously described [15] custom SABiosciences PCR Array (Qiagen) was created to include a list of 84 genes generated from literature to evaluate estrogen receptor signaling, breast and ductal morphogenesis, cellular growth and differentiation, proliferation, tumor progression and epithelial to mesenchymal transition as well as 5 housekeeping genes (S1 Table). Genomic DNA contamination and reverse transcription controls were included on the plate, and genomic controls showed no amplification. Samples were prepared per manufacturer’s instructions, and were added to 384-well plates using an epMotion 5075 automated pipettor (Eppendorf). Plates were run on an ABI 7900HT machine using cycling conditions recommended by the manufacturer.
Statistical Analysis
The raw PCR cycle (Ct) values were imported into the R statistical environment [18]. Raw PCR cycles were normalized (dCt) using the SLqPCR package [19] to optimize selection of house-keeping genes.
For comparison of E2 treated samples to respective vehicle controls and comparison of normalized E2-treated 2D and 3D samples, dCt were used to construct a linear model of adjusted Ct values using the LIMMA package in R [20]. The empirical Bayes statistic was used and genes with an adjusted p-value significance for multiple comparisons (q-value) or less than 0.05 and absolute fold chance of greater then 1.5 were selected.
Cluster Analysis
Fold change values for all significantly changed genes across time and culture method were log transformed and imported into Cluster [21], and clustered using Euclidean distance and complete linkage analysis. Values were imported into Treeview (reference) and visualized.
Results
When compared to vehicle-treated controls, MCF-7 monolayer cultures exposed to 1nM estradiol (E2) for 4 hours exhibit significant changes in expression of 19 transcripts as summarized in Table 1. E2 increased expression of insulin-like growth factor binding protein 4 (IGFBP4), progesterone receptor (PGR), V-Myc avian myelocytomatosis viral oncogene homolog (MYC), transforming growth factor alpha (TGFA) and FBJ murine osteosarcoma viral oncogene homolog (FOS). Treatment of MCF-7 monolayer cultures with E2 for 4 hours also increased expression of growth response to estrogen breast cancer (GREB1), PDZ-domain containing 1 (PDZK1), cyclin D1 (CCND1), snail family zinc finger 1 (SNAI1), transforming growth factor beta 3 (TGFB3) and xbox-binding protein 1 (XBP1). E2 treatment also increased expression of vascular endothelial growth factor alpha (VEGFA), amphiregulin (AREG), and wingless-type MMTV integration site family member 4 (WNT4). Expression of BCL2-like 1 (BCL2L1), Erb-B2 receptor tyrosine kinase 2 (ERBB2twist family transcription factor 1 (TWIST1), g-protein coupled estrogen receptor (GPER) and cytochrome P450 1A1, (CYP1A1) were significantly decreased following exposure to estradiol. Complete gene expression results are reported in S2 Table.
Table 1.
