Abstract
Antagonism of pro-inflammatory transcription factors by monomeric glucocorticoid receptor (GR) has long been viewed as central to glucocorticoid (GC) efficacy. However, the mechanisms and targets through which GCs exert therapeutic effects in diseases such as asthma remain incompletely understood. We previously defined a surprising cooperative interaction between GR and NF-κB that enhanced expression of A20 (TNFAIP3), a potent inhibitor of NF-κB. Here we extend this observation to establish that A20 is required for maximal cytokine repression by GCs. To ascertain the global extent of GR and NF-κB cooperation, we determined genome-wide occupancy of GR, the p65 subunit of NF-κB, and RNA polymerase II in airway epithelial cells treated with dexamethasone, TNF, or both using chromatin immunoprecipitation followed by deep sequencing. We found that GR recruits p65 to dimeric GR binding sites across the genome and discovered additional regulatory elements in which GR-p65 cooperation augments gene expression. GR targets regulated by this mechanism include key anti-inflammatory and injury response genes such as SERPINA1, which encodes α1 antitrypsin, and FOXP4, an inhibitor of mucus production. Although dexamethasone treatment reduced RNA polymerase II occupancy of TNF targets such as IL8 and TNFAIP2, we were unable to correlate specific binding sequences for GR or occupancy patterns with repressive effects on transcription. Our results suggest that cooperative anti-inflammatory gene regulation by GR and p65 contributes to GC efficacy, whereas tethering interactions between GR and p65 are not universally required for GC-based gene repression.
Keywords: asthma, ChIP-sequencing (ChIP-seq), chromatin immunoprecipitation (ChiP), glucocorticoid receptor, NF-κB (NF-KB), transcription enhancer, A20, SERPINA1, airway
Introduction
Glucocorticoids (GCs)3 are the primary controller therapy for asthma (1) and are also widely used to treat chronic obstructive pulmonary disease (2, 3). GCs function by binding to the glucocorticoid receptor (GR, NR3C1), a ubiquitously expressed nuclear receptor transcription factor. Upon ligand binding, GR translocates to the nucleus where it regulates transcription (4, 5). Direct association of GR with DNA through canonical GR binding sites is implicated in transcriptional induction by GCs, whereas interactions between GR and inflammatory transcription factor complexes such as NF-κB, in a process that may not require direct association of GR with DNA, has been suggested to result in transcriptional repression (6). So-called transrepression (i.e. repressive interactions between GR and inflammatory regulators) was long considered to be the primary mechanism through which GR represses inflammation in many cell types (7, 8). More recent studies, however, have suggested that far greater mechanistic complexity underpins the anti-inflammatory properties of GCs (9, 10). For example, genome-wide analysis of GR interactions with DNA (i.e. chromatin-immunoprecipitation of GR followed by deep sequencing, or ChIP-seq) indicated that interactions between GR and inflammatory transcription factors do not necessarily lead to repressive regulatory outcomes (11–14). Moreover, an increasing number of GR-induced genes are now recognized as contributing to inflammatory repression by GCs (15–17). It is also clear that GR actions are subordinate to cellular and chromatin context (18–22), with wide variations in both GR binding and activity occurring between cell types and as a consequence of exposure to different cytokines and growth factors (12, 23).
One of the major targets of GCs in obstructive airway diseases such as asthma is the airway epithelium (24). The airway epithelium engages in complex pathophysiologic cross-talk with immune cells (25, 26), which are themselves GC targets, and infectious and environmental stimuli (27–29), ultimately leading to airway inflammation and symptoms in susceptible patients (30). It is well established that GCs act via GR in cultured airway epithelial cells to repress inflammatory responses to a range of clinically relevant inflammatory stimuli, including IL1B, TNF, and rhinoviral infection (31, 32). It has also been shown that GCs directly modify human airway epithelial gene expression in vivo (33, 34), with GC treatment linked to increased mRNA levels of the canonical GR target FKBP5 and decreased expression of CLCA1, SERPINB2, and POSTN (periostin), whose levels in asthma are correlated with clinical responses to GCs (35). However, the mechanistic basis for GR-mediated repression of gene expression in airway epithelial cells remains incompletely understood, and the relative role of transrepression versus induction of anti-inflammatory genes by GCs in repressing airway inflammation has yet to be established.
To further understand the mechanisms of GR signaling in the airway epithelium, we previously studied the effects of dexamethasone (dex, a potent synthetic GC) on the expression of several pro- and anti-inflammatory targets of TNF signaling in Beas-2B cells (36), which are an immortalized human bronchial epithelial cell line. We showed that, in comparison to robust repression of cytokine mRNA levels, GCs spare or augment the expression of negative feedback regulators of NF-κB, such as TNFAIP3 (frequently referred to as A20). Furthermore, we identified inductive synergy between GR and NF-κB at a novel enhancer within the TNFAIP3 locus and proposed a model in which cooperation between GR and NF-κB at anti-inflammatory loci is central to the therapeutic effects of GCs. Subsequent studies by other researchers have supported this notion, with co-induction of IRAK3 (frequently referred to as IRAKM) and Sphk1 by GR and inflammatory stimuli linked to anti-inflammatory effects of GCs in various models of lung disease (37, 38). Thus, cooperative gene induction mediated by convergence of activated GR and inflammatory signals at specific enhancers appears to contribute to the anti-inflammatory properties of GCs. However, the overall pattern of cross-talk between GR and NF-κB in airway epithelial cells and whether GR and NF-κB cooperate at additional loci to repress inflammation has yet to be established.
In this study we extended our prior work on GR cross-talk with NF-κB to determine whether GCs selectively spare or augment the expression of anti-inflammatory targets of TNF in primary human tracheobronchial epithelial cells and whether TNFAIP3 suppresses TLR3-mediated cytokine induction. We applied siRNA knockdown of TNFAIP3 to determine whether this GR-p65 co-regulated target is required for maximal repression of cytokine expression by GCs. Furthermore, we expanded our studies of interactions between GR and NF-κB to encompass integrated ChIP-seq studies of GR, the p65 subunit of NF-κB, and RNA polymerase II (RNAPII) occupancy in Beas-2B cells. Our results provide a framework for understanding the molecular basis of GC action in the airway epithelium.
Experimental Procedures
Cell Culture and Reagents
With approval from the Institutional Review Board at National Jewish Health, de-identified organ donor lungs deemed unsuitable for transplantation were used as a source for normal human tracheobronchial (i.e. primary airway) epithelial cells, which were obtained as previously described (39). Thawed airway epithelial cells at passage 1 were cultured and expanded in bronchial epithelial cell growth medium (BEGM) with supplements (Lonza) in 5% CO2 at 37 °C using 60-mm tissue culture dishes coated with collagen until 90% confluence was achieved (40). Beas-2B cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM; Corning) containing l-glutamine and 4.5 g/liter glucose and supplemented with 10% fetal bovine serum (Fisher) and 1% penicillin/streptomycin (Corning). Dex (D1756), obtained from Sigma, was dissolved in sterile 100% ethanol and used at a final concentration of 100 nm. Recombinant human tumor necrosis factor-α (TNF; PHC3015L), purchased from Life Technologies, was diluted in 1× Dulbecco's phosphate-buffered saline containing 0.2% fetal bovine serum and used at a final concentration of 20 ng/ml. Polyinosine-polycytidylic acid (poly(I:C)) was purchased from Invivogen. Human rhinovirus serotype 1A (HRV-1A; ATCC) was propagated in H1-HeLa cells (CRL-1958; ATCC), purified, and titrated as described previously (41). Adenoviral expression constructs for TNFAIP3 (Ad-TNFAIP3) and GFP (Ad-GFP as control) have been previously described (36) and were prepared by Welgen. ON-TARGETplus SMARTpool siRNA targeting TNFAIP3 (si-TNFAIP3; L-009919-00-0005), GR (si-NR3C1; L-003424-00-0005), and a non-targeting control (si-Ctrl: D-001810-10-20) from Dharmacon were used for TNFAIP3 and GR knockdown studies. Antibodies against TNFAIP3 (ab13597) and β-actin (ab75186) were purchased from Abcam. Pan-TEAD antibody (D3F7L, 13295) was obtained from Cell Signaling Technology. Anti-GAPDH antibody (sc-25778) was purchased from Santa Cruz Biotechnology. Secondary antibodies ECL sheep anti-mouse IgG-HRP (95017-332) and ECL donkey anti-rabbit IgG-HRP (95017-330) were purchased from VWR. Anti-NFKB p65 (C-20, sc-372x) from Santa Cruz Biotechnology, anti-RNA polymerase II (8WG16, 920101) from BioLegend, and anti-GR (IA-1, a generous gift from Dr. Miles Pufall) antibodies were used for chromatin immunoprecipitation (ChIP).
Plasmids
The pFSTL3, pTNFAIP3, and pABR plasmids were generated in the pGL3-Promoter vector. PCR primers were designed to amplify putative GR-p65 cooperative binding regions of FSTL3, TNFAIP3, and ABR identified through the ChIP-Seq data. Amplified PCR product from FSTL3 and TNFAIP3 was introduced into pCR 2.1 TOPO (Life Technologies) and subsequently cloned into the pGL3-Promoter vector using KpnI/BglII. PCR product from ABR was directly cloned into the pGL3-Promoter using NheI/BglII. The cloning primers are provided in supplemental Table S1. Transfections were performed, and luciferase activity was assayed as previously described (42).
Adenoviral Transduction
Primary airway epithelial cells at passage 2 were seeded into 12-well culture plates at 1 × 105 cells/well. At ∼80% confluence, cells were infected with Ad-TNFAIP3 or Ad-GFP (control) adenovirus at a multiplicity of infection of 50 for 17 h and then allowed to recover in fresh medium for 30 h. Cells were then treated with poly(I:C) (2.5 μg/ml) or infected with HRV-1A (TCID50 = 107) for 24 h. After treatment, the cells were harvested with TRIzol for RNA, and changes in gene expression were assayed by qRT-PCR as described previously (42). The supernatants were assayed for IL8 protein using human IL8 enzyme-linked immunosorbent assay (ELISA) kit (R&D systems).
siRNA Transfection
Beas-2B cells were seeded in 6-well dishes and transfected with si-TNFAIP3 or si-Ctrl using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer's protocol. Approximately 48 h after transfection cells were treated with vehicle, dex, TNF, or TNF + dex for 4 or 24 h and harvested for RNA and protein. IL8 protein levels in the supernatants from cells treated for 24 h was assayed by ELISA (R&D Systems). For GR knockdown experiments, cells were seeded in 6-well dishes or 10-cm dishes and transfected with si-GR or si-Ctrl. After 48 h, cells in 6-well dishes were treated with vehicle or dex for 4 h and harvested for RNA and protein. Cells in 10-cm dishes were treated for 1 h, and chromatin immunoprecipitation was performed. Knockdown was confirmed using Western blotting.
