Abstract
The zinc finger homeobox 3 (ZFHX3, also named ATBF1 for AT motif binding factor 1) is a transcription factor that suppresses prostatic carcinogenesis and induces neuronal differentiation. It also interacts with estrogen receptor α to inhibit cell proliferation and regulate pubertal mammary gland development in mice. In the present study, we examined whether and how Zfhx3 regulates lactogenic differentiation in mouse mammary glands. At different stages of mammary gland development, Zfhx3 protein was expressed at varying levels, with the highest level at lactation. In the HC11 mouse mammary epithelial cell line, an in vitro model of lactogenesis, knockdown of Zfhx3 attenuated prolactin-induced β-casein expression and morphological changes, indicators of lactogenic differentiation. In mouse mammary tissue, knock-out of Zfhx3 interrupted lactogenesis, resulting in underdeveloped glands with much smaller and fewer alveoli, reduced β-casein expression, accumulation of large cytoplasmic lipid droplets in luminal cells after parturition, and failure in lactation. Mechanistically, Zfhx3 maintained the expression of Prlr (prolactin receptor) and Prlr-Jak2-Stat5 signaling activity, whereas knockdown and knock-out of Zfhx3 in HC11 cells and mammary tissues, respectively, decreased Prlr expression, Stat5 phosphorylation, and the expression of Prlr-Jak2-Stat5 target genes. These findings indicate that Zfhx3 plays an essential role in proper lactogenic development in mammary glands, at least in part by maintaining Prlr expression and Prlr-Jak2-Stat5 signaling activity.
Keywords: development, homeobox, mammary gland, peptide hormone, prolactin, transcription factor, STAT5 phosphorylation, ZFHX3/ATBF1, lactogenic differentiation, prolactin receptor (PRLR)
Introduction
Development of the mammary gland occurs in connection with sexual development and reproduction and is regulated by multiple hormones (e.g. estrogen, progesterone, and prolactin) and growth factors (EGF, FGF, insulin-like growth factor, etc.). It can be divided into six distinct stages: embryonic, prepubertal, pubertal (the linear phase), pregnancy, lactation, and involution (the cyclic phase) (1, 2). Unlike most other organs, 5 of the 6 stages of mammary gland development occur postnatally, providing an ideal model for studying genes in both normal development and neoplastic progression. Mammary glands retain plasticity for undergoing the cyclic phase, which implies the existence of specific molecules that are able to integrate a variety of signals from hormones and growth factors. For example, the STAT genes have been demonstrated to play roles in both hormone response and growth factor signaling (3). At present, a few factors have been identified and characterized for their roles in mammary gland development in response to hormonal signaling, but many more remain to be discovered (4, 5).
The homeobox gene family contains the homeobox sequence that encodes for the homeodomain, a DNA-binding domain about 60 amino acids long. More than 200 homeodomain-containing proteins have been identified and characterized in a variety of species, most of which act as transcription factors in a wide range of critical activities during normal development and tumorigenesis (6). Several homeobox genes are expressed in mammary epithelial cells, and their functions as regulators of mammary gland development have been established using genetically modified mice (7, 8). For example, loss of Msx2, Hoxc6, Hoxa9, and Pax2 in the mammary gland leads to a series of defects during mammary gland development, including failures in side branching and lobulo-alveolar development (8, 9).
Zinc finger homeobox 3 (ZFHX3), also named ATBF1 for AT motif binding factor 1, is a large transcription factor with 23 zinc finger and 4 homeodomains (10, 11). Very few homeobox genes have more than 1 homeobox (12), and the existence of 4 homeoboxes in ZFHX3 suggests that it has a dynamic function in biological processes (13). For example, ZFHX3 is necessary for neuronal and myogenic differentiation in cell culture models (11, 14–16), and deletion of Zfhx3 in mice causes developmental defects, interrupts epithelial homeostasis, and induces neoplastic morphology in mouse prostates (17–19). In mouse mammary glands, we previously demonstrated that Zfhx3 mRNA expression varies at different stages during development, reaching the highest level at lactation (20), and that Zfhx3 regulates pubertal mammary gland development (20). In addition, both estrogen and progesterone, two hormones essential for normal mammary gland development, induce or enhance the transcription of ZFHX3 in human and mouse mammary epithelial cells (21, 22), although estrogen also causes protein degradation of ZFHX3 when too much estrogen is present (21). Taken together with the observation that deletion of Zfhx3 in mouse prostates alters the transcription level of Prlr (17), a key regulator of lactogenic differentiation in the mammary gland, we hypothesize that ZFHX3 is more relevant to lactogenic differentiation during mammary gland development.
In this study, we examined whether and how Zfhx3 regulates lactogenic development in the mammary gland using both a cell culture model and mouse model. In the HC11 mouse mammary epithelial cell line, where prolactin induces morphological and molecular changes indicative of lactogenic differentiation, knockdown of Zfhx3 expression attenuated the effect of prolactin. Consistently, deletion of Zfhx3 in mouse mammary glands prevented proper alveologenesis and lactogenic differentiation. Mechanistically, the effect of Zfhx3 was mediated at least in part by regulating the Prlr-Jak2-Stat5 signaling axis.
Experimental Procedures
Mice
Wild type C57BL/6 mice were purchased from the Academy of Military Medical Sciences (Beijing, China), and breeding was carried out following standard procedures for the collection of mammary tissues at different developmental stages. Breeding, genotyping, and preparation of mice with MMTV-Cre-mediated mammary-specific knock-out of Zfhx3 were previously described (20). All mice were fed with pathogen-free food and water and were closely monitored and humanely euthanized. At least four mice were used for each genotype at a time point for different analyses, including whole mount analysis, immunohistochemical staining, and real time PCR. All mouse experiments were approved by the Institutional Animal Care and Use Committee at College of Life Sciences of Nankai University.
Cell Culture
The human embryonic kidney cell line 293T and human breast cancer cell line MCF-7 were cultured in DMEM (Invitrogen) supplemented with 10% FBS. Human breast cancer cell lines MDA-MB-231 and T-47D were cultured in RPMI 1640 medium supplemented with 10% FBS. For prolactin treatment, T-47D cells were first incubated in serum-free medium overnight and then treated with 500 ng/ml recombinant human prolactin (R&D Systems) for indicated times.
In Vitro Lactogenic Differentiation of HC11 Cells
Mouse mammary epithelial cell line HC11 was maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS, 1× GlutaMAX (Invitrogen), 5 μg/ml insulin (Roche), and 10 ng/ml epidermal growth factor (EGF, R&D). To induce cell differentiation, HC11 cells were grown to confluence, incubated in medium deprived of EGF for 24 h, and then treated for 5 days with a combination of lactogenic hormones including 1 μm dexamethasone (Sigma), 5 μg/ml insulin, and 5 μg/ml recombinant mouse prolactin (R&D), referred to as DIP.2 The mammospheres were photographed by phase contrast microscopy using a Olympus CKX41SF inverted microscope (Olympus, Tokyo, Japan) with a Canon EOS 600D digital camera (Canon, Tokyo, Japan).
Lentivirus Production and Cell Transduction
MISSIONTM shRNAs targeting human ZFHX3 and mouse Zfhx3 in pLKO.1-puro plasmid and empty vector control plasmid were purchased from Sigma. The insert sequences for these shRNAs are listed in Table 1. Lentiviral particles were produced in 293T cells by cotransfecting pLKO.1 with pMD2.G and psPAX2 plasmids using the FuGENE 6 transfection reagent (Promega, Beijing, China) according to the manufacturer's protocol. On the day after transfection, the medium was replaced, and the cells were incubated for an additional 2 days before viral supernatant was collected. Human T-47D or mouse HC11 cells were seeded in 6-well culture plates and grown to ∼70% confluency, at which time the culture medium was replaced with fresh medium containing 8 μg/ml of Polybrene (Sigma), and 1 ml of lentiviral solution was added dropwise. 6–12 h after viral infection, the virus-containing medium was replaced with fresh medium again, and on the following day 2 μg/ml of puromycin (Sigma) was added to the medium for 3–5 days to select cells stably expressing shRNAs. RNA was extracted, and protein lysate was prepared for analysis. Cells stably expressing the desired shRNAs were maintained in medium containing 1 μg/ml of puromycin.