| Gene Name | Abbreviation | Fold Change | q value |
|---|---|---|---|
| Insulin-Like Growth Factor Binding Protein 4 | IGFBP4 | 5.78 | <0.0001 |
| Progesterone Receptor | PGR | 4.59 | <0.0001 |
| V-Myc Avian Myelocytomatosis Viral Oncogene Homolog | MYC | 3.72 | <0.0001 |
| Transforming growth factor, alpha | TGFA | 3.61 | 0.0001 |
| FBJ Murine Osteosarcoma Viral Oncogene Homolog | FOS | 3.35 | 0.0001 |
| Growth response to estrogen, breast cancer 1 | GREB1 | 3.23 | 0.0001 |
| PDZ domain containing 1 | PDZK1 | 3.17 | <0.0001 |
| Cyclin D1 | CCND1 | 2.75 | 0.0002 |
| Snail Family Zinc Finger 1, | SNAI1 | 2.60 | 0.0052 |
| Tranforming growth factor beta 3 | TGFB3 | 2.50 | 0.0004 |
| Xbox binding protein 1 | XBP1 | 2.10 | 0.0011 |
| vascular endotheilal growth factor alpha | VEGFA | 2.01 | 0.0089 |
| Amphiregulin | AREG | 1.84 | 0.0207 |
| Wingless-Type MMTV Integration Site Family, Member 4 | WNT4 | 1.68 | 0.0123 |
| BCL2-like 1 | BCL2L1 | -1.58 | 0.0306 |
| Erb-B2 Receptor Tyrosine Kinase 2 | ERBB2 | -1.78 | 0.0454 |
| Twist Family BHLH Transcription Factor 1 | TWIST1 | -2.10 | 0.0023 |
| G-protein coupled estrogen receptor | GPER | -3.06 | <0.0001 |
| cytochrome P450 1A1 | CYP1A1 | -3.27 | <0.0001 |
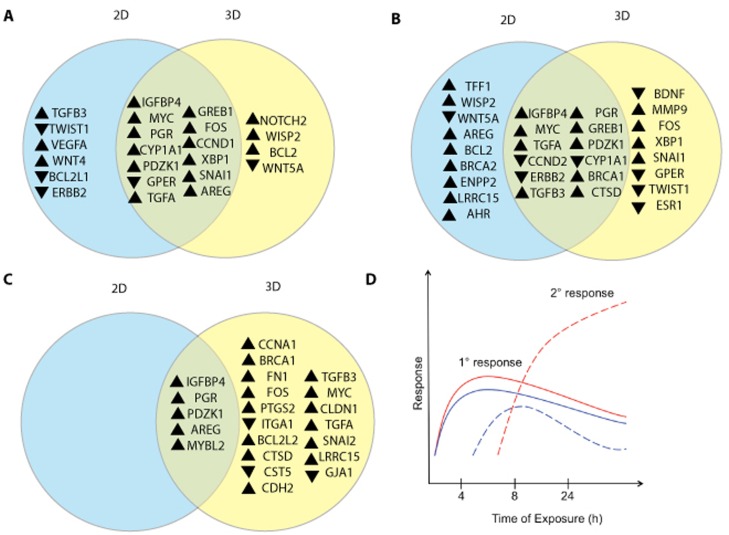
Exposure of 3D MCF-7 microtissues to estradiol for 4 hours altered expression of 17 transcripts (Table 2), resulting in an increase in expression of PGR, WNT1-inducible signaling protein 2 (WISP2), IGFBP4 and GREB1. E2 exposure also increased expression of PDZK1, MYC, CCND1, TGFA and AREG. E2-treated MCF-7 microtissues demonstrated increased expression of SNAI1, XBP1, FOS, notch 2 (NOTCH2) and B-cell lymphoma 2 (BCL2). MCF-7 microtissues treated with 1nM E2 for 8 hours displayed decreased expression of GPER, CYP1A1 and wingless-type MMTV integration site family member 5A (WNT5A). All gene expression results are reported in S3 Table. Overlapping and unique expression changes in 2D and 3D cultures exposed to estradiol are reported in Fig 1A.
Table 2.
| Gene Name | Abbreviation | Fold Change | q value |
|---|---|---|---|
| Progesterone receptor | PGR | 10.49 | <0.0001 |
| WNT1 Inducible Signaling Pathway Protein | WISP2 | 8.37 | 0.0126 |
| insulin-like growth factor binding protein 4 | IGFBP4 | 5.33 | <0.0001 |
| growth response to estrogen, breast cancer 1 | GREB1 | 5.18 | <0.0001 |
| PDZ containing domain 1 | PDZK1 | 5.00 | <0.0001 |
| V-Myc Avian Myelocytomatosis Viral Oncogene Homolog | MYC | 3.39 | <0.0001 |
| cyclin D1 | CCND1 | 3.31 | <0.0001 |
| transforming growth facotr alpha | TGFA | 2.95 | 0.0002 |
| amphiregulin | AREG | 2.57 | 0.0012 |
| Snail Family Zinc Finger 1, | SNAI1 | 2.40 | 0.0061 |
| xbox binding protein 1 | XBP1 | 2.07 | 0.0017 |
| FBJ Murine Osteosarcoma Viral Oncogene Homolog | FOS | 2.02 | 0.0049 |
| Notch 2 | NOTCH2 | 1.97 | 0.0063 |
| B-cell lymphoma 2 | BCL2 | 1.57 | 0.0431 |
| g-protein coupled estrogen receptor | GPER | -2.14 | 0.0009 |
| Cytochrome P450 1A1 | CYP1A1 | -3.78 | <0.0001 |
| Wingless-Type MMTV Integration Site Family, Member 5A | WNT5A | -4.26 | 0.0457 |
Fig 1. Gene expression of 2D and 3D cultures exposed to 1nM E2 for 4, 8 and 24 hours.