Western Blotting
Beas-2B cells were lysed with radioimmunoprecipitation assay buffer containing 1× protease inhibitor mixture (Thermo Scientific). Approximately 30–50 μg of total protein were separated by SDS-PAGE and transferred onto PVDF membranes (Immun-blot; Bio-Rad). Membranes were immunoblotted with antibodies against proteins of interest, and bands were visualized using ECL Prime detection system from GE Healthcare.
RNA Preparation and Analysis
Primary Airway epithelial cells at passage 2 were plated on 12-well plates at 1 × 105 cells/well and grown to 80% confluence. Beas-2B cells were plated on 6-well plates at 3.5 × 105 cells/well and grown to 90% confluence. Cells were then treated with vehicle, TNF, dex, or TNF + dex for 4 h and then lysed in TRIzol (Life Technologies). RNA was purified using the PureLink RNA Mini kit from Life Technologies. Reverse transcription of RNA, qRT-PCR, and gene expression analyses were performed as previously described (42). Primers are listed in supplemental Table S1. Percent repression, where indicated, was calculated as ([cycle number change (relative to vehicle) for TNF treatment] − [cycle number change (relative to vehicle) for TNF + dex treatment]) ÷ [relative cycle number change for TNF treatment]. Percent repression for IL8 protein levels was determined similarly.
ChIP
Beas-2B cells at 100% confluence were treated with vehicle, dex, TNF, or TNF + dex in fresh medium for 1 h. ChIP and qPCR analysis of purified ChIP DNA was performed as previously described except that the chromatin was fragmented at high power for 34 cycles (1 cycle = 30 s on, 30 s off) in a Diagenode Bioruptor. Enrichment of the protein of interest at a putative target region was calculated on a log2 scale and was determined by comparing the Ct values of the specific target region with the geometric mean of the Ct values of three negative control regions. For the siRNA knockdown ChIP experiments, enrichment was normalized relative to input, as the Ct values for the internal negative control regions were too high to be reliable. All ChIP-qPCR experiments were performed in biological quadruplicate and repeated two or more times. Primers are listed in supplemental Table S1.
ChIP Sequencing
ChIP-seq and analysis was performed essentially as described (43). Briefly, a minimum of 1 ng of DNA from ChIP samples was used to prepare libraries (Nugen Ovation Ultralow System V2 1-16 Part no. 0344). Samples were run on an Illumina HiSeq using 1 × 50-bp end reads. FASTQ sequence file quality was assessed (Babraham Bioinformatics). Sequences were mapped to the human genome (hg19) using Bowtie2 (44). Peak calling was performed using MACS2 software with a narrow peak range set at 300 (45). Using the HOMER group, peaks were converted to the BigWig format and uploaded into the UCSC Genome Browser using the custom track feature. Custom tracks annotated on the UCSC Genome Browser Feb. 200 (GRCh37/hg19) Assembly are freely available upon request. Using an R implementation, the consistency of replicated sample peaks was assessed with the Irreproducible Discovery Rate algorithm (46). Differential binding analysis was performed using an R Bioconductor package DBChIP (47) and annotated using ChIPseeker (48). To facilitate comparisons with ChIP-seq data we hope to generate in the future using patient derived airway epithelial cells, only autosomal binding regions were used to define differential binding and subsequent analysis. All ChIP-seq data has been deposited in the Gene Expression Omnibus (GSE79803). An overview of this experiment was previously published in abstract form (49).
ChIP-seq Quality Control
Standard initial analysis and quality control metrics indicated a base call accuracy >99.9% for 96% of base reads. The average read number per sample was ∼30 million, well above the 10-million minimal read number proposed by the ENCODE consortium (50). R-values between ChIP samples for GR and p65 ChIPs were computed. The technical duplicate samples generally showed high concordance, with r values of ∼0.9 or greater, exemplified by an r value of 0.95 for the two dex-treated GR ChIP samples. An exception to the robust correlations between technical duplicates occurred with the two TNF-treated p65 ChIP samples (r = 0.82). This was subsequently found to reflect a suboptimal p65 ChIP in one of the two discordant samples; this sample was excluded from subsequent analysis. Thus, a total of 19 samples were used for analysis, with 9 of 10 conditions represented in technical duplicate.
Statistical Considerations and Experimental Uncertainty
Two tailed t tests were performed to compare selected ChIP-qPCR, qRT-PCR, ELISA, and luciferase data with significance level sets at p < 0.05. As t test performance is sensitive to normality and variance assumptions at the relatively low numbers used for comparisons in this work, results of statistical tests were confirmed with non-parametric tests, which do not require an assumption of normality or equal variance. siRNA knockdown of GR followed by ChIP was performed on quadruplicate samples on two occasions. Luciferase assays were performed on quadruplicate samples on two occasions for the ABR construct; for the TNFAIP3 and FSTL3 constructs, two independent clones of each construct were tested in quadruplicate on two occasions. qPCR analysis of primary airway epithelial cells was performed on quadruplicate samples on two or three occasions. TNFAIP3 knockdown and subsequent qRT-PCR was performed on quadruplicate samples on two occasions after 4 and 24 h of dex treatment. Secreted IL8 levels with siRNA treatment were measured from quadruplicate samples in two independent experiments. ChIP-qPCR validation of ChIP-seq data were performed in quadruplicate on two or three occasions. ChIP-seq was performed and analyzed using duplicate samples, with the exception of the TNF-treated p65 ChIP samples, as described above. qRT-PCR analysis of gene expression responses to dex, TNF, and dex + TNF in Beas-2B cells was performed in quadruplicate on two occasions. In all cases, data between independent replicates of each experiment were qualitatively similar and thus indicative of experimental reproducibility. Error bars in figures represent S.D.
Results
Glucocorticoids Selectively Spare Expression of TNF-induced Anti-inflammatory Targets in Primary Airway Epithelial Cells
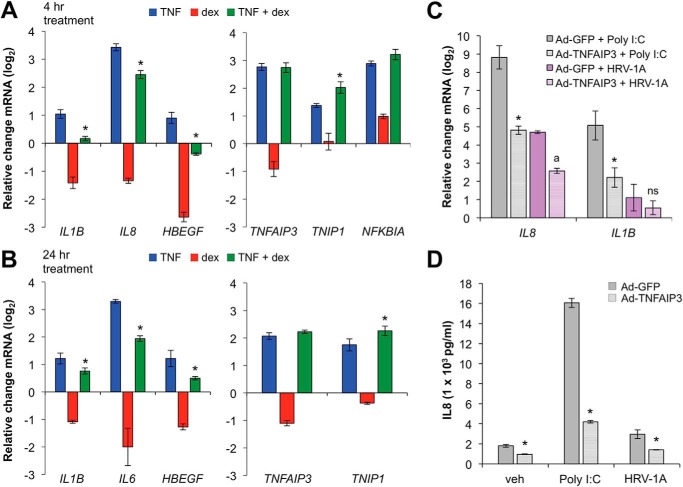
To explore whether GCs differentially regulate pro- and anti-inflammatory targets of TNF in primary airway cells, submerged primary airway epithelial cells were cultured with vehicle, TNF, dex, or a combination of TNF and dex for 4 h or 24 h. Consistent with our previous Beas-2B results (36), induction of pro-inflammatory targets such as IL1B, IL8, and HBEGF by TNF was strongly suppressed by dex co-treatment (left panels, Fig. 1, A and B), whereas expression of TNFAIP3, TNIP1, and NFKBIA, which are known to exert negative feedback control of inflammation in various contexts, was either spared or enhanced (right panels, Fig. 1, A and B). The group difference in the effect of dex on the set of pro- versus anti-inflammatory genes, as assessed using non-parametric rank-sum testing, was highly significant (p < 0.005). Moreover, adenoviral driven overexpression of TNFAIP3 abrogated the inductive effects of both rhinoviral infection and treatment with poly(I:C), a synthetic TLR3 agonist, on IL1B and IL8 mRNA expression (Fig. 1C) and secreted IL8 (Fig. 1D). Thus, similar to our previous results in Beas-2B cells, GCs differentially regulate pro- and anti-inflammatory targets of TNF in primary airway epithelial cells, and TNFAIP3 potently suppresses inflammatory cytokine induction.
FIGURE 1.
Glucocorticoids spare expression of TNF targets that mediate cytokine repression in airway epithelial cells. A and B, primary airway epithelial cells were treated with TNF (20 ng/ml), dex (100 nm), or TNF + dex as indicated for 4 h (A) or 24 h (B), and expression of pro (left)- and anti (right)-inflammatory genes was assayed using qRT-PCR. Bars indicate mean CT values on a log2 scale (±S.D.) relative to vehicle-treated controls. C, primary airway epithelial cells were transduced with adenoviral expression constructs for TNFAIP3 (Ad-TNFAIP3) or GFP (Ad-GFP; control) before treatment with poly(I:C) (2.5 μg/ml) or infection with HRV-1A (TCID50 = 107) for 24 h, after which IL8 and IL1B mRNA levels were assayed using qRT-PCR. Bars indicate mean CT values (±S.D.) relative to Ad-GFP + vehicle-treated controls. D, supernatants from C were used to measure secreted IL8 protein levels using ELISA. Statistical comparisons for each panel are as follows. A and B, *, p ≤ 0.05 versus TNF treatment (blue) for the same gene. C, *, p ≤ 0.05 compared with Ad-GFP + poly(I:C) for each gene; a, p ≤ 0.05 compared with Ad-GFP + HRV-1A for each gene. D, *, p ≤ 0.05 compared with Ad-GFP for each treatment as indicated. ns, not significant.
