Abstract
Biofilm formation is important for infection by many pathogens. Bordetella bronchiseptica causes respiratory tract infections in mammals and forms biofilm structures in nasal epithelium of infected mice. We previously demonstrated that cyclic di-GMP is involved in biofilm formation in B. bronchiseptica. In the present work, based on their previously reported function in Pseudomonas fluorescens, we identified three genes in the B. bronchiseptica genome likely involved in c-di-GMP-dependent biofilm formation: brtA, lapD and lapG. Genetic analysis confirmed a role for BrtA, LapD and LapG in biofilm formation using microtiter plate assays, as well as scanning electron and fluorescent microscopy to analyze the phenotypes of mutants lacking these proteins. In vitro and in vivo studies showed that the protease LapG of B. bronchiseptica cleaves the N-terminal domain of BrtA, as well as the LapA protein of P. fluorescens, indicating functional conservation between these species. Furthermore, while BrtA and LapG appear to have little or no impact on colonization in a mouse model of infection, a B. bronchiseptica strain lacking the LapG protease has a significantly higher rate of inducing a severe disease outcome compared to the wild type. These findings support a role for c-di-GMP acting through BrtA/LapD/LapG to modulate biofilm formation, as well as impact pathogenesis, by B. bronchiseptica
Introduction
Bordetella bronchiseptica is a Gram-negative bacterium that causes respiratory tract infections in mammals, atrophic rhinitis in pigs, kennel cough in dogs and snuffles in rabbits [1]. B. bronchiseptica has a variety of virulence factors that allow host infection. Each factor, such as pertactin, filamentous hemagglutinin, adenylate cyclase, the type three secretory system and lipopolysaccharide are likely to perform specific functions required for successful colonization [2–6].
The ability of Bordetella spp. to form biofilms has been reported in previous work, including the ability of B. bronchiseptica to form biofilms on abiotic surfaces regulated by the two-component system BvgAS [7,8]. As has been found for other biofilm-forming organisms, extracellular DNA (eDNA) and exopolysaccharide are important for biofilm formation by B. bronchiseptica [9,10]. Particularly in Bordetella, BvgAS-regulated factors, including the filamentous hemagglutinin and adenylate cyclase, may also participate in biofilm formation [7,11]. Static growth in intermediate nicotinic acid concentrations, a regulator of the BvgAS system, resulted in the best conditions for promoting biofilm formation [8]. Studies from our group and others indicate that biofilm is enhanced in the intermediate phase [7,12], however, there are conflicting findings regarding BvgAS-dependent biofilm regulation [8]. Furthermore, transcriptome analysis has shown that more than 33% of the B. bronchiseptica genes are differentially regulated during biofilm formation compared to planktonic culture [13]. Thus, further studies are needed to elucidate all factors affecting biofilm formation.
Sloan and colleagues observed biofilm-like structures in vivo in the nasal epithelium of B. bronchiseptica infected mice, and these communities expressed a polysaccharide essential for in vivo biofilm development [9]. The absence of polysaccharides, key factor for in vitro biofilm formation, also impaired infection suggesting that biofilm formation may also participate in host-pathogen interactions [9]. Thus, biofilm formation may play an important role in host-B. bronchiseptica interactions. Recently, we showed that bis-(3′-5′)-cyclic-dimeric guanosine monophosphate (c-di-GMP) regulates motility and biofilm formation in B. bronchiseptica [12]. C-di-GMP is a bacterial second messenger known to regulate a variety of cellular processes including biofilm formation, motility and virulence of bacterial pathogens [14,15]. Like in other bacteria where c-di-GMP-related functions have been studied, high c-di-GMP levels in B. bronchiseptica correlated with an enhanced biofilm formation phenotype [12]. However, in this previous study we did not uncover the mechanism by which c-di-GMP enhanced biofilm formation in B. bronchiseptica.
Here we confirm the role for the Bvg-regulated adhesin BrtA, a putative homolog of the well-characterized LapA adhesin of Pseudomonas fluorescens, in biofilm formation by B. bronchiseptica, as has recently been reported by Nishikawa and colleagues [16]. Furthermore, we show that the c-di-GMP receptor LapD and the LapD-regulated protease LapG also participate in biofilm formation by B. bronchiseptica, likely via the control of BrtA localization to the cell surface as has been reported for the LapA adhesin of P. fluorescens. We also present data that the appropriate control of BrtA via LapG appears to be important for modulating pathogenesis in a mouse model of infection. Thus, this work describes the basis of c-di-GMP-mediated control of biofilm formation in B. bronchiseptica, both in vivo and in vitro.
Materials and Methods
Ethics Statement
This study was performed in strict accordance with animal use protocols approved by Faculty of Sciences, National University of La Plata, Institutional Animal Care and Use Committee (IACUC), protocol number 007-00-15. All animals were properly anesthetized with isofluorane for bacterial inoculation, monitored daily and euthanized with isofluorane overdose if they met any early removal criteria (lethargy, hunched posture, or ruffled coat) to limit suffering.
Microbiological Methods
For routine culture, B. bronchiseptica strains were grown on Bordet Gengou agar (BGA) (Difco) supplemented with 15% (vol/vol) defibrinated fresh sheep blood (BGA medium) at 36°C for 48 h, and replated on the same medium for 24 h. Liquid cultures were grown in Stainer-Scholte (SS) [17] medium at 36°C and 160 rpm. When appropriate BGA or SS was supplemented with kanamycin (80 μg ml-1), streptomyicin (200 μg ml-1) or gentamycin (50 μg ml-1). P. fluorescens and E. coli were grown in lysogeny broth (LB) [18] at 30°C and 37°C, respectively. When appropriate, antibiotics were added to the medium at the following concentrations: E. coli, 10 μg ml-1 gentamycin; P. fluorescens, 30 μg ml-1 gentamycin.
Replicative plasmids were introduced to E. coli and B. bronchiseptica by electroporation using standard techniques. Non-replicating plasmids were introduced into B. bronchiseptica by conjugation. The yeast strain InvSc1 (Saccharomyces cerevisiae; Invitrogen), was routinely cultured on YPD medium. When selecting for plasmids carrying the URA3 gene, yeast were grown on YNB with complete supplemental mixture minus uracil.
Plasmid and Strain Construction
Strains and plasmids were constructed using standard molecular biology techniques and are listed in S1 Table. Oligonucleotides used in this study are listed in S2 Table. Detailed descriptions of strain and plasmid construction procedures can be found in the S1 Text.
