Abstract
Cohesin, a multi-subunit protein complex involved in chromosome organization, is frequently mutated or aberrantly expressed in cancer. Multiple functions of cohesin, including cell division and gene expression, highlight its potential as a novel therapeutic target. The SMC3 subunit of cohesin is acetylated (ac) during S phase to establish cohesion between replicated chromosomes. Following anaphase, ac-SMC3 is deacetylated by HDAC8. Reversal of SMC3 acetylation is imperative for recycling cohesin so that it can be reloaded in interphase for both non-mitotic and mitotic functions. We blocked deacetylation of ac-SMC3 using an HDAC8-specific inhibitor PCI-34051 in MCF7 breast cancer cells, and examined the effects on transcription of cohesin-dependent genes that respond to estrogen. HDAC8 inhibition led to accumulation of ac-SMC3 as expected, but surprisingly, had no influence on the transcription of estrogen-responsive genes that are altered by siRNA targeting of RAD21 or SMC3. Knockdown of RAD21 altered estrogen receptor α (ER) recruitment at SOX4 and IL20, and affected transcription of these genes, while HDAC8 inhibition did not. Rather, inhibition of HDAC8 delayed cell cycle progression, suppressed proliferation and induced apoptosis in a concentration-dependent manner. We conclude that HDAC8 inhibition does not change the estrogen-specific transcriptional role of cohesin in MCF7 cells, but instead, compromises cell cycle progression and cell survival. Our results argue that candidate inhibitors of cohesin function may differ in their effects depending on the cellular genotype and should be thoroughly tested for predicted effects on cohesin's mechanistic roles.
Keywords: breast cancer, cell cycle, estrogen receptor, gene expression, histone deacetylase inhibitor (HDAC inhibitor) (HDI), HDAC8, SMC3, cohesin
Introduction
The cohesin complex is a chromatin-associated multi-subunit protein comprised of two SMC (structural maintenance of chromosomes, SMC1A and SMC3)2 and two non-SMC subunits (RAD21, STAG1/2). Cohesin's canonical function establishes cohesion between replicated sister chromatids, thus ensuring their precise segregation at anaphase (1, 2). Research over the last one and a half decades has revealed that cohesin has additional cohesion-independent functions in interphase nuclei, such as chromatin organization and transcription regulation (3, 4).
Recently, cancer genome sequencing projects have revealed that genes encoding cohesin subunits are frequently mutated in several different types of cancer, with particularly high frequency in acute myeloid leukemia (AML) (5–9). Recent mouse models indicate that cohesin contributes to leukemia progression likely through controlling transcription and genome organization, rather than through chromosome separation (10–12). In breast cancer, the cohesin subunit RAD21 is overexpressed and confers poor prognosis (13, 14). We previously showed that cohesin mediates transcription of the MYC gene (15–17) and modulates the transcription of estrogen-dependent genes in MCF7 breast cancer cells (18). The emergence of cohesin as an important contributor to cancer has generated interest in the development of therapeutics that compromise cohesin function.
SMC3 acetylation and deacetylation is essential for cohesin function (19–22). The SMC3 subunit is acetylated on Lys-105 and Lys-106 by ESCO1/2 during S phase, and acetylation is reversed by the Class I HDAC, HDAC8, at telophase (22–25). Deacetylation of SMC3 is crucial for recycling of cohesin for use in subsequent cell cycles (22, 24, 26). Deardorff et al. (25) showed that mutation or depletion of HDAC8 led to the accumulation of ac-SMC3, reduced chromatin-bound cohesin, and resulted in genome-wide cohesin-mediated transcriptional dysregulation. Depletion of HDAC8 affected transcriptional regulation in HeLa cells without markedly disrupting cycling of these cells. Chemical inhibition of HDAC8 was deemed equivalent to blocking its expression using siRNA (25). In addition to ac-SMC3, acetylated p53 is a target of HDAC8. In inv(16)+ AML, chemical inhibition of HDAC8 prevented p53 deacetylation, leading to restoration of p53-induced apoptosis of leukemia-initiating cells (28).
PCI-34051 was found to be a highly selective and potent inhibitor of HDAC8 in 2008, where it was shown to induce apoptosis in T cell-derived but not solid tumor cell lines (29). Since then, many studies have used PCI-34051 in various cell lines and tissues to functionally characterize the biological role of HDAC8 (30–33). Two recent studies showed that PCI-34051 is relatively specific for HDAC8's SMC3 deacetylase activity (34, 35). Based on these findings, PCI-34051 has the potential to be a cohesin inhibitor.
Previously, we showed that cohesin is essential for normal transcription of a subset of estrogen-responsive genes in MCF7 breast cancer cells, via DNA looping and the recruitment of estrogen receptor α (ER) (15, 18). Here, we sought to determine if HDAC8 inhibition leading to accumulated ac-SMC3 blocks the estrogen-specific transcriptional role of cohesin in MCF7 breast cancer cells. We found that HDAC8 inhibition impedes cell growth, but is not functionally equivalent to RAD21 or SMC3 siRNA-based abrogation of cohesin. Our results suggest that the effectiveness of targeting cohesin's transcription regulatory function via inhibition of HDAC8 might depend on cell type.
Experimental Procedures
Cell Culture
MCF7 cells (ATCC HTB-22) of low passage number (p9-p15) were cultured in Dulbecco's Modified Eagle Medium (DMEM, Life Technologies or Sigma) supplemented with 10% fetal bovine serum (FBS) in a humidified cell culture incubator at 37 °C and 5% CO2. Hormone deprivation, 17-β-estradiol stimulation and RAD21 depletion using siRNA in MCF7 cells were carried as previously described (15, 18). To deplete SMC3 in MCF7 cells, we used the ON-TARGET plus siRNA smart pool LU-006834-00-0005 (GE Dharmacon). The non-targeting control siRNA smart pool D-001810-10-05 (GE Dharmacon) was used as a negative control. SMC3 and control siRNAs were used at final concentrations of 20 nm. Cells were reverse-transfected using Lipofectamine RNAiMAX (Life Technologies) following 24 h of hormone depletion. After transfection, cells were cultured in hormone-depleted conditions for a further 48 h prior to estradiol stimulation.
Estrogen deprivation for PCI-34051 experiments was carried out as for the siRNA transfections, except that the siRNA was replaced by 48 h PCI-34051 treatment (Cayman Chemical) and for controls we used dimethyl sulfoxide (DMSO) solvent.
RNA Isolation and Quantitative PCR
Total RNA was isolated, reverse-transcribed into cDNA, and analyzed by quantitative PCR (qRT-PCR) as described previously (15, 18). Two reference genes, GAPDH and CYCLOPHILIN, were used for normalization of gene expression. Primer sequences for expression analysis have been described previously (15, 18).
Cell Cycle Synchronization
For G1-S phase cell synchronization experiments, MCF7 cells (3 × 105 cells per well of a 6-well plate) were cultured in full growth medium and allowed to grow to 25–30% confluence. Double thymidine block was carried out as follows: 18 h incubation with 2 mm thymidine, 9 h release and then 17 h of incubation with 2 mm thymidine. Following the second block, cells were released and treated with PCI-34051 as indicated.