TNFAIP3 Is Required for Maximal Repression of TNF-induced Pro-inflammatory Targets by Dex
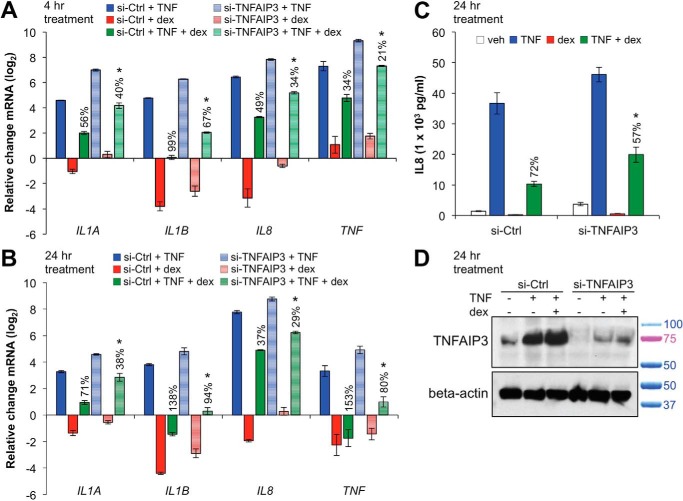
Whereas the effect of TNFAIP3 overexpression on cytokine induction indicates that TNFAIP3 represses inflammatory gene expression, these data do not establish a direct link between TNFAIP3 and the anti-inflammatory effects of GCs. Therefore, to determine whether TNFAIP3 contributes to cytokine repression by GCs, we reduced TNFAIP3 protein expression using siRNA-mediated gene knockdown in Beas-2B cells. We selected this cell type for these experiments and for the remainder of this work as Beas-2B cells exhibit patterns of anti-inflammatory gene expression that are similar to primary cells but are more amenable to molecular manipulation. After siRNA transfection with control (si-Ctrl) or anti-TNFAIP3 siRNA (si-TNFAIP3), cells were treated with TNF, dex, or TNF + dex for 4 or 24 h. As shown in Fig. 2, A and B, in comparison to si-Ctrl, transfection with si-TNFAIP3 led to increased expression of pro-inflammatory genes with TNF treatment (shaded blue versus solid blue bars) and decreased percent repression by dex (solid green versus shaded green bars). The -fold repression (log2) with TNF + dex co-treatment compared with TNF alone for IL8 and IL1A was also significantly less (p < 0.05) with si-TNFAIP3 versus si-Ctrl at 24 h. Furthermore, dex-mediated repression of secreted IL8 was significantly attenuated by transfection with si-TNFAIP3 in comparison to control (Fig. 2C). We confirmed by Western blot that si-TNFAIP3 transfection reduced TNFAIP3 protein levels (Fig. 2D). Taken together, these data indicate that TNFAIP3 is required for maximal repressive effects of ligand-activated GR on cytokine expression.
FIGURE 2.
TNFAIP3 is required for maximal repression of cytokines by glucocorticoids. A and B, Beas-2B cells were transfected with siRNA against TNFAIP3 (si-TNFAIP3) or control (si-Ctrl) and then treated with TNF, dex, or TNF + dex for 4 or 24 h. Transcript levels of indicated proinflammatory genes were assessed by qRT-PCR and are presented as the mean CT values (±S.D.) relative to si-Ctrl + vehicle-treated controls. For a specific gene, 100% repression is defined as TNF + dex treatment resulting in base-line expression levels in comparison to TNF treatment under the same conditions. Percent repression for each assayed gene is indicated in the figure above the TNF + dex columns. C, cells were transfected as described above and treated for 24 h with TNF, dex, or TNF + dex as indicated. IL8 levels in the supernatant were measured using ELISA, and percent repression is indicated. D, Western blots for TNFAIP3 and β-actin (loading control) proteins confirming knockdown by si-TNFAIP3 in the experiment described in C. Relative positions of standard weight markers in kilodaltons are as indicated to the right of the blot. Statistical comparisons for each panel are as follows: A and B, *, p ≤ 0.05 versus si-Ctrl with TNF + dex treatment for the same gene (shaded green versus solid green bars); C, *, p ≤ 0.05 versus si-Ctrl with TNF + dex treatment.
Genome-wide Analysis of GR, p65, and RNAPII Occupancy in Beas-2B Cells Using ChIP-Seq
Our data implicate selective regulation of pro- versus anti-inflammatory targets of TNF by GCs and cooperation between GR and p65 at genes such as TNFAIP3 as important for GC efficacy in the airway epithelium. We, therefore, sought to determine how GR and the p65 subunit of the NF-κB complex interact with DNA on a genome-wide basis and how occupancy of these two factors correlates with recruitment of RNAPII at both pro- and anti-inflammatory genes. To accomplish this, we performed ChIP-seq for GR, p65, and RNAPII in Beas-2B cells treated for 1 h with vehicle (GR, p65, RNAPII), dex (GR, RNAPII), TNF (p65, RNAPII), or both dex and TNF (GR, p65, RNAPII). Initial peak calling and quality control are detailed under “Experimental Procedures.” Integrated analyses of the resulting datasets are described below.
Recruitment of RNAPII and Binding Site Enrichment Associated with GR Occupancy
Our initial analysis of the biologic implications of these data focused on occupancy of GR after treatment with dex. To identify high confidence peaks where GR occupancy was regulated by dex, we performed differential binding analysis comparing GR peaks within the dex-treatment datasets to vehicle control. With a false discovery rate (FDR) of 0.05 (51), 5612 peaks with differential GR occupancy associated with dex treatment in comparison to vehicle were identified (supplemental Table S2) from a total of 60,680 MACS defined GR peaks common to both dex-treated ChIP samples. The distribution of these differentially occupied GR binding peaks with respect to annotated transcriptional start sites is similar to prior reports (13), with ∼25% of sites mapping to the promoter or first exon, ∼30% of sites mapping to the first intron, and ∼40% of binding sites mapping to intergenic regions.
Based on average read number, >75% of the set of 5612 high confidence dex-regulated GR binding regions showed dex-induced GR occupancy. This is consistent with the canonical activity of GCs in inducing nuclear localization of GR and subsequent DNA binding (52). Peaks indicative of non-canonical interactions between GR and DNA in which ligand reduced GR occupancy, a finding of unclear biologic significance that was recently reported by another group (53), were also identified.
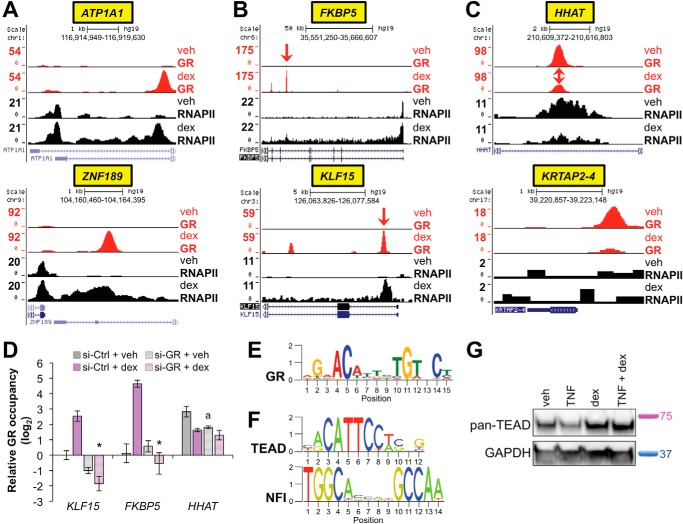
We visualized strongly dex-induced peaks in the UCSC genome browser (54). This was performed in conjunction with the RNAPII occupancy data, in which 1411 peaks were differentially occupied (FDR < 0.05) by RNAPII with dex in comparison to vehicle (supplemental Table S3). This integrated analysis allowed us to unequivocally define genes such as ATP1A and ZNF189 as direct targets of GR (Fig. 3A). We also confirmed that canonical GR binding sites and target genes, such as FKBP5 and KLF15 (42, 55), were represented within these data (Fig. 3B). Examples of peaks that are present in the absence of dex are also depicted (Fig. 3C).
FIGURE 3.
ChIP-seq for GR and RNAPII establishes genome-wide sites of GR activity and associated transcriptional consequences in airway epithelial cells. A–C, GR (red) and RNAPII (black) ChIP-seq peaks visualized in the UCSC Genome Browser from dex- versus vehicle (veh)-treated Beas-2B samples as indicated. Peak heights reflect normalized factor occupancy (reads per million reads, see the vertical scale on the left of each panel). Gene symbols are highlighted in a yellow box on the bottom, and corresponding chromosomal locations are provided on the top of each panel. A, representative examples of genes identified as novel direct transcriptional targets of GR. B, confirmation of GR occupancy at well established genomic binding sites within FKBP5 (top) and KLF15 (bottom). Red arrows indicate approximate positions targeted by GR ChIP-qPCR primers tested in D. C, representative examples of genes exhibiting GR occupancy in the absence of dex. D, ChIP-qPCR for GR, as performed on Beas-2B cells transfected with siRNA against GR (si-GR) or control and treated with dex or vehicle for 1 h. *, p ≤ 0.05 for KLF15 and FKBP5 compared with si-Ctrl + dex (compare shaded purple to solid purple bars); a, p ≤ 0.05 for HHAT compared with si-Ctrl + veh (compare shaded gray to the solid gray bar). E, GR binding logo identified though de novo binding motif analysis using MEME-ChIP to analyze regions differentially bound by GR in dex- versus veh-treated samples. F, binding motifs for TEAD and NFI family transcription factors, which were identified by CentriMo as centrally enriched within the differentially occupied GR binding regions described in E. G, Western blots for TEAD family members and GAPDH (loading control) in Beas-2B cells treated as indicated for 4 h. Relative positions of standard mass markers in kilodaltons are as indicated to the right of the blot.
siRNA knockdown of GR followed by ChIP-qPCR confirmed that dex-induced peaks within the FKBP5 and KLF15 loci reflect GR occupancy (Fig. 3D). The GR peak within HHAT that was present in the absence of dex was also modestly reduced by GR knockdown (Fig. 3D). We conclude that our ChIP-seq data for GR and RNAPII comprehensively delineates genomic sites of GR activity and associated transcriptional consequences in the Beas-2B airway epithelial cell line model. These data are publically available through the GEO repository; custom tracks annotated in the UCSC genome Browser are available upon request.
We next sought to determine whether specific DNA sequences were enriched within the sets of GR genomic binding regions. To accomplish this, we used MEME-ChIP to interrogate 301-base pair regions centered at each of the 5612 dex-regulated GR binding regions (56). This analysis resulted in unbiased identification of a close match for the consensus GR binding sequence (Fig. 3E). The Centrimo tool within the MEME suite (57) identified central enrichment for matches to the consensus GR binding site within 2837/5612, or 51%, of the sequences, a percentage that is consistent with those from other genome-wide studies of GR occupancy (55). Central enrichment was also detected for several additional motifs including AP-1 (∼38% of sequences), CEBP (∼23%), TEAD (∼69%; Fig. 3F), and NFI (∼17%; Fig. 3F) family members. Western blot analysis using a pan-TEAD antibody showed substantial expression of TEAD family members in this cell type (Fig. 3G), supporting a role in Beas-2B cells for TEAD family cross-talk with GR, as reported in other cell types (58). The complete MEME-ChIP analysis is available in the supplemental File S1. These data confirm enrichment for the consensus binding sequence for dimeric GR and suggest new potential GR interacting partners within airway epithelial cells.