TABLE 1.
Sequences of shRNAs against mouse Zfhx3/human ZFHX3 in Sigma PLKO.1 vector
| Name | Sequence | Sigma ID |
|---|---|---|
| shZfhx3-8 | CCGGGCCAGGAAGAATTACGAGAATCTCGAGATTCTCGTAATTCTTCCTGGCTTTTTG | TRCN0000075408 |
| shZfhx3-9 | CCGGCCCGGACAATAAGCCGTTTAACTCGAGTTAAACGGCTTATTGTCCGGGTTTTTG | TRCN0000075409 |
| shZfhx3-11 | CCGGCCGGAAGAAGTTAGCGGATATCTCGAGATATCCGCTAACTTCTTCCGGTTTTTG | TRCN0000075411 |
| shZfhx3-12 | CCGGGCCATGCTCTTAGACTGTGATCTCGAGATCACAGTCTAAGAGCATGGCTTTTTG | TRCN0000075412 |
| shZFHX3-2 | CCGGGCGATGCTCTTAGACTGTGATCTCGAGATCACAGTCTAAGAGCATCGCTTTTT | TRCN0000013561 |
| shZFHX3-3 | CCGGGCCAGGAAGAATTATGAGAATCTCGAGATTCTCATAATTCTTCCTGGCTTTTT | TRCN0000013558 |
| shZFHX3-5 | CCGGCCCTTTAGTTTCCACAGCTAACTCGAGTTAGCTGTGGAAACTAAAGGGTTTTT | TRCN0000013560 |
Reverse Transcription and Real Time PCR
Total RNA was used for cDNA synthesis with the Moloney murine leukemia virus reverse transcriptase system (Promega). RT-PCR was performed on the Mastercycler ep Realplex real time PCR system (Eppendorf, Hamburg, Germany) using the SYBR premix Ex Taq (TaKaRa Bio Inc., Tianjin, China). Human GAPDH or mouse β-actin gene was used as the internal control in real time PCR. Quantification of gene expression levels was determined using the comparative ▵▵CT method. Primer sequences used in real time PCR for each gene are listed in Table 2 and were from the PrimerBank (23) or the indicated references.
TABLE 2.
Primer sequences for real time PCR
| Gene | Sequence (5′-3′), forward/reverse | Reference |
|---|---|---|
| hZFHX3 | TGTTCCAGATCGAGATGGGAAT/CTTTCCCAGATCCTCTGAGGTTT | Ref. 20 |
| hGAPDH | GGTGGTCTCCTCTGACTTCAACA/GTTGCTGTAGCCAAATTCGTTGT | Ref. 20 |
| mZfhx3 | AGAGCAAGAGGGCAGCGTCATC/CGGTTCACGTCAGCGTTGCTATAC | Ref. 19 |
| mCsn2 | ACATCCAGCCTATTGCTCAAC/GAGGGGCATCTGTTTGTGCT | Ref. 23 |
| mCtgf | GGGCCTCTTCTGCGATTTC/ATCCAGGCAAGTGCATTGGTA | Ref. 23 |
| mPrlr total | GTGGAATCCTGGGTCAGATG/GGGCCACTGGTTTTGTAGTC | Ref. 80 |
| mPrlr long | ATAAAAGGATTTGATACTCATCTGCTAGAG/TGTCATCCACTTCCAAGAACTCC | Ref. 80 |
| mId2 | CGACCCGATGAGTCTGCTCTA/GACGATAGTGGGATGCGAGTC | Ref. 23 |
| mElf5 | TGGACTCCGTAACCCATAGCACCT/ATTGCTTAAGGGCTGATGGCATCG | Ref. 81 |
| mLmo4 | CCCAAGGCAACGTGTATCAT/GGGCTGTGGGTCTATCATGT | Ref. 75 |
| mCebpb | AACTCTCTGCTTCTCCCTCTG/AAGCCCGTAGGAACATCTTT | Ref. 82 |
| mActin | GATCTGGCACCACACCTTCT/GGGGTGTTGAAGGTCTCAAA | Ref. 19 |
Immunoblotting
Cell lysate preparation and Western blotting were performed as previously described (20). Briefly, cells or tissues were lysed using radioimmune precipitation assay buffer supplemented with protease inhibitor (cOmplete, Roche) and phosphatase inhibitor (PhosStop, Roche). Protein concentrations were determined using a Pierce BCA protein assay kit (Thermo Fisher) according to the manufacturer's protocol. Cell lysates containing equal amounts of proteins were subjected to SDS-PAGE, transferred to a nitrocellulose membrane (PALL, Beijing, China), incubated with antibodies, and visualized using WesternBright ECL (Advansta, CA). Actin or tubulin was used as a loading control. For the detection of Zfhx3 protein, which has an estimated molecular mass of 406 kDa, 4% SDS-PAGE was run at 100 V for 3.5 h. The spectra multicolor high range protein marker (Thermos Fisher, catalog no. 26625) was used in those 4% gels, and the estimated molecular mass of Zfhx3 (406 kDa) is indicated in relevant figure panels because the highest band in the molecular mass marker (300 kDa) was still too distant from the Zfhx3 band. For the detection of other proteins, 8–12% gels were used with the PageRuler Plus Prestained protein marker (Thermo Fisher, catalog no. 26616), and molecular mass markers in kDa are indicated in applicable figure panels. The antibodies and dilution factors used in this study are listed in Table 3.
TABLE 3.
Antibodies, dilution factors, and antigen retrieval buffer used in Western blotting (WB) and/or IHC staining
| Protein | ID (lot number) | Application and dilution | Antigen retrieval buffer |
|---|---|---|---|
| ZFHX3 | Anti-ZFHX3 (1–160 amino acids) (27) | WB (1:800) | |
| ZFHX3 | Anti-ZFHX3 (2107–2147 amino acids) (27) | IHC (1:1000) | Citrate, pH 6 |
| STAT5 | sc-835 (G0111, Santa Cruz) | WB (1:500) | |
| p-STAT5(Y694) | #9351 (Lot 7 of Ref 06/2012, Cell Signaling Technology) | WB (1:1000); IHC (1:1000) | Citrate, pH 6 |
| PRLR | sc-20992 (J1910, Santa Cruz) | WB (1:1000); IHC (1:200) | Tris-EDTA, pH 9 |
| CSN2 | sc-17969 (C1605, Santa Cruz) | WB (1:200); IHC (1:200) | Citrate, pH 6 |
| Tubulin | sc-23949 (B0408, Santa Cruz) | WB (1:3000) | |
| Actin | A2228 (Sigma) | WB (1:5000) |
Whole Mount Preparation and Histological Analysis of Mammary Glands
Whole mount analysis of mouse mammary glands was performed as previously described (20) except that whole mount glands were stained with carmine alum instead of hematoxylin. Briefly, the fourth abdominal mammary gland on the left was dissected at lactation day 2, placed between two glass slides and spread by placing weights on top of the slide for 10 min, followed by fixing in 3.7% formalin overnight. Tissues were hydrated, stained with carmine alum, and cleared with xylene. For histological analysis, formalin-fixed tissues were processed for sectioning, and stained with hematoxylin and eosin (H&E).