MCF-7 2D and 3D cultures were treated with 1nM E2 for 4 hours and E2 treatment of 2D cultures altered expression of 19 transcripts (blue), 14 of which were also altered in 3D cultures (A). Four transcripts were significantly altered in 3D microtissues and not in monolayer cultures (yellow) (A). Monolayer MCF-7 cultures treated with E2 for 8 hours exhibited significant gene expression changes in in 21 transcripts (blue), while 3D microtissues treated with E2 display altered expression of 20 transcripts (yellow) (B). Twelve transcripts were significantly altered in both 2D and 3D cultures (B). E2 treatment altered expression of 5 transcripts in 2D cultures (blue), while 3D cultures exhibited altered expression of 22 transcripts (yellow) following 24 hours of estradiol exposure (C). Five transcripts were significantly altered in both 2D and 3D cultures (C). Responses in 2D and 3D MCF-7 culture differ following exposure to estradiol. In both 2D and 3D cultures, estrogenic responses to estradiol are similar, and involve altered expression of typical estrogen-responsive genes (D). At 8 hours of exposure, secondary responses diverge in 2D and 3D systems, with differences persisting at 24 hours (D).
Monolayer MCF-7 cultures treated with 1nM E2 for 8 hours display altered expression of 21 transcripts (Table 3), with increased expression of ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENPP2), PGR, AREG, GREB1, IGFBP4, PDZK1 and LRCC15 compared to vehicle-treated controls. Expression of breast cancer 1 (BRCA1), breast cancer 2 (BRCA2), cathepsin D (CTSD), BCL2, and WISP2 were significantly increased. TFF1, MYC, Aryl hydrocarbon receptor (AHR), TGFA and TGFB3 expression were significantly increased. E2 treatment of monolayer cultures for 8 hours resulted in statistically significant decreased expression of ERBB2, CYP1A1, CCND2 and WNT5A compared to vehicle-treated controls. All gene expression results following E2 exposure are included in S4 Table.
Table 3.