Ethanol Fixation for Cell Cycle Analysis
Floating and adherent cells were harvested by adding phosphate-buffered saline (PBS), 0.1% EDTA solution, and centrifuged at 2000 rpm for 5 min at 4 °C. Cell pellets were washed in PBS, 1% FBS solution, resuspended in ice-cold PBS, and fixed in 96% ethanol overnight at 4 °C.
Propidium Iodide Staining and Flow Cytometry
Fixed cells were pelleted by centrifugation at 2000 rpm for 5 min at 4 °C and washed in PBS, 1% FBS solution. Cells were pelleted, resuspended in propidium iodide (10 μg/ml) with RNase A (250 μg/ml), and incubated at 37 °C for 30 min. Samples were analyzed on a Beckman Coulter Gallios Flow Cytometer (Beckman Coulter), and cell cycle profiles were generated using the acquisition interface software. Data were analyzed using the FlowJo software (version 9.7, Tree Star).
Antibodies
Primary antibodies used are as follows: anti-RAD21 (Ab992, Abcam), ER (HC-20, sc-543, Santa Cruz Biotechnology), total-SMC3 (D47B5, Cell Signaling Technology), ac-SMC3 (Custom antibody kindly donated by Dr. Katsuhiko Shirahige, University of Tokyo, Japan), γ-tubulin (T5326, Sigma), and α-tubulin (T6199, Sigma).
Quantitative Chromatin Immunoprecipitation (ChIP)
ChIP for RAD21 and ER was performed as described previously (15, 18). Primer sequences were: SOX4 primer 1 forward: 5′-CGCTAGGAAATGACCCGAGA-3′; SOX4 primer 1 reverse: 5′-TTCAGTTTGACCGTGAACCC-3′; SOX4 primer 2 forward: 5′-ATTCCAATTTATTTCCTCCCTGT-3′; SOX4 primer 2 reverse: 5′-GCTTAAGCTCGCCAAGGATT-3′; SOX4 primer 3 forward: 5′-GGCTTGTAACGCTGGCTTAG-3′; SOX4 primer 3 reverse: 5′-CCCGTTGTTGCAATTACAGTT-3′ and IL20 primer 1 forward: 5′-CACCCAGGAGTGCCTGACTA-3′; IL20 primer 1 reverse: 5′-GCAAGACGTGATGGGCAAT-3′.
Immunoblotting
Proteins were quantified using the PierceTMBCA protein assay kit, and equal amounts of protein (20–60 μg) were loaded onto SDS-PAGE gels. Immunoblotting analyses were carried out as described previously using the LI-COR Odyssey® infrared detection system and quantified using LI-COR Image Studio software (18). Primary antibodies used were: total-SMC3 (1:1000), ac-SMC3 (1:500), ER (1:1000), and γ-tubulin (1:5000).
Immunofluorescence and Confocal Microscopy
MCF7 cells were grown on sterile coverslips (6.25 × 104 cells per well of a 24-well plate). Cells were fixed in 4% paraformaldehyde (Fisher Chemical) solution for 10 min at room temperature and permeabilized with 0.3% (for α-tubulin) or 0.5% (for total and ac-SMC3) Triton X-100 (Sigma) for 10 min at room temperature. Cells were blocked in PBS containing 10% FBS for 1–1.5 h. Primary antibodies were diluted in blocking buffer, and incubation was carried out at room temperature for 1.5 h (α-tubulin, 1:500) or overnight at 4 °C (total SMC3, 1:200 and ac-SMC3 1:100). Cells were washed with PBS and incubated with the secondary antibodies and 1 μg/ml of Hoechst 33342 at room temperature for 1 h. Cells were then washed and mounted onto microscope slides using ProLong Gold Anti-fade (Life Technology) mounting medium. Immunostained cells were visualized on a Nikon C2 confocal microscope; z-series images were taken using the NIS elements software, digitally zoomed, z-stacked, and processed using FIJI software. Secondary antibodies used were: anti-mouse Alexa Fluor 488 (1:2000, Life Tecnologies), anti-rabbit Alexa Fluor 568 (1:2000, Life Technologies).
MTT Assay
Cells were seeded in quadruplet into 96-well plates at a density of 9900 cells for days 1 and 3, 6250 cells for day 5 and 4000 cells for day 7 incubation times. The next day, cells were treated with varying concentrations of PCI-34051. On the day of the assay, 0.25 mg/ml of MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium) was added to each well, and plates were incubated at 37 °C for 3–4 h. Culture medium was then removed, and reaction was stopped with 100 μl of DMSO. Absorbance was read at 570 nm on a microtiter plate reader (BioRad).
IncuCyte and YOYO-1 Dye Assays
Cells were seeded and dosed as above. After PCI-34051 addition, plates were immediately placed in the IncuCyte FLR chamber. Using the IncuCyte FLR's 10× objective, images were acquired every 3 h. At the end of each treatment period, kinetic growth curves were generated, and cell proliferation was quantified using IncuCyte's confluence-based automated analysis software.
YOYO-1 dye assay (Life Technologies) was used to measure cell viability following PCI-34051 treatment. Cells were seeded as described above and the next day, YOYO-1 (1:10,000 dilution) was added to the culture medium, simultaneously with PCI-34051, and fluorescence was monitored in real time on the IncuCyte FLR and quantified using the IncuCyte software.
Statistical Analyses
GraphPad Prism (version 6, GraphPad software Inc.) was used to graph and analyze the data.
Results
HDAC8 Inhibition Leads to Accumulation of Acetylated SMC3 in MCF7 Cells
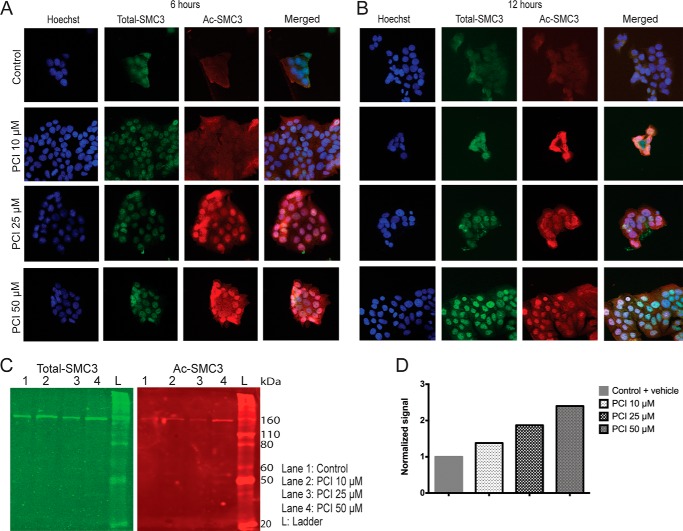
Previously, 25 μm of PCI-34051 was shown to inhibit HDAC8 and block SMC3 deacetylation in HeLa cells (25). To determine whether a similar effect occurs in MCF7 breast cancer cells, we used immunofluorescence with specific antibodies to examine the status of total SMC3 and ac-SMC3 post-PCI-34051 treatment. MCF7 cells were synchronized in the G1-S phase and treated with 10, 25, and 50 μm of PCI-34051 or vehicle control for 6 or 12 h (Fig. 1, A and B). At both time points and at all concentrations, PCI-34051 caused an accumulation of acetylated SMC3 compared with vehicle controls, whereas total SMC3 remained unchanged. Immunoblot analysis on protein lysates from asynchronized MCF7 cells treated with PCI-34051 for 48 (Fig. 1, C and D) or 72 (data not shown) hours showed a dose-dependent increase in acetylated SMC3 relative to total SMC3. Levels of acetylated SMC3 increased by ∼1.5-fold with 10 μm PCI-34015, ∼2-fold with 25 μm, and ∼2.5-fold with 50 μm. The increase in acetylated SMC3 indicates that, consistent with previous observations in HeLa cells, PCI-34051 blocks HDAC8-mediated deacetylation of SMC3.
