Abstract
The cyclic di-GMP (c-di-GMP) second messenger represents a signaling system that regulates many bacterial behaviors and is of key importance for driving the lifestyle switch between motile loner cells and biofilm formers. This review provides an up-to-date compendium of c-di-GMP pathways connected to biofilm formation, biofilm-associated motilities, and other functionalities in the ubiquitous and opportunistic human pathogen Pseudomonas aeruginosa. This bacterium is frequently adopted as a model organism to study bacterial biofilm formation. Importantly, its versatility and adaptation capabilities are linked with a broad range of complex regulatory networks, including a large set of genes involved in c-di-GMP biosynthesis, degradation, and transmission.
Keywords: antibiotic resistance, biofilm, cyclic di-GMP (c-di-GMP), Pseudomonas aeruginosa (P. aeruginosa), signaling
Introduction
Bacteria can live as planktonic cells exploring aqueous environments or as a sessile biofilm community. The switch from planktonic to sessile occurs when, under propitious conditions, individual cells encounter a surface and undergo a series of dramatic physiological, metabolic, and phenotypic changes. Among these changes are the slowdown of metabolic activities and the production of an extracellular matrix, a complex mixture of exopolysaccharides, proteins, and nucleic acids (1). In the case of pathogens, the two bacterial lifestyles also differ in terms of virulence factor production and infection strategies. Although planktonic cells cause fulminant acute infections, the formation of a biofilm correlates with deep-rooted chronic infections and resistance to both phagocytosis and antimicrobial agents (2).
Cyclic di-GMP (c-di-GMP)3 is recognized as an intracellular signaling molecule coordinating the “lifestyle transition” from motility to sessility and vice versa (i.e. dispersion) (3). The correlation between high c-di-GMP concentration in the cell and biofilm formation or between low c-di-GMP levels and motility has been demonstrated in several bacteria species, e.g. Escherichia coli, Pseudomonas aeruginosa, and Salmonella enterica serovar Typhimurium (4). P. aeruginosa biofilms are estimated to contain on average 75–110 pmol of c-di-GMP per mg of total cell extract, whereas planktonic cells contain less than 30 pmol mg−1 (5). This concept is widely accepted but does not include the multiplicity of c-di-GMP transmission cascades operating during biofilm. Biofilm determinants modulated by c-di-GMP range from flagella rotation to type IV pili retraction, exopolysaccharide production, surface adhesin expression, antimicrobial resistance and other stress responses, secondary metabolite production, and biofilm dispersion (3). How do we reconcile the global effect of the intracellular c-di-GMP concentration on stimulating the biofilm lifestyle with the discrete actions of c-di-GMP on biofilm formation? Biofilm formation is considered as a developmental process that includes attachment to and movement on the surface, formation of microcolonies, maturation, and ultimately dispersal (1, 6, 7). It is proposed that cells use c-di-GMP as a checkpoint to proceed through the distinct stages of biofilm development until they fully commit to the biofilm lifestyle, although they may still be offered the choice to revert the decision at any time (3, 8).
The c-di-GMP Metabolism
The levels of c-di-GMP in the cell are modified by the rate of its synthesis and degradation. The molecule is synthesized from two molecules of GTP by enzymes called diguanylate cyclases (DGCs) and is degraded into 5′-phosphoguanylyl-(3′-5′)-guanosine (pGpG) and/or GMP by phosphodiesterases (PDEs) (Fig. 1A). Using bioinformatics, biochemical, and structural approaches, the catalytic domains of DGCs and PDEs have been identified and characterized: the former carrying a GGDEF active site motif, and the latter carrying either EAL or HD-GYP domains (9, 10). These domains can stand alone in a protein or can be present in association with receiver or transmission domains, suggesting a modulation of their enzymatic activity in response to external/internal signals, whereas several have multiple hydrophobic segments, suggesting membrane localization (Fig. 1B). This indicates a possible post-translational regulation of DGCs and PDEs that may segregate their activity temporally or spatially. Moreover, GGDEF and EAL domains can both be present in the same protein. In these so-called “hybrid” proteins, either only one of the two domains is catalytically active, the other having acquired a regulatory function, or a third regulatory domain is present, probably disjoining the activity of the GGDEF and EAL domains (11, 12). Recently, examples of proteins with dual DGC and PDE activities have been described, shedding some light on this “biochemical conundrum” (13–15). In P. aeruginosa, the GGDEF and the EAL domains of MucR are activated differently so that in planktonic cells, MucR functions as a DGC and as a positive regulator of alginate biosynthesis, whereas in biofilms, it functions as a PDE and is a positive regulator of biofilm dispersal induced by nitric oxide or glutamate (16).
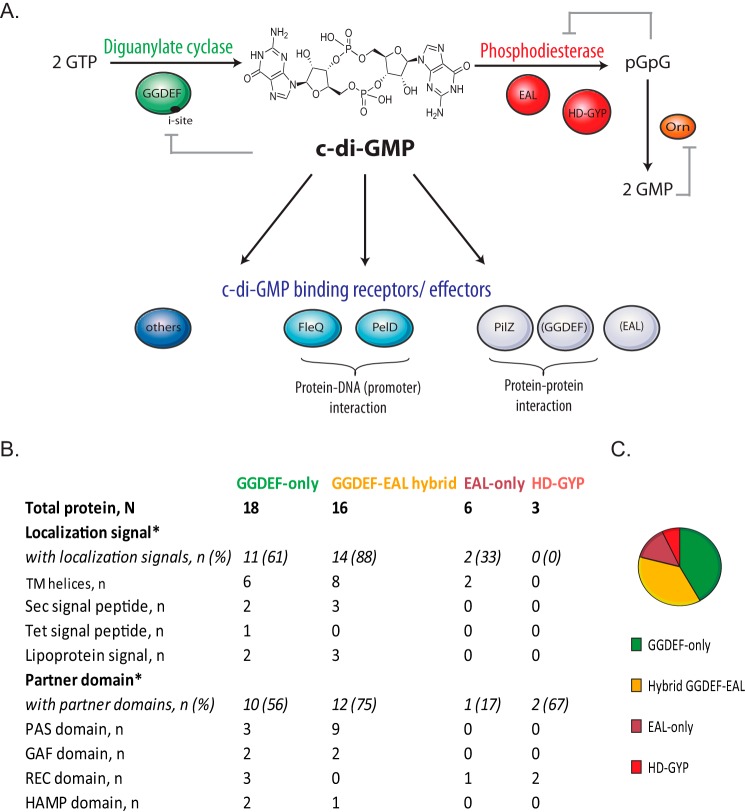
FIGURE 1.