| Gene Name | Abbreviation | Fold Change | q value |
|---|---|---|---|
| Ectonucleotide Pyrophosphatase/ Phosphodiesterase 2 | ENPP2 | 15.49 | 0.0063 |
| Progesterone receptor | PGR | 5.63 | <0.0001 |
| Amphiregulin | AREG | 3.99 | 0.0026 |
| Growth response to estrogen, breast cancer 1 | GREB1 | 3.83 | <0.0001 |
| insulin-like growth factor binding protein 4 | IGFBP4 | 3.33 | 0.0004 |
| PDZ containg domain 1 | PDZK1 | 3.31 | <0.0001 |
| Leucine-rich repeat containing 15 | LRRC15 | 2.71 | 0.0106 |
| Breast cancer 1 | BRCA1 | 2.37 | 0.0001 |
| Breast cancer 2 | BRCA2 | 2.13 | 0.0047 |
| Cathepsin D | CTSD | 2.05 | 0.0002 |
| B-cell lymphoma 2 | BCL2 | 1.93 | 0.0034 |
| WNT1 Inducible Signaling Pathway Protein | WISP2 | 1.92 | 0.0001 |
| Trefoil factor 1 | TFF1 | 1.80 | <0.0001 |
| V-Myc Avian Myelocytomatosis Viral Oncogene Homolog | MYC | 1.72 | 0.0016 |
| Aryl-hydrocarbon receptor | AHR | 1.67 | 0.0498 |
| Transforming growth factor alpha | TGFA | 1.55 | 0.0052 |
| Transforming growth factor beta 3 | TGFB3 | 1.53 | 0.0108 |
| Notch 2 | NOTCH2 | 1.40 | 0.0059 |
| Erb-B2 Receptor Tyrosine Kinase 2 | ERBB2 | -1.75 | 0.0095 |
| Cytochrome P450 1A1 | CYP1A1 | -2.10 | 0.0001 |
| Cyclin D2 | CCND2 | -2.32 | 0.0090 |
| Wingless-Type MMTV Integration Site Family, Member 5A | WNT5A | -5.29 | 0.0001 |
Exposure of MCF-7 microtissues to 1nM E2 for 8 hours resulted in significantly altered expression of 20 transcripts, compared to vehicle-treated controls (Table 4). Estradiol induced expression of PGR, GREB1, IGFBP4, PDZK1, TGFB3 and MYC. Expression of TGFA, SNAI1, CTSD, FOS, BRCA1 and XBP1 were significantly increased by E2 treatment. Complete gene expression results can be found in S5 Table. Differences and similarities in expression are summarized in Fig 1B.
Table 4.
| Gene Name | Abbreviation | Fold Change | q value |
|---|---|---|---|
| Progesterone receptor | PGR | 8.52 | <0.0001 |
| growth response to estrogen, breast cancer 1 | GREB1 | 7.05 | <0.0001 |
| insulin-like growth factor binding protein 4 | IGFBP4 | 6.10 | <0.0001 |
| PDZ containing domain 1 | PDZK1 | 5.43 | <0.0001 |
| Transforming growth factor beta 3 | TGFB3 | 3.09 | <0.0001 |
| V-Myc Avian Myelocytomatosis Viral Oncogene Homolog | MYC | 2.99 | <0.0001 |
| transforming growth factor alpha | TGFA | 2.38 | <0.0001 |
| Snail Family Zinc Finger 1, | SNAI1 | 1.99 | 0.0006 |
| Cathepsin D | CTSD | 1.88 | 0.0004 |
| FBJ Murine Osteosarcoma Viral Oncogene Homolog | FOS | 1.88 | 0.0002 |
| Breast cancer 1 | BRCA1 | 1.72 | 0.0031 |
| xbox binding protein 1 | XBP1 | 1.62 | 0.0003 |
| Estrogen receptor alpha | ESR1 | -1.50 | 0.0031 |
| Twist Family BHLH Transcription Factor 1 | TWIST1 | -1.70 | 0.0021 |
| g-protein coupled receptor 1 | GPER | -1.71 | 0.0006 |
| Matrix metallopeptidase 9 | MMP9 | -1.75 | 0.0001 |
| Erb-B2 Receptor Tyrosine Kinase 2 | ERBB2 | -2.05 | 0.0020 |
| Cytochrome P450 1A1 | CYP1A1 | -2.22 | <0.0001 |
| Cyclin D2 | CCND2 | -2.86 | 0.0026 |
| Brain-derived neurotrophic factor | BDNF | -4.34 | 0.0001 |
MCF-7 monolayers exposed to 1nM E2 for 24 hours displayed increased expression of 5 transcripts (Table 5), including IGFBP4, PGR, PDZK1, AREG and MYBL2. Complete results are reported in S6 Table.