FIGURE 1.
HDAC8 inhibitor PCI-34051 causes accumulation of acetylated (ac)-SMC3 in MCF7 cells. A and B, representative immunofluorescence analysis of total- and ac-SMC3 in G1-S synchronized MCF7 cells treated with the indicated concentrations of PCI-34051 for 6 and 12 h, respectively. MCF7 cells grown in complete medium were synchronized to G1-S phase by double thymidine block and released in the presence of different concentrations of PCI-34051 for 6 or 12 h. Cells were stained for DNA (Hoechst-H33342, blue) and with antibodies detecting total-SMC3 (green) and ac-SMC3 (red). Cells were analyzed by Nikon C2 confocal microscope; images were taken at 20× magnification (z-series) using the NIS elements software, z-stacked, and processed using FIJI. C, immunoblots of total and ac-SMC3 cells following treatment with PCI-34051. Asynchronous MCF7 cells grown in complete medium were incubated with different concentrations of PCI-34051 for 48 h. Total cell lysates were prepared, and levels of total- and ac-SMC3 were analyzed by SDS-PAGE and immunoblotting. Total-SMC3 (green) and ac-SMC3 (red) were immune-detected on the same membrane using the two-color Odyssey® infrared immunoblotting system. D, bar plot showing the quantification of ac-SMC3 relative to total-SMC3 (n = 1).
HDAC8 Inhibition Does Not Affect ER-driven Transcription of Cohesin-sensitive Genes in MCF7 Cells
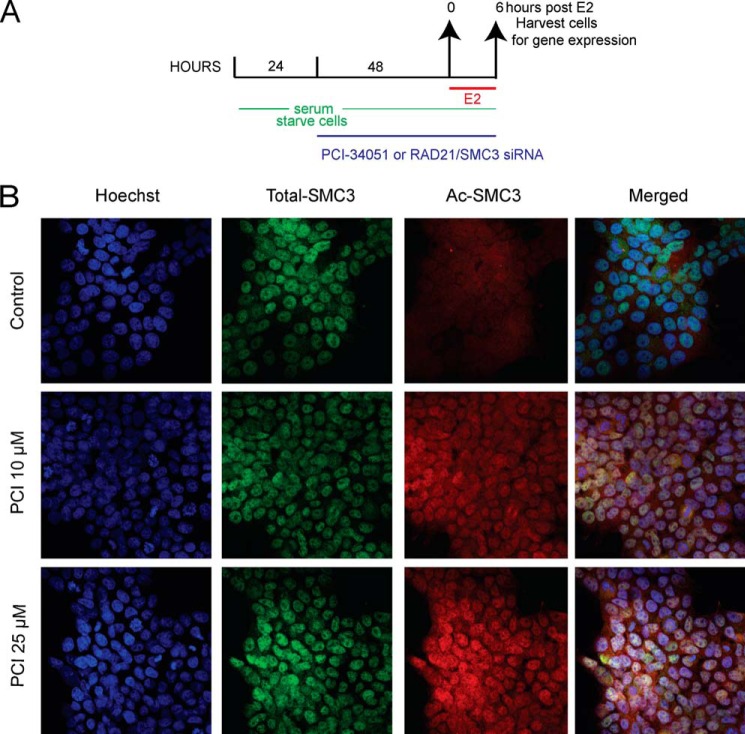
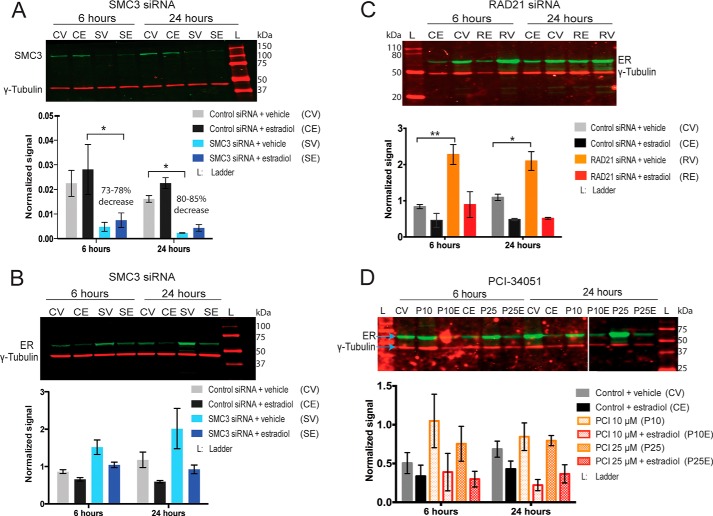
We previously had identified ER target genes that were regulated by cohesin in breast cancer cells (15, 18). To determine whether HDAC8 inhibition alters cohesin's gene regulatory function, we compared the expression of 10 ER-regulated genes following knockdown of the cohesin subunits RAD21 or SMC3 with PCI-34051 treatment, following 6 h of estradiol stimulation in MCF7 cells. Immunostaining of estradiol-starved MCF7 cells treated with 10 and 25 μm PCI-34051 confirmed robust accumulation of ac-SMC3, while no changes were visible for total SMC3 (Fig. 2). We previously described ∼70% depletion of RAD21 protein levels by siRNA (15, 18), and the same conditions were used here (data not shown). Depletion of SMC3 was confirmed by qRT-PCR (data not shown) and immunoblotting with anti-SMC3 antibody showed a decrease in protein levels by 73–78% (Fig. 3A).
FIGURE 2.
Accumulation of ac-SMC3 in estrogen-starved MCF7 cells upon HDAC8 inhibition. A, schematic of the experimental design used for immunofluorescence, transcriptional, and immunoblotting analyses of PCI-34051-treated or RAD21-depleted MCF7 cells grown in estrogen-starved medium. Prior to estradiol stimulation, cells were estrogen-starved for 3 days and were treated with RAD21 or SMC3 siRNA or PCI-34051 (10 and 25 μm) for 48 h. E2: estradiol. B, representative immunofluorescence images from immunostainings with total- and ac-SMC3 in estrogen-starved and PCI-34051-treated MCF7 cells. Estrogen-deprived MCF7 cells were treated with 10 or 25 μm of PCI-34051 for 48 h. Cells were fixed, permeabilized, and stained with Hoechst H33342 (blue, for DNA) and with antibodies against total-SMC3 (green) and ac-SMC3 (red). Cells were analyzed by Nikon C2 confocal microscope; images were taken at 20× magnification (z-series) using the NIS elements software, z-stacked, and processed using FIJI.