Biofilm Assays
Biofilm formation assays using static cultures were performed as described previously [12] from overnight cultures inoculated into SS liquid medium. Briefly, the culture was pipetted into glass tubes or wells of a sterile 96-well U bottom microtiter plate (polyvinylchloride, PVC) and incubated statically at 37°C. Nicotinic acid was added as indicated. After 24 hours, unless otherwise indicated, planktonic bacteria were removed and the attached cells were stained with 0.1% crystal violet (CV) solution. The stain was dissolved by adding 120 μl of 33% acetic acid solution. One hundred microliters of the dissolved stain solution was transferred to a new, flat-bottom microplate and then quantified by measuring OD at 595 nm.
Scanning Electron Microscopy
Scanning electron microscopy pictures were obtained as described previously by Sisti et al. [12]. Briefly, Bordetella strains were cultured statically on glass coverslips partially submerged vertically in plastic tubes such that an air-liquid interface was established on the coverslip. After 24 h of incubation, the coverslip was removed and washed with sterile PBS, and the bacteria were fixed with 2.5% glutaraldehyde in PBS. Samples were dehydrated in a graded ethanol series (20, 50, 70, 90 and 100% for 60 min each), subjected to critical point drying using liquid carbon dioxide (EMITECH, K850) and sputter coated with gold (SPI Supplies). The surface topographies of the biofilm in A panels were visualized with a scanning electron microscope (ESEM FEI QUANTA 200), and detected with SDD (EDAX Apollo 40) camera. The surface topographies in B panels were visualized with a scanning electron microscope (Philips SEM 505), and the images were processed with the Image Soft Imaging System ADDA II.
RNA Extraction for RT-PCR Studies
RNA extraction and RT-PCR studies were performed as described previously by our group [12]. RNA preparation from bacteria was performed using the Illustra RNAspin kit (GE, USA), and quantification of RNA was performed using a ND-1,000 NanoDrop spectrophotometer at 260 nm. Measurements of A260/280 were used to determine the purity of the RNA. The synthesis of cDNA was performed with a Reverse Transcription System kit (Promega) according to the manufacturer’s protocol using random primers. One microgram of RNA was used for each sample. The reaction was incubated at room temperature for 10 min, and reverse transcription was performed in a thermal cycler at 42°C for 15 min and 95°C for 5 min for a total of 35 cycles. PCR was performed with Kappa Taq from Biosystems. Samples that had not undergone the reverse transcription process were used as controls for the absence of genomic DNA in RT-PCR experiments. Primers employed are summarized in S2 Table, including the primers designed to amplify the intergenic region between the lapA and lapG genes. PCR analysis of internal fragment of recA gene, which is constitutively expressed, was used as control.
LapA Localization
For Western and dot blot assays of the LapA protein, we used strains with internal HA-tag engineered into the sequence of the chromosomal lapA gene. Culture conditions, preparation of clarified cell extracts, and analysis of samples for LapA localization were performed as described previously without modification [19]. Detection of cell surface LapA by dot blotting was also performed as previously described [19,20].
LapG Activity Assays
Bacterial cultures were grown overnight in LB, and clarified cell extracts were subsequently prepared by sonication (4×10 s on ice) in resuspension buffer, followed by centrifugation 12 min at 15,000 × g. As described previously [19], to assess cleavage of N-terminal domain of LapA (N-term-LapA), cell extracts E. coli carrying the pMini-LapA plasmid were mixed 1:1 with cell extracts from strains with and without a LapG-expressing plasmid, followed by incubation at room temperature for 30 min. When indicated, purified P. fluorescens LapG was added (750 ng of LapG; est. purity: 50%).
Murine Respiratory Infection Model
Female BALB/c mice (originally from National Institute of Health (USA) and maintained at Faculty of Veterinary, National University of La Plata) 3 to 4 weeks of age were used as a model of in vivo respiratory infection by B. bronchiseptica. Bacteria grown on BGA medium were resuspended and adjusted to approximately 107 CFU ml-1 in PBS. Fifty microliters of bacterial suspension was delivered intranasally to each mouse via an air displacement pipette. At different times post-inoculation, three to four mice from each group were euthanized and their lungs and nose were removed aseptically. Tissues were homogenized in PBS, and appropriate dilutions were plated onto BGA medium to determine the number of viable bacteria present in the lungs and nose. Results presented in figures are from two independent experiments performed with at least four mice per strain per time point. Counts from euthanized mice were excluded from calculations, as described in more detail in the Results section.
Statistical Analysis
All the results were compared by analysis of variance (ANOVA) followed by the Tukey test using Infostat software (Cordoba National University).
Results
Genes Coding for Lap-Like Proteins Are Present and Expressed in B. bronchiseptica
We searched the B. bronchiseptica genome for genes coding for proteins with properties similar to the LapA protein of P. fluorescens, including a type I secretion signal, a Von Willebrand factor type A domain (vWFA), an overall large size, a domain with sequence repeats, as well as lapG- and lapD-like genes found in close proximity to the candidate LapA-like protein(s). We performed a BLASTP search using ~1200 C-term amino acids of P. fluorescens LapA as the query against the available B. bronchiseptica RB50 genome. This portion of LapA includes the vWFA domain and type I secretion signal. The search identified a protein with ~30% identity to the query sequence; the identified protein contained a C-term domain of a putative hemolysin. Inspection of the complete amino acid sequence identified a protein with LapA-like properties in B. bronchiseptica—BB_RS05895.
The BB_RS05895 ORF was recently described by Nishikawa and co-workers and named brtA [16], thus we use that nomenclature here. The B. bronchiseptica RB50 BrtA protein is predicted to be 3345 amino acids in length, with eight repeat regions. Each repeat region is ~200 amino acids long and is composed of distinct regions designated the CADG and VCBS (Dystroglycan-type cadherin-like domains and Vibrio, Colwellia, Bradyrhizobium, and Shewanella repeats superfamily respectively) domains based on conserved amino acid sequence motifs (Fig 1, S1 Fig). Interestingly, the number of repeats in BrtA vary among B. bronchiseptica strains. As described in S3 Table, the number of repeats varies between 2 and 15 with a median of 3 repeats. These differences in repeat length may impact host tropism, however no correlation was observed between isolate source and number of repeated regions (S3 Table). It is important to note that many draft genomes have incomplete BrtA sequences, likely due to high frequency of repeated sequences in this protein. Hence data regarding number of CADG-IDR1-VCBS-IDR2 repeats in BrtA homologs is not available for all genomes.