Integrated Analysis of p65 and RNAPII Occupancy with TNF Treatment
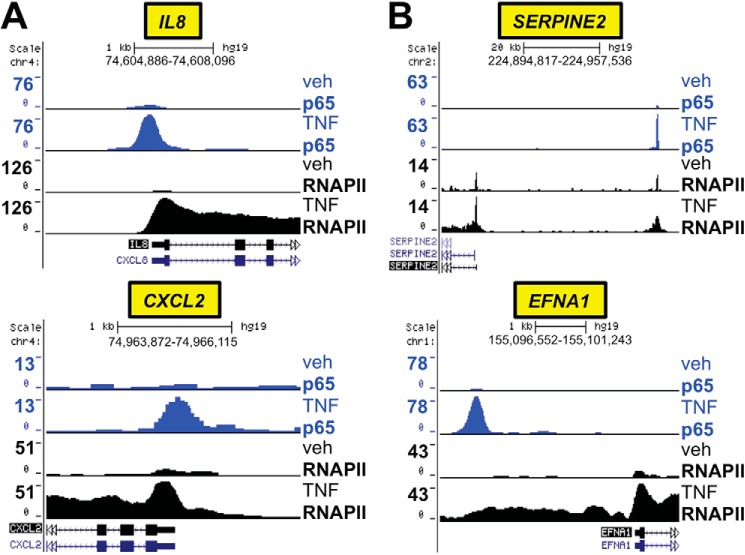
We applied a similar analysis pipeline to the p65 ChIP-seq data. Differential binding analysis comparing TNF treatment to vehicle identified 4863 sites with TNF-regulated p65 occupancy with an FDR of < 0.05 (supplemental Table S4) from a total of 52,501 p65 peaks in the TNF-treated sample. Coordinated visualization of the p65 and RNAPII ChIP-seq data established that genes such as IL8 and CXCL2 are regulated directly by TNF-activated p65 in association with robust recruitment of RNAPII (Fig. 4A). Targets of NF-κB that have been identified in other cell types, such as SERPINE2 and EFNA1 (59, 60), were also confirmed to be directly regulated by p65 in Beas-2B cells, in concert with RNAPII recruitment (Fig. 4B). The dataset for differential RNAPII occupancy (542 peaks, FDR < 0.05) with TNF treatment in comparison to vehicle is available in supplemental Table S5.
FIGURE 4.
Coordinated analysis of p65 and RNAPII ChIP-seq defines direct transcriptional targets of NF-κB in Beas-2B cells. A and B, p65 (blue) and RNAPII (black) ChIP-seq peaks in TNF- versus vehicle (veh)-treated Beas-2B samples, as detailed for Fig. 3. A, representative examples of genes exhibiting increased p65 binding and robust RNAPII recruitment with TNF treatment in Beas-2B cells. B, verification of p65 and RNAPII recruitment to genes reported in other cell types to be direct transcriptional targets of NF-κB.
Application of the Centrimo tool within the MEME suite showed central enrichment for matches to the NF-κB binding consensus at ∼62% of sequences. In addition, central enrichment for other transcription factor families, including ETS-like factor binding sites (51%) and TEAD family members (19%), were identified within the set of p65 binding regions. The complete file generated by MEME-ChIP for this dataset is available in supplemental File S2. Thus, similar to our results for GR, the ChIP-seq data for p65 and RNAPII with TNF treatment provides a genome-wide map of sites of p65 action and identifies potential interactions between p65 and other transcription factor families in airway epithelial cells.
GR Occupancy Patterns and Repression of TNF Target Genes
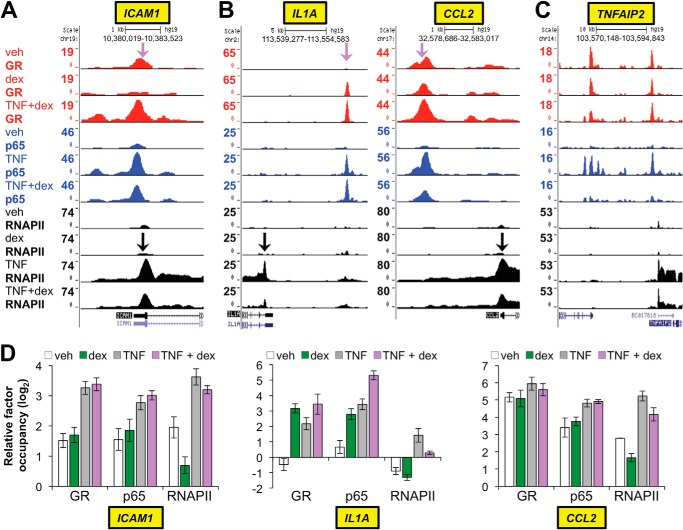
We next focused on analyzing cross-talk between TNF and GC signaling, which was a primary goal of the ChIP-seq experiment. Similar to the above analysis for the individual GR-dex and p65-TNF datasets, we utilized a combination of genome-wide differential binding analysis and visualization of individual loci regulated by GR and p65 within the UCSC genome browser. Differential binding analysis for GR comparing dex treatment alone with TNF + dex co-treatment identified 831 sites (FDR < 0.05) with differential GR binding between the two conditions (supplemental Table S6). This indicates that a comparatively small fraction (<20%) of the ∼5600 dex-regulated sites of GR occupancy (FDR < 0.05) was altered significantly by TNF co-treatment. Whereas this is consistent with the high r values (∼0.92) we calculated between the dex and TNF + dex GR ChIP-seq datasets, these data strongly suggest that TNF-mediated redirection of GR binding, as would be expected with tethering interactions between genomic p65 and GR, is not a dominant determinant of GR occupancy in the setting of dex + TNF co-treatment. Furthermore, >⅓ of the GR binding regions whose occupancy was increased with TNF had significant occupancy (i.e. >15 sequence reads; data are shown in supplemental Table S6) with dex treatment alone, indicating that locus-specific features mediate GR occupancy in these regions, in addition to any potential contribution of GR-p65 tethering. We also observed surprising heterogeneity of GR and p65 occupancy patterns at loci in which dex treatment resulted in reduced recruitment of RNAPII by TNF, as illustrated in Fig. 5. Specifically, at the ICAM1 locus, GR occupancy was only detected with dex + TNF co-treatment and mapped to the same genomic location as a strong p65 peak (Fig. 5A). In contrast, at the IL1A and CCL2 loci, although GR occupancy was again highest with TNF + dex co-treatment and mapped to a region of TNF-induced p65 occupancy, significant GR binding within the IL1A and CCL2 loci was present with dex treatment alone (Fig. 5B). A third pattern occurred at the TNFAIP2 locus in which, despite a substantial decrease in TNF-induced RNAPII occupancy with dex treatment, no appreciable change in GR binding with dex + TNF in comparison to dex or vehicle alone was observed (Fig. 5C). The SERPINB2 locus, which is repressed by GCs in human airways clinically (34), exhibited a pattern that was similar to TNFAIP2 (data not shown). Taken together, the complexity of GR-p65 occupancy patterns associated with GC-mediated transcriptional repression is incompatible with simple tethering models.
FIGURE 5.
GR binding patterns associated with repression of TNF-induced pro-inflammatory genes. A–C, GR (red), p65 (blue), and RNAPII (black) ChIP-seq peaks in Beas-2B samples treated as indicated on the far left and visualized in the UCSC Genome Browser. Purple arrows indicate approximate positions targeted by both GR and p65 ChIP-qPCR primers tested in D, whereas black arrows indicate regions targeted by RNAPII ChIP-qPCR primers. A, example of a gene at which GR occupancy was detected only with TNF + dex co-treatment. B, genes like IL1A (left) and CCL2 (right) exhibited significant GR binding with dex alone that was further enhanced by TNF + dex co-treatment. C, in contrast to A and B, TNFAIP2 exemplifies genes showing substantial reductions in TNF-induced RNAPII recruitment with dex co-treatment but no notable difference in GR binding between vehicle (veh)-, dex-, or TNF + dex-treated samples. D, independent validation of the GR, p65, and RNAPII binding patterns described in A–C using ChIP-qPCR. Bars represent the mean (±S.D.) relative factor occupancy on a log2 scale, determined by comparing CT values at the target region to the geometric mean of CT values at three negative control regions.
We noticed that there was detectable GR occupancy with vehicle treatment alone at putative tethering sites within the ICAM1 and CCL2 loci where GR occupancy with TNF + dex treatment was higher than GR occupancy with dex treatment alone (Fig. 5). Furthermore in our previous work, TNF treatment resulted in significant GR recruitment to chromatin at the IL1B locus in the absence of dex (36). These data suggest that recruitment of GR to sites of p65 occupancy may not require exogenous ligand but instead may be due to effects of TNF alone. We, therefore, used ChIP-qPCR to evaluate GR occupancy at sites within the ICAM1, CCL2, and IL1A loci after TNF treatment in comparison to treatment with TNF + dex. At the same time we validated the GR, p65, and RNAPII ChIP-seq data for these loci using ChIP-qPCR on independent samples (Fig. 5D). The resultant data generally mirrored the peak intensities we observed in the ChIP-seq datasets. In addition, TNF treatment increased GR occupancy at these loci in the absence of exogenous ligand. These results provide further evidence that treatment with dex mediates repression through mechanisms that are not necessarily associated with changes in GR occupancy at repressed loci.