FIGURE 3.
Effects of cohesin depletion or HDAC8 inhibition on ER protein levels in MCF7 cells. Estrogen-starved MCF7 cells were transfected with control non-targeting or SMC3 or RAD21 siRNA, or were treated with 10 or 25 μm of PCI-34051 for 48 h, then treated with 100 nm estradiol for 6 and 24 h. A, SMC3 siRNA decreases SMC3 protein levels in MCF7 cells. B–D, ER protein levels following SMC3 siRNA (B), RAD21 siRNA (C), and PCI-34051 treatment (D) in MCF7 cells. Immunoblots showing total-SMC3 and ER were simultaneously probed for γ-tubulin, and detection was carried out using the two-color Odyssey® infrared system. Shown are representative blots of overlay of γ-tubulin (red) with either total-SMC3 or ER (green). Bar graphs represent levels of the indicated proteins normalized to γ-tubulin. Errors are the mean ± S.E. of 2–3 independent biological replicates. Significant difference for SMC3 levels was determined by paired ratio t test and for ER levels via two-way ANOVA. *, p ≤ 0.05; **, p ≤ 0.01.
Depletion of SMC3 or RAD21 increased basal levels of ER protein (Fig. 3, B and C), however this was only significant in RAD21-depleted cells. Upon estradiol stimulation in RAD21 siRNA conditions, ER protein levels returned to control conditions and no significant differences were observed. The small increase in ER protein levels upon SMC3 depletion was still apparent with estradiol stimulation but the change was not significant. Treatment with 10 and 25 μm PCI-34051 also increased basal levels of ER, although not significantly, and levels returned to control conditions upon estradiol stimulation (Fig. 3D). Therefore, levels of ER are not rate-limiting for the transcription of estrogen responsive genes.
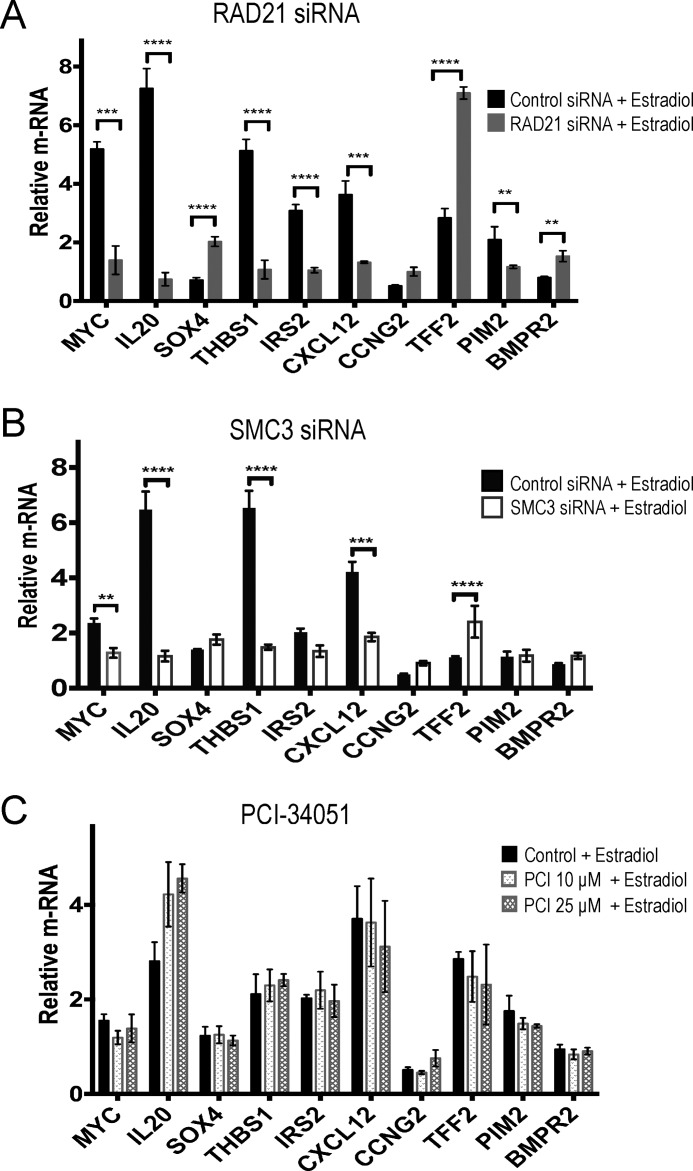
Previous microarray analysis identified MYC, IL20, SOX4, THBS1, IRS2, CXCL12, CCNG2, TFF2, PIM2, and BMPR2 as having altered expression in response to estrogen upon RAD21 depletion (18). We found that altered expression of these genes upon RAD21 knockdown was largely recapitulated by SMC3 knockdown in estradiol-stimulated MCF7 cells (Fig. 4, A and B). The results suggest that RAD21 or SMC3 depletion have similar effects on transcription, as might be expected if the cohesin complex as a whole is essential for transcription. In contrast, HDAC8 inhibition by PCI-34051 did not alter gene transcription in the presence of estradiol (Fig. 4C) despite eliciting increased levels of ac-SMC3 (Fig. 2). Together, the results show that HDAC8 inhibition by PCI-34051 in MCF7 cells does not alter ER-associated transcription, unlike depletion of cohesin subunits.
FIGURE 4.
HDAC8 inhibition does not alter regulation of cohesin-sensitive ER target genes. Estrogen-starved MCF7 cells were transfected with control non-targeting or RAD21 siRNA (A) or SMC3 siRNA (B), or were treated with 10 or 25 μm of PCI-34051 (C) for 48 h, then treated with 100 nm estradiol for 6 h. Transcript levels of the indicated genes were quantitated by qRT-PCR and are expressed relative to the mean of two reference genes, CYCLOPHILIN and GAPDH. Error bars represent the mean ± S.E. of three independent biological replicates for each treatment type. Significance was determined using two-way ANOVA. **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
HDAC8 Inhibition Does Not Significantly Alter Chromatin-bound Cohesin or ER
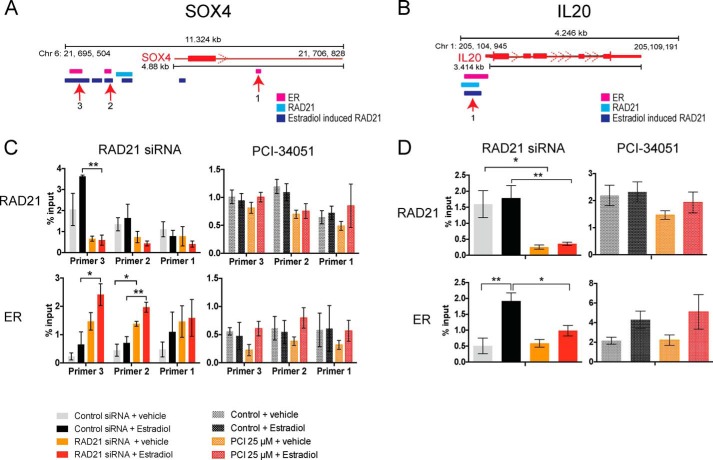
With the exception of CCNG2, the cohesin-regulated ER target genes identified above have cohesin and ER binding sites within 15 kb of the transcription start site (TSS) (15, 18), implying they can be regulated directly by cohesin and ER. We used quantitative chromatin immunoprecipitation (qChIP) to determine if HDAC8 inhibition alters cohesin and ER binding. Estrogen-starved MCF7 cells were RAD21-depleted or treated with PCI-34051 for 48 h then stimulated with estradiol for 45 min (Fig. 5). Recruitment of RAD21 and ER was analyzed at SOX4 (Fig. 5A) and IL20 (Fig. 5B), genes at which transcription increased and decreased respectively, upon RAD21 siRNA depletion.