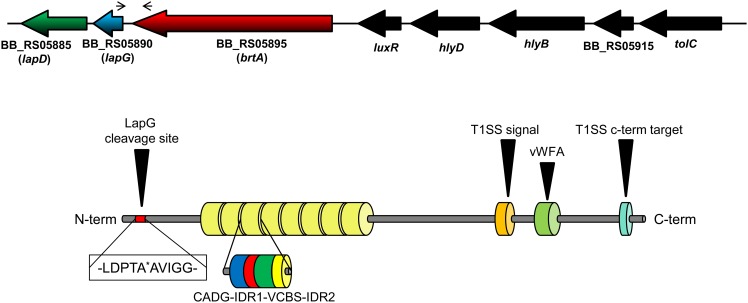
Fig 1. B. bronchiseptica LapA domain organization.
Diagram showing the organization of the lapD, lapG and brtA genes in B. bronchiseptica (top) as well as the domain organization of the BrtA protein (bottom). The narrow arrows indicate the location of the primers used in the RT-PCR experiment. The thick, black arrow indicates the specific cleavage site for LapG. The type I secretion signal (T1SS) and T1SS target domains of BrtA are also shown (orange and cyan, respectively), as is the von Willebrand Factor A (vWFA) domain (light green). The repeat regions are indicated by the 8 yellow barrels. Domains found in repeated regions are also indicated: CADG domain (blue), VCBS domain (green) and IDR (interdomain region, red and yellow). The CADG and VCBS domain names are based on core conserved amino acid.
While BrtA architecture is similar to the LapA protein of P. fluorescens and the RtxA protein of Legionella pneumophila [21], the sequence of the repeat regions is distinct. Nishikawa and colleagues showed that the BrtA protein localizes to the surface of B. bronchiseptica, and using a brtA mutant strain, these workers also showed that BrtA is required for the adhesion to an abiotic (polystyrene) substratum but not to rat alveolar type 2 cells [16]. Upstream of BB_RS05895 we found BB_RS05890 and BB_RS05885, which show sequence similarity to the P. fluorescens LapG (48% identity) and LapD (30% identity) proteins, respectively. Based on their sequence as well as functional conservation to the P. fluorescens proteins (as described below), we decided to name BB_RS05890 and BB_RS05885 as lapG and lapD, respectively. We also identified a canonical predicted cleavage site for LapG (an Ala-Ala motif) in the N-terminal portion of BrtA (Fig 1) [21], indicating that BrtA cell surface localization is likely controlled by the LapG protease, a hypothesis we test further below.
The gene organization in B. bronchiseptica suggested the lapD, lapG and brtA genes are part of an operon. Intergenic regions of 81 bp and 16 bp were found between the end of the brtA gene and start of the lapG gene and between the lapG and lapD genes, respectively. To test whether these genes are co-transcribed, we performed RT-PCR using primers that amplified the intergenic region between the brtA and lapG genes (Fig 1). Results presented in Fig 2A show that the brtA and lapG genes appear to be co-transcribed. Moreover, these genes appear to be transcribed in both virulent and avirulent phases, as indicated by the detection of the PCR product across a range of nicotinic acid (NA) concentrations.
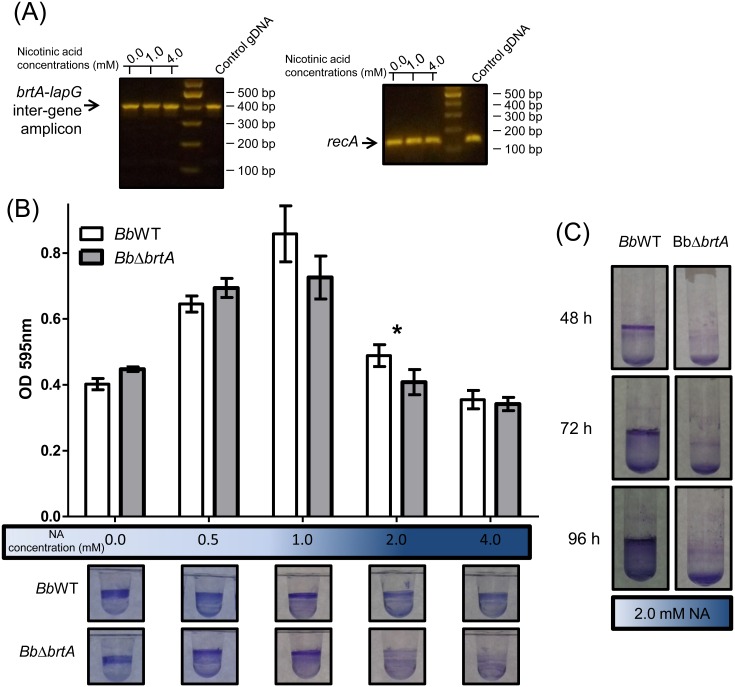
Fig 2. Loss of BrtA in B. bronchiseptica negatively impacts biofilm formation in vitro.
(A) RT-PCR using B. bronchiseptica cDNA as template to amplify intergenic regions between the lapA and lapG genes. The recA amplicon was used as a positive control for the constitutively expressed recA gene. cDNA obtained from wild type B. bronchiseptica grown in different NA concentrations was employed in these assays. Note: the RT-PCR assays used are not quantitative, but are useful for detecting the presence/absence of a transcript. (B) Biofilm development by the BbWT and BbΔbrtA strains cultured in SS medium either alone or supplemented with NA at the indicated concentration. Biofilm formation was assessed by PVC microtiter plate assays as described in the Materials and Methods. *, indicates a significant difference between the wild type and mutant with same concentration of NA, p<0.01. After incubation in static conditions, the medium was removed, the bacteria were stained with CV, and the extent of biofilm formation quantified. (C) Biofilm formation by the BbWT and BbΔbrtA strains assessed in SS supplemented with NA 2.0 mM in glass tubes.
We compared the amino acid sequence of the B. bronchiseptica LapD- and LapG-like proteins. LapD of B. bronchiseptica has cytosolic GGDEF and EAL domains that have been shown to be critical to the function of the LapD protein of P. fluorescens [19]. The B. bronchiseptica LapG-like protein is 48% identical to its P. fluorescens counterpart; sequence analysis shows that all the amino acids described by Chatterjee et al. as important for protease activity, as well as for binding to LapD and calcium [20] are conserved between the LapG proteins of B. bronchiseptica and P. fluorescens.