p65 Is Recruited to Binding Sites for Dimeric GR
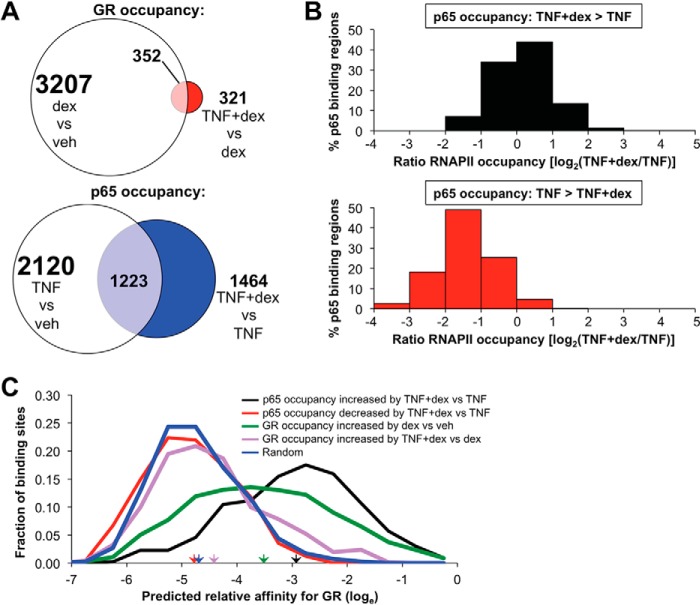
In contrast to the relatively modest effects of TNF treatment on GR occupancy (831 differentially occupied peaks of ∼5600 dex-regulated GR peaks), differential binding analysis of p65 occupancy with TNF + dex co-treatment compared with TNF alone identified 3597 peaks that were differentially occupied by p65 (FDR < 0.05) between the two conditions (supplemental Table S7). Thus, in comparison to the effect of TNF on GR occupancy, ligand-activated GR appears to alter p65 occupancy at both a larger percentage of binding peaks and in association with many more unique genes IDs (Fig. 6A; note that many genes contain multiple binding peaks, so the number of gene IDs depicted by the Venn diagrams are less than the total number of peaks). Although many of the p65 binding regions in which occupancy differed with TNF + dex co-treatment in comparison to TNF alone had relatively low read numbers, 1083 differentially bound p65 peaks were identified after filtering for sites with higher occupancy (i.e. >15 reads per peak). Of these binding regions, p65 occupancy was increased by TNF + dex treatment in comparison to TNF alone at 658 peaks and decreased at 425 peaks, with 400 peaks having a 4-fold or greater increase in occupancy with dex and 241 peaks exhibiting a 4-fold or greater reduction in occupancy when dex was combined with TNF. The set of sites in which p65 occupancy was enhanced by dex were evenly associated with generally modest increases and decreases in local RNAPII occupancy with dex + TNF treatment in comparison to TNF alone, whereas sites in which p65 occupancy was reduced with dex treatment were almost universally associated with reductions in RNAPII occupancy in comparison to TNF treatment (Fig. 6B). Thus, activation of GR signaling substantially modifies the p65 cistrome in association with both increases and decreases in RNAPII occupancy.
FIGURE 6.
Recruitment and exclusion of p65 by dex is associated with distinct effects on RNAPII occupancy and differential enrichment for dimeric GR binding sites. A, scaled Venn diagram depicting the effect of the addition of TNF on dex-regulated occupancy of GR (top) in comparison to the effect of dex on TNF-regulated p65 occupancy. Numbers represent unique genes associated with sites of occupancy. B, distribution of relative local RNAPII occupancy (TNF + dex compared with TNF alone, expressed as log2 of the occupancy ratio) at sites in which TNF + dex increased p65 occupancy (top) or decreased p65 occupancy (bottom) relative to TNF treatment alone. C, frequency distributions of maximal predicted GR binding affinities (on a loge scale) within p65 binding regions in which p65 occupancy was increased (black) or decreased (red) by TNF + dex co-treatment versus TNF alone, within GR binding regions in which GR recruitment was increased with dex versus veh (green) or increased with TNF + dex versus dex alone (purple). The blue line shows the distribution of maximal calculated affinities within randomly shuffled sequences from the data set corresponding to the black line. Colored arrowheads indicate mean predicted GR binding affinities within corresponding subsets.
Whether GR interactions with p65 are associated with canonical binding sites for dimeric GR or with a distinct class of sequences termed negative glucocorticoid response elements (GREs) is controversial. Therefore, we determined whether there was enrichment for dimeric GR binding sequences within p65 binding regions in which p65 occupancy was altered with dex treatment. To accomplish this, we used a position weight matrix for interactions between dimeric GR and DNA that we previously created and validated (43), in conjunction with the Patser program, which employs position weight matrices to estimate maximal binding site energies for transcription factors within specific DNA sequences (61). This system was applied to identify the maximal predicted binding affinity for dimeric GR within 301-bp fragments centered on p65 binding peaks that exhibited a 4-fold or greater increase in read numbers when dex was added to TNF treatment and a minimum of 15 reads (total of 400 sequences, supplemental Table S8). In addition, we calculated the maximal GR binding affinity within each of the p65 binding regions with 25% or less occupancy with dex + TNF treatment compared with TNF alone (total of 241 sequences with a 15-read minimum with TNF treatment; supplemental Table S9). For comparison, we also determined the peak predicted affinity for GR within 1) each of the GR binding regions that exhibited greater occupancy with TNF + dex in comparison to dex alone, 2) each of the dex-regulated GR binding regions with sequence reads of 15 or greater, and 3) within each of the 400 sequences with dex-enhanced p65 binding after subjecting them to random shuffling (a control). Remarkably, as shown in Fig. 6C, the predicted maximal GR binding affinities within p65 binding regions that show increased p65 occupancy with dex + TNF in comparison to TNF are substantially greater than predicted affinities within dex-induced GR binding peaks (Fig. 6C, compare black and green lines). Moreover, the average GR occupancy (based on ChIP-seq counts) within these 400 p65 binding regions sequences with dex treatment alone was ∼220, versus a mean GR occupancy of 12 with vehicle alone. This strongly implicates dex-regulated dimeric GR as contributing to increased p65 occupancy at numerous loci across the genome.
In contrast to these findings for sites in which dex treatment enhanced p65 occupancy, GR binding affinity within regions in which p65 occupancy was reduced by dex (Fig. 6C, red line) had an almost indistinguishable distribution from shuffled DNA (Fig. 6C, blue line). MEME-ChIP analysis also failed to reveal enrichment for GR binding motifs or so called negative GREs (supplemental File S3) within sites in which dex treatment reduced p65 occupancy. Moreover, average GR occupancy within these p65 binding regions was not substantially increased with TNF + dex (average of 12 reads) or dex treatment (average of 10 reads) relative to vehicle treatment (average of 19 reads). Taken together, these results established a clear role for ligand-induced dimeric GR in enhancing p65 occupancy, whereas sites in which dex treatment resulted in reductions in p65 binding and RNAPII recruitment did not appear to require ligand-induced GR occupancy nor the presence of recognized binding sites for GR.
Novel Targets of Cooperation between GR and p65
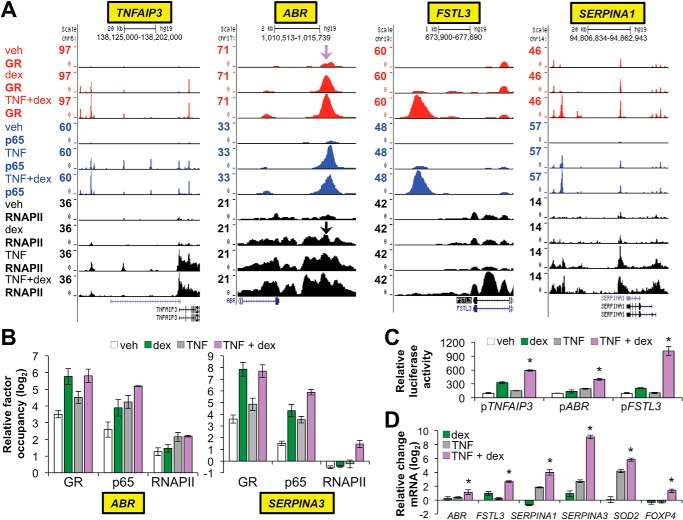
We had previously defined cooperative regulation by p65 and GR of the anti-inflammatory gene, TNFAIP3, through an intronic response element containing binding sites for dimeric GR and p65. Moreover, our ChIP-seq data identified a second region upstream of TNFAIP3 with reciprocal robustly enhanced recruitment of GR by TNF and p65 by dex (Fig. 7A). The patterns of occupancy for these TNFAIP3-associated regulatory elements contrasts factor binding at repressed pro-inflammatory genes, such as ICAM1 and CCL2, in which p65 and RNAPII occupancy was significantly curtailed by dex. Accordingly, we hypothesized that sites in which RNAPII occupancy was enhanced by dex + TNF co-treatment in comparison to treatment with TNF or dex alone may tag novel targets of GR-p65 cooperation that potentially contribute to inflammatory resolution and injury repair in the airway. To test this hypothesis, we performed differential binding analysis for RNAPII occupancy after TNF + dex treatment in comparison to vehicle (supplemental Table S10). Of 4460 differentially occupied sites (FDR < 0.05), which encompass both non-coding regulatory regions and coding regions, 1162 (26%) exhibited maximal average occupancy with TNF + dex as opposed to either treatment alone. This suggests that enhanced RNAPII occupancy is an important regulatory outcome of GR and p65 cross-talk.
FIGURE 7.
Novel targets of cooperative regulation by GR and p65. A, GR (red), p65 (blue), and RNAPII (black) ChIP-seq peaks in Beas-2B samples treated as indicated on the far left and visualized in the UCSC Genome Browser at the TNFAIP3, ABR, FSTL3, and SERPINA1 loci. The purple arrow indicates the approximate location of ChIP primers used to validate GR and p65 occupancy (see panel B) upstream of ABR; black arrows indicate the location of the ChIP primers used to validate RNAPII occupancy. B, ChIP-qPCR analysis of GR, p65, and RNAPII occupancy within the ABR and SERPINA3 loci. Bars represent mean (±S.D.) relative factor occupancy on a log2 scale, as detailed for Fig. 5. RNAPII and GR/p65 ChIP-qPCR primers for SERPINA3 targeted the transcription start site and a region ∼10 kb upstream of the transcription start site, respectively. C, relative luciferase activity of the indicated reporter constructs after transfection into Beas-2B cells and treatment with vehicle, dex, TNF, or TNF + dex for 8 h. *, p ≤ 0.05 compared with the other treatments for each reporter. D, relative mRNA levels of the indicated genes in Beas-2B cells treated with vehicle, dex, TNF, or TNF + dex for 4 h. *, p ≤ 0.05 versus treatment with dex or TNF + dex for each gene as indicated.
To determine whether enhanced RNAPII occupancy with TNF + dex in comparison to TNF or dex alone was associated with potentially beneficial gene expression responses to GCs, we used the UCSC genome browser to visualize loci with evidence of likely GR-p65 cooperation based on occupancy patterns for RNAPII, GR, and p65. Whereas occupied sites could not always be clearly linked to cooperative regulatory responses within coding regions, we nevertheless identified a set of regulatory elements that appear to control the expression of novel anti-inflammatory targets of GR-p65 cooperation. Included in this set were: SERPINA1 (Fig. 7A), the gene that encodes for α1 antitrypsin, a major genetic cause of emphysema in humans (62); EDN2, whose disruption causes emphysema in mice (63); SOD2, which protects the airway epithelium from oxidative damage (64, 65); SERPINA3, which encodes the α1 anti-chymotrypsin gene, a potent inhibitor of mast cell proteases that is implicated in asthma pathogenesis (66, 67); FOXP4, which represses goblet cell differentiation and mucus secretion (68); and ABR (Fig. 7A), a RAC inhibitor whose deficiency worsens airway hyperresponsiveness and inflammation in murine allergic asthma models (69). Cooperative regulation of TNFAIP3 and IRAK3 was also reconfirmed, and several genes with less well described biologic functions, such as FSTL3 (Fig. 7A), DENND3, and GRAMD3, were also identified as controlled through a GR-p65 cooperative regulatory regime.