FIGURE 5.
HDAC8 inhibition does not affect ER binding at candidate genes in MCF7 cells. Estrogen-deprived MCF7 cells were transfected with control/RAD21 siRNA or treated with 25 μm of PCI-34051 for 48 h and then treated with vehicle or 100 nm of estradiol for 45 min. Schematics of the SOX4 gene (A) and IL20 gene (B) showing RAD21, estradiol-induced RAD21, and ER binding, mapped on the UCSC genome browser (2006 NCBI36/hg18 version) using previously published ChIP-seq data in MCF7 cells. Numbers represent positions of the ChIP primers used to amplify various sites. Bar plots represent qChIP analyses at SOX4 gene (C) and at IL20 gene (D) using RAD21 and ER antibodies in RAD21 siRNA or PCI-34051-treated MCF7 cells. Binding is shown relative to input chromatin. Error bars are ±S.E. of four/three independent biological replicates for the knockdown and PCI-34051 experiments, respectively. Significance was determined using two-way ANOVA. *, p ≤ 0.05; **, p ≤ 0.01.
As expected, siRAD21 diminished RAD21 binding at cognate sites upstream of SOX4 and IL20. Depletion of RAD21 also altered ER binding. At SOX4 (where RAD21 depletion enhanced transcription), ER binding increased at two upstream locations (Fig. 5, A and B). In contrast, at IL20 (down-regulated by RAD21 depletion), ER binding decreased at a site near the TSS (Fig. 5, C and D). In contrast, HDAC8 inhibition decreased RAD21 binding only slightly (but not significantly) at specific sites upstream of SOX4 and IL20, while ER binding remained unaltered (Fig. 5, C and D). Consistent with the lack of transcriptional changes (Fig. 4), HDAC8 inhibition did not significantly reduce chromatin-bound cohesin and had no effect on ER recruitment to gene targets.
HDAC8 Inhibition Delays Cell Cycle Progression and Compromises Viability of MCF7 Cells
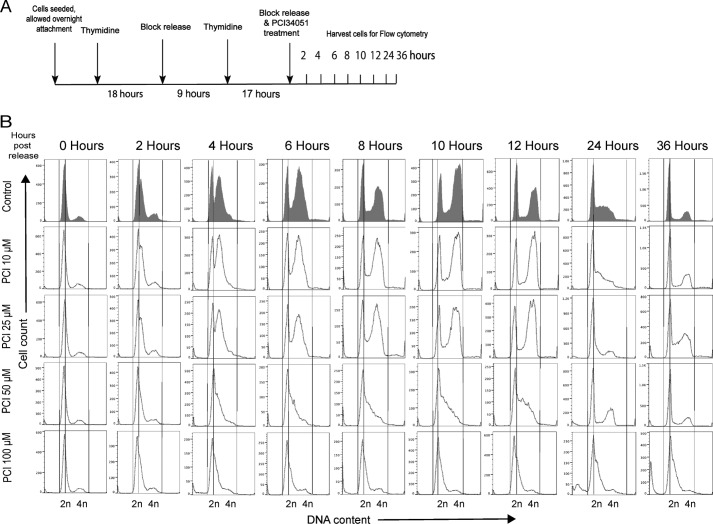
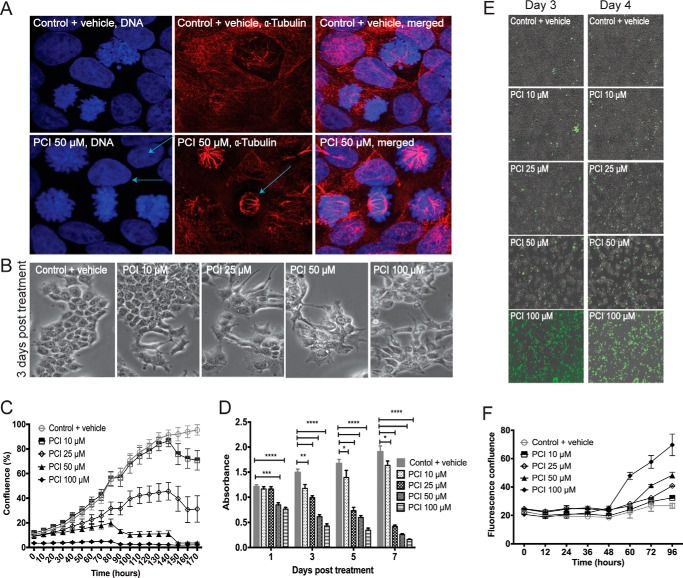
Despite the lack of effects on cohesin-mediated transcription, growth of MCF7 cells was sensitive to HDAC8 inhibition. To better understand the effects of HDAC8 inhibition on cell cycle progression, we conducted flow cytometry on synchronized cells (Fig. 6A). Control cells entered early S phase 2 h after release of the thymidine block, and traversed S phase, G2, and mitosis at 12 h as a synchronous cohort (Fig. 6B). By 24 h, cells began to re-enter the next cell cycle. At 10 μm PCI-34051, the transit time through S and G2+M was similar to the control, but there was a notable delay at 24 h as fewer cells re-entered the next cycle This delay was marked at 25 μm PCI-34051. At higher doses cells barely made it into S phase. PCI-34051 treatment caused a similar cell cycle delay in non-cancerous breast epithelial MCF10A cells and in T47D ER-positive breast cancer cells, with delay being more profound in T47D (data not shown). Thus, the main effect of PCI-34051 is to slow progression through the G1 phase of the cell cycle, with potency dependent on cell type. Mitosis was not compromised, as mitotic figures were normal as revealed by α-tubulin and DAPI staining (Fig. 7A), even at the 50 μm dose of PCI-34051.
FIGURE 6.