BrtA Is Important for Biofilm Formation in a Surface-Dependent Manner
Recently BrtA was described as an avirulence factor involved in biofilm formation in B. bronchiseptica RB50 [16]. To confirm its role in biofilm formation, we deleted the brtA gene of B. bronchiseptica 9.73H+. The ability to form a biofilm was evaluated by the crystal violet (CV) biofilm assay using a 96-well polyvinylchloride (PVC) microtiter plates as described in the Material and Methods. We determined the amount of biofilm biomass formed as a function of NA concentration; this chemical serves as an environmental modulator of BvgAS activity [22]. As shown in Fig 2B, we observed a modest but significant reduction in the amount of biofilm formed by a mutant lacking the BrtA protein (BbΔbrtA) at 2 mM NA. In contrast, biofilm formation by the BbΔbrtA strain was substantially reduced at 2 mM NA when biofilm formation was assessed on a hydrophilic surface (glass, Fig 2C). Taken together, our data indicate that BrtA differentially contributes to biofilm formation on different surfaces.
B. bronchiseptica LapG Is a Protease that Cleaves the N-Terminus of BrtA
Newell and co-workers previously described the P. fluorescens mutant PfΔlapG, a strain with a clean deletion of the lapG gene and expressing a fully functional, HA-tagged variant of the LapA adhesin [20]. The lack of LapG in P. fluorescens results in a strain that forms a hyper-biofilm due to the enhanced levels of LapA on the surface of the cell [20].
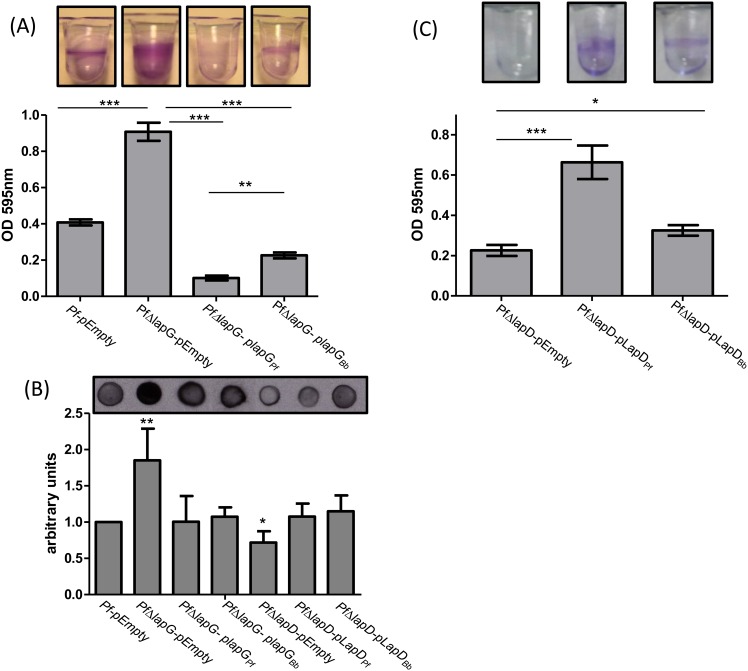
We hypothesized that if the LapG-like protein of B. bronchiseptica shared function with LapG of P. fluorescens, expression of the B. bronchiseptica LapG protein would reduce the biofilm formed by the PfΔlapG mutant strain. The B. bronchiseptica lapG-like gene was cloned into a broad host vector, and this construct and the vector control were introduced into the wild-type P. fluorescens or the PfΔlapG mutant strain. Expression of the B. bronchiseptica LapG protein in the PfΔlapG strain significantly (p< 0.01) reduced the biofilm formed compared to the same mutant carrying pEmpty control vector (Fig 3A). Expression of the P. fluorescens LapG protein in the PfΔlapG mutant served as a positive control for this assay.
Fig 3. B. bronchiseptica LapG and LapD complement their respective P. fluorescens mutations.
(A) Biofilm formation by the P. fluorescens ΔlapG mutant (PfΔlapG) carrying plasmids expressing the B. bronchiseptica or P. fluorescens lapG genes was determined by the CV biofilm assay. (B) Cell surface levels of LapA as measured by dot blot. Shown is a representative dot blot assay (top) and quantification of the pixel density (n = 6 ± SD; bottom) to assess the level of cell surface LapA of the P. fluorescens strains expressing LapG or LapD, as well as indicated control strains. *, ** or *** indicate significant differences between strains, p<0.05, p<0.01 and p<0.005 respectively. (C) Biofilm formation by the P. fluorescens ΔlapD mutant (PfΔlapD) carrying plasmids expressing the B. bronchiseptica or P. fluorescens lapD genes was determined by the CV biofilm assay. *, ** or *** indicate significant differences between strains, p<0.05, p<0.01 and p<0.005 respectively.
To confirm that the reduction in biofilm formation caused by the expression of the B. bronchiseptica LapG protein in the PfΔlapG mutant was due to a reduction in LapA on the cell surface, we performed the reported dot blot assay [19] to quantify cell-surface localized LapA protein in P. fluorescens. LapA was indeed reduced on the bacterial cell surface when B. bronchiseptica LapG was expressed in the PfΔlapG mutant (Fig 3B, first 4 bars), suggesting that LapG of B. bronchiseptica is indeed a protease that can target P. fluorescens LapA as a substrate.