Immunohistochemistry
Immunohistochemical staining was performed on sections of formalin-fixed, paraffin-embedded tissues. Briefly, tissue sections were first deparaffinized and rehydrated following standard procedure. Antigen retrieval was carried out by heating in sodium citrate buffer or Tris-EDTA buffer using a pressure cooker for 3 min at full pressure. Sections were incubated with primary antibodies at 4 °C overnight and then with the HRP solution and the DAB-chromogen. All sections were counterstained in hematoxylin, dehydrated in ethanol, cleared in xylene, and covered with a coverslip with Eukitt quick-hardening mounting medium (Sigma). Antibodies, dilution factors, and antigen retrieval buffers are listed in Table 3.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 5 software (GraphPad, San Diego, CA). A two-tailed Student's t test was used to determine statistical differences between two groups, and one-way analysis of variance was used to compare three or more groups. p values less than 0.05 were considered statistically significant.
Results
Expression of Zfhx3 Protein during Mammary Gland Development
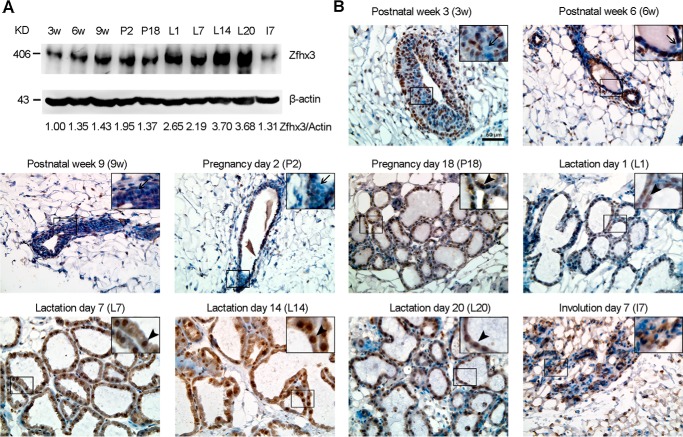
We previously measured Zfhx3 mRNA expression at different stages of mammary gland development and found that the Zfhx3 mRNA level varies at different stages, being the highest in late pregnancy and lactation (20). Here we determined Zfhx3 protein expression. Mammary glands were collected from mice at postnatal weeks 3, 6, and 9; pregnancy days 2 and 18; lactation days 1, 7, 14, and 20; and involution day 7. Western blotting demonstrated that the Zfhx3 protein level increased from weeks 3 to 9 after birth, further increased to an intermediate level during pregnancy, increased to and maintained a maximum level during lactation, and then dramatically reduced during involution (Fig. 1A). Even during lactation, the Zfhx3 protein level kept increasing, being higher at days 14 and 20 than at days 1 and 7 (Fig. 1A). This pattern of expression is almost identical to that for Zfhx3 mRNA expression (20).
FIGURE 1.
Dynamic expression of Zfhx3 protein at different stages of postnatal mammary gland development. A, detection of Zfhx3 protein by Western blotting in C57BL/6 female mouse mammary tissues at indicated developmental stages. β-Actin served as the loading control. 3w, postnatal week 3; 6w, postnatal week 6; 9w, postnatal week 9; P2, pregnancy day 2; P18, pregnancy day 18; L1, lactation day 1; L7, lactation day 7; L14, lactation day 14; L20, lactation day 20; I7, involution day 7. The data are representative of three mice per group. Band intensities were quantified using the ImageJ program, and ratios of Zfhx3/β-actin, normalized to that of postnatal week 3, are shown at the bottom. B, IHC staining in mammary glands of C57BL/6 female mice at indicated developmental stages. Arrows show Zfhx3-negative body cells in the terminal end bud (postnatal week 3, prepuberty) or Zfhx3-negative ductal luminal cells at postnatal weeks 6 and 9 (puberty) and pregnancy day 2 (early pregnancy); arrowheads show mature alveolar luminal (Zfhx3-positive) cells in pregnancy day 18 (late pregnancy) and lactation days 1, 7, 14, and 20 (lactation). A representative image from three mice is shown for each stage. Scale bar, 50 μm.
We also performed IHC staining to detect Zfhx3 protein in the mammary gland at each developmental stage (Fig. 1B). We observed that Zfhx3 was expressed in both the epithelial cells and some types of stromal cells and was mainly localized in the nucleus. In a typical terminal end bud structure at postnatal week 3 (prepuberty) mammary gland, we could see two morphologically distinct cell types: the single-layered cap cells (outer layer, giving rise to basal cells) and multiple-layered body cells (centrally located, giving rise to luminal cells) (2). We observed that a significant portion of body cells in the terminal end bud were Zfhx3-negative (indicated by arrows in Fig. 1). During the pubertal and the early pregnancy stages when ductal morphogenesis occurs, we observed that some of the ductal luminal cells were also Zfhx3-negative (indicated by arrows in Fig. 1), but in contrast nearly all the mature alveolar luminal cells (indicated by arrowheads in Fig. 1) from the late pregnancy stage to the lactation stage were Zfhx3-positive, and at day 7 of involution Zfhx3 expression dramatically decreased. This pattern of expression strongly suggests that Zfhx3 may play a role in alveologenesis and lactogenic hormone-induced differentiation of mammary epithelial cells at lactation.
Zfhx3 Is Necessary for Prolactin-induced Lactogenic Differentiation in HC11 Mammary Epithelial Cells in Culture
We first used the HC11 cell line model to test Zfhx3 function in lactogenic differentiation induced by lactogenic hormones. HC11 is a clone of the COMMA-1D mouse mammary epithelial cell line (24), derived from mammary tissue of BALB/c mice in the middle of pregnancy (25). Treatment with prolactin in the presence of insulin and glucocorticoid hormone dexamethasone (the DIP treatment) makes cultured HC11 cells produce milk protein β-casein, a marker of lactogenic differentiation (26), and form mammospheres in high density, making this cell line a widely used in vitro model of mammary gland differentiation in the context of prolactin and glucocorticoid stimulation (24).
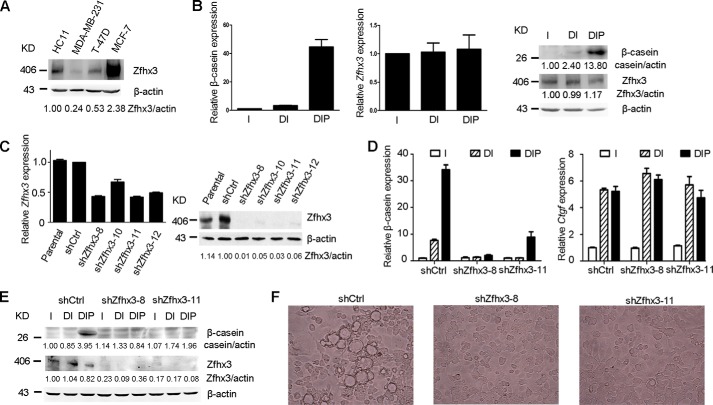
First, we measured the expression of Zfhx3 in HC11 cells. Compared with three human breast cancer cell lines with low (MDA-MB-231), intermediate (T-47D), and high (MCF-7) levels of ZFHX3 (27), HC11 cells showed an expression level higher than T-47D cells but lower than MCF-7 cells, as detected by Western blotting (Fig. 2A). We then grew HC11 cells to confluence, incubated cells in EGF-depleted medium for 24 h, and treated them with a combination of lactogenic hormones including DIP to induce β-casein expression. Consistent with previous studies (28), an induction of β-casein was detected at both the mRNA and protein levels as expected (Fig. 2B, left and right panels). Meanwhile, hormone treatment did not alter Zfhx3 expression at either the mRNA or protein level (Fig. 2B, middle and right panels). Immunofluorescence staining showed that Zfhx3 was localized in the nucleus regardless of hormone treatment (data not shown). These results validated the HC11 model for Zfhx3 study.
FIGURE 2.