To provide additional validation that expression of ABR and SERPINA3 is controlled by cooperative interactions between GR and p65, we performed ChIP-qPCR validation of the ChIP-seq findings for ABR and SERPINA3, which confirmed that GR, p65, and RNAPII recruitment was equal to or maximal with combined TNF + dex treatment in comparison to either agent alone (Fig. 7B). We also cloned the presumptive cooperative GR-p65 binding regions within ABR, FSTL3, and the novel distal element for TNFAIP3 into the pGL3-Promoter vector. These three potential GR-p65 response elements each contain at least one match for the consensus binding sequences for both GR and p65 (supplemental Fig. S1). The resultant reporters were transfected into Beas-2B cells, which were subsequently treated with TNF, dex, or TNF + dex and harvested for luciferase assays. For each reporter, we found that combined treatment with TNF + dex resulted in higher luciferase activity than treatment with either dex or TNF alone (Fig. 7C), with the FSTL3 response element exhibiting particularly robust synergistic induction. We also used qRT-PCR to verify that increased RNAPII occupancy with TNF + dex co-treatment in comparison to dex or TNF alone was reflective of the expression levels for ABR, FSTL3, SERPINA1, SERPINA3, SOD2, and FOXP4 under these treatment conditions. As shown in Fig. 7D, the mRNA levels for each of these genes were maximal with TNF + dex co-treatment. Taken together, these data suggest that cooperation between GR and p65 increases the expression of numerous genes linked to repressing airway epithelial inflammation and lung disease.
Discussion
Whereas the classical notion that GCs exert therapeutic effects by causing monomeric GR to repress inflammatory transcription factors through physical tethering is now generally recognized as inadequate (9), substantial questions regarding mechanism of inflammatory repression by GCs remain unanswered. Here, in airway epithelial cells, we have shown that a subset of interactions between GR and p65 result in enhanced p65 occupancy and/or RNAPII recruitment, frequently in association with binding sites that are energetically favorable for interactions with dimeric GR. Moreover, cooperative interactions between GR and p65 appear to maintain or augment the expression of a number of genes with known or likely anti-inflammatory properties in the lung, including SERPINA1, SERPINA3, FOXP4, ABR, and SOD2, in addition to A20/TNFAIP3, which we demonstrate is required for full suppressive function of GCs. We also uncovered GC-mediated repression of TNF-induced loci, such as TNFAIP2, in which no substantial changes in GR occupancy were observed with either TNF + dex co-treatment or dex treatment alone in comparison to vehicle, indicating that cis-acting ligand-induced GR occupancy is not a universal requirement for significant reductions in RNAPII and p65 occupancy. Taken together, our data support a model in which cooperative regulation of anti-inflammatory genes by GR and p65 is a major mechanism underlying the therapeutic effects of GCs in the airway.
Interactions between GCs and inflammatory stimuli have been previously studied on a genome-wide basis in other cell types (70), including at least two publications on overlap between the GR and p65 cistromes that identified various possible regulatory consequences of GR-p65 interactions (12, 13). However, these prior studies did not specifically recognize cooperative regulation of anti-inflammatory targets by GR and p65 as a potential mechanism underlying the therapeutic effect of GCs. Instead, several studies, including our own publication on regulation of TNFAIP3 by GR and p65 and subsequent work by other groups on Sphk1 and IRAK3 (37, 38), have established that this principal applies on a case-by-case basis to these individual genes. In the work presented here, we have extended these findings using genome-wide methodology to identify numerous additional genes that are regulated through this paradigm. Furthermore, we have verified GR-p65 cooperation by reporter assays at three previously uncharacterized response elements. We have also provided evidence for widespread recruitment of RNAPII by dimeric GR and p65, which thus appears to be a major regulatory outcome for interactions between these two factors. Although the role of the newly defined targets of GR-p65 cooperation in mediating the therapeutic effects of GCs in airway-centered diseases remains to be experimentally confirmed, the established GC-independent functions of the genes highlighted in Fig. 7 strongly implicate GR-p65 cooperation as an important mechanism underlying GC efficacy.
The complexity of regulatory outcomes and downstream transcription factor occupancy patterns associated with simultaneous activation of TNF and GC signaling may serve to obfuscate potential sites of GR-p65 cooperation. For example, in addition to the cooperative intronic response element within TNFAIP3 we defined previously and the new upstream element highlighted in this work, the TNFAIP3 locus contains several p65 binding regions that did not appear to be targets of GR-p65 cooperation, as dex treatment resulted in reduced p65 occupancy at these sites (see Fig. 7). The activity of these p65 response elements that did not cooperate with GR were likely repressed through the effects of TNFAIP3 itself, with additional negative regulators of NF-κB that were induced by GCs, such as NFKBIA and DUSP1 (71–73), potentially playing a similar repressive role. Thus, enhancers subject to GR-p65 cooperation may only serve to maintain or limit dex-mediated reductions in RNAPII occupancy at loci that are regulated through multiple p65 binding regions. Defining regulatory outcomes of co-occupied elements simply based on gross changes in RNAPII occupancy within the closest linked coding region, an approach used in other cistrome-based studies of GR (13), is therefore unlikely to correctly assign the regulatory contribution of specific GR-p65 response elements. Local transcription measured by GRO-seq at enhancers may be a more sensitive way to determine specifically whether an enhancer is co-induced by GR and NF-κB (74, 75).
Controversy exists regarding the role of specific sequences in contributing to GR-mediated repression. For example, it has been argued that GR is capable of binding so-called negative glucocorticoid response elements that mediate gene repression (76). Others, however, have found no evidence for negative GREs and suggest instead that repression can occur through standard dimeric GR binding sites (12). It has also recently been argued that under physiologic levels of hormone, GR binds to potentially activating “half-sites” near inflammatory genes as a monomer, with exogenous ligand causing evacuation of these sites and redistribution of RNAPII to targets of dimeric GR-mediated gene induction, resulting indirectly in repression through competition for both GR and RNAPII (53). Our data, however, do not provide strong support for any of these models serving as a unifying basis for GR-mediated repression. None of the data sets we interrogated with MEME-ChIP showed enrichment for negative GREs. Similarly, we found minimal evidence for dimeric GR binding sites within either the set of p65 binding regions in which dex treatment reduced occupancy nor within the set of GR binding regions in which TNF treatment enhanced GR binding. We also observed a range of possible occupancy patterns for GR in association with dex-repressed genes, including no obvious changes in GR occupancy between vehicle, dex, and dex + TNF treatment at the TNFAIP2 locus despite marked decreases in RNAPII occupancy with dex + TNF in comparison to TNF treatment. This alteration in RNAPII levels within the TNFAIP2 coding region is unlikely to be secondary to dex-activated GR recruiting limiting amounts of RNAPII to other genomic regions, as many genes showed no change in RNAPII levels after dex treatment. Based on the GR binding pattern, we cannot exclude the possibility that repression of TNFAIP2 is due entirely to the activity of genes that are induced by GR, such as TNFAIP3/A20. Moreover, as ICAM1 and CCL2 exhibited declines in RNAPII that were comparable with those we observed at TNFAIP2, the association between repression and GR occupancy at these loci in airway epithelial cells is not entirely clear. Rather than active recruitment of repressors, it is conceivable that a primary role for GR in these regions is to increase the on/off rate and disassembly of p65-containing activating complexes (77). Support for this versus other models of GR activity within promoter-proximal p65-occupied regions will require additional experimentation.
What is the significance of GR occupancy under basal conditions (i.e. occupancy in the absence of supplemental dex treatment)? We previously attributed this phenomenon, which has been observed in earlier studies by us and other groups (36, 53, 55, 78, 79), to the activity of glucocorticoid-like molecules presumed to be present at low levels within the culture medium. However, several aspects of our ChIP-seq data are incompatible with this notion. First, there was little to no GR occupancy detected under basal culture conditions at binding sites within the KLF15 (Fig. 3B) and PER1 loci, which others have shown are robustly occupied by GR after exposure to low nanomolar concentrations of supplemental ligand (80). Second, whereas GR occupancy was markedly reduced at some sites after dex treatment, other sites had dramatically increased occupancy, and still other regions exhibited minimal changes with the addition of supplemental ligand (see Figs. 3C and 5, A–C). Thus, dex treatment does not simply increase occupancy engendered by possible glucocorticoid-like molecules in the culture medium but instead induces qualitatively different interactions between GR and chromatin. We envision three remaining possible explanations. First, although GR knockdown resulted in a ∼50% reduction in GR occupancy within HHAT (Fig. 3D), which exhibited a robust GR peak in the absence of dex (Fig. 3C), it remains theoretically possible that our vehicle treatment ChIP data reflects interactions between the GR antibody we used and a non-GR protein. Alternatively, a ligand that, in comparison to dex and other synthetic GR agonists, confers markedly distinct biologic properties to GR could be present in the medium. A third explanation is that Beas-2B cells may, as a result of alternate splicing or post-translational modification, harbor at least two species of GR with different biochemical characteristics. Stochastic alternate folding could also result in multiple GR protein moieties, some of which may be incompatible with the formation of apoGR, the canonical unliganded GR species that is retained in the cytoplasm through interactions with chaperones such as HSP90 in the absence of hormone (77). We are in the process of testing each of these not necessarily mutually exclusive possibilities experimentally. The biologic consequences of presumptive GR occupancy in the absence of supplemental ligand also remain to be determined.
Although they have been in clinical use for >50 years, GCs are still by far the most effective drug class for treating many inflammatory diseases. As such, the mechanisms through which they exert their potent therapeutic effects are likely to represent novel pharmacologic targets and encompass pathways whose disruption engenders resistance to GC-based therapies, which complicates subsets of most generally GC-responsive diseases. In this work we have presented strong evidence that cooperative regulation of negative feedback control of inflammation by GR and p65 is a key mechanistic underpinning of therapeutic responses to GCs in airway epithelial cells. A logical extension of our data is that cooperation with other transcription factors, such as STAT family members, may contribute to GC efficacy in other inflammatory contexts. In that regard, reminiscent of our results for cistrome-based interactions between GR and p65, STAT3 engages in complex cross-talk with GR that is incompatible with a simple tethering-based trans-repression model (11). Moreover, our ChIP-seq data (supplemental Tables S2 and S3) revealed robust recruitment of GR and RNAPII to the PTPN1 locus. PTPN1 is a direct target of the STAT family that represses STAT-dependent responses to cytokines, including the asthma-associated cytokine, IL4 (11, 81). We thus speculate that GR cooperatively regulates negative feedback responses to diverse inflammatory signals, providing a general mechanistic explanation for the broad potency of GC-based therapies in treating a multitude of immune-mediated diseases.