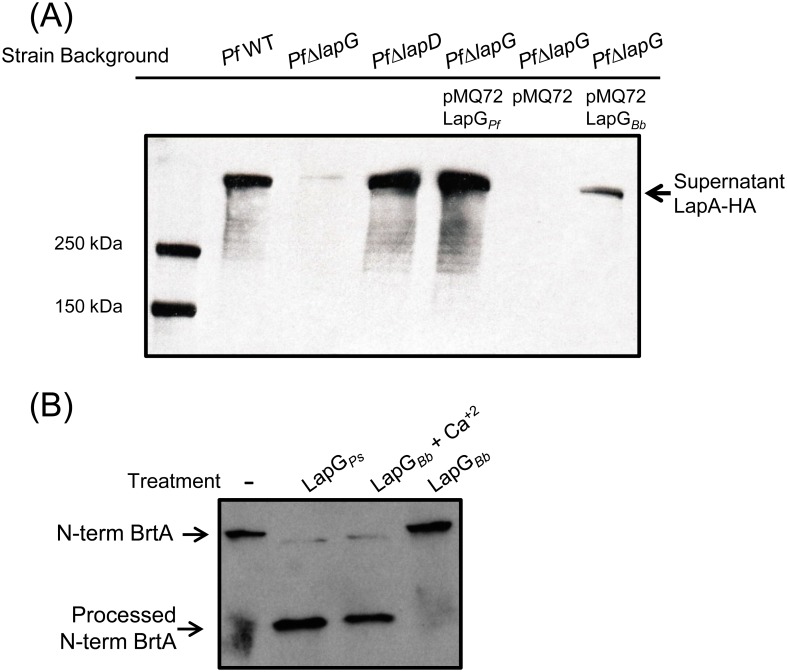
To confirm that B. bronchiseptica LapG is a protease that cleaves BrtA we performed biochemical assays using two different approaches. In the experiment shown in Fig 4A, the indicated P. fluorescens strain was transformed with a vector control (pMQ72), a plasmid carrying the LapG protease from P. fluorescens (LapGPf) as a positive control, or B. bronchiseptica (LapGBb). In these assays, we assess LapG activity by measuring the amount of LapA protein of P. fluorescens released into the supernatant; detecting supernatant LapA requires a functional LapG protease. The source of LapA in these studies is the chromosomally-encoded LapA modified with an HA-tag to facilitate its detection. As expected, in the wild type P. fluorescens strain (PfWT, Fig 4A, lane 2), LapA protein can be detected in the supernatant but no LapA is detected in the P. fluorescens strain lacking LapG (PfΔlapG, Fig 4A, lane 3). As an additional control, we see abundant LapA in the supernatant in a lapD mutant strain (PfΔlapD, Fig 4A, lane 4); a lapD mutation results in high, unregulated LapG activity, and thus serves as an additional positive control for LapA release into the supernatant.
Fig 4. B. bronchiseptica LapG cleaves P. fluorescens LapA and B. bronchiseptica BrtA.
(A) Cleavage of LapA-HA from the surface of P. fluorescens cells with B. bronchiseptica LapG. Western blot with anti-HA antibodies were used to detect LapA-HA of P. fluorescens released into the supernatant fraction for each indicated strain. (B) Cleavage of N-terminal fragment of BrtA by B. bronchiseptica LapG in vitro. Samples correspond to reaction mixture of E. coli lysate expressing indicated protein or empty vector with E. coli lysate expressing the N-terminus of BrtA (N-term BrtA) incubated at room temperature for 1h. Western blot with anti-HA antibodies was used to detect N-term BrtA.
Our data indicate that B. bronchiseptica LapG can indeed substitute for the LapG protein of P. fluorescens as indicated by its ability to release LapA into the supernatant (Fig 4A, lane 7), albeit to a lesser extent than the P. fluorescens LapG protein (Fig 4A, lane 5). In the P. fluorescens strain lacking LapG (PfΔlapG) carrying the pMQ72 vector (lane 6), as expected, no LapA was detected in the supernatant. These data are consistent with the hypothesis that B. bronchiseptica LapG is a protease that can target cell-surface adhesins like LapA.
To extend the finding in Fig 4A to an in vitro system, an E. coli strain expressing B. bronchiseptica LapG was grown, concentrated and sonicated to generate a crude cell-free extract, as described in the Materials and Methods. Crude cell free extracts of E. coli containing the HA-tagged, N-term fragment of B. bronchiseptica BrtA (N-term BrtA, amino acids 1–330) were also generated. The crude extract containing B. bronchiseptica LapG was mixed in a 1:1 ratio with the extract containing the N-terminal fragment of B. bronchiseptica BrtA, incubated at room temperature for 1 hr, then analyzed by Western blotting using an anti-HA-tag antibody to detect processing of BrtA. As a positive control, LapG from P. fluorescens was shown to process N-term BrtA, and B. bronchiseptica LapG was also able to cleave the N-terminal fragment of B. bronchiseptica BrtA (Fig 4B). Interestingly, B. bronchiseptica LapG only cleaved BrtA in presence of 10 mM calcium. This finding is consistent with previous work showing that the LapG protein of P. fluorescens is a calcium-dependent enzyme [23]. Interestingly, Nishikawa and colleagues observed that treatment of B. bronchiseptica with EGTA, a calcium chelator, reduced biofilm formation; this reduction in biofilm formation may be due to the need for calcium to promote proper folding of some secreted proteins [24,25].
B. bronchiseptica LapD Appears Partially Functional in P. fluorescens
The B. bronchiseptica lapD-like gene was cloned in a broad host vector to evaluate its ability to complement the phenotype of a lapD mutant of P. fluorescens (PfΔlapD). The PfΔlapD strain shows reduced biofilm formation and low LapA levels on the bacterial surface compared with the wild type [19]. When the B. bronchiseptica lapD gene was expressed in the PfΔlapD strain, partial complementation was observed in CV biofilm assay (Fig 3C). The level of cell surface LapA in the strain carrying the B. bronchiseptica LapD was increased to an extent similar to that observed when the P. fluorescens lapD gene was introduced into this strain (Fig 3B). We do not understand why the increase in cell surface LapA to wild-type levels in the strain carrying B. bronchiseptica LapD does not also result in an increase in biofilm formation to levels equivalent to the wild-type strain.
The Deletion or Over-Expression of LapG Alters Biofilm Formation by B. bronchiseptica
Previous studies of P. fluorescens showed that unregulated LapG expression results in increased cleavage of LapA and the release of this adhesin into the supernatant, results in reduced biofilm formation. In contrast, deleting the lapG gene of P. fluorescens results in enhanced levels of LapA on the cell surface and increased biofilm formation [20]. Thus, we assessed the effect of expressing LapG from a plasmid and deleting the gene from the chromosome on biofilm formation by B. bronchiseptica as a function of NA, an in vitro regulator of biofilm formation.
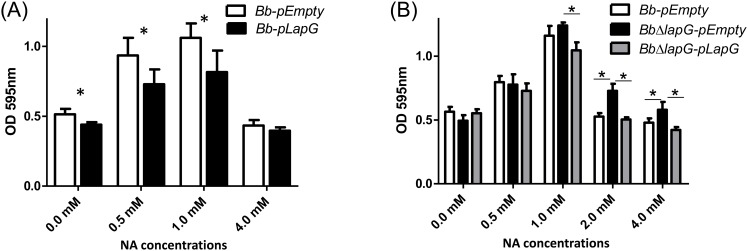
We hypothesized that expression of B. bronchiseptica LapG under a constitutive promoter would decrease biofilm formation in B. bronchiseptica. As expected, LapG overexpression resulted in significantly (p<0.001) reduced biofilm formation (Fig 5A) but only at low concentrations of NA added 0, 0.5 and 1 mM. At the highest concentration of NA tested (4.0 mM), where the extent of biofilm formation was the lowest, expression of the B. bronchiseptica LapG did not further reduce biofilm formation.