Knockdown of Zfhx3 impairs prolactin-induced differentiation of the mouse HC11 mammary epithelial cells. A, detection of Zfhx3 protein expression by Western blotting in HC11 and three human breast cancer cell lines that show low, moderate, and high expression of ZFHX3, with β-actin as the loading control. B, detection of the expression of β-casein and Zfhx3 in lactogenic hormone-treated HC11 cells through RT-PCR (left and middle panels) and Western blotting (right panel). C, lentiviral expression of shRNAs against Zfhx3 (shZfhx3-8, -10, -11, and -12) knocked down Zfhx3 expression in HC11 cells, as detected by RT-PCR (left panel) and Western blotting (right panel). Parental HC11 cells were used as a positive control. D and E, knockdown of Zfhx3 attenuated prolactin-induced β-casein expression while not affecting the expression of Ctgf, a target gene of dexamethasone but not prolactin, as detected by RT-PCR (D) and Western blotting (E). F, prolactin-induced bubble-like structures, an established indicator of lactogenic hormone-induced differentiation, disappeared upon the knockdown of Zfhx3 in HC11 cells. Cells in B and D–F were treated with insulin (I), insulin plus dexamethasone (DI), or DIP for 5 days; in F only the DIP groups are shown. All Western blots were quantified using the ImageJ program. Ctrl, control.
We then knocked down Zfhx3 expression by lentivirus-mediated stable expression of shRNAs against Zfhx3 in HC11 cells. All four shRNAs induced significant knockdown of Zfhx3, as confirmed by real time PCR for Zfhx3 mRNA and Western blotting for its protein (Fig. 2C). Two of the shRNAs, shZfhx3-8 and shZfhx3-11, were more effective in Zfhx3 knockdown and thus were used for further experimentation. At day 5 of DIP treatment, β-casein mRNA (Fig. 2D, left panel) and protein (Fig. 2E) were significantly induced in the control shRNA group, whereas knockdown of Zfhx3 almost abolished the induction of β-casein (Fig. 2, D and E). Meanwhile, the connective tissue growth factor gene (Ctgf), which enhances lactogenic differentiation and only responds to dexamethasone but not to prolactin in its induction (29, 30), did not show an expression change upon the knockdown of Zfhx3 (Fig. 2D, right panel). Parallel to β-casein induction, DIP-treated HC11 cells formed mammospheres during differentiation at high density in culture (Fig. 2F, left panel) as previously reported (31), but the sphere formation was interrupted by Zfhx3 knockdown (Fig. 2F, middle and right panel). These results suggest that Zfhx3 is necessary for DIP to induce lactogenic differentiation in HC11 cells, and the effect only involves the prolactin signaling, not the glucocorticoid signaling.
Deletion of Zfhx3 Attenuates Alveologenesis and Lactogenic Differentiation in Mouse Mammary Glands
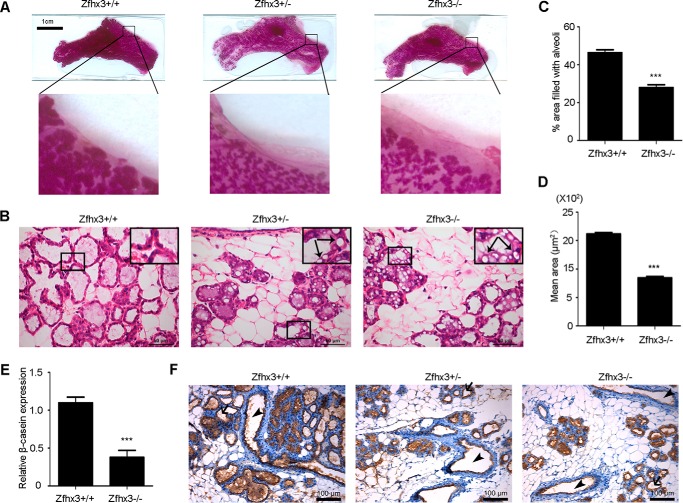
To more directly test the necessity of Zfhx3 for lactogenic differentiation, we bred mice with floxed Zfhx3 alleles to MMTV-Cre mice in which the Cre recombinase is specifically expressed in mammary epithelial cells under the MMTV promoter, as described in our previous study (20). 9-week-old MMTV-Cre/Zfhx3+/+, MMTV-Cre/Zfhx3f/+, and MMTV-Cre/Zfhx3f/f female mice were mated and sacrificed at lactation day 2. Whole mount analysis showed that Zfhx3 deletion led to defects in alveologenesis, as indicated by reduced alveolar density and size (Fig. 3A). H&E staining of tissue sections demonstrated that Zfhx3 deletion caused the accumulation of large cytoplasmic lipid droplets in the luminal layer (Fig. 3B), which has been established as an indicator of failure in secretory activation in multiple lines of genetically engineered mice (32). We estimated the proportion of areas occupied by alveoli and the mean area of a single lumen in H&E-stained tissue sections using the ImageJ program. A significant decrease was detected in both parameters when Zfhx3 was deleted (Fig. 3, C and D).
FIGURE 3.
Deletion of Zfhx3 interferes with the lactogenic development of mouse mammary gland. A and B, underdeveloped mammary glands in Zfhx3 depleted females at lactation day 2, as seen in images of mammary glands from whole mount preparation (A) and H&E staining of tissue sections (B). Arrows in B indicate large cytoplasmic lipid droplets in the luminal epithelial cells of Zfhx3-deleted mammary glands. C, quantification of the total area filled by lactating alveoli in the H&E-stained sections using ImageJ. At least four mice were analyzed, and for each section, at least 20 images (×20) were used to analyze the area occupied by alveoli. D, quantification of the mean area of a single alveoli in the mammary glands using ImageJ. A total of at least 1000 alveoli were analyzed per genotype. For C and D, the data are means ± S.E. E, expression of β-casein by real time PCR; the data are means ± S.E. F, IHC staining of β-casein at lactation in the mammary glands; arrows show empty alveolar lumen, and arrowheads show empty ducts. Statistical significance was calculated by two-tailed Student's t test. ***, p < 0.001.
We also measured the milk protein gene β-casein expression in these mice. The mRNA level of β-casein was significantly reduced by Zfhx3 deletion, as detected by real time PCR (Fig. 3E), consistent with the findings from HC11 cells. The protein expression of β-casein, as detected by IHC staining, was hardly detectable in some alveolar lumen or the mammary ducts, and the staining intensity was indistinguishable between wild type mice and those with Zfhx3 deletion (Fig. 3F), which could be due to quick emptying of the glands, as reported for IHC staining of the WAP milk protein in the Stat5a knock-out mammary glands at lactation day 1 (33). Lobulo-alveolar hypoplasia was evident in all four mice with homozygous deletion of Zfhx3.
Deletion of one Zfhx3 allele caused an intermediate abnormality in the size and density of alveoli, accumulation of cytoplasmic lipid droplets (Fig. 3B, middle panel). In addition, on day 2 after birth, although all litters from wild type mothers survived, most litters from mice with one Zfhx3 allele and all from Zfhx3-null mice died, likely because of dehydration. These findings indicate that Zfhx3 is required for proper development, maintenance, and function of structurally organized secretory lobulo-alveolar units in mammary glands.
Zfhx3 Is Required for Prolactin-induced Tyrosine Phosphorylation of Stat5
During lactogenic differentiation, prolactin binds to its receptor (PRLR), which then recruits the JAK2 kinase to phosphorylate STAT5 at Tyr694/699 to transduce the signal into the nucleus, where lactogenic genes including milk protein β-casein are induced (34–37). We therefore analyzed the phosphorylation status of Stat5 to determine whether Zfhx3 regulates alveologenesis and lactogenic differentiation by the canonical PRLR-JAK2-STAT5 signaling pathway.