PCI-34051 delays cell cycle progression in a concentration-dependent manner. MCF7 cells were synchronized to G1-S phase by double thymidine block and released in the presence of different concentrations of PCI-34051 for the indicated time points. Cells were fixed, stained with propidium iodide, and cell cycle distributions were assessed by flow cytometry. Data were processed using the FlowJo software (version 9.7). A, schematic of the experiment design used for synchronizing cells to G1-S phase by double thymidine block and release. B, histograms of flow cytometry analyses showing fluorescence intensity on the x axis and cell counts on the y axis. DNA content analyses of vehicle-treated control cells and PCI-34051-treated cells are presented from 0, 2, 4, 6, 8, 10, 12, 24, and 36 h, post-treatment. Lines are shown for ease of comparison of gated cell populations between G1 and S/G2-M phases of vehicle-treated control and PCI-34051-treated cells.
FIGURE 7.
PCI-34051 reduces MCF7 cell growth and viability. A, confocal immunofluorescence analysis showing nuclear morphology and tubulin organization of PCI-34051-treated MCF7 cells. G1-S synchronized MCF7 cells were treated with 50 μm PCI-34051 for 24 h. Cells were then fixed, permeabilized, and stained with Hoechst H33342 (for DNA, blue) and for anti- α-tubulin (red). Arrows point to a metaphase cell with tubulin staining showing the mitotic spindle. B, images showing phenotype of PCI-34051-treated MCF7 cells. MCF7 cells were treated with varying concentrations of PCI-34051 (as indicated) for 3 days and visualized on an IX71 Olympus inverted microscope at 20× magnification. Images were taken using the Olympus DP71 camera and DC software, digitally zoomed and processed using Image J. C, confluence-based IncuCyte assessment of cell growth in MCF7 cells treated with varying concentrations of PCI-34051. Percentage of cell confluence is plotted on the y axis with treatment times on the x axis. Error bars are ± S.E. of two independent experiments. D, MTT assessment of proliferation of MCF7 cells treated with different concentration of PCI-34051 for various time periods. Bar graphs represent absorbance readings ± S.E. of two independent experiments. Significance was determined using two-way ANOVA. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001. E, phase contrast images of YOYO-1 fluorescent dye (green) staining in MCF7 cells 3 and 4 days post-treatment with different concentrations of PCI-34051. Images were taken on the IncuCyte FLR at ×10 magnification in real time. F, graph showing assessment of cell viability using YOYO-1 fluorescent nucleic acid dye in MCF7 treated with varying concentrations of PCI-34051. Non-viability was quantified by plotting fluorescence confluence versus time using the IncuCyte FLR object counting algorithm. Error bars represent ± S.E. of two independent experiments.
A sub-G1 population, indicative of apoptotic cell death, was observed after 24 h with the 50 and 100 μm PCI-34051 (Fig. 6B). Morphological examination showed that PCI-34051 doses of 50 μm and above resulted in alteration of the characteristic MCF7 cell shape (Fig. 7B). HDAC8 inhibition with PCI-34051 led to a dose-dependent decrease in cell confluence (Fig. 7C) and cellular metabolism as measured by MTT assay (Fig. 7D). Cells lost viability after 48 h of drug treatment (Fig. 7, E and F). Our results show that HDAC8 inhibition compromises growth and survival of MCF7 breast cancer cells.
Discussion
Cohesin shows promise as therapeutic target in cancers where it has a known role in tumor biology, such as AML (8–12). The lack of chromosome abnormalities in many cancers with cohesin alterations raises the intriguing possibility that cohesin contributes to cancer through its role in transcription (3, 6, 7). In MCF7 breast cancer cells, cohesin binds throughout the genome in combination with ER, leading to the idea that cohesin is essential for estrogen-dependent transcription (15, 18, 36, 37). In a previous study, we found that cohesin is an important transcriptional modulator of a subset of estrogen-dependent genes (18). Therefore, inhibition of cohesin in ER-positive breast cancer could have therapeutic potential through blockade of transcriptional response to estrogen.
HDAC8 was previously shown to be the specific deacetylase for SMC3 and therefore important for SMC3 recycling, which is assumed to be essential for all cohesin functions. Indeed, humans with mutations in HDAC8 fall within the spectrum of the human developmental disorder, Cornelia de Lange Syndrome (CdLS) (25). CdLS is caused by mutations in the cohesin loader NIPBL (around 50–60%) with a subset having lesions in cohesin subunits or HDAC8 (∼5%) (38, 39). The consensus is that developmental anomalies in CdLS are due to global problems with gene transcription (25, 40–42). Therefore, HDAC8 likely has a role in gene transcription, in a developmental context, through its role in deacetylation of SMC3.
In the present study, we tested the idea that HDAC8 inhibition could cause similar transcriptional changes to cohesin subunit knockdown in MCF7 cells. Surprisingly, we found that although MCF7 cells accumulated ac-SMC3 following HDAC8 inhibition, transcription in response to estrogen was unaffected. Chromatin-associated cohesin (RAD21) was only slightly reduced at two candidate gene loci, SOX4 and IL20 on a background of increased ac-SMC3. This minor decrease is consistent with the global 17% decrease in RAD21 binding in HeLa cells that have reduced HDAC8 activity (25). Furthermore, recruitment of ER remained unaltered, which contrasts with its altered binding in RAD21-depleted cells. Instead, HDAC8 inhibition delayed transit through the G1 phase of the cell cycle, consistent with a predicted defect in cohesin reloading because of accumulation of ac-SMC3.
We observed a cytostatic effect of HDAC8 inhibition, particularly at higher concentrations of PCI-34051, which eventually compromised survival of MCF7 cells. A recent study showed that HDAC8 is also responsible for the deacetylation of p53. In inv(16)+ leukemia cells, inhibition of p53 deacetylation reactivated p53, and induced apoptosis (28). It is possible that the cell cycle delay and growth arrest we observed in MCF7 cells is also mediated by altered p53 acetylation rather than solely from accumulated ac-SMC3.
It is possible that cell type may be important for HDAC8 inhibition to have appreciable effects on cohesin and its transcription function. In T47D ER-positive breast cancer cells with mutant p53, we observed minor changes in expression of a small number of genes following PCI-34051 treatment that reflected RAD21 or SMC3 knockdown. However, T47D cells also exhibited severe cell cycle delay following PCI-34051 treatment such that we could not exclude indirect effects of cell cycle arrest on transcription (data not shown). Differences in transcriptional responses between cell types could also be cohesion dose-dependent (12).
Mutations in genes of the cohesin subunits have been identified in a range of cancers (7), and are not generally associated with aneuploidy (43). Evidence suggests that cohesin functions to establish chromatin interactions that influence contact between gene promoters and regulatory elements (18, 27, 44–47). In hematopoietic stem cells, haploinsufficiency of cohesin alters transcription and induces leukemia (10–12), suggesting that in cancers, the primary role of cohesin might be transcriptional. Our findings in this study call for caution in the development of drug design strategies for therapeutic targeting of cohesin, since not all tumor types may respond in the same way, and targeting cohesin transcriptional function may not have the anticipated outcome. Future screening strategies for cohesin-inhibitory candidates may require the inclusion of multiple platforms and robust biochemical testing for predicted function.