Fig 5. LapG regulates biofilm formation by B. bronchiseptica.
(A) Biofilm formation in different NA concentrations by the wild type carrying a vector control (Bb-pEmpty) or a plasmid over-expressing B. bronchiseptica LapG (Bb-lapG). *, indicates a significant difference versus the Bb-pEmpty in same NA concentration, p<0.001. (B) Biofilm formation by the wild-type strain carrying a vector control (Bb-pEmpty), a strain with a deletion of the lapG gene carrying a vector control (BbΔlapG-pEmpty), and the BbΔlapG mutant complemented with a wild-type copy of the B. bronchiseptica lapG gene (BbΔlapG-pLapG). Biofilm formation was assessed using the microtiter plate assays as described in the Materials and Methods. *, indicates a significant difference in the indicated comparisons, p<0.001.
Based on the work in P. fluorescens, we would expect a B. bronchiseptica strain with a deletion of the lapG gene to develop very robust biofilms compared to the wild type. As expected, the B. bronchiseptica strain lacking the lapG gene (BbΔlapG) showed more biofilm formation compared to wild type at the higher concentrations of NA tested, and this defect could be complemented in trans by a wild-type copy of LapGBb (Fig 5B). This phenotype was observed on either a PVC (Fig 5B) or glass surface (not shown).
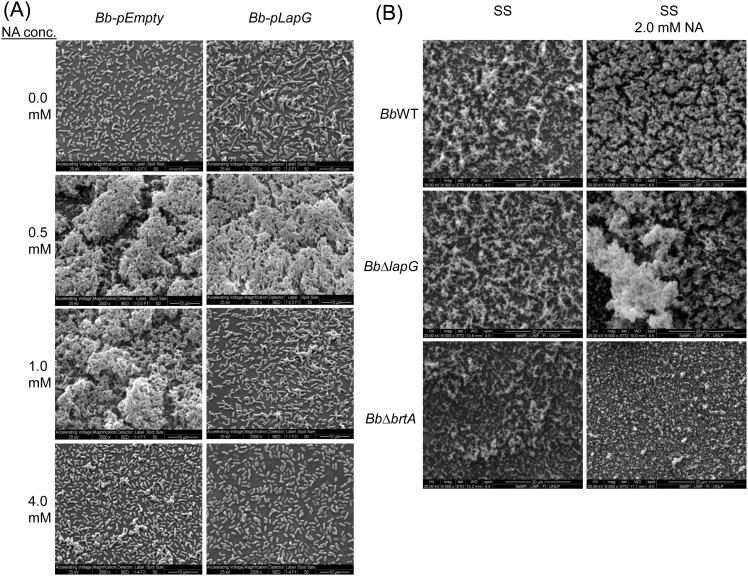
The observations in Fig 5 using the microtiter plate assay were confirmed by SEM analysis of biofilm structures formed in the air-liquid interface using glass as an abiotic surface. Fig 6 shows representative SEM images of the biofilms formed by strains over-expressing LapG compared to a vector control (Fig 6A) and in a strain lacking the lapG gene compared to the wild type (Fig 6B). As a control, deletion of the brtA gene reduced (but did not eliminate) the biofilm formed under these conditions (Fig 6B). Similar results were observed when fluorescence microscopy was employed (S2 Fig).
Fig 6. SEM images of B. bronchiseptica biofilms.
(A) The strains Bb-pEmpty (left column of panels) or Bb-pLapG (right column of panels) were grown on vertically submerged coverslips in SS medium alone or supplemented with indicated NA concentrations. After 24 h of growth, biofilms formed at the air–liquid interface were visualized by SEM. (B) Indicated strains were grown on vertically submerged coverslips in SS medium alone (left column of panels) or SS supplemented with 2.0 mM NA (right column of panels).
LapG May Contribute to Infection In Vivo
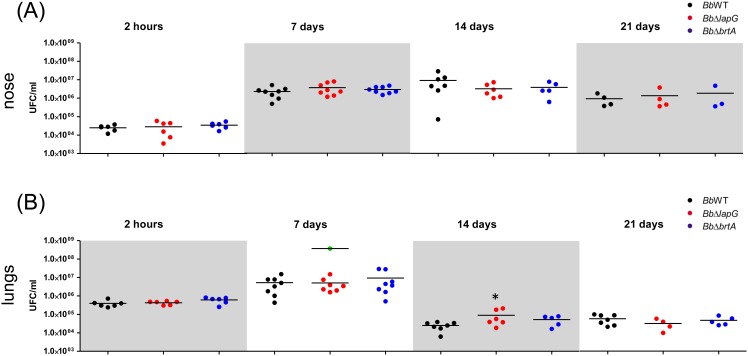
Biofilm formation on epithelial cells in the nose is thought to be a key early step in Bordetella infection [9]. Given the role of BrtA and LapG for in vitro biofilm formation, we hypothesized that these proteins might also play a role during colonization in vivo. We intranasally inoculated suspensions of the wild type or mutant B. bronchiseptica strains into BALB/c mice, and the infection progress was evaluated for 21 days in the nose and lungs (Fig 7).
Fig 7. B. bronchiseptica infection in a mouse model.
Comparison of B. bronchiseptica bacterial burden in the mouse nose (A) and lungs (B) were determined as described in the Materials and Methods after the indicated days post-infection. Bacteria were intranasally inoculated into external nares with an air displacement pipette (5 x 105 CFU in 50 μl). Green dots correspond to CFU from euthanized mice infected with BbΔlapG. * indicates a significant difference compared to the CFU determined in mice infected with wild type strain, p<0.01.
Mice were infected with wild type B. bronchiseptica as previously described [4]. Wild type bacteria persisted in lung and nose mice after 21 days post infection. A typical B. bronchiseptica infection was observed; lung burden increased during the first 7 days and this microbe was gradually reduced across the remainder of the experiment. Our analysis indicates that BrtA does not contribute to bacterial burden at any stage during infection in either body site evaluated (Fig 7). For the strain lacking the lapG gene (BbΔlapG), there was a small but statistically significant increase in bacterial burden at 14 days for those mice that survived until the end of the experiment, but no significant difference was detected at any other time point (Fig 7).