Molecular basis of c-di-GMP signaling in P. aeruginosa. A, c-di-GMP is synthesized by diguanylate cyclases (green) that carry GGDEF domains and degraded by phosphodiesterases (red) that carry either EAL or HD-GYP domains. EAL phosphodiesterases linearize c-di-GMP into pGpG, which is successively hydrolyzed into 2 GMP molecules primarily by the oligoribonuclease Orn (orange) (34, 35). HD-GYP-phosphodiesterases are proposed to perform both steps of the c-di-GMP degradation process (31). Feedback inhibition mechanisms are illustrated by gray lines. In the cell, c-di-GMP regulates cellular processes at different levels (transcriptional, post-transcriptional, and post-translational). The diversity of c-di-GMP-binding receptors and effectors (blue) is the key of the c-di-GMP pleiotropic mechanisms. B, spatial localization signals and partner domain occurrence for GGDEF, EAL, and HD-GYP proteins of P. aeruginosa. Table based on the work of Seshasayee et al. (17) *: The sets of proteins corresponding to each of the category are not mutually exclusive. Organization of classes is in agreement as described previously (17). TM helices, transmembrane helices. C, pie chart illustrating numerical proportion of GGDEF, EAL, and HD-GYP proteins in P. aeruginosa.
Large-scale genome sequencing led to the discovery that GGDEF- and EAL-containing proteins are nearly ubiquitous in the bacterial kingdom and that bacterial genomes contain multiple copies of genes encoding GGDEF, EAL, or HD-GYP domain-containing proteins (17). A census of all the GGDEF, EAL, and HD-GYP domains in bacterial genomes is available at http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html (18). The abundance of DGCs and PDEs in a genome may be correlated to the number of complex cellular functions linked with c-di-GMP signaling and to the diversity of possible signals coordinating these functions. The P. aeruginosa genome encodes one of the highest numbers of DGCs and PDEs: 18 GGDEF, 5 EAL, 16 GGDEF/EAL, and 3 HD-GYP predicted proteins (supplemental Table S1).
DGCs: GGDEF Domain Proteins
DGCs function as homodimers. The GGDEF catalytic site is placed at the dimer interface and is involved in the binding of two molecules of GTP and in their conversion into c-di-GMP, with Mg2+ as cofactor. Five amino acids upstream of the GGDEF active site is the inhibitory site (I-site) RXXD, where the feedback inhibition of the cyclase activity occurs. Binding of c-di-GMP at the I-site prevents the formation of enzymatically active DGC dimers (19). The first experimental demonstration of a DGC activity comes from the work on PleD, a response regulator in Caulobacter crescentus (20). Nowadays the PleD activity is well defined together with its receiver (REC) domain and the phosphorylation-induced dimerization. In P. aeruginosa, the first biochemical characterization of a DGC stems from the work on WspR, which contains a REC-GGDEF domain organization (supplemental Table S1). The DGC was named after its regulatory role on the P. aeruginosa wrinkly spreader phenotype that is correlated with a thick biofilm due to an increased production of exopolysaccharides (21). The control of WspR activity occurs by three different routes that are proposed to occur sub-sequentially. First, upon sensing growth on the surface, the Wsp signal transduction complex phosphorylates WspR and triggers c-di-GMP synthesis (21, 22). In turn, the WspR phosphorylation triggers subcellular WspR oligomerization and cluster formation, which further increases the DGC activity (23). Finally, the feedback inhibition of WspR activity occurs by c-di-GMP binding at the I-site (24). The mechanisms of WspR regulation are supported by structural studies, which revealed that, in solution, the protein can exist in three stable forms: a globular dimer (active), a tetramer (more active), and an elongated dimer (less active due to c-di-GMP binding) (25, 26).
PDEs: EAL or HD-GYP Domain Proteins
The EAL domain hydrolyzes c-di-GMP into linear pGpG (Fig. 1). Contrary to DGCs, the EAL activity of PDEs seems to be independent of protein oligomerization, whereas it is dependent on binding metal ions (requiring Mg2+ or Mn2+ and inhibited by Ca2+ and Zn2+) (27). The glutamate residue (E) in the EAL signature motif is essential, whereas a change of the alanine residue (A) into tyrosine or valine (ETL and EVL) still sustains the enzymatic activity. In P. aeruginosa, the CheY-EAL domain protein RocR was identified as a response regulator in the RocSAR signaling system (28). This system is composed of a membrane sensor RocS1 and two response regulators, RocA1 and RocR. RocR activity is triggered by phosphorylation at the CheY domain, and the protein competes with RocA1 for the phosphoryl transfer from the RocS1 sensor. Overall, the Roc system regulates biofilm formation and virulence genes expression (cup fimbriae gene clusters and type III secretion system genes) (28, 29).
HD-GYP domain-containing proteins belong to the HD superfamily of metal-dependent phosphohydrolases (11). This enzyme hydrolyzes c-di-GMP in a two-step reaction, producing as a final product two molecules of GMP (Fig. 1). Contrary to GGDEF and EAL proteins, this class of enzyme is not ubiquitous in bacteria, but still widely distributed (18). The first biochemical studies on HD-GYP proteins were conducted on the RpfG PDE from Xanthomonas campestris (30). In P. aeruginosa, two of the three HD-GYP proteins (PA4108, PA4781, and PA2572) were shown to have a PDE activity in vivo and in vitro (supplemental Table S1) (31, 32). The structure of PA4781 has been resolved, showing that PA4781 preferentially binds to pGpG over c-di-GMP, and the low rate in hydrolyzing c-di-GMP brought into question its primary work as a genuine PDE (33). Interestingly, pGpG is also a signaling molecule, and it is proposed as a possible alternative to c-di-GMP in certain conditions (3, 31). Finally, the 3′-5′exoribonuclease Orn has been identified in P. aeruginosa as primarily responsible for the pGpG cleavage into two GMP molecules (34, 35).
Discrete Role of DGCs and PDEs on P. aeruginosa Biofilm Formation and during Infection
Besides WspR and RocR, described previously, other DGCs and PDEs have been reported as key players in P. aeruginosa biofilm formation. Careful examination of dgc and pde mutant phenotypes, combined with epistasis analysis, pointed at specific features about the role of, for example, SadC and RoeA (DGC) or BifA (PDE) (supplemental Table S1). This resulted in a more global understanding of their relative importance at different stages of the biofilm development process (36, 37). In Fig. 2, we illustrate this concept by including all the P. aeruginosa DGCs and PDEs that have been in one way or another associated with biofilm formation. At least five DGCs have been described to specifically control the transition from planktonic to surface-associated growth: WspR, SadC, RoeA, SiaD, and YfiN/TpbB (21, 36–39). Instead, the GcbA and NicD DGCs or the DipA (Pch), RbdA, and NbdA PDEs have been linked to biofilm dispersal (5, 40–44).