Author Contributions
J. A. H., T. D., J. A., and A. W. B. designed the study and analyzed the data; T. D. performed cell culture, quantitative PCR, ChIP, and cell biology experiments involving PCI-34051 and siRAD21. J. A. performed ChIP and siSMC3 experiments; T. D., J. A., A. W. B., and J. A. H. wrote the paper. All authors reviewed and approved the final version of the manuscript.
Acknowledgment
We thank Katie Young for assistance with flow cytometry.
This work was supported by Genesis Oncology Trust, Cancer Society of New Zealand (11-18), and Health Research Council of NZ (HRC) (10-873, 15-229) grants (to J. A. H.), along with HRC Grants 13-61, 14-219, and 15-500) (to A. W. B.). The authors declare that they have no conflicts of interest with the contents of this article.
- SMC
- structural maintenance of chromosomes
- AML
- acute myeloid leukemia
- HDI
- histone deacetylase inhibitor
- HDAC
- histone deacetylase
- CdLS
- Cornelia de Lange Syndrome
- ER
- estrogen receptor
- TSS
- transcription start site.
References
- 1.Nasmyth K., and Haering C. H. (2009) Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 [DOI] [PubMed] [Google Scholar]
- 2.Peters J. M., Tedeschi A., and Schmitz J. (2008) The cohesin complex and its roles in chromosome biology. Genes Dev. 22, 3089–3114 [DOI] [PubMed] [Google Scholar]
- 3.Losada A. (2014) Cohesin in cancer: chromosome segregation and beyond. Nat. Rev. Cancer 14, 389–393 [DOI] [PubMed] [Google Scholar]
- 4.Remeseiro S., Cuadrado A., and Losada A. (2013) Cohesin in development and disease. Development 140, 3715–3718 [DOI] [PubMed] [Google Scholar]
- 5.Kon A., Shih L. Y., Minamino M., Sanada M., Shiraishi Y., Nagata Y., Yoshida K., Okuno Y., Bando M., Nakato R., Ishikawa S., Sato-Otsubo A., Nagae G., Nishimoto A., Haferlach C., et al. (2013) Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 45, 1232–1237 [DOI] [PubMed] [Google Scholar]
- 6.Leeke B., Marsman J., O'Sullivan J. M., and Horsfield J. A. (2014) Cohesin mutations in myeloid malignancies: underlying mechanisms. Exp. Hematol. Oncol. 3, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leiserson M. D. M., Vandin F., Wu H.-T., Dobson J. R., Eldridge J. V., Thomas J. L., Papoutsaki A., Kim Y., Niu B., McLellan M., Lawrence M. S., Gonzalez-Perez A., Tamborero D., Cheng Y., Ryslik G. A., Lopez-Bigas N., Getz G., Ding L., and Raphael B. J. (2015) Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 47, 106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thol F., Bollin R., Gehlhaar M., Walter C., Dugas M., Suchanek K. J., Kirchner A., Huang L., Chaturvedi A., Wichmann M., Wiehlmann L., Shahswar R., Damm F., Gohring G., Schlegelberger B., et al. (2014) Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood 123, 914–920 [DOI] [PubMed] [Google Scholar]
- 9.Thota S., and Viny A. D. (2014) Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood 124, 1790–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazumdar C., Shen Y., Xavy S., Zhao F., Reinisch A., Li R., Corces M. R., Flynn R. A., Buenrostro J. D., Chan S. M., Thomas D., Koenig J. L., Hong W.-J., Chang H. Y., and Majeti R. (2015) Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell 17, 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullenders J., Aranda-Orgilles B., Lhoumaud P., Keller M., Pae J., Wang K., Kayembe C., Rocha P. P., Raviram R., Gong Y., Premsrirut P. K., Tsirigos A., Bonneau R., Skok J. A., Cimmino L., Hoehn D., and Aifantis I. (2015) Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J. Exp. Med. 212, 1833–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viny A. D., Ott C. J., Spitzer B., Rivas M., Meydan C., Papalexi E., Yelin D., Shank K., Reyes J., Chiu A., Romin Y., Boyko V., Thota S., Maciejewski J. P., Melnick A., Bradner J. E., and Levine R. L. (2015) Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J. Exp. Med. 212, 1819–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atienza J. M., Roth R. B., Rosette C., Smylie K. J., Kammerer S., Rehbock J., Ekblom J., and Denissenko M. F. (2005) Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol. Cancer Ther. 4, 361–368 [DOI] [PubMed] [Google Scholar]
- 14.Xu H., Yan M., Patra J., Natrajan R., Yan Y., Swagemakers S., Tomaszewski J. M., Verschoor S., Millar E. K., van der Spek P., Reis-Filho J. S., Ramsay R. G., O'Toole S. A., McNeil C. M., Sutherland R. L., McKay M. J., and Fox S. B. (2011) Enhanced RAD21 cohesin expression confers poor prognosis and resistance to chemotherapy in high grade luminal, basal and HER2 breast cancers. Breast Cancer Res. 13, R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McEwan M. V., Eccles M. R., and Horsfield J. A. (2012) Cohesin is required for activation of MYC by estradiol. PLoS ONE 7, e49160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes J. M., Bentley F. K., Print C. G., Dorsett D., Misulovin Z., Dickinson E. J., Crosier K. E., Crosier P. S., and Horsfield J. A. (2010) Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev. Biol. 344, 637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhodes J. M., McEwan M., and Horsfield J. A. (2011) Gene regulation by cohesin in cancer: is the ring an unexpected party to proliferation? Mol. Cancer Res. 9, 1587–1607 [DOI] [PubMed] [Google Scholar]
- 18.Antony J., Dasgupta T., Rhodes J. M., McEwan M. V., Print C. G., O'Sullivan J M., and Horsfield J. A. (2014) Cohesin modulates transcription of estrogen-responsive genes. Biochim. Biophys. Acta 24, 00311–00313 [DOI] [PubMed] [Google Scholar]
- 19.Zhang J., Shi X., Li Y., Kim B. J., Jia J., Huang Z., Yang T., Fu X., Jung S. Y., Wang Y., Zhang P., Kim S. T., Pan X., and Qin J. (2008) Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 31, 143–151 [DOI] [PubMed] [Google Scholar]
- 20.Unal E., Heidinger-Pauli J. M., Kim W., Guacci V., Onn I., Gygi S. P., and Koshland D. E. (2008) A molecular determinant for the establishment of sister chromatid cohesion. Science 321, 566–569 [DOI] [PubMed] [Google Scholar]
- 21.Rolef Ben-Shahar T., Heeger S., Lehane C., East P., Flynn H., Skehel M., and Uhlmann F. (2008) Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321, 563–566 [DOI] [PubMed] [Google Scholar]
- 22.Beckouët F., Hu B., Roig M. B., Sutani T., Komata M., Uluocak P., Katis V. L., Shirahige K., and Nasmyth K. (2010) An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol. Cell 39, 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borges V., Lehane C., Lopez-Serra L., Flynn H., Skehel M., Rolef Ben-Shahar T., and Uhlmann F. (2010) Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol. Cell 39, 677–688 [DOI] [PubMed] [Google Scholar]