Interestingly, across all experiments, 25% of BbΔlapG infected mice presented symptoms of illness (indicated by ruffled fur, labored breathing, and diminished responsiveness) and were euthanized to avoid suffering (S3 Fig). Lungs of these euthanized mice carrying the BbΔlapG strain had on average 3 x 108 CFU ml-1, a significant increase of 1–2 log over the wild type (7 x 106 CFU ml-1) and the mice carrying the BbΔlapG mutant that survived until the end of the experiment. These data indicated the possibility that loss of LapG function by B. bronchiseptica could contribute to enhanced virulence, at least in some circumstances.
Discussion
In this work we analyze the BrtA protein of B. bronchiseptica, a predicted adhesin based on its similarity in domain structure to the LapA adhesin of P. fluorescens. We confirm the recent finding of Nishikawa and colleagues [16] that BrtA participates in biofilm formation by B. bronchiseptica, and build on this recent study by identifying and characterizing homologs of the LapD and LapG proteins of P. fluorescens. In P. fluorescens, LapD and LapG participate in the regulation of cell surface localization of the LapA protein; we show that the B. bronchiseptica LapD and LapG proteins likely play a similar role for BrtA. We also demonstrate that loss of BrtA results in a mild biofilm defect on plastic, and a much more severe defect in biofilm formation on glass, a hydrophilic substrate. BrtA, while having limited sequence similarity to the LapA protein of P. fluorescens shares a number of conserved features, including T1SS and vWFA domains, and the conserved Ala-Ala motif near its N-terminus that serves as the site of proteolysis in LapA for the LapG protein of P. fluorescens.
In P. fluorescens, the localization of the LapA protein to the surface of the cell is postranscriptionally regulated by LapG. LapG is a periplasmic protease that targets the N-terminus of LapA and cleaves this adhesin at a conserved Ala-Ala motif, resulting in release of LapA into the supernatant. Loss of LapA results in a reduction in biofilm formation by P. fluorescens. The LapD protein of P. fluorescens, in response to c-di-GMP levels in the cytoplasm, regulates LapG’s ability to target LapA by sequestering LapG via a protein-protein interaction mechanism [20,26]. The studies presented here indicate that the LapD and LapG proteins of B. bronchiseptica likely play a similar role in the context of BrtA. Our in vivo and in vitro studies, including complementation assays, support the conclusion that LapG is a protease that targets the N-terminal domain of BrtA for cleavage, thereby negatively impacting biofilm formation. Similarly, our studies implicate LapD in the control of LapG in B. bronchiseptica.
An interesting observation from our SEM and fluorescence microscopy studies was that in a mutant lacking BrtA, biofilm formation was reduced but not eliminated. This observation suggested that there may be at least one other adhesin contributing to biofilm formation on abiotic surfaces. Previous reports implicated FHA and the adenylate cyclase enzyme as involved in biofilm formation by Bordetella [7,11], further work should be directed at assessing the role of these proteins (or others) in the context of a strain lacking BrtA. Interestingly, BrtA appeared to play no detectable role in disease progression in a mouse model of infection; there was no change in bacterial burden in the nose or lungs of mice for up to 21 days post-infection in the brtA mutant strain compared to the wild type. This finding is consistent with the work of Nishikawa and colleagues who showed that a brtA mutant of B. bronchiseptica showed no defect in a model of rat nasal septum and tracheal colonization. Given that BrtA plays a role in biofilm formation on abiotic surfaces, we propose that a major role for this adhesion might be in environmental persistence as the microbe transits between hosts.
A strain of B. bronchiseptica lacking LapG function, which presumably has increased cell-surface BrtA based on our in vitro studies, when infecting mice, yielded two distinct phenotypes. In 75% of the animals, infection was maintained for 21 days with a bacterial burden similar to the wild type, with the exception of a small but significant increase in bacterial numbers in lungs at day 14 post-infection. However, 25% of the mice infected with the B. bronchiseptica lapG mutant showed increased disease symptoms that resulted in the need to euthanize these animals typically between days 4–9. These data may indicate that BrtA, when present in larger levels on the cell surface can enhance virulence, at least in some animals. However, given the lack of impact upon loss of BrtA function in this infection model, it is also possible that LapG may have (at least) one other target in B. bronchiseptica. Distinguishing between these possibilities should be explored in future studies.
Overall, we have established that the LapD/LapG system of this pathogen likely functions in an analogous manner to the well-described system in P. fluorescens. In B. bronchiseptica, the target of the LapD/LapG system appears to be BrtA, a LapA-like protein that has a clear role in biofilm formation on abiotic surfaces. The role of the LapD/LapG system in a mouse model of infection is less clear—additional investigation will be required to dissect the role (if any) of BrtA and the LapD/LapG system in host-pathogen interactions.
Supporting Information
A consensus sequence among the eight repeat regions in BrtA is shown.
(TIF)
Bordetella strains were cultured on glass coverslips as described for the SEM studies. After 12 hr of incubation the coverslips were washed with sterile PBS, and the bacteria were fixed with 4% formaldehyde in PBS. The samples were observed in a Nikon Ti-U Fluorescence Microscope, and the images were obtained with a Nikon Digital Sight DS Ri1 camera using the Nis-Elements software. White bars indicate 100 μm.
(TIF)
Kaplan Meier survival curve is represented for mice infected with BbWT (blue line) or BbΔlapG (red line). The difference between the survival curves was analyzed by the log-rank test (z = 1.98; p = 0.047 with 95% confidence).
(PDF)
(DOCX)
(DOCX)
(PDF)
(DOCX)
Acknowledgments
We thank Peggy Cotter for providing plasmids pGFLIP and pTNS3. We thank Paula Giménez, Silvana Tongiani, Luciana Cayuela, Juan Guzmán and Abel Bortolameotti for their excellent technical assistance. We thank to Ulises Mancini for technical assistance on S3 Table. We acknowledge the Laboratorio de Investigaciones de Metalurgia Física (LIMF) service (Facultad de Ingeniería–UNLP) and the Servicio de Microscopía Electrónica de Barrido (CINDECA-CONICET-UNLP) for SEM analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by NIH grant R01-AI097307 and NSF grant MCB-9984521 to GAO, and International Cooperation grants (2012 and 2013) from CONICET to FS and JF, PICT 2013-0092, PÏP N° 0092 and PIP N° 0486 to FS and JF. NA was supported by CONICET doctoral fellowship.