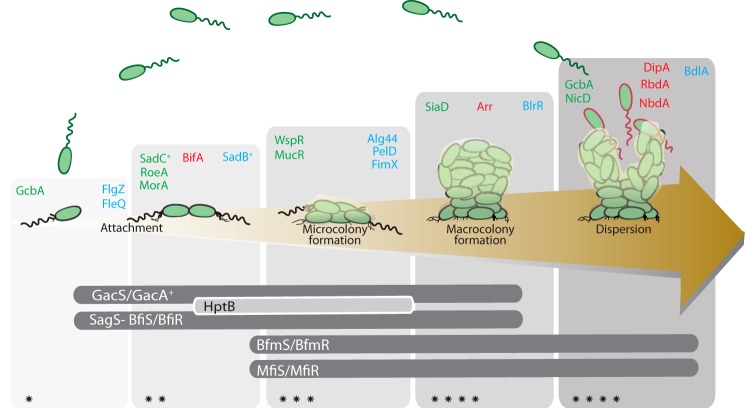
FIGURE 2.
Coordinated action of c-di-GMP signaling pathways and two-component system cascades in the control of P. aeruginosa biofilm development. In the laboratory, biofilm formation is shown to be a cyclic process that initiates with attachment to the surface of planktonic bacteria (first reversible and then irreversible). A bacteria microcolony is subsequently formed, which evolves into a mature mushroom-shaped macrocolony until the biofilm-associated cells disperse to resume again a planktonic lifestyle. Planktonic, biofilm, and dispersed cells possess distinct physiological stages (green, black, and red outline, respectively) (1, 7). The upper panel illustrates DGC (green), PDE (red), and c-di-GMP receptors/effectors (blue) and the developmental stage in which they are proposed to act. Specific references to each DGC/PDE/effector are available in supplemental Tables S1 and S2. The lower panel illustrates biofilm stage-specific two-component regulatory systems (45). The gradient of the gray panels in the background of the figure indicates increasing intracellular c-di-GMP levels (also indicated with *, **, ***, and ****).
The sequential intervention of these enzymes reveals that c-di-GMP pathways are well coordinated, organized, insulated, and tuned by global regulatory networks (45). These networks repress or activate distinct c-di-GMP pathways in a defined temporal window. In P. aeruginosa, this concept is supported through several examples such as the connection between c-di-GMP signaling and the Gac/Rsm cascade for the control of biofilm formation (Fig. 3), between c-di-GMP signaling and the SagS pathway for the regulation of biofilm antimicrobial resistance, or between c-di-GMP signaling and the Las-mediated quorum-sensing system for the control of biofilm formation and collective motilities (44, 46–48).
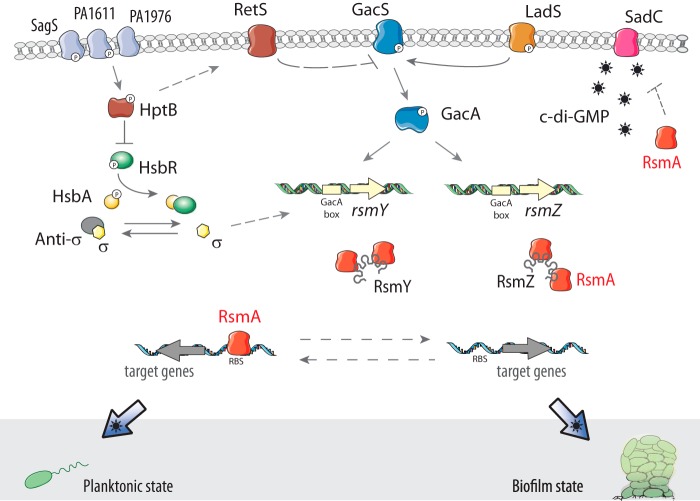
FIGURE 3.
The Gac/Rsm cascade in P. aeruginosa is genetically linked to c-di-GMP through SadC. The GacS/GacA two-component system is promoting the expression of two small regulatory RNAs, RsmY and RsmZ, which sequester the translational repressor RsmA. Titration of RsmA induces the production of sessile and biofilm determinants, whereas free RsmA leads to a planktonic and more virulent lifestyle (45, 99). Several additional regulators modulate the Gac/Rsm system, such as the two hybrid sensors RetS and LadS, as well as the histidine phosphotransfer protein HptB and other pathways. The elevated concentration of c-di-GMP in a hyperbiofilm-forming retS mutant was the first hint of the link between the Gac/Rsm and the c-di-GMP pathways (100). Later on, the molecular details of the link were elucidated: SadC, a DGC whose production is repressed by RsmA, is a central player for the Gac/Rsm regulation of biofilm formation (46). It appears therefore evident that the c-di-GMP signaling network and the Gsc/Rsm cascade are not independent to each other and that they are both instrumental for a proper development of the biofilm.
P. aeruginosa is predominant in chronic infection of cystic fibrosis patients, where the bacterium persists for many years, creating life-threatening lung damage. Over the course of long-term infections, P. aeruginosa undergoes extensive genetic and phenotypic adaptation to the lung environment, resulting in a less virulent state with increased production of biofilm (49). A consequence of the P. aeruginosa adaptation to the lungs is its phenotypic heterogeneity, e.g. the mucoid or the small colony variant (SCV) phenotype (50). In general, SCV colonies appear small, slow growing, and more resistant to several classes of antibiotics, with an increased production of exopolysaccharides and high c-di-GMP levels (50, 51). The c-di-GMP signaling has been proposed to be instrumental for SCV formation because overexpression/activation of DGC such as WspR or YfiN (TbpB) induces the SCV phenotype, whereas mutations in the wsp and yfi systems were identified in SCVs isolated from cystic fibrosis patients. YfiN is a membrane-anchored DGC, which up-regulates the pel and psl exopolysaccharide operons (39), whereas its activity is repressed by the YfiR periplasmic protein (52). YfiB is an outer membrane lipoprotein and an antagonist of YfiR (53). Finally, exposure to sub-inhibitory concentration of antibiotic triggers SCVs formation (54, 55), and in the case of kanamycin, this effect is linked to c-di-GMP via the PvrR PDE (55).