Table 5.
| Gene Name | Abbreviation | Fold Change | q value |
|---|---|---|---|
| Insulin-Like Growth Factor Binding Protein 4 | IGFBP4 | 3.68 | 0.0002 |
| Progesterone Receptor | PGR | 6.98 | 0.0017 |
| PDZ domain containing 1 | PDZK1 | 7.27 | 0.0030 |
| Amphiregulin | AREG | 3.84 | 0.0184 |
| V-Myb Avian Myeloblastosis Viral Oncogene Homolog-Like 2 | MYBL2 | 2.47 | 0.0468 |
Exposure to E2 for 24 hours significantly altered expression of 22 transcripts in MCF-7 microtissues (Table 6). Expression of BCL2-like 2 (BCL2L2), cystatin D (CST5), PDZK1, AREG, leucine rich repeat containing 15 (LRRC15) and IGFBP4 was significantly increased following estradiol exposure. CTSD, PGR, prostaglandin synthase 2 (PTGS2), snail family zinc finger 2 (SNAI2) and claudin 1 (CLDN1) expression was significantly increased in MCF-7 microtissues treated with E2 for 24 hours. Increases in expression of TGFB3, MYC, cyclin A1 (CCNA1), MYBL2, TGFA, N-cadherin (CDH2), fibronectin (FN1), BRCA1 and FOS were observed in E2-treated microtissues (Table 6). Expression of gap junction 1 (GJA1) and integrin alpha 1 (ITGA1) was significantly decreased in E2-treated microtissues. Complete gene expression results are reported in S7 Table. Monolayer and microtissue cultures treated with E2 for 24 hours displayed large differences in gene expression (Fig 1C).
Table 6.
| Gene Name | Abbreviation | Fold Change | q value |
|---|---|---|---|
| Insulin-Like Growth Factor Binding Protein 4 | IGFBP4 | 3.35 | 0.0002 |
| PDZ domain containing 1 | PDZK1 | 4.49 | 0.0004 |
| Amphiregulin | AREG | 4.01 | 0.0006 |
| Progesterone Receptor | PGR | 3.08 | 0.0010 |
| Transforming growth factor beta 3 | TGFB3 | 2.35 | 0.0015 |
| V-Myc Avian Myelocytomatosis Viral Oncogene Homolog | MYC | 2.05 | 0.0026 |
| Claudin 1 | CLDN1 | 2.54 | 0.0042 |
| V-Myb Avian Myeloblastosis Viral Oncogene Homolog-Like 2 | MYBL2 | 1.87 | 0.0049 |
| transforming growth factor alpha | TGFA | 1.84 | 0.0070 |
| Snail Family Zinc Finger 2 | SNAI2 | 2.85 | 0.0072 |
| Leucine rich repeat containing 15 | LRRC15 | 3.66 | 0.0073 |
| Gap Junction 1 | GJA1 | -2.18 | 0.0079 |
| Cyclin A1 | CCNA1 | 2.02 | 0.0081 |
| Breast cancer 1 | BRCA1 | 1.65 | 0.0084 |
| Fibronectin 1 | FN1 | 1.66 | 0.0103 |
| FBJ Murine Osteosarcoma Viral Oncogene Homolog | FOS | 1.55 | 0.0105 |
| Prostaglandin synthase 2 | PTGS2 | 2.97 | 0.0117 |
| Integrin, alpha 1 | ITGA1 | 4.07 | 0.0147 |
| BCL-like 2 | BCL2L2 | 34.16 | 0.0192 |
| Cathepsin D | CTSD | 3.25 | 0.0194 |
| Cystatin D | CST5 | 6.95 | 0.0197 |
| N-cadherin | CDH2 | 1.82 | 0.0311 |
Discussion
When compared to 2D monolayer MCF-7 cultures, MCF-7 microtissues display differences in gene expression following exposure to estradiol. The current study indicates that in both 2D and 3D cultures exposed to estradiol, genes that are typically used as indicators of estrogenic response are upregulated by estradiol. These genes include PGR, PDZK1 [22], and IGFBP4 [23, 24], which are all increased at 4, 8, and 24 hours of exposure in 2D and 3D cultures, though the degree of fold change is greater in 3D microtissues. The similar induction of transcription of other well-known estrogen responsive genes including GREB1 [22], AREG [25], MYC and inhibition of CYP1A1 demonstrates that in terms of estrogenic activity, MCF-7 monolayers and 3D microtissues respond similarly. Following hierarchical clustering analysis, these genes cluster together and exhibit similar patterns in 2D and 3D cultures over time (Fig 1D). The primary response encompasses changes in expression of genes that contain estrogen response elements, and is consistent with previous studies that demonstrate the binding of ERs and transcription of target genes following exposure to estradiol [22].