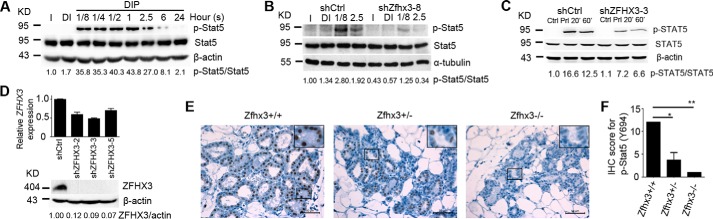
In the HC11 cell system, treatment with prolactin for 7.5 min clearly induced Stat5 phosphorylation as expected, and the phosphorylation was still detectable after 2.5 h of treatment and hardly seen after 24 h (Fig. 4A). When Zfhx3 was knocked down in HC11 cells, prolactin-induced Stat5 phosphorylation was dramatically decreased, whereas total Stat5 showed similar expression levels (Fig. 4B). In the T-47D human breast cancer cell line, which expresses a higher level of PRLR (38), prolactin treatment also significantly induced STAT5 phosphorylation as in HC11 cells (Fig. 4C). Stable knockdown of ZFHX3 expression using shRNA in T-47D cells, which was confirmed by real time PCR and Western blotting (Fig. 4D), compromised the phosphorylation of STAT5 (Fig. 4C). Similar results were obtained using a different shRNA(s) in both HC11 and T-47D cells (data not shown). These results showed that Zfhx3 is necessary for STAT5 phosphorylation not only in the normal mammary epithelial cells but also in breast cancer cell lines, indicating that ZFHX3 protein may play a role in both normal development and tumorigenesis of the breast.
FIGURE 4.
Zfhx3 is necessary for prolactin-induced phosphorylation of STAT5. A, parental HC11 cells were treated with 5 μg/ml prolactin in a time-dependent manner and Stat5 phosphorylation at tyrosine 694 (p-Y694-Stat5) was detected by Western blotting. B, mouse HC11 mammary epithelial cells with stable knockdown of Zfhx3 (shZfhx3-8) or control (shCtrl) were treated with insulin (I), insulin plus dexamethasone (DI), or DIP for 7.5 min or 2.5 h and then subjected to Western blotting to detect p-Y694-Stat5 and total Stat5. C and D, stable knockdown of ZFHX3 in human breast cancer cell line T-47D also reduced prolactin (Prl)-induced STAT5 phosphorylation, as detected by Western blotting. Knockdown efficiency was confirmed by RT-PCR and Western blotting (D). E and F, IHC staining of p-Y694-Stat5 in lactation day 2 mammary glands (E). IHC score is shown as means ± S.E. (F) and was produced by multiplying the percentage of positive cells (scored as 0 for negative, 1 for < 10%, 2 for 11–50%, 3 for 50–80%, and 4 for > 80%) with the intensity of staining (0 for negative, 1 for weak, 2 for moderate, and 3 for strong). Statistical significance was calculated by one-way analysis of variance followed by Bonferroni's test. *, p < 0.05; **, p < 0.01. All Western blots were quantified using the ImageJ program.
In mouse mammary tissues at lactation day 2 with different Zfhx3 deletion status, IHC staining demonstrated that although most epithelial cells had intense staining for phospho-Stat5 (Fig. 4E, left panel), homozygous deletion of Zfhx3 almost eliminated the staining (Fig. 4E, right panel). Deletion of one Zfhx3 allele showed an intermediate effect in both the p-Stat5 staining intensity and the number of p-Stat5-positive cells (Fig. 4E, middle panel). The IHC staining index (IHC score) for p-Stat5, which was produced by multiplying the percentage of positive cells (scored as 0 for negative, 1 for <10%, 2 for 11–50%, 3 for 50–80%, and 4 for >80%) with the intensity of staining (0 for negative, 1 for weak, 2 for moderate, and 3 for strong), was significantly lower in Zfhx3-null tissue than in normal tissue (Fig. 4F). These observations suggest that Zfhx3 plays a role in the activation of Stat5 by prolactin-Prlr signaling.
Zfhx3 Plays a Role in the Expression of Prlr
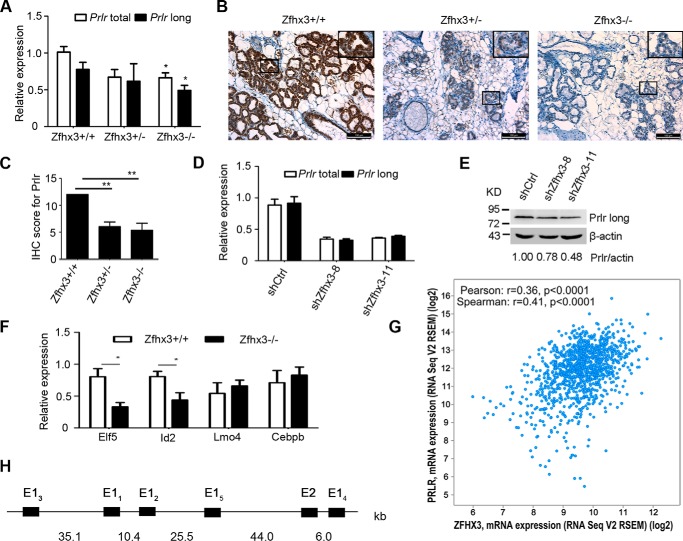
In a previous work, Yan et al. (39) carried out ChIP-seq experiments using ChIP grade antibodies of more than 100 transcription factors, including ZFHX3. Reanalyzing their data using the ChIP-Enrich tool (40) with annotation of all peaks to the nearest transcription start sites, we found that a number of potential target genes of ZFHX3, including PRLR, played crucial roles in mammary gland development (data not shown). We therefore tested whether ZFHX3 regulates PRLR expression to modulate prolactin-PRLR signaling. We found that in mouse mammary glands at lactation day 2, deletion of Zfhx3 decreased the Prlr mRNA level, as detected by real time PCR (Fig. 5A). The decrease in Prlr mRNA level by Zfhx3 deletion was already significant at postnatal week 9 (data not shown), suggesting that Prlr down-regulation is independent of lactogenesis. We also detected Prlr protein in mouse mammary tissues by IHC staining and found that deletion of Zfhx3 also down-regulated Prlr protein at lactation day 2 (Fig. 5, B and C). Down-regulation of Prlr was also induced by the knockdown of Zfhx3 in HC11 cells at both the mRNA level (Fig. 5D) and the protein level (Fig. 5E). Consistent with the down-regulation of Prlr, the expression of two established players downstream of prolactin-Prlr signaling, Elf5 and Id2, was also down-regulated by Zfhx3 knock-out in mouse mammary glands at lactation day 2 (Fig. 5F), although the expression of two other prolactin signaling molecules, Lmo4 and Cebpb, was not altered by Zfhx3 deletion (Fig. 5F). Using the cBioPortal web tool (41, 42), we analyzed the expression of ZFHX3 and PRLR mRNAs in 1100 breast cancer samples (TCGA, Provisional), and found a positive correlation between the expression of the two genes (Fig. 5G). These results indicate that ZFHX3 plays a crucial role in the expression of PRLR in mammary epithelial cells, which could mediate the effect of Zfhx3 on PRLR-JAK2-STAT5 signaling and mammary gland development.
FIGURE 5.
Zfhx3 is needed for the expression of Prlr in mouse mammary epithelial cells. A–C, deletion of Zfhx3 down-regulates Prlr expression, as detected by RT-PCR analysis for Prlr mRNA (total and long form, A) or by IHC staining for Prlr protein (B) at day 2 of lactation. C, IHC score is shown as means ± S.E., statistical significance was calculated by one-way analysis of variance followed by Bonferroni's test; **, p < 0.01). D and E, detection of Prlr expression in HC11 cells with stable knockdown of Zfhx3 by RT-PCR (total and long form, D) or Western blotting (long form, E), band intensities in Western blot were quantified using the ImageJ program. F, Zfhx3-mediated gene regulation in mammary alveolar development. Gene expression analysis of mammary glands from MMTV-Cre/Zfhx3+/+ and MMTV-Cre/Zfhx3f/f mice at day 2 of lactation. At least four mice per genotype were used; the data are means ± S.E. Statistical significance was calculated by two-tailed Student's t test using GraphPad Prism 5; *, p < 0.05. G, the mRNA expression data of ZFHX3 and PRLR in breast invasive carcinoma (TCGA, Provisional) were obtained using cBioPortal. The case set contained 1100 tumor samples. Pearson or Spearman correlation coefficients (r) and two-tailed p value calculations to determine the significance of the correlations were calculated using GraphPad Prism 5. H, the 5′ region of mouse Prlr and the location of five alternative first exons (represented by black boxes). E2, exon 2.