Molecular Mechanisms of c-di-GMP Regulation
The regulation of cellular functions by c-di-GMP occurs at multiple levels, including (i) allosteric regulation of an enzyme activity or protein function, (ii) regulation of gene expression through modulation of a transcription factor, and (iii) regulation of gene expression by direct interaction with noncoding RNA molecules (riboswitches). The molecular bricks by which c-di-GMP builds these regulatory connections are constituted by an array of different c-di-GMP-binding receptors or c-di-GMP effector molecules. We define here c-di-GMP receptors as those molecules that detect c-di-GMP levels in the cell and consequently translate the information into the activation of a specific cellular response/signaling pathway. Instead, c-di-GMP effectors are defined as proteins whose activity changes allosterically upon c-di-GMP binding and consequently regulate a defined interacting target protein. A list of identified c-di-GMP receptors/effectors in P. aeruginosa is presented in supplemental Table S2. Among the known c-di-GMP-binding motifs, we include inactive GGDEF, EAL, HD-GYP domains, PilZ domains, and other less characterized examples (11, 56).
In P. aeruginosa, PelD is a c-di-GMP receptor whose expression and binding to c-di-GMP are required for Pel polysaccharide production (57). PelD is an inner membrane protein with a GAF domain and a degenerated GGDEF domain with a conserved I-site (supplemental Table S2). The binding of c-di-GMP to PelD occurs at the I-site (57). How the binding stimulates Pel production and/or secretion remains unclear. One can speculate that the c-di-GMP-bound form of PelD interacts with the Pel machinery in a way that induces conformational changes which stimulate exopolysaccharide transport (58, 59).
PilZ domains contain two conserved motifs: an RXXXR motif with two conserved arginine residues surrounding one of the c-di-GMP guanine and a DXSXXG motif that surrounds the other guanine (60). Alg44 is a membrane-associated protein with a cytoplasmic PilZ domain. This protein binds c-di-GMP and is required for P. aeruginosa alginate production (61, 62).
Although inactive DGCs, PDEs, and PilZ domains can be recognized in silico, other effectors are challenging to identify using bioinformatics prediction. A number of transcriptional regulators have been identified as c-di-GMP receptors. In P. aeruginosa, FleQ is an enhancer-binding protein that at low levels of c-di-GMP is the master activator of flagellar gene expression (63). Homologs of FleQ are present in all Pseudomonas species and in many flagellated gamma-proteobacteria (64). FleQ does not possess a PilZ domain, but c-di-GMP competitively inhibits FleQ ATPase activity by interacting with the ATP-binding site (65). At high levels of intracellular c-di-GMP, the binding of the molecule to FleQ converts its function as a repressor of the pel, psl, and cdr genes, involved in production of exopolysaccharides and adhesins, into an activator (66). Another c-di-GMP-responsive transcriptional regulator of P. aeruginosa is BrlR (67). BrlR participates in the resistance of biofilm cells to antimicrobial agents by increasing the expression of genes encoding multidrug efflux pumps (68, 69). Interestingly, BrlR has a stronger binding affinity for c-di-GMP than FleQ (as characterized by a Kd of 2.2 μm and of 15–20 μm respectively; supplemental Table S2), which suggests that BrlR activation occurs at lower c-di-GMP levels and at earlier stages in the biofilm development process as compared with FleQ (67). In general, determination of the affinity constants of the different receptors or effectors for c-di-GMP can be considered as useful information to determine at which global levels of c-di-GMP they are activated and by extension within which physiological window they act. Finally, c-di-GMP could also act as a competitive inhibitor for certain enzymes capable of catabolizing ATP, such as the FliI flagellar ATPase (70).
The hunt for identifying new c-di-GMP-binding proteins is ongoing, and both a priori and a posteriori (or targeted) approaches are being employed. A priori approaches are based on affinity pulldown assays using c-di-GMP-conjugated Sepharose resin, biotin, or a tripartite c-di-GMP capture compound to enrich c-di-GMP-binding proteins from whole cell lysates (71–73). The differential radial capillary action of ligand assay (DRaCALA) is also used to systematically screen protein expression libraries for their c-di-GMP binding activity(74). Alternatively, the a posteriori approaches are “educated guesses,” in which gene products functionally associated with c-di-GMP-regulated processes are tested for c-di-GMP binding via several biochemical assays, among them DRaCALA, isothermal titration calorimetry, and a peptide array approach (74–76).
The Specificity of c-di-GMP Signaling
A pioneering analysis of all GGDEF and EAL domain-containing proteins from two P. aeruginosa strains (PAO1 and PA14), using transposon mutant libraries or strains overexpressing dgc/pde genes, revealed that DGCs or PDEs are not redundant and have a different impact on biofilm formation or cytotoxicity (77). Several plausible explanations are proposed for the partial loss or gain of a specific phenotype when deleting a dgc or a pde gene. One is that DGCs and PDEs are differentially controlled at the level of gene expression or enzyme activity and therefore could have a distinct impact on the global pool of c-di-GMP. Another is related to the degree of c-di-GMP signaling specificity and the existence of local c-di-GMP pools in the cell.
c-di-GMP is a small molecule and presumably diffuses freely in the bacterial cytoplasm. In such a context, all DGCs and PDEs may affect the pool of c-di-GMP uniformly throughout the cell. The degree of c-di-GMP-mediated responses is then possibly determined by the binding affinity of c-di-GMP for different effectors, which in turn leads to various outputs and phenotypes.
The low specificity model does not clash with the idea of a temporal sequestration of DGCs and PDEs. Temporal sequestration is reached by modulation of dgc or pde gene expression at a defined time period, in response to environmental or cellular alterations through functional association to specific regulatory networks. In P. aeruginosa, for example, a case can be made for the repression of SadC by the Gac/Rsm cascade (46), the nutrient-induced activation of the NicD/BdlA/DipA cascade (5), or the presence of Wsp and Yfi multi-protein complexes that control WspR and YfiN DGCs activity, respectively (21, 39, 53).
An alternative hypothesis that may result in highly specific signaling is that each individual DGC and PDE regulates only a subset of c-di-GMP-regulated behaviors. The way this may be achieved is via molecular mechanisms that sequester the signal (c-di-GMP pool) in multi-protein complexes or at distinct cellular sites. An example is the PleD polar sequestration during cell division in C. crescentus (20), the YcgR flagellar motor control in E. coli and Salmonella (78, 79), the PilZ-FimXEAL-c-di-GMP complex of Xanthomonas citri (80), the c-di-GMP dependent localization mechanism of LapA in Pseudomonas fluorescens (81), or the WspR subcellular clustering in P. aeruginosa (23). Interesting lessons on signaling molecule compartmentalization can be taken from cAMP signaling studies in eukaryotes, where the creation of cAMP compartments is achieved mainly by localization of PDEs (82).