Interestingly, transcription of other genes in 2D and 3D MCF-7 cultures is not similar following E2 exposure. As early as 4 hours of E2 exposure there are differences in expression of many markers between 2D and 3D cultures, though the majority of significantly altered genes are changed in both 2D and 3D cultures exposed to E2. Over time, the number of genes that are significantly altered in 2D or 3D cultures increases, while the number of genes shared between the two decreases, indicating a divergence of the response in each system. Hierarchical clustering demonstrated that genes expressed in 3D microtissues and not in 2D cluster separately from typical estrogen-responsive genes (Fig 1D). Expression of downstream genes involved in cell cycle progression, cell-cell communication and cell adhesion is differentially altered in monolayer and 3D cultures after 24 hours of exposure. The large differences in expression of cell adhesion markers at later time points is likely due to the differences in cellular differentiation between 2D and 3D cultures that have been previously described [15]. This difference is most striking at 24 hours of estradiol exposure, where 2D cultures display changes in only 5 genes, while 3D cultures have 22 transcripts significantly changed. A number of the 22 genes altered by estradiol exposure in 3D cultures are involved with cell adhesion (FN1, ITGA1, CST5, CDH2, CLDN1, SNAI2 and GJA1) These changes in cell adhesion molecules were not detected in E2-treated monolayers in the present study. In vitro and in vivo studies have demonstrated that exposure to E2 increases epithelial-mesenchymal transition (EMT), increasing the invasive potential and mobility of cells [26, 27]. In the present study, increases in expression of the EMT transcriptional regulators TWIST1 [28] and SNAI1 [29] are detected as early as 8 hours. Both SNAI1 and TWIST initiate EMT, which results in degradation of cell-cell contacts, loss of cellular polarity and increased invasive potential [30]. Following those increases in expression at 8 hours, at 24 hours increased expression of several classical EMT markers was detected, including FN1, CLDN1 and SNAI2. Previous work has demonstrated that CLDN1 expression is capable of inducing EMT [31], while FN1 and SNAI2 are highly expressed in mesenchymal-like cells [32, 33]. Coupled with the increased expression of mesenchymal markers was the decreased expression of epithelial markers ITGA1 and GJA1 in MCF-7 microtissues treated with E2 for 24 hours, indicating that MCF-7 cells cultured in 3D begin to undergo EMT. Interestingly, in this study, MCF-7 monolayers did not demonstrate any significant changes in expression of EMT-related genes following E2 exposure. It is clear from the current study that 2D and 3D MCF-7 cultures respond differently to E2, underlining the importance of selecting a relevant cell system.
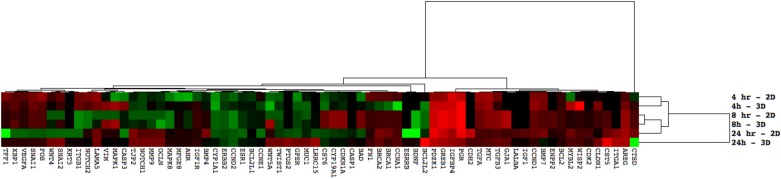
Overall, the current study indicates that when examining 2D and 3D cultures for estrogenic responses by assessing the expression of target genes, levels of expression of classical estrogen-induced genes are similar at early time points. However, at later time points, the estrogenic responses diverge and result in activation of different targets in each culture system (Fig 2). In this analysis, both 2D and 3D 4 hour cultures clustered together, with the 8 hour 2D, 8 hour 3D and 24 hour 2D samples clustering together. The 24 hour 3D sample clustered separately from all treatments, further emphasizing the large differences observed in 2D and 3D culture systems. Expression of downstream genes involved in cell cycle progression, cell-cell communication and cell adhesion are significantly altered in 3D cultures and not 2D cultures. The large differences in expression of cell adhesion markers at later time points is likely due to the differences in cellular differentiation between 2D and 3D cultures that have been previously described [15]. This observation further supports the need to develop more physiologically relevant, human cell based models for toxicity testing, as underlying differences in gene and protein expression in various systems can lead to altered interpretation of results.