Author Contributions
V. K. designed, performed, and interpreted experiments and co-wrote the manuscript. S. K. S. designed, performed, and interpreted the experiments and co-wrote the manuscript. M. O. A. performed and interpreted experiments and reviewed the manuscript. R. B. performed primary airway epithelial experiments and reviewed the manuscript. H. W. C. designed primary airway epithelial experiments and reviewed the manuscript. T. L. P. analyzed and interpreted ChIP-seq data and reviewed the manuscript. A. N. G. conceived the project, designed and interpreted experiments, and co-wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Katrina Diener for expert technical assistance and Robert Newton for critical review of the manuscript. ChIP sequencing was performed at the University of Colorado Cancer Center, which is supported by National Institutes of Health Grant P30-CA046934 (NCI).
This work was supported by NHLBI, National Institutes of Health Grant R01HL109557 (to A. N. G.). This work was also supported by institutional funds from National Jewish Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Files S1–S3, Tables S1–S10, and Fig. S1.
- GC
- glucocorticoid
- GR
- glucocorticoid receptor
- dex
- dexamethasone
- seq
- sequencing
- RNAPII
- RNA polymerase II
- HRV-1A
- human rhinovirus serotype 1A
- TCID50
- 50% tissue culture infective dose
- qRT
- quantitative real-time
- FDR
- false discovery rate
- qPCR
- quantitative PCR.
References
- 1.Bateman E. D., Hurd S. S., Barnes P. J., Bousquet J., Drazen J. M., FitzGerald M., Gibson P., Ohta K., O'Byrne P., Pedersen S. E., Pizzichini E., Sullivan S. D., Wenzel S. E., and Zar H. J. (2008) Global strategy for asthma management and prevention: GINA executive summary. Eur. Respir. J. 31, 143–178 [DOI] [PubMed] [Google Scholar]
- 2.Pelaia G., Muzzio C. C., Vatrella A., Maselli R., Magnoni M. S., and Rizzi A. (2015) Pharmacological basis and scientific rationale underlying the targeted use of inhaled corticosteroid/long-acting β2-adrenergic agonist combinations in chronic obstructive pulmonary disease treatment. Expert. Opin. Pharmacother. 16, 2009–2021 [DOI] [PubMed] [Google Scholar]
- 3.MacIntyre N. R. (2006) Corticosteroid therapy and chronic obstructive pulmonary disease. Respir. Care 51, 289–296 [PubMed] [Google Scholar]
- 4.Meijsing S. H. (2015) Mechanisms of glucocorticoid-regulated gene transcription. Adv. Exp. Med. Biol. 872, 59–81 [DOI] [PubMed] [Google Scholar]
- 5.Nicolaides N. C., Galata Z., Kino T., Chrousos G. P., and Charmandari E. (2010) The human glucocorticoid receptor: molecular basis of biologic function. Steroids 75, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMaster A., and Ray D. W. (2008) Drug insight: selective agonists and antagonists of the glucocorticoid receptor. Nat. Clin. Pract. Endocrinol. Metab. 4, 91–101 [DOI] [PubMed] [Google Scholar]
- 7.De Bosscher K., Vanden Berghe W., and Haegeman G. (2003) The interplay between the glucocorticoid receptor and nuclear factor-κB or activator protein-1: molecular mechanisms for gene repression. Endocr. Rev. 24, 488–522 [DOI] [PubMed] [Google Scholar]
- 8.Schäcke H., Berger M., Rehwinkel H., and Asadullah K. (2007) Selective glucocorticoid receptor agonists (SEGRAs): novel ligands with an improved therapeutic index. Mol. Cell Endocrinol. 275, 109–117 [DOI] [PubMed] [Google Scholar]
- 9.Clark A. R., and Belvisi M. G. (2012) Maps and legends: the quest for dissociated ligands of the glucocorticoid receptor. Pharmacol. Ther. 134, 54–67 [DOI] [PubMed] [Google Scholar]
- 10.King E. M., Chivers J. E., Rider C. F., Minnich A., Giembycz M. A., and Newton R. (2013) Glucocorticoid repression of inflammatory gene expression shows differential responsiveness by transactivation- and transrepression-dependent mechanisms. PLoS ONE 8, e53936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langlais D., Couture C., Balsalobre A., and Drouin J. (2012) The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol. Cell 47, 38–49 [DOI] [PubMed] [Google Scholar]
- 12.Uhlenhaut N. H., Barish G. D., Yu R. T., Downes M., Karunasiri M., Liddle C., Schwalie P., Hübner N., and Evans R. M. (2013) Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell 49, 158–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao N. A., McCalman M. T., Moulos P., Francoijs K. J., Chatziioannou A., Kolisis F. N., Alexis M. N., Mitsiou D. J., and Stunnenberg H. G. (2011) Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res. 21, 1404–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biddie S. C., John S., Sabo P. J., Thurman R. E., Johnson T. A., Schiltz R. L., Miranda T. B., Sung M. H., Trump S., Lightman S. L., Vinson C., Stamatoyannopoulos J. A., and Hager G. L. (2011) Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell 43, 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandevyver S., Dejager L., Tuckermann J., and Libert C. (2013) New insights into the anti-inflammatory mechanisms of glucocorticoids: an emerging role for glucocorticoid receptor-mediated transactivation. Endocrinology 154, 993–1007 [DOI] [PubMed] [Google Scholar]
- 16.Holden N. S., George T., Rider C. F., Chandrasekhar A., Shah S., Kaur M., Johnson M., Siderovski D. P., Leigh R., Giembycz M. A., and Newton R. (2014) Induction of regulator of G-protein signaling 2 expression by long-acting β2-adrenoceptor agonists and glucocorticoids in human airway epithelial cells. J. Pharmacol. Exp. Ther. 348, 12–24 [DOI] [PubMed] [Google Scholar]
- 17.Vandevyver S., Dejager L., Van Bogaert T., Kleyman A., Liu Y., Tuckermann J., and Libert C. (2012) Glucocorticoid receptor dimerization induces MKP1 to protect against TNF-induced inflammation. J. Clin. Invest. 122, 2130–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gertz J., Savic D., Varley K. E., Partridge E. C., Safi A., Jain P., Cooper G. M., Reddy T. E., Crawford G. E., and Myers R. M. (2013) Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol. Cell 52, 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.So A. Y., Chaivorapol C., Bolton E. C., Li H., and Yamamoto K. R. (2007) Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 3, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roesler W. J., and Park E. A. (1998) Hormone response units: one plus one equals more than two. Mol. Cell. Biochem. 178, 1–8 [DOI] [PubMed] [Google Scholar]
- 21.Stafford J. M., Waltner-Law M., and Granner D. K. (2001) Role of accessory factors and steroid receptor coactivator 1 in the regulation of phosphoenolpyruvate carboxykinase gene transcription by glucocorticoids. J. Biol. Chem. 276, 3811–3819 [DOI] [PubMed] [Google Scholar]
- 22.John S., Sabo P. J., Thurman R. E., Sung M. H., Biddie S. C., Johnson T. A., Hager G. L., and Stamatoyannopoulos J. A. (2011) Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43, 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert W. M., Xu C. F., Neubert T. A., Chao M. V., Garabedian M. J., and Jeanneteau F. D. (2013) Brain-derived neurotrophic factor signaling rewrites the glucocorticoid transcriptome via glucocorticoid receptor phosphorylation. Mol. Cell. Biol. 33, 3700–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerber A. N. (2015) Glucocorticoids and the Lung. Adv. Exp. Med. Biol. 872, 279–298 [DOI] [PubMed] [Google Scholar]
- 25.Kraft M., Martin R. J., Lazarus S. C., Fahy J. V., Boushey H. A., Lemanske R. F. Jr., Szefler S. J., and Asthma Clinical Research, N. (2003) Airway tissue mast cells in persistent asthma: predictor of treatment failure when patients discontinue inhaled corticosteroids. Chest 124, 42–50 [DOI] [PubMed] [Google Scholar]
- 26.Walsh G. M., Sexton D. W., and Blaylock M. G. (2003) Corticosteroids, eosinophils and bronchial epithelial cells: new insights into the resolution of inflammation in asthma. J. Endocrinol. 178, 37–43 [DOI] [PubMed] [Google Scholar]
- 27.Hallstrand T. S., Hackett T. L., Altemeier W. A., Matute-Bello G., Hansbro P. M., and Knight D. A. (2014) Airway epithelial regulation of pulmonary immune homeostasis and inflammation. Clin. Immunol. 151, 1–15 [DOI] [PubMed] [Google Scholar]
- 28.Hiemstra P. S., McCray P. B. Jr., and Bals R. (2015) The innate immune function of airway epithelial cells in inflammatory lung disease. Eur. Respir. J. 45, 1150–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Q., and Chu H. W. (2009) Role of infections in the induction and development of asthma: genetic and inflammatory drivers. Expert Rev. Clin. Immunol. 5, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez F. D., and Vercelli D. (2013) Asthma. Lancet 382, 1360–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards M. R., Johnson M. W., and Johnston S. L. (2006) Combination therapy: synergistic suppression of virus-induced chemokines in airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 34, 616–624 [DOI] [PubMed] [Google Scholar]
- 32.Zhang N., Truong-Tran Q. A., Tancowny B., Harris K. E., and Schleimer R. P. (2007) Glucocorticoids enhance or spare innate immunity: effects in airway epithelium are mediated by CCAAT/enhancer binding proteins. J. Immunol. 179, 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly M. M., King E. M., Rider C. F., Gwozd C., Holden N. S., Eddleston J., Zuraw B., Leigh R., O'Byrne P. M., and Newton R. (2012) Corticosteroid-induced gene expression in allergen-challenged asthmatic subjects taking inhaled budesonide. Br. J. Pharmacol. 165, 1737–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodruff P. G., Boushey H. A., Dolganov G. M., Barker C. S., Yang Y. H., Donnelly S., Ellwanger A., Sidhu S. S., Dao-Pick T. P., Pantoja C., Erle D. J., Yamamoto K. R., and Fahy J. V. (2007) Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc. Natl. Acad. Sci. U.S.A. 104, 15858–15863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhakta N. R., Solberg O. D., Nguyen C. P., Nguyen C. N., Arron J. R., Fahy J. V., and Woodruff P. G. (2013) A qPCR-based metric of Th2 airway inflammation in asthma. Clin. Transl. Allergy 3, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altonsy M. O., Sasse S. K., Phang T. L., and Gerber A. N. (2014) Context-dependent cooperation between nuclear factor κB (NF-κB) and the glucocorticoid receptor at a TNFAIP3 intronic enhancer: a mechanism to maintain negative feedback control of inflammation. J. Biol. Chem. 289, 8231–8239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyata M., Lee J. Y., Susuki-Miyata S., Wang W. Y., Xu H., Kai H., Kobayashi K. S., Flavell R. A., and Li J. D. (2015) Glucocorticoids suppress inflammation via the up-regulation of negative regulator IRAK-M. Nat. Commun. 