Discussion
Postnatal mammary gland development is a multistage process involving a sequence of morphological and molecular changes (4, 32). The transition from pregnancy to lactation activates the secretion of all the nutrients required for the newborns, including milk proteins, lactose, and milk fat. Like other stages of mammary gland development, alveologenesis and lactogenic differentiation are controlled by hormones such as progesterone and prolactin and key transcription factors (4, 32). In this study, we performed histological and molecular analyses using in vitro and in vivo models to establish the Zfhx3 transcription factor as a crucial regulator of normal alveologenesis and lactogenic differentiation. We further demonstrated that Zfhx3 functions by maintaining Prlr-Jak2-Stat5 signaling activity.
We first demonstrated that the Zfhx3 protein level varied at different stages of mammary gland development and reached a peak in late pregnancy and lactation (Fig. 1), which is consistent with the mRNA level of Zfhx3 reported previously (20). Based on transcriptomic analysis of non-human primate breast tissues at prepubertal, adolescent, adult luteal, pregnant, lactating, and postmenopausal stages, four distinct patterns of gene expression were noticed during reproductive life, including the juvenile pattern, adult pattern 1, adult pattern 2, and the lactational pattern (43). The pattern of Zfhx3 expression fits between the adult pattern 1 and the lactational pattern, and genes with either pattern of expression likely play a role in prolactin signaling (43). This appears to be the case for Zfhx3.
Our functional studies indicated the essential role of Zfhx3 in prolactin signaling and lactogenic differentiation. Prevention of Zfhx3 expression compromised alveologenesis and lactogenic differentiation. In the mouse HC11 cell line, a well established and widely used in vitro model of lactogenic differentiation, knockdown of Zfhx3 compromised prolactin-induced cell differentiation including β-casein expression (Fig. 2). More directly, when Zfhx3 was deleted in mouse mammary glands, the number and size of alveoli decreased, milk protein secretion was attenuated, and litters died after birth, likely because of dehydration (Fig. 3). Zfhx3 is essential for proper alveologenesis and lactogenic differentiation.
It has been established that lactation involves the expression of numerous genes and the secretion of their gene products in the milk (43, 44). Our previous study demonstrated that deletion of Zfhx3 in mouse prostates dysregulates a large number of genes, and those encoding for secretory and membrane proteins are the most affected gene groups (17). Using β-casein as a representative of milk proteins, we found that knockdown in HC11 cells and knock-out in mammary tissues indeed interrupted β-casein expression (Figs. 2 and 3), further indicating that Zfhx3 is necessary for lactogenic differentiation and the production of milk.
Mechanistically, our findings suggest that Zfhx3 regulates alveolar and lactogenic differentiation by maintaining prolactin-Prlr signaling activity, which is the single most potent regulator of alveologenesis and lactogenic differentiation (45, 46). In this signaling pathway, binding of prolactin to PRLR activates JAK2 kinase, which then phosphorylates the STAT5 transcription factor, leading to the up-regulation of the Elf5 transcription factor and the expression of genes underlying alveolar and lactogenic differentiation (33, 47–51). We found that Zfhx3 was not only needed for the expression of the milk protein β-casein (Figs. 2 and 3), an indicator of the PRLR-STAT5 signaling axis, it was also necessary for the phosphorylation of Stat5 in both mouse mammary tissues and cultured HC11 mouse mammary epithelial cells (Fig. 4). Furthermore, we found that Zfhx3 was required for the expression of Prlr. For example, deletion of Zfhx3 in mouse mammary tissues and knockdown of Zfhx3 in cultured HC11 mouse mammary epithelial cells significantly down-regulated Prlr expression at both the mRNA and protein levels (Fig. 5). Consistently, a significant correlation was detected between mRNA levels of ZFHX3 and PRLR in hundreds of human breast cancer samples (Fig. 5G). Therefore, maintaining PRLR expression and PRLR-JAK2-STAT5 signaling activity could be the major mechanism by which ZFHX3 regulates alveolar and lactogenic development of the mammary gland.
The promoter of PRLR is complicated, and so is its transcriptional regulation. For example, in addition to the abundantly expressed and widely conserved exon 1 among different species, the PRLR gene has multiple additional first exons because of alternative splicing in different tissues (52). Different tissues can have different 5′-regions of the Prlr gene (53). Human PRLR also has multiple first exons that employ distinct mechanisms for controlling PRLR transcription (54). In mice, five alternative first exons have been identified for Prlr, including E11, E12, E13, E14, and E15 (Fig. 5H) (55). Little is known about how the putative promoters upstream of these first exons drive Prlr transcription and what factors bind to the promoter sequences. We cloned the putative promoters of the five first exons and measured their promoter activities using dual luciferase reporter assay. Except for the E15 promoter, which showed a slight change in promoter activity, none of the other four putative promoters showed detectable changes when Zfhx3 was knocked down in HC11 cells (data not shown). It remains to be determined whether Zfhx3 directly binds to Prlr promoter, and if so where it binds. Porcine Prlr promoter, which is the best studied so far among different species for the potential use of Prlr as a marker for reproductive performance (56–59), has 12 alternative first exons (52). Similar to porcine Prlr, the 5′ region of mouse Prlr is also quite large, spanning more than 120 kb (Fig. 5H). It is thus possible that mouse Prlr has more alternative first exons. Furthermore, whereas STAT5 can bind to an interferon-γ-activated sequence (GAS) in the PRLR gene to regulate its transcription (60), it remains to be determined whether ZFHX3 interacts with STAT5 on the same promoter region. Currently we are in the process of examining how ZFHX3 regulates the transcription of PRLR in mammary epithelial cells.
Regulation of the PRLR-JAK2-STAT5 signaling by ZFHX3 could play an important role in human breast cancer, because the level of phosphorylated STAT5 gradually decreases during breast carcinogenesis, whereas the total expression level of STAT5 does not appear to change. The decrease of phosphorylated STAT5 is correlated with poorer patient survival and predicts both poor clinical outcome and increased risk of anti-estrogen therapy failure (61–65). PRLR has been shown to suppress epithelial-mesenchymal transition and invasiveness of breast cancer cells (66), and PRLR expression is significantly down-regulated in invasive breast cancer, the down-regulation being associated with lymph node metastasis, lower tumor grade, and metastasis-free survival in breast cancer patients (67). Consistent with the findings of p-STAT5 and PRLR expression, the ZFHX3 mRNA level has also been shown to be frequently down-regulated in human breast cancer (68), which is likely mediated by hemizygous deletion (69), and again down-regulation was correlated with a worse patient survival (68). Indeed, a tumor suppressor function has been established for ZFHX3 in prostate cancer, because mutation of ZFHX3 is frequent in human prostate cancer (70, 71), and deletion of Zfhx3 in mouse prostates induced neoplastic lesions (17). These findings suggest that ZFHX3-mediated PRLR-JAK2-STAT5 activity plays a suppressive role in breast cancer development. Currently we are testing this hypothesis by investigating whether Zfhx3 deletion induces neoplastic lesions in mouse mammary glands.
In addition to Prlr, other molecules could also mediate the effect of Zfhx3 in alveolar and lactogenic development of the mammary gland. For example, deletion of Zfhx3 in mouse mammary glands led to the down-regulation of Elf5 and Id2 (Fig. 5F), both of which have established roles in mammary gland development. Id2 mediates the function of RANKL in the survival and proliferation of mammary epithelial cells (72, 73), and deletion of Id2 leads to lactation defects in mice (74). The Elf5 transcription factor has a similar effect on mammary gland development as Zfhx3, and deletion of Elf5 leads to failure of Stat5 activation and defects in alveologenesis (75). Elf5 has also been demonstrated to regulate mammary gland stem/progenitor cell fate (76), although such a role has not been examined for Zfhx3. Both Elf5 and Zfhx3 are transcription factors, but whether they directly interact on gene promoters remains to be clarified. Recently Zfhx3 was also shown to modify circadian function by directly activating circadian genes Cry2, Per1, and Per2 (77). Interestingly, both Per1 and Per2 have been implicated in the development and differentiation of mouse mammary gland (78, 79). Therefore, it is more likely that Zfhx3 regulates alveologenesis and lactogenic differentiation by interacting with or regulating the expression of other factors.