Fig 2. Euclidean hierarchical clustering of all significantly altered genes over time.
All fold change values for significantly changed transcripts were analyzed using hierarchical clustering. Transcripts clustered into 2 large groups, aligning with the observation that classical estrogen responsive genes (ex PGR, GREB, AREG) are similarly expressed in 2D and 3D cultures, but secondary responses are different (ex VIM, OCLN). Green is decreased expression, red indicates increased expression.
Supporting Information
(XLS)
Values determined relative to vehicle-treated controls, positive fold change indicates increased expression.
(XLSX)
Positive fold change indicates increased expression, values compared to time-matched, vehicle-treated controls.
(XLSX)
Positive fold change indicates increased expression, relative to vehicle-treated controls.
(XLSX)
Fold change relative to vehicle-treated controls.
(XLSX)
Fold change values relative to vehicle-treated controls.
(XLSX)
Fold change relative to vehicle-treated controls.
(XLSX)
Acknowledgments
Major support for this research was provided NIEHS Training Grant T32 ES007272, Unilever, and the generous gift of Donna McGraw Weiss ‘89 and Jason Weiss, with additional support by NIH RO1 ES 020750. The authors would like to thank Dr. Edward Dere for assistance with data analysis and visualization.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Major support for this research was provided by the National Institute of Environmental Health Sciences (NIEHS) Training Grant T32 ES007272, Unilever, and the generous gift of Donna McGraw Weiss ‘89 and Jason Weiss, with additional support by NIH RO1 ES 020750. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The authors would like to thank Dr. Edward Dere for assistance with data analysis and visualization.
References
- 1.Chambers A, Krewski D, Birkett N, Plunkett L, Hertzberg R, Danzeisen R, et al. An exposure-response curve for copper excess and deficiency. Journal of toxicology and environmental health Part B, Critical reviews. 2010;13(7–8):546–78. 10.1080/10937404.2010.538657 . [DOI] [PubMed] [Google Scholar]
- 2.Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition, (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Three-dimensional culture models of normal and malignant breast epithelial cells, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell-cell interaction and morphological polarity., (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures, (2003). [DOI] [PubMed] [Google Scholar]
- 7.Modelling glandular epithelial cancers in three-dimensional cultures, (2005). [DOI] [PubMed] [Google Scholar]
- 8.Use of three-dimensional basement membrane cultures to model oncogene-induced changes in mammary epithelial morphogenesis., (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fibroblast biology in three-dimensional collagen matrices. (2003). [DOI] [PubMed] [Google Scholar]
- 10.Hirst SJ, Twort CH, Lee TH. Differential effects of extracellular matrix proteins on human airway smooth muscle cell proliferation and phenotype. American journal of respiratory cell and molecular biology. 2000;23(3):335–44. Epub 2000/09/06. 10.1165/ajrcmb.23.3.3990 . [DOI] [PubMed] [Google Scholar]
- 11.Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(19):9064–8. Epub 1992/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen-Ngoc KV, Cheung KJ, Brenot A, Shamir ER, Gray RS, Hines WC, et al. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(39):E2595–604. Epub 2012/08/28. 10.1073/pnas.1212834109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Napolitano AP, Dean DM, Man AJ, Youssef J, Ho DN, Rago AP, et al. Scaffold-free three-dimensional cell culture utilizing micromolded nonadhesive hydrogels. BioTechniques 2007. p. 494-6–500. [DOI] [PubMed] [Google Scholar]
- 14.Dynamics of the Self-Assembly of Complex Cellular Aggregates on Micromolded Nonadhesive Hydrogels, (2007). [DOI] [PubMed] [Google Scholar]