It becomes obvious that understanding regulatory mechanisms of DGCs and PDEs is not as simple as measuring global c-di-GMP levels in the cell, and c-di-GMP-dependent control involves highly complex and tightly regulated signaling systems. Low and high signaling specificity could not be mutually exclusive. In the context of c-di-GMP regulation of localized structural machineries, such as flagella or type IV pili, it is reasonable to think that the maintenance of a local c-di-GMP pool would guarantee a more rapid and efficient control of their activity (78–80). Instead, for the overall development of a biofilm, the global c-di-GMP pool may guarantee coordination and cross-talking between multiple pathways (Fig. 2).
Emerging Challenges in c-di-GMP Signaling Research
Novel and original observations on c-di-GMP signaling in P. aeruginosa have recently emerged and have raised new fundamental and challenging questions.
Heterogeneity of c-di-GMP Levels in Individual Cells
A FRET-based biosensor has been recently constructed and an asymmetrical distribution of c-di-GMP was observed during P. aeruginosa and C. crescentus cell division (83). The concept of a bimodal distribution of c-di-GMP in C. crescentus was not surprising, given its asymmetric cell cycle and the PleD/TipF/PopA localization and activity (8). In the case of P. aeruginosa, this observation was more unexpected, as the bacterium produces morphologically similar progeny. Along this line, the same group showed that a specific PDE (named Pch and previously identified as DipA) modulates motility by localizing at the flagellated cell pole. The enzyme is thus asymmetrically partitioned upon cell division to generate c-di-GMP heterogeneity (84). Phenotypic heterogeneity in a population of genetically identical cells has been demonstrated in many bacterial species, particularly for biofilm-forming bacteria. An example is the bistable expression of the biofilm master regulator CsgD in Salmonella (85), with CsgD connected to a complex c-di-GMP-dependent regulatory network. Therefore, c-di-GMP might be instrumental for survival and persistence within a changing environment by creating a phenotypic heterogeneous clonal population.
Cross-talk between Second Messengers
Although c-di-GMP is the second messenger associated with biofilm and chronic infection, cAMP has been shown as being a hallmark for P. aeruginosa virulence (i.e. acute infection) (86). The dichotomy between these two second messengers is suggested by the observation that increasing c-di-GMP levels, via activation of WspR and YfiN, consequently decreases cAMP levels via an unknown mechanism (87). Interestingly, in the biofilm state, cAMP and c-di-GMP are observed to be spatially organized. Indeed, bacterial cells carrying a cAMP reporter display only little activity in flow chamber-grown biofilm except for cells in the outer layer, whereas a c-di-GMP reporter is overall more active, especially at the bottom of the biofilm and in the middle of microcolonies. Further evidence of a connection between cAMP and c-di-GMP is given by the cAMP-dependent regulation of the minor pilin gene pilY1, which seems to activate a signaling cascade causing the increase of c-di-GMP levels during P. aeruginosa transition from reversible to irreversible attachment (88). This cross-talk concept is likely to be further expanded and might involve other small molecules such c-di-AMP or ppGpp (89). A P. aeruginosa strain lacking (p)ppGpp is sensitive to multiple classes of antibiotics and is defective in biofilm formation (90). The connection between c-di-GMP and (p)ppGpp has been recently proposed in Mycobacterium smegmatis, where both signaling molecules may be involved in the metabolism of glycopeptidolipids and polar lipids, leading to an increase of the bacterium antibiotic resistance (91).
c-di-GMP Regulation of Antimicrobial Resistance
Cells in a biofilm can be up to 1000 times less susceptible to antimicrobial agents than planktonic cells (92). The reasons for the biofilm tolerance are multiple, including slow growth or the presence of an extracellular matrix (93, 94). By regulating biofilm, c-di-GMP signaling can therefore also influence the antimicrobial resistance of the bacterium. Recently, new c-di-GMP-related mechanisms have been described to contribute to P. aeruginosa antibiotic resistance, independently from biofilm formation. A pel mutant strain with high c-di-GMP levels (overexpression of the PA5487 DGC) has a higher fitness in the presence of imipenem as compared with the same strain with low c-di-GMP levels (PvrR PDE overexpression) (95). Sub-inhibitory concentrations of aminoglycosides induce biofilm formation in terms of biomass but are not linked to exopolysaccharide production. The PDE Arr has been demonstrated to be necessary for such a response (96). Finally, lowering c-di-GMP levels in P. aeruginosa by engineering a sagS deletion renders the bacterium more susceptible to antibiotics, whereas this strain is still capable of forming proper biofilms (47, 48). Furthermore, upon overexpression of the AdcA DGC, resistance to antibiotics is restored to wild type levels (48).
Overall, the possibility to fight against biofilm formation, antimicrobial resistance, and chronic infections by manipulating and subverting c-di-GMP signaling is an interesting therapeutic challenge (97, 98). The targets are multiple and give the opportunity to intervene at a global level by targeting DGCs or PDEs, or to be more clinical by aiming at specific receptors/effectors and thus inhibit specific pathways.
Final Remarks and Future Perspectives
P. aeruginosa has come to be a remarkable model organism for bacterial pathogenesis (2, 55, 93). Nowadays a wide variety of technical tools are available for researchers who intend to study this microorganism. The significant progresses that have been made in understanding c-di-GMP-regulated phenotypes in P. aeruginosa could therefore be applicable to other bacteria that are relatively less easy to manipulate in the laboratory.
Importantly, despite this progress, many questions about c-di-GMP mechanisms of action remain unanswered. The basics of c-di-GMP metabolism have been elucidated, and we understand most of the enzymology behind its synthesis and degradation. However, the detailed mechanisms through which c-di-GMP operates, and in particular the process of specific transmission, remain obscure. Identification of new c-di-GMP receptors/effectors surely helps researchers in making better connections between c-di-GMP signaling and functional output. Now, have we identified all the players and their role in the c-di-GMP contest? Surely not! In the case of the c-di-GMP regulation of exopolysaccharide production/secretion in P. aeruginosa, for example, although a good number of involved DGCs/PDEs/effectors have been identified, e.g. PelD or Alg44, how they act on the associated molecular mechanism(s) remains to be deciphered.