In summary, we found in this study that the Zfhx3 protein level varied during mammary gland development, reaching the highest level in late pregnancy and lactation. Knockdown of Zfhx3 in HC11 mammary epithelial cells and deletion of Zfhx3 in mouse mammary tissues attenuated prolactin-induced alveologenesis and lactogenic differentiation, as indicated by morphological and molecular alterations. Mechanistically, knockdown and knock-out of Zfhx3 decreased the expression level of Prlr, Elf5, Id2, and phosphorylated Stat5. These findings suggest that Zfhx3 plays an essential role in proper development of the mammary gland in pregnancy and lactation by maintaining the activity of the PRLR-JAK2-STAT5 signaling pathway.
Author Contributions
D. Z., M. L., L.F., and J.-T. D. conceived the project. D. Z., G. M., T. N., and J.-T. D. designed all experiments and analyzed all data. D. Z., G. M., X. Z., Y. H., X. H., L. F., X.-Y. D., and Q. Z. prepared and performed experiments. D. Z. and J.-T. D. wrote the manuscript.
Acknowledgments
We sincerely thank Dr. Bernd Groner for kindly providing the HC11 cells and Dr. Tamas Nagy for kindly providing MMTV-Cre mice in pure C57BL/6 background. We thank TCGA project organizers, as well as all study participants. We thank Dr. Zhengmao Zhu, Dr. Xinpei Ci, Dr. Baotong Zhang, Ang Luo, Huanhuan Zhao, Qiao Wu, and Shiying Zhang for help and support throughout the study.
This work was supported by Grant 81472464 from the National Natural Science Foundation of China. The authors declare that they have no conflicts of interest with the contents of this article.
- DIP
- dexamethasone, insulin, and prolactin
- H&E
- hematoxylin and eosin
- IHC
- immunohistochemical
- PRLR
- prolactin receptor.
References
- 1.Hennighausen L., and Robinson G. W. (2001) Signaling pathways in mammary gland development. Dev. Cell 1, 467–475 [DOI] [PubMed] [Google Scholar]
- 2.Hennighausen L., and Robinson G. W. (2005) Information networks in the mammary gland. Nat. Rev. Mol. Cell Biol. 6, 715–725 [DOI] [PubMed] [Google Scholar]
- 3.Silva C. M., and Shupnik M. A. (2007) Integration of steroid and growth factor pathways in breast cancer: focus on signal transducers and activators of transcription and their potential role in resistance. Mol. Endocrinol. 21, 1499–1512 [DOI] [PubMed] [Google Scholar]
- 4.Brisken C., and O'Malley B. (2010) Hormone action in the mammary gland. Cold Spring Harb. Perspect. Biol. 2, a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hynes N. E., and Watson C. J. (2010) Mammary gland growth factors: roles in normal development and in cancer. Cold Spring Harb. Perspect. Biol. 2, a003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abate-Shen C. (2002) Deregulated homeobox gene expression in cancer: cause or consequence? Nat. Rev. Cancer 2, 777–785 [DOI] [PubMed] [Google Scholar]
- 7.Duverger O., and Morasso M. I. (2008) Role of homeobox genes in the patterning, specification, and differentiation of ectodermal appendages in mammals. J. Cell Physiol. 216, 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis M. T. (2000) Homeobox genes in mammary gland development and neoplasia. Breast Cancer Res. 2, 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H., and Sukumar S. (2003) Role of homeobox genes in normal mammary gland development and breast tumorigenesis. J. Mammary Gland Biol. Neoplasia 8, 159–175 [DOI] [PubMed] [Google Scholar]
- 10.Morinaga T., Yasuda H., Hashimoto T., Higashio K., and Tamaoki T. (1991) A human α-fetoprotein enhancer-binding protein, ATBF1, contains four homeodomains and seventeen zinc fingers. Mol. Cell Biol. 11, 6041–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miura Y., Tam T., Ido A., Morinaga T., Miki T., Hashimoto T., and Tamaoki T. (1995) Cloning and characterization of an ATBF1 isoform that expresses in a neuronal differentiation-dependent manner. J. Biol. Chem. 270, 26840–26848 [DOI] [PubMed] [Google Scholar]
- 12.Holland P. W., Booth H. A., and Bruford E. A. (2007) Classification and nomenclature of all human homeobox genes. BMC Biol. 5, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuda H., Mizuno A., Tamaoki T., and Morinaga T. (1994) ATBF1, a multiple-homeodomain zinc finger protein, selectively down-regulates AT-rich elements of the human α-fetoprotein gene. Mol. Cell Biol. 14, 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung C. G., Kim H. J., Kawaguchi M., Khanna K. K., Hida H., Asai K., Nishino H., and Miura Y. (2005) Homeotic factor ATBF1 induces the cell cycle arrest associated with neuronal differentiation. Development 132, 5137–5145 [DOI] [PubMed] [Google Scholar]
- 15.Kataoka H., Joh T., Miura Y., Tamaoki T., Senoo K., Ohara H., Nomura T., Tada T., Asai K., Kato T., and Itoh M. (2000) AT motif binding factor 1-A (ATBF1-A) negatively regulates transcription of the aminopeptidase N gene in the crypt-villus axis of small intestine. Biochem. Biophys. Res. Commun. 267, 91–95 [DOI] [PubMed] [Google Scholar]
- 16.Berry F. B., Miura Y., Mihara K., Kaspar P., Sakata N., Hashimoto-Tamaoki T., and Tamaoki T. (2001) Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J. Biol. Chem. 276, 25057–25065 [DOI] [PubMed] [Google Scholar]
- 17.Sun X., Fu X., Li J., Xing C., Frierson H. F., Wu H., Ding X., Ju T., Cummings R. D., and Dong J. T. (2014) Deletion of Atbf1/Zfhx3 in mouse prostate causes neoplastic lesions, likely by attenuation of membrane and secretory proteins and multiple signaling pathways. Neoplasia 16, 377–389 Cover article [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun X., Xing C., Fu X., Li J., Zhang B., Frierson H. F. Jr., and Dong J. T. (2015) Additive effect of Zfhx3/Atbf1 and Pten deletion on mouse prostatic tumorigenesis. J. Genet. Genomics 42, 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun X., Fu X., Li J., Xing C., Martin D. W., Zhang H. H., Chen Z., and Dong J. T. (2012) Heterozygous deletion of Atbf1 by the Cre-loxP system in mice causes preweaning mortality. Genesis 50, 819–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M., Fu X., Ma G., Sun X., Dong X., Nagy T., Xing C., Li J., and Dong J. T. (2012) Atbf1 regulates pubertal mammary gland development likely by inhibiting the pro-proliferative function of estrogen-ER signaling. PLoS One 7, e51283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong X. Y., Guo P., Sun X., Li Q., and Dong J. T. (2011) Estrogen up-regulates ATBF1 transcription but causes its protein degradation in estrogen receptor-α-positive breast cancer cells. J. Biol. Chem. 286, 13879–13890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M., Zhao D., Ma G., Zhang B., Fu X., Zhu Z., Fu L., Sun X., and Dong J. T. (2013) Upregulation of ATBF1 by progesterone-PR signaling and its functional implication in mammary epithelial cells. Biochem. Biophys. Res. Commun. 430, 358–363 [DOI] [PubMed] [Google Scholar]