- 15.Vantangoli MM, Madnick SJ, Huse SM, Weston P, Boekelheide K. MCF-7 Human Breast Cancer Cells Form Differentiated Microtissues in Scaffold-Free Hydrogels. PloS one. 2015;10(8):e0135426 10.1371/journal.pone.0135426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A human cell line from a pleural effusion derived from a breast carcinoma., (1973). [DOI] [PubMed] [Google Scholar]
- 17.Napolitano AP, Dean DM, Man AJ, Youssef J, Ho DN, Rago AP, et al. Scaffold-free three-dimensional cell culture utilizing micromolded nonadhesive hydrogels. BioTechniques. 2007;43(4):494-6–500. [DOI] [PubMed] [Google Scholar]
- 18.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 19.Kohl M. SLqPCR: Functions for analysis of real-time quantitative PCR data at SIRS-Lab GmbH. SIRS-Lab GmbH, Jena, Germany; 2007. [Google Scholar]
- 20.Smyth G. Limma: linear models for microarray data In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. p. 397–420. [Google Scholar]
- 21.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(25):14863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh MG, Thompson DA, Weigel RJ. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer research. 2000;60(22):6367–75. . [PubMed] [Google Scholar]
- 23.Vendrell JA, Magnino F, Danis E, Duchesne MJ, Pinloche S, Pons M, et al. Estrogen regulation in human breast cancer cells of new downstream gene targets involved in estrogen metabolism, cell proliferation and cell transformation. Journal of molecular endocrinology. 2004;32(2):397–414. Epub 2004/04/10. . [DOI] [PubMed] [Google Scholar]
- 24.Safe S. Transcriptional activation of genes by 17 beta-estradiol through estrogen receptor-Sp1 interactions. Vitamins and hormones. 2001;62:231–52. Epub 2001/05/11. . [DOI] [PubMed] [Google Scholar]
- 25.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144(10):4562–74. Epub 2003/09/10. 10.1210/en.2003-0567 . [DOI] [PubMed] [Google Scholar]
- 26.Park SH, Cheung LW, Wong AS, Leung PC. Estrogen regulates Snail and Slug in the down-regulation of E-cadherin and induces metastatic potential of ovarian cancer cells through estrogen receptor alpha. Molecular endocrinology. 2008;22(9):2085–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherbakov AM, Andreeva OE, Shatskaya VA, Krasil'nikov MA. The relationships between snail1 and estrogen receptor signaling in breast cancer cells. Journal of cellular biochemistry. 2012;113(6):2147–55. Epub 2012/02/07. 10.1002/jcb.24087 . [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–39. Epub 2004/06/24. 10.1016/j.cell.2004.06.006 . [DOI] [PubMed] [Google Scholar]
- 29.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nature cell biology. 2000;2(2):84–9. Epub 2000/02/03. 10.1038/35000034 . [DOI] [PubMed] [Google Scholar]
- 30.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Current opinion in cell biology. 2005;17(5):548–58. [DOI] [PubMed] [Google Scholar]
- 31.Suh Y, Yoon CH, Kim RK, Lim EJ, Oh YS, Hwang SG, et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene. 2013;32(41):4873–82. [DOI] [PubMed] [Google Scholar]
- 32.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–15. Epub 2008/05/20. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer research. 2002;62(6):1613–8. . [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
Values determined relative to vehicle-treated controls, positive fold change indicates increased expression.
(XLSX)
Positive fold change indicates increased expression, values compared to time-matched, vehicle-treated controls.
(XLSX)
Positive fold change indicates increased expression, relative to vehicle-treated controls.
(XLSX)
Fold change relative to vehicle-treated controls.
(XLSX)
Fold change values relative to vehicle-treated controls.
(XLSX)
Fold change relative to vehicle-treated controls.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.