Supplementary Material
Acknowledgment
We thank Ronan McCarthy for critical reading of the manuscript.
This work was supported by Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/L007959/1 (to A. F.). This work was also supported by an Imperial College Antimicrobial Research Collaborative (ARC) Early Career Research Fellowship (to M. V.). This is the third article in the Thematic Minireview series “Biofilms.” The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1 and S2.
- c-di-GMP
- cyclic-di-GMP
- DGC
- diguanylate cyclase
- PDE
- phosphodiesterase
- I-site
- inhibitory site
- SCV
- small colony variant
- guanosine pGpG
- 5′-phosphoguanylyl-(3′-5′)-guanosine
- ppGpp
- guanosine tetraphosphate
- (p)ppGpp
- tetra- or pentaphosphate guanosine.
References
- 1.O'Toole G., Kaplan H. B., and Kolter R. (2000) Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L., Costerton J. W., and Stoodley P. (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 [DOI] [PubMed] [Google Scholar]
- 3.Römling U., Galperin M. Y., and Gomelsky M. (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simm R., Morr M., Kader A., Nimtz M., and Römling U. (2004) GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53, 1123–1134 [DOI] [PubMed] [Google Scholar]
- 5.Basu Roy A., and Sauer K. (2014) Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol. Microbiol. 94, 771–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauer K., Camper A. K., Ehrlich G. D., Costerton J. W., and Davies D. G. (2002) Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chua S. L., Liu Y., Yam J. K., Chen Y., Vejborg R. M., Tan B. G., Kjelleberg S., Tolker-Nielsen T., Givskov M., and Yang L. (2014) Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 5, 4462. [DOI] [PubMed] [Google Scholar]
- 8.Hengge R. (2009) Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273 [DOI] [PubMed] [Google Scholar]
- 9.Ryjenkov D. A., Tarutina M., Moskvin O. V., and Gomelsky M. (2005) Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187, 1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt A. J., Ryjenkov D. A., and Gomelsky M. (2005) The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187, 4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou S. H., and Galperin M. Y. (2016) Diversity of c-di-GMP-binding proteins and mechanisms. J. Bacteriol. 198, 32–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galperin M. Y. (2010) Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 13, 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feirer N., Xu J., Allen K. D., Koestler B. J., Bruger E. L., Waters C. M., White R. H., and Fuqua C. (2015) A Pterin-dependent signaling pathway regulates a dual-function diguanylate cyclase-phosphodiesterase controlling surface attachment in Agrobacterium tumefaciens. MBio 6, e00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarutina M., Ryjenkov D. A., and Gomelsky M. (2006) An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 281, 34751–34758 [DOI] [PubMed] [Google Scholar]
- 15.Boles B. R., and McCarter L. L. (2002) Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 184, 5946–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hay I. D., Remminghorst U., and Rehm B. H. (2009) MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 75, 1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seshasayee A. S., Fraser G. M., and Luscombe N. M. (2010) Comparative genomics of cyclic-di-GMP signalling in bacteria: post-translational regulation and catalytic activity. Nucleic Acids Res. 38, 5970–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galperin M. Y. (2005) A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen M. W., Kotaka M., Vonrhein C., Bricogne G., Rao F., Chuah M. L., Svergun D., Schneider G., Liang Z. X., and Lescar J. (2012) Structural insights into the regulatory mechanism of the response regulator RocR from Pseudomonas aeruginosa in cyclic Di-GMP signaling. J. Bacteriol. 194, 4837–4846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paul R., Weiser S., Amiot N. C., Chan C., Schirmer T., Giese B., and Jenal U. (2004) Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18, 715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickman J. W., Tifrea D. F., and Harwood C. S. (2005) A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U.S.A. 102, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Güvener Z. T., and Harwood C. S. (2007) Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66, 1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huangyutitham V., Güvener Z. T., and Harwood C. S. (2013) Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. MBio 4, e00242–00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De N., Pirruccello M., Krasteva P. V., Bae N., Raghavan R. V., and Sondermann H. (2008) Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 6, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De N., Navarro M. V., Wang Q., Krasteva P. V., and Sondermann H. (2010) Biophysical assays for protein interactions in the Wsp sensory system and biofilm formation. Methods Enzymol. 471, 161–184 [DOI] [PubMed] [Google Scholar]
- 26.Malone J. G., Williams R., Christen M., Jenal U., Spiers A. J., and Rainey P. B. (2007) The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology 153, 980–994 [DOI] [PubMed] [Google Scholar]
- 27.Tchigvintsev A., Xu X., Singer A., Chang C., Brown G., Proudfoot M., Cui H., Flick R., Anderson W. F., Joachimiak A., Galperin M. Y., Savchenko A., and Yakunin A. F. (2010) Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol. 402, 524–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulasekara H. D., Ventre I., Kulasekara B. R., Lazdunski A., Filloux A., and Lory S. (2005) A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55, 368–380 [DOI] [PubMed] [Google Scholar]
- 29.Rao F., Yang Y., Qi Y., and Liang Z. X. (2008) Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190, 3622–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryan R. P., Fouhy Y., Lucey J. F., Crossman L. C., Spiro S., He Y. W., Zhang L. H., Heeb S., Cámara M., Williams P., and Dow J. M. (2006) Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U.S.A. 103, 6712–6717 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Stelitano V., Giardina G., Paiardini A., Castiglione N., Cutruzzolà F., and Rinaldo S. (2013) C-di-GMP hydrolysis by Pseudomonas aeruginosa HD-GYP phosphodiesterases: analysis of the reaction mechanism and novel roles for pGpG. PLoS ONE 8, e74920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan R. P., Lucey J., O'Donovan K., McCarthy Y., Yang L., Tolker-Nielsen T., and Dow J. M. (2009) HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ. Microbiol. 11, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 33.Rinaldo S., Paiardini A., Stelitano V., Brunotti P., Cervoni L., Fernicola S., Protano C., Vitali M., Cutruzzolà F., and Giardina G. (2015) Structural basis of functional diversification of the HD-GYP domain revealed by the Pseudomonas aeruginosa PA4781 protein, which displays an unselective bimetallic binding site. J. Bacteriol. 197, 1525–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orr M. W., Donaldson G. P., Severin G. B., Wang J., Sintim H. O., Waters C. M., and Lee V. T. (2015) Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc. Natl. Acad. Sci. U.S.A. 112, E5048–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen D., Mechold U., Nevenzal H., Yarmiyhu Y., Randall T. E., Bay D. C., Rich J. D., Parsek M. R., Kaever V., Harrison J. J., and Banin E. (2015) Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 112, 11359–11364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernier S. P., Ha D. G., Khan W., Merritt J. H., and O'Toole G. A. (2011) Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Res. Microbiol. 162, 680–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merritt J. H., Ha D. G., Cowles K. N., Lu W., Morales D. K., Rabinowitz J., Gitai Z., and O'Toole G. A. (2010) Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. MBio 1, e00183–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klebensberger J., Birkenmaier A., Geffers R., Kjelleberg S., and Philipp B. (2009) SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa. Environ. Microbiol. 11, 3073–3086 [DOI] [PubMed] [Google Scholar]