References
- 1.Goodnow RA. Biology of Bordetella bronchiseptica. Microbiol Rev. 1980;44: 722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harvill ET, Cotter PA, Yuk MH, Miller JF. Probing the function of Bordetella bronchiseptica adenylate cyclase toxin by manipulating host immunity. Infect Immun. 1999;67: 1493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvill ET, Preston A, Cotter PA, Allen AG, Maskell DJ, Miller JF. Multiple roles for Bordetella lipopolysaccharide molecules during respiratory tract infection. Infect Immun. 2000;68: 6720–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sisti F, Fernández J, Rodríguez ME, Lagares A, Guiso N, Hozbor DF. In vitro and in vivo characterization of a Bordetella bronchiseptica mutant strain with a deep rough lipopolysaccharide structure. Infect Immun. 2002;70: 1791–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skinner JA, Reissinger A, Shen H, Yuk MH. Bordetella type III secretion and adenylate cyclase toxin synergize to drive dendritic cells into a semimature state. J Immunol. 2004;173: 1934–1940. [DOI] [PubMed] [Google Scholar]
- 6.Inatsuka CS, Julio SM, Cotter PA. Bordetella filamentous hemagglutinin plays a critical role in immunomodulation, suggesting a mechanism for host specificity. Proc Natl Acad Sci U S A. 2005;102: 18578–18583. 10.1073/pnas.0507910102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irie Y, Mattoo S, Yuk MH. The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J Bacteriol. 2004;186: 5692–5698. 10.1128/JB.186.17.5692-5698.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mishra M, Parise G, Jackson KD, Wozniak DJ, Deora R. The BvgAS signal transduction system regulates biofilm development in Bordetella. J Bacteriol. 2005;187: 1474–1484. 10.1128/JB.187.4.1474-1484.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sloan GP, Love CF, Sukumar N, Mishra M, Deora R. The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J Bacteriol. 2007;189: 8270–8276. 10.1128/JB.00785-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conover MS, Mishra M, Deora R. Extracellular DNA is essential for maintaining Bordetella biofilm integrity on abiotic surfaces and in the upper respiratory tract of mice. PLoS One. 2011;6: e16861 10.1371/journal.pone.0016861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serra DO, Conover MS, Arnal L, Sloan GP, Rodriguez ME, Yantorno OM, et al. FHA-mediated cell-substrate and cell-cell adhesions are critical for Bordetella pertussis biofilm formation on abiotic surfaces and in the mouse nose and the trachea. PLoS One. 2011;6: e28811 10.1371/journal.pone.0028811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sisti F, Ha D-G, O’Toole GA, Hozbor D, Fernández J. Cyclic-di-GMP signalling regulates motility and biofilm formation in Bordetella bronchiseptica. Microbiology. 2013;159: 869–79. 10.1099/mic.0.064345-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson TL, Conover MS, Deora R. Transcriptome profiling reveals stage-specific production and requirement of flagella during biofilm development in Bordetella bronchiseptica. PLoS One. 2012;7: e49166 10.1371/journal.pone.0049166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whiteley CG, Lee D-J. Bacterial diguanylate cyclases: structure, function and mechanism in exopolysaccharide biofilm development. Biotechnol Adv. 33: 124–41. 10.1016/j.biotechadv.2014.11.010 [DOI] [PubMed] [Google Scholar]
- 15.Ryan RP. Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiology. 2013;159: 1286–97. 10.1099/mic.0.068189-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikawa S, Shinzawa N, Nakamura K, Ishigaki K, Abe H, Horiguchi Y. The bvg-repressed gene brtA, encoding biofilm-associated surface adhesin, is expressed during host infection by Bordetella bronchiseptica. Microbiol Immunol. 2016;60: 93–105. 10.1111/1348-0421.12356 [DOI] [PubMed] [Google Scholar]
- 17.Stainer DW, Scholte MJ. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol. 1970;63: 211–220. 10.1099/00221287-63-2-211 [DOI] [PubMed] [Google Scholar]
- 18.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. American Society for Microbiology (ASM); 1951;62: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newell PD, Monds RD, O’Toole GA. LapD is a bis-(3’,5')-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A. 2009;106: 3461–6. 10.1073/pnas.0808933106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newell PD, Boyd CD, Sondermann H, O’Toole GA. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 2011;9: e1000587 10.1371/journal.pbio.1000587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatterjee D, Boyd CD, O’Toole GA, Sondermann H. Structural characterization of a conserved, calcium-dependent periplasmic protease from Legionella pneumophila. J Bacteriol. 2012;194: 4415–25. 10.1128/JB.00640-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akerley BJ, Monack DM, Falkow S, Miller JF. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J Bacteriol. 1992;174: 980–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd CD, Chatterjee D, Sondermann H, O’Toole GA. LapG, required for modulating biofilm formation by Pseudomonas fluorescens Pf0-1, is a calcium-dependent protease. J Bacteriol. 2012;194: 4406–14. 10.1128/JB.00642-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakás L, Veiga MP, Soloaga A, Ostolaza H, Goñi FM. Calcium-dependent conformation of E. coli alpha-haemolysin. Implications for the mechanism of membrane insertion and lysis. Biochim Biophys Acta. 1998;1368: 225–34. [DOI] [PubMed] [Google Scholar]
- 25.Subrini O, Sotomayor-Pérez AC, Hessel A, Spiaczka-Karst J, Selwa E, Sapay N, et al. Characterization of a membrane-active peptide from the Bordetella pertussis CyaA toxin. J Biol Chem. 2013;288: 32585–32598. 10.1074/jbc.M113.508838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarro MVAS, Newell PD, Krasteva P V, Chatterjee D, Madden DR, O’Toole GA, et al. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol. 2011;9: e1000588 10.1371/journal.pbio.1000588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A consensus sequence among the eight repeat regions in BrtA is shown.
(TIF)
Bordetella strains were cultured on glass coverslips as described for the SEM studies. After 12 hr of incubation the coverslips were washed with sterile PBS, and the bacteria were fixed with 4% formaldehyde in PBS. The samples were observed in a Nikon Ti-U Fluorescence Microscope, and the images were obtained with a Nikon Digital Sight DS Ri1 camera using the Nis-Elements software. White bars indicate 100 μm.
(TIF)
Kaplan Meier survival curve is represented for mice infected with BbWT (blue line) or BbΔlapG (red line). The difference between the survival curves was analyzed by the log-rank test (z = 1.98; p = 0.047 with 95% confidence).
(PDF)
(DOCX)
(DOCX)
(PDF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.