6, 6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vettorazzi S., Bode C., Dejager L., Frappart L., Shelest E., Klaßen C., Tasdogan A., Reichardt H. M., Libert C., Schneider M., Weih F., Henriette Uhlenhaut N., David J. P., Gräler M., Kleiman A., and Tuckermann J. P. (2015) Glucocorticoids limit acute lung inflammation in concert with inflammatory stimuli by induction of SphK1. Nat. Commun. 6, 7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Q., Jiang D., Smith S., Thaikoottathil J., Martin R. J., Bowler R. P., and Chu H. W. (2012) IL-13 dampens human airway epithelial innate immunity through induction of IL-1 receptor-associated kinase M. J. Allergy Clin. Immunol. 129, 825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berman R., Huang C., Jiang D., Finigan J. H., Wu Q., and Chu H. W. (2014) MUC18 differentially regulates pro-inflammatory and anti-viral responses in human airway epithelial cells. J. Clin. Cell Immunol. 5, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Q., van Dyk L. F., Jiang D., Dakhama A., Li L., White S. R., Gross A., and Chu H. W. (2013) Interleukin-1 receptor-associated kinase M (IRAK-M) promotes human rhinovirus infection in lung epithelial cells via the autophagic pathway. Virology 446, 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasse S. K., Mailloux C. M., Barczak A. J., Wang Q., Altonsy M. O., Jain M. K., Haldar S. M., and Gerber A. N. (2013) The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol. Cell. Biol. 33, 2104–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasse S. K., Zuo Z., Kadiyala V., Zhang L., Pufall M. A., Jain M. K., Phang T. L., Stormo G. D., and Gerber A. N. (2015) Response element composition governs correlations between binding site affinity and transcription in glucocorticoid receptor feed-forward loops. J. Biol. Chem. 290, 19756–19769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B., and Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu T. (2014) Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol. Biol. 1150, 81–95 [DOI] [PubMed] [Google Scholar]
- 46.Li Q., Brown J. B., Huang H., and Bickel P. J. (2011) Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 [Google Scholar]
- 47.Liang K., and Keles S. (2012) Detecting differential binding of transcription factors with ChIP-seq. Bioinformatics 28, 121–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu G., Wang L. G., and He Q. Y. (2015) ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 [DOI] [PubMed] [Google Scholar]
- 49.Kadiyala V., Sasse S. K., Altonsy M. O., Phang T. L., and Gerber A. N. (2016) Cistrome analysis of glucocorticoid receptor activity in bronchial epithelial cells defines novel mechanisms of steroid efficacy. Ann. Am. Thorac. Soc. 13, S103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.ENCODE Project Consortium (2011) A user's guide to the encyclopedia of DNA elements (ENCODE). PLos Biol. 9, e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57, 289–300 [Google Scholar]
- 52.Sacta M. A., Chinenov Y., and Rogatsky I. (2016) Glucocorticoid signaling: an update from a genomic perspective. Annu. Rev. Physiol. 78, 155–180 [DOI] [PubMed] [Google Scholar]
- 53.Lim H. W., Uhlenhaut N. H., Rauch A., Weiner J., Hübner S., Hübner N., Won K. J., Lazar M. A., Tuckermann J., and Steger D. J. (2015) Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 25, 836–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raney B. J., Dreszer T. R., Barber G. P., Clawson H., Fujita P. A., Wang T., Nguyen N., Paten B., Zweig A. S., Karolchik D., and Kent W. J. (2014) Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics 30, 1003–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reddy T. E., Pauli F., Sprouse R. O., Neff N. F., Newberry K. M., Garabedian M. J., and Myers R. M. (2009) Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 19, 2163–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Machanick P., and Bailey T. L. (2011) MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., and Noble W. S. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Starick S. R., Ibn-Salem J., Jurk M., Hernandez C., Love M. I., Chung H. R., Vingron M., Thomas-Chollier M., and Meijsing S. H. (2015) ChIP-exo signal associated with DNA-binding motifs provides insight into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res. 25, 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki S., Singhirunnusorn P., Nakano H., Doi T., Saiki I., and Sakurai H. (2006) Identification of TNF-α-responsive NF-κB p65-binding element in the distal promoter of the mouse serine protease inhibitor SerpinE2. FEBS Lett. 580, 3257–3262 [DOI] [PubMed] [Google Scholar]
- 60.Deregowski V., Delhalle S., Benoit V., Bours V., and Merville M. P. (2002) Identification of cytokine-induced nuclear factor-κB target genes in ovarian and breast cancer cells. Biochem. Pharmacol. 64, 873–881 [DOI] [PubMed] [Google Scholar]
- 61.Hertz G. Z., and Stormo G. D. (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15, 563–577 [DOI] [PubMed] [Google Scholar]
- 62.Thun G. A., Imboden M., Ferrarotti I., Kumar A., Obeidat M., Zorzetto M., Haun M., Curjuric I., Couto Alves A., Jackson V. E., Albrecht E., Ried J. S., Teumer A., Lopez L. M., Huffman J. E. et al. (2013) Causal and synthetic associations of variants in the SERPINA gene cluster with α1-antitrypsin serum levels. PLoS Genet. 9, e1003585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang I., Bramall A. N., Baynash A. G., Rattner A., Rakheja D., Post M., Joza S., McKerlie C., Stewart D. J., McInnes R. R., and Yanagisawa M. (2013) Endothelin-2 deficiency causes growth retardation, hypothermia, and emphysema in mice. J. Clin. Invest. 123, 2643–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siedlinski M., van Diemen C. C., Postma D. S., Vonk J. M., and Boezen H. M. (2009) Superoxide dismutases, lung function and bronchial responsiveness in a general population. Eur. Respir. J. 33, 986–992 [DOI] [PubMed] [Google Scholar]
- 65.Hosakote Y. M., Jantzi P. D., Esham D. L., Spratt H., Kurosky A., Casola A., and Garofalo R. P. (2011) Viral-mediated inhibition of antioxidant enzymes contributes to the pathogenesis of severe respiratory syncytial virus bronchiolitis. Am. J. Respir. Crit. Care Med. 183, 1550–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horvath A. J., Irving J. A., Rossjohn J., Law R. H., Bottomley S. P., Quinsey N. S., Pike R. N., Coughlin P. B., and Whisstock J. C. (2005) The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins. J. Biol. Chem. 280, 43168–43178 [DOI] [PubMed] [Google Scholar]
- 67.Bradding P., and Arthur G. (2016) Mast cells in asthma: state of the art. Clin. Exp. Allergy 46, 194–263 [DOI] [PubMed] [Google Scholar]
- 68.Li S., Wang Y., Zhang Y., Lu M. M., DeMayo F. J., Dekker J. D., Tucker P. W., and Morrisey E. E. (2012) Foxp1/4 control epithelial cell fate during lung development and regeneration through regulation of anterior gradient 2. Development 139, 2500–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gong D., Fei F., Lim M., Yu M., Groffen J., and Heisterkamp N. (2013) Abr, a negative regulator of Rac, attenuates cockroach allergen-induced asthma in a mouse model. J. Immunol. 191, 4514–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lannan E. A., Galliher-Beckley A. J., Scoltock A. B., and Cidlowski J. A. (2012) Proinflammatory actions of glucocorticoids: glucocorticoids and TNFα coregulate gene expression in vitro and in vivo. Endocrinology 153, 3701–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shipp L. E., Lee J. V., Yu C. Y., Pufall M., Zhang P., Scott D. K., and Wang J. C. (2010) Transcriptional regulation of human dual specificity protein phosphatase 1 (DUSP1) gene by glucocorticoids. PLoS ONE 5, e13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Auphan N., DiDonato J. A., Rosette C., Helmberg A., and Karin M. (1995) Immunosuppression by glucocorticoids: inhibition of NF-κB activity through induction of IκB synthesis. Science 270, 286–290 [DOI] [PubMed] [Google Scholar]
- 73.Scheinman R. I., Cogswell P. C., Lofquist A. K., and Baldwin A. S. Jr. (1995) Role of transcriptional activation of IκBα in mediation of immunosuppression by glucocorticoids. Science 270, 283–286 [DOI] [PubMed] [Google Scholar]
- 74.Azofeifa J., Allen M., Lladser M., and Dowell R. (2016) An annotation agnostic algorithm for detecting nascent RNA transcripts in GRO-seq. IEEE/ACM Trans. Comput. Biol. Bioinform., 10.1109/TCBB.2016.2520919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Allen M. A., Andrysik Z., Dengler V. L., Mellert H. S., Guarnieri A., Freeman J. A., Sullivan K. D., Galbraith M. D., Luo X., Kraus W. L., Dowell R. D., and Espinosa J. M. (2014) Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife 3, e02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Surjit M., Ganti K. P., Mukherji A., Ye T., Hua G., Metzger D., Li M., and Chambon P. (2011) Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 145, 224–241 [DOI] [PubMed] [Google Scholar]
- 77.Freeman B. C., and Yamamoto K. R. (2002) Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296, 2232–2235 [DOI] [PubMed] [Google Scholar]
- 78.Bouazza B., Debba-Pavard M., Amrani Y., Isaacs L., O'Connell D., Ahamed S., Formella D., and Tliba O. (2014) Basal p38 mitogen-activated protein kinase regulates unliganded glucocorticoid receptor function in airway smooth muscle cells. Am. J. Respir. Cell Mol. Biol. 50, 301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu A., Josephson M. B., Diener B. L., Nino G., Xu S., Paranjape C., Orange J. S., and Grunstein M. M. (2013) Pro-asthmatic cytokines regulate unliganded and ligand-dependent glucocorticoid receptor signaling in airway smooth muscle. PLoS ONE 8, e60452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reddy T. E., Gertz J., Crawford G. E., Garabedian M. J., and Myers R. M. (2012) The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol. Cell. Biol. 32, 3756–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu X., Malumbres R., Shields B., Jiang X., Sarosiek K. A., Natkunam Y., Tiganis T., and Lossos I. S. (2008) PTP1B is a negative regulator of interleukin 4-induced STAT6 signaling. Blood 112, 4098–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.