- 39.Malone J. G., Jaeger T., Spangler C., Ritz D., Spang A., Arrieumerlou C., Kaever V., Landmann R., and Jenal U. (2010) YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 6, e1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrova O. E., Cherny K. E., and Sauer K. (2014) The Pseudomonas aeruginosa diguanylate cyclase GcbA, a homolog of P. fluorescens GcbA, promotes initial attachment to surfaces, but not biofilm formation, via regulation of motility. J. Bacteriol. 196, 2827–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy A. B., Petrova O. E., and Sauer K. (2012) The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 194, 2904–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.An S., Wu J., and Zhang L. H. (2010) Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl. Environ. Microbiol. 76, 8160–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y., Heine S., Entian M., Sauer K., and Frankenberg-Dinkel N. (2013) NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J. Bacteriol. 195, 3531–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueda A., and Wood T. K. (2009) Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 5, e1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikkelsen H., Sivaneson M., and Filloux A. (2011) Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ. Microbiol. 13, 1666–1681 [DOI] [PubMed] [Google Scholar]
- 46.Moscoso J. A., Jaeger T., Valentini M., Hui K., Jenal U., and Filloux A. (2014) The diguanylate cyclase SadC is a central player in Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 196, 4081–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta K., Marques C. N., Petrova O. E., and Sauer K. (2013) Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. J. Bacteriol. 195, 4975–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta K., Liao J., Petrova O. E., Cherny K. E., and Sauer K. (2014) Elevated levels of the second messenger c-di-GMP contribute to antimicrobial resistance of Pseudomonas aeruginosa. Mol. Microbiol. 92, 488–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marvig R. L., Sommer L. M., Molin S., and Johansen H. K. (2015) Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 47, 57–64 [DOI] [PubMed] [Google Scholar]
- 50.Häussler S., Tümmler B., Weissbrodt H., Rohde M., and Steinmetz I. (1999) Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29, 621–625 [DOI] [PubMed] [Google Scholar]
- 51.Smith E. E., Buckley D. G., Wu Z., Saenphimmachak C., Hoffman L. R., D'Argenio D. A., Miller S. I., Ramsey B. W., Speert D. P., Moskowitz S. M., Burns J. L., Kaul R., and Olson M. V. (2006) Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U.S.A. 103, 8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li S., Li T., Xu Y., Zhang Q., Zhang W., Che S., Liu R., Wang Y., and Bartlam M. (2015) Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1. Sci. Rep. 5, 16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malone J. G., Jaeger T., Manfredi P., Dötsch A., Blanka A., Bos R., Cornelis G. R., Häussler S., and Jenal U. (2012) The YfiBNR signal transduction mechanism reveals novel targets for the evolution of persistent Pseudomonas aeruginosa in cystic fibrosis airways. PLoS Pathog. 8, e1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barclay M. L., Begg E. J., Chambers S. T., Thornley P. E., Pattemore P. K., and Grimwood K. (1996) Adaptive resistance to tobramycin in Pseudomonas aeruginosa lung infection in cystic fibrosis. J. Antimicrob. Chemother. 37, 1155–1164 [DOI] [PubMed] [Google Scholar]
- 55.Drenkard E., and Ausubel F. M. (2002) Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416, 740–743 [DOI] [PubMed] [Google Scholar]
- 56.Hengge R. (2010) Cyclic-di-GMP reaches out into the bacterial RNA world. Sci. Signal. 3, pe44. [DOI] [PubMed] [Google Scholar]
- 57.Lee V. T., Matewish J. M., Kessler J. L., Hyodo M., Hayakawa Y., and Lory S. (2007) A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65, 1474–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z., Chen J. H., Hao Y., and Nair S. K. (2012) Structures of the PelD cyclic diguanylate effector involved in pellicle formation in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 287, 30191–30204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitney J. C., Colvin K. M., Marmont L. S., Robinson H., Parsek M. R., and Howell P. L. (2012) Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J. Biol. Chem. 287, 23582–23593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amikam D., and Galperin M. Y. (2006) PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22, 3–6 [DOI] [PubMed] [Google Scholar]
- 61.Merighi M., Lee V. T., Hyodo M., Hayakawa Y., and Lory S. (2007) The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65, 876–895 [DOI] [PubMed] [Google Scholar]
- 62.Oglesby L. L., Jain S., and Ohman D. E. (2008) Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology 154, 1605–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hickman J. W., and Harwood C. S. (2008) Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69, 376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baraquet C., and Harwood C. S. (2013) Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc. Natl. Acad. Sci. U.S.A. 110, 18478–18483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su T., Liu S., Wang K., Chi K., Zhu D., Wei T., Huang Y., Guo L., Hu W., Xu S., Lin Z., and Gu L. (2015) The REC domain mediated dimerization is critical for FleQ from Pseudomonas aeruginosa to function as a c-di-GMP receptor and flagella gene regulator. J. Struct. Biol. 192, 1–13 [DOI] [PubMed] [Google Scholar]