- 24.Xiong B., Lu S., and Gerton J. L. (2010) Hos1 is a lysine deacetylase for the Smc3 subunit of cohesin. Curr. Biol. 20, 1660–1665 [DOI] [PubMed] [Google Scholar]
- 25.Deardorff M. A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., Saitoh K., Komata M., Katou Y., Clark D., Cole K. E., De Baere E., Decroos C., Di Donato N., Ernst S. et al. (2012) HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 489, 313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Remeseiro S., and Losada A. (2013) Cohesin, a chromatin engagement ring. Curr. Opin. Cell Biol. 25, 63–71 [DOI] [PubMed] [Google Scholar]
- 27.Sofueva S., Yaffe E., Chan W. C., Georgopoulou D., Vietri Rudan M., Mira-Bontenbal H., Pollard S. M., Schroth G. P., Tanay A., and Hadjur S. (2013) Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 32, 3119–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi J., Singh S., Hua W.-K., Cai Q., Chao S.-W., Li L., Liu H., Ho Y., McDonald T., Lin A., Marcucci G., Bhatia R., Huang W.-J., Chang C.-I., and Kuo Y.-H. (2015) HDAC8 inhibition specifically targets Inv(16) acute myeloid leukemic stem cells by restoring p53 acetylation. Cell Stem Cell 17, 597–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balasubramanian S., Ramos J., Luo W., Sirisawad M., Verner E., and Buggy J. J. (2008) A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia 22, 1026–1034 [DOI] [PubMed] [Google Scholar]
- 30.Sleiman S. F., Olson D. E., Bourassa M. W., Karuppagounder S. S., Zhang Y. L., Gale J., Wagner F. F., Basso M., Coppola G., Pinto J. T., Holson E. B., and Ratan R. R. (2014) Hydroxamic acid-based histone deacetylase (HDAC) inhibitors can mediate neuroprotection independent of HDAC inhibition. J. Neurosci. 34, 14328–14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez G., Bill K. L., Bid H. K., Braggio D., Constantino D., Prudner B., Zewdu A., Batte K., Lev D., and Pollock R. E. (2015) HDAC8, A potential therapeutic target for the treatment of malignant peripheral nerve sheath tumors (MPNST). PLoS ONE 10, e0133302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C., Eessalu T. E., Barth V. N., Mitch C. H., Wagner F. F., Hong Y., Neelamegam R., Schroeder F. A., Holson E. B., Haggarty S. J., and Hooker J. M. (2013) Design, synthesis, and evaluation of hydroxamic acid-based molecular probes for in vivo imaging of histone deacetylase (HDAC) in brain. Am. J. Nucl. Med. Mol. Imaging 4, 29–38 [PMC free article] [PubMed] [Google Scholar]
- 33.Ha S. D., Han C. Y., Reid C., and Kim S. O. (2014) HDAC8-mediated epigenetic reprogramming plays a key role in resistance to anthrax lethal toxin-induced pyroptosis in macrophages. J. Immunol. 193, 1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schölz C., Weinert B. T., Wagner S. A., Beli P., Miyake Y., Qi J., Jensen L. J., Streicher W., McCarthy A. R., Westwood N. J., Lain S., Cox J., Matthias P., Mann M., Bradner J. E., and Choudhary C. (2015) Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat. Biotechnol. 33, 415–423 [DOI] [PubMed] [Google Scholar]
- 35.Olson D. E., Udeshi N. D., Wolfson N. A., Pitcairn C. A., Sullivan E. D., Jaffe J. D., Svinkina T., Natoli T., Lu X., Paulk J., McCarren P., Wagner F. F., Barker D., Howe E., Lazzaro F., Gale J. P., Zhang Y. L., Subramanian A., Fierke C. A., Carr S. A., and Holson E. B. (2014) An unbiased approach to identify endogenous substrates of “histone” deacetylase 8. ACS Chem. Biol. 9, 2210–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fullwood M. J., Liu M. H., Pan Y. F., Liu J., Xu H., Mohamed Y. B., Orlov Y. L., Velkov S., Ho A., Mei P. H., Chew E. G. Y., Huang P. Y. H., Welboren W.-J., Han Y., Ooi H. S. et al. (2009) An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt D., Schwalie P. C., Ross-Innes C. S., Hurtado A., Brown G. D., Carroll J. S., Flicek P., and Odom D. T. (2010) A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 20, 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaiser F. J., Ansari M., Braunholz D., Concepción Gil-Rodríguez M., Decroos C., Wilde J. J., Fincher C. T., Kaur M., Bando M., Amor D. J., Atwal P. S., Bahlo M., Bowman C. M., Bradley J. J., Brunner H. G. et al. (2014) Loss-of-function HDAC8 mutations cause a phenotypic spectrum of Cornelia de Lange syndrome-like features, ocular hypertelorism, large fontanelle and X-linked inheritance. Hum. Mol. Genet. 23, 2888–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mannini L., Cucco F., Quarantotti V., Krantz I. D., and Musio A. (2013) Mutation spectrum and genotype-phenotype correlation in Cornelia de Lange syndrome. Hum. Mutat. 34, 1589–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horsfield J. A., Print C. G., and Mönnich M. (2012) Diverse developmental disorders from the one ring: distinct molecular pathways underlie the cohesinopathies. Front. Genet. 3, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan B., Pehlivan D., Karaca E., Patel N., Charng W. L., Gambin T., Gonzaga-Jauregui C., Sutton V. R., Yesil G., Bozdogan S. T., Tos T., Koparir A., Koparir E., Beck C. R., Gu S. et al. (2015) Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J. Clin. Investig. 125, 636–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuster K., Leeke B., Meier M., Wang Y., Newman T., Burgess S., and Horsfield J. A. (2015) A neural crest origin for cohesinopathy heart defects. Hum. Mol. Genet. 24, 7005–7016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solomon D. A., Kim J.-S., and Waldman T. (2014) Cohesin gene mutations in tumorigenesis: from discovery to clinical significance. BMB Rep. 47, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., van Berkum N. L., Ebmeier C. C., Goossens J., Rahl P. B., Levine S. S., Taatjes D. J., Dekker J., and Young R. A. (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marsman J., O'Neill A. C., Kao B. R., Rhodes J. M., Meier M., Antony J., Mönnich M., and Horsfield J. A. (2014) Cohesin and CTCF differentially regulate spatiotemporal runx1 expression during zebrafish development. Biochim. Biophys. Acta 1839, 50–61 [DOI] [PubMed] [Google Scholar]
- 46.Merkenschlager M., and Odom D. T. (2013) CTCF and cohesin: linking gene regulatory elements with their targets. Cell 152, 1285–1297 [DOI] [PubMed] [Google Scholar]
- 47.Seitan V. C., Faure A. J., Zhan Y., McCord R. P., Lajoie B. R., Ing-Simmons E., Lenhard B., Giorgetti L., Heard E., Fisher A. G., Flicek P., Dekker J., and Merkenschlager M. (2013) Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 23, 2066–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]