- 66.Baraquet C., Murakami K., Parsek M. R., and Harwood C. S. (2012) The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 40, 7207–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chambers J. R., Liao J., Schurr M. J., and Sauer K. (2014) BrlR from Pseudomonas aeruginosa is a c-di-GMP-responsive transcription factor. Mol. Microbiol. 92, 471–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liao J., and Sauer K. (2012) The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J. Bacteriol. 194, 4823–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liao J., Schurr M. J., and Sauer K. (2013) The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms. J. Bacteriol. 195, 3352–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trampari E., Stevenson C. E., Little R. H., Wilhelm T., Lawson D. M., and Malone J. G. (2015) Bacterial rotary export ATPases are allosterically regulated by the nucleotide second messenger cyclic-di-GMP. J. Biol. Chem. 290, 24470–24483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Düvel J., Bertinetti D., Möller S., Schwede F., Morr M., Wissing J., Radamm L., Zimmermann B., Genieser H. G., Jänsch L., Herberg F. W., and Häussler S. (2012) A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J. Microbiol. Methods 88, 229–236 [DOI] [PubMed] [Google Scholar]
- 72.Laventie B. J., Nesper J., Ahrne E., Glatter T., Schmidt A., and Jenal U. (2015) Capture compound mass spectrometry: a powerful tool to identify novel c-di-GMP effector proteins. J. Vis. Exp. 10.3791/51404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nesper J., Reinders A., Glatter T., Schmidt A., and Jenal U. (2012) A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J. Proteomics 75, 4874–4878 [DOI] [PubMed] [Google Scholar]
- 74.Roelofs K. G., Wang J., Sintim H. O., and Lee V. T. (2011) Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc. Natl. Acad. Sci. U.S.A. 108, 15528–15533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Düvel J., Bense S., Möller S., Bertinetti D., Schwede F., Morr M., Eckweiler D., Genieser H. G., Jänsch L., Herberg F. W., Frank R., and Häussler S. (2016) Application of synthetic peptide arrays to uncover c-di-GMP binding motifs. J. Bacteriol. 198, 138–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whitney J. C., Whitfield G. B., Marmont L. S., Yip P., Neculai A. M., Lobsanov Y. D., Robinson H., Ohman D. E., and Howell P. L. (2015) Dimeric c-di-GMP is required for post-translational regulation of alginate production in Pseudomonas aeruginosa. J. Biol. Chem. 290, 12451–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kulasakara H., Lee V., Brencic A., Liberati N., Urbach J., Miyata S., Lee D. G., Neely A. N., Hyodo M., Hayakawa Y., Ausubel F. M., and Lory S. (2006) Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U.S.A. 103, 2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boehm A., Kaiser M., Li H., Spangler C., Kasper C. A., Ackermann M., Kaever V., Sourjik V., Roth V., and Jenal U. (2010) Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141, 107–116 [DOI] [PubMed] [Google Scholar]
- 79.Paul K., Nieto V., Carlquist W. C., Blair D. F., and Harshey R. M. (2010) The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38, 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guzzo C. R., Dunger G., Salinas R. K., and Farah C. S. (2013) Structure of the PilZ-FimXEAL-c-di-GMP complex responsible for the regulation of bacterial type IV pilus biogenesis. J. Mol. Biol. 425, 2174–2197 [DOI] [PubMed] [Google Scholar]
- 81.Newell P. D., Yoshioka S., Hvorecny K. L., Monds R. D., and O'Toole G. A. (2011) Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0–1. J. Bacteriol. 193, 4685–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baillie G. S. (2009) Compartmentalized signalling: spatial regulation of cAMP by the action of compartmentalized phosphodiesterases. FEBS J. 276, 1790–1799 [DOI] [PubMed] [Google Scholar]
- 83.Christen M., Kulasekara H. D., Christen B., Kulasekara B. R., Hoffman L. R., and Miller S. I. (2010) Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328, 1295–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kulasekara B. R., Kamischke C., Kulasekara H. D., Christen M., Wiggins P. A., and Miller S. I. (2013) c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. Elife 2, e01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grantcharova N., Peters V., Monteiro C., Zakikhany K., and Römling U. (2010) Bistable expression of CsgD in biofilm development of Salmonella enterica serovar typhimurium. J. Bacteriol. 192, 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolfgang M. C., Lee V. T., Gilmore M. E., and Lory S. (2003) Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4, 253–263 [DOI] [PubMed] [Google Scholar]
- 87.Almblad H., Harrison J. J., Rybtke M., Groizeleau J., Givskov M., Parsek M. R., and Tolker-Nielsen T. (2015) The cyclic AMP-Vfr signaling pathway in Pseudomonas aeruginosa is inhibited by cyclic Di-GMP. J. Bacteriol. 197, 2190–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luo Y., Zhao K., Baker A. E., Kuchma S. L., Coggan K. A., Wolfgang M. C., Wong G. C., and O'Toole G. A. (2015) A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. MBio 6, e02456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kalia D., Merey G., Nakayama S., Zheng Y., Zhou J., Luo Y., Guo M., Roembke B. T., and Sintim H. O. (2013) Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42, 305–341 [DOI] [PubMed] [Google Scholar]
- 90.Nguyen D., Joshi-Datar A., Lepine F., Bauerle E., Olakanmi O., Beer K., McKay G., Siehnel R., Schafhauser J., Wang Y., Britigan B. E., and Singh P. K. (2011) Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gupta K. R., Kasetty S., and Chatterji D. (2015) Novel functions of (p)ppGpp and cyclic di-GMP in mycobacterial physiology revealed by phenotype microarray analysis of wild-type and isogenic strains of Mycobacterium smegmatis. Appl. Environ. Microbiol. 81, 2571–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nickel J. C., Ruseska I., Wright J. B., and Costerton J. W. (1985) Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob. Agents Chemother. 27, 619–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Drenkard E. (2003) Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5, 1213–1219 [DOI] [PubMed] [Google Scholar]
- 94.Mah T. F., and O'Toole G. A. (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39 [DOI] [PubMed] [Google Scholar]
- 95.Nicastro G. G., Kaihami G. H., Pereira T. O., Meireles D. A., Groleau M. C., Déziel E., and Baldini R. L. (2014) Cyclic-di-GMP levels affect Pseudomonas aeruginosa fitness in the presence of imipenem. Environ. Microbiol. 16, 1321–1333 [PubMed] [Google Scholar]
- 96.Hoffman L. R., D'Argenio D. A., MacCoss M. J., Zhang Z., Jones R. A., and Miller S. I. (2005) Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436, 1171–1175 [DOI] [PubMed] [Google Scholar]
- 97.Christensen L. D., van Gennip M., Rybtke M. T., Wu H., Chiang W. C., Alhede M., Høiby N., Nielsen T. E., Givskov M., and Tolker-Nielsen T. (2013) Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic Di-GMP level in the bacteria. Infect. Immun. 81, 2705–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lieberman O. J., Orr M. W., Wang Y., and Lee V. T. (2014) High-throughput screening using the differential radial capillary action of ligand assay identifies ebselen as an inhibitor of diguanylate cyclases. ACS Chem. Biol. 9, 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lapouge K., Schubert M., Allain F. H., and Haas D. (2008) Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67, 241–253 [DOI] [PubMed] [Google Scholar]
- 100.Moscoso J. A., Mikkelsen H., Heeb S., Williams P., and Filloux A. (2011) The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13, 3128–3138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.