- 23.Spandidos A., Wang X., Wang H., and Seed B. (2010) PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38, D792–D799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ball R. K., Friis R. R., Schoenenberger C. A., Doppler W., and Groner B. (1988) Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J. 7, 2089–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danielson K. G., Oborn C. J., Durban E. M., Butel J. S., and Medina D. (1984) Epithelial mouse mammary cell line exhibiting normal morphogenesis in vivo and functional differentiation in vitro. Proc. Natl. Acad. Sci. U.S.A. 81, 3756–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier V. S., and Groner B. (1994) The nuclear factor YY1 participates in repression of the beta-casein gene promoter in mammary epithelial cells and is counteracted by mammary gland factor during lactogenic hormone induction. Mol. Cell Biol. 14, 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong X. Y., Sun X., Guo P., Li Q., Sasahara M., Ishii Y., and Dong J. T. (2010) ATBF1 inhibits ER function by selectively competing with AIB1 for binding to ER in ER-positive breast cancer cells. J. Biol. Chem. 285, 32801–32809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doppler W., Groner B., and Ball R. K. (1989) Prolactin and glucocorticoid hormones synergistically induce expression of transfected rat beta-casein gene promoter constructs in a mammary epithelial cell line. Proc. Natl. Acad. Sci. U.S.A. 86, 104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dammeier J., Beer H. D., Brauchle M., and Werner S. (1998) Dexamethasone is a novel potent inducer of connective tissue growth factor expression. Implications for glucocorticoid therapy. J. Biol. Chem. 273, 18185–18190 [DOI] [PubMed] [Google Scholar]
- 30.Morrison B. L., Jose C. C., and Cutler M. L. (2010) Connective tissue growth factor (CTGF/CCN2) enhances lactogenic differentiation of mammary epithelial cells via integrin-mediated cell adhesion. BMC Cell Biol. 11, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison B., and Cutler M. L. (2009) Mouse mammary epithelial cells form mammospheres during lactogenic differentiation. J. Vis. Exp. e1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson S. M., Rudolph M. C., McManaman J. L., and Neville M. C. (2007) Key stages in mammary gland development: secretory activation in the mammary gland: it's not just about milk protein synthesis! Breast Cancer Res. 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Robinson G. W., Wagner K.-U., Garrett L., Wynshaw-Boris A., and Hennighausen L. (1997) Stat5a is mandatory for adult mammary gland development and lactogenesis. Gene. Dev. 11, 179–186 [DOI] [PubMed] [Google Scholar]
- 34.Hynes N. E., Cella N., and Wartmann M. (1997) Prolactin mediated intracellular signaling in mammary epithelial cells. J. Mammary Gland Biol. Neoplasia 2, 19–27 [DOI] [PubMed] [Google Scholar]
- 35.Watson C. J., and Burdon T. G. (1996) Prolactin signal transduction mechanisms in the mammary gland: the role of the Jak/Stat pathway. Rev. Reprod. 1, 1–5 [DOI] [PubMed] [Google Scholar]
- 36.Brooks C. L. (2012) Molecular mechanisms of prolactin and its receptor. Endocr. Rev. 33, 504–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly P. A., Bachelot A., Kedzia C., Hennighausen L., Ormandy C. J., Kopchick J. J., and Binart N. (2002) The role of prolactin and growth hormone in mammary gland development. Mol. Cell Endocrinol. 197, 127–131 [DOI] [PubMed] [Google Scholar]
- 38.Peirce S. K., Chen W. Y., and Chen W. Y. (2001) Quantification of prolactin receptor mRNA in multiple human tissues and cancer cell lines by real time RT-PCR. J. Endocrinol. 171, R1–4 [DOI] [PubMed] [Google Scholar]
- 39.Yan J., Enge M., Whitington T., Dave K., Liu J., Sur I., Schmierer B., Jolma A., Kivioja T., Taipale M., and Taipale J. (2013) Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154, 801–813 [DOI] [PubMed] [Google Scholar]
- 40.Welch R. P., Lee C., Imbriano P. M., Patil S., Weymouth T. E., Smith R. A., Scott L. J., and Sartor M. A. (2014) ChIP-Enrich: gene set enrichment testing for ChIP-seq data. Nucleic Acids Res. 42, e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao J. J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y. C., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stute P., Sielker S., Wood C. E., Register T. C., Lees C. J., Dewi F. N., Williams J. K., Wagner J. D., Stefenelli U., and Cline J. M. (2012) Life stage differences in mammary gland gene expression profile in non-human primates. Breast Cancer Res. Treat 133, 617–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paten A. M., Duncan E. J., Pain S. J., Peterson S. W., Kenyon P. R., Blair H. T., and Dearden P. K. (2015) Functional development of the adult ovine mammary gland: insights from gene expression profiling. BMC Genomics 16, 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brisken C., and Rajaram R. D. (2006) Alveolar and lactogenic differentiation. J. Mammary Gland Biol. Neoplasia 11, 239–248 [DOI] [PubMed] [Google Scholar]
- 46.Ben-Jonathan N., LaPensee C. R., and LaPensee E. W. (2008) What can we learn from rodents about prolactin in humans? Endocr. Rev. 29, 1–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oakes S. R., Naylor M. J., Asselin-Labat M. L., Blazek K. D., Gardiner-Garden M., Hilton H. N., Kazlauskas M., Pritchard M. A., Chodosh L. A., Pfeffer P. L., Lindeman G. J., Visvader J. E., and Ormandy C. J. (2008) The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Gene Dev 22, 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horseman N. D. (1999) Prolactin and mammary gland development. J. Mammary Gland Biol. 4, 79–88 [DOI] [PubMed] [Google Scholar]
- 49.Miyoshi K., Shillingford J. M., Smith G. H., Grimm S. L., Wagner K.-U., Oka T., Rosen J. M., Robinson G. W., and Hennighausen L. (2001) Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J. Cell Biol. 155, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brisken C. (2002) Hormonal control of alveolar development and its implications for breast carcinogenesis. J. Mammary Gland Biol. Neoplasia 7, 39–48 [DOI] [PubMed] [Google Scholar]
- 51.Harris J., Stanford P. M., Sutherland K., Oakes S. R., Naylor M. J., Robertson F. G., Blazek K. D., Kazlauskas M., Hilton H. N., Wittlin S., Alexander W. S., Lindeman G. J., Visvader J. E., and Ormandy C. J. (2006) Socs2 and elf5 mediate prolactin-induced mammary gland development. Mol. Endocrinol. 20, 1177–1187 [DOI] [PubMed] [Google Scholar]
- 52.Schennink A., Trott J. F., Manjarin R., Lemay D. G., Freking B. A., and Hovey R. C. (2015) Comparative genomics reveals tissue-specific regulation of prolactin receptor gene expression. J. Mol. Endocrinol. 54, 1–15 [DOI] [PubMed] [Google Scholar]
- 53.Galsgaard E. D., Nielsen J. H., and Møldrup A. (1999) Regulation of prolactin receptor (PRLR) gene expression in insulin-producing cells. Prolactin and growth hormone activate one of the rat prlr gene promoters via STAT5a and STAT5b. J. Biol. Chem. 274, 18686–18692 [DOI] [PubMed] [Google Scholar]
- 54.Hu Z. Z., Zhuang L., Meng J., Tsai-Morris C. H., and Dufau M. L. (2002) Complex 5′ genomic structure of the human prolactin receptor: multiple alternative exons 1 and promoter utilization. Endocrinology 143, 2139–2142 [DOI] [PubMed] [Google Scholar]
- 55.Tabata H., Kobayashi M., Ikeda J. H., Nakao N., Saito T. R., and Tanaka M. (2012) Characterization of multiple first exons in murine prolactin receptor gene and the effect of prolactin on their expression in the choroid plexus. J. Mol. Endocrinol. 48, 169–176 [DOI] [PubMed] [Google Scholar]
- 56.Feng F.-Y., Li X.-J., Lu G., Gao J.-W., and Ren G.-Z. (2011) Relationship of genetic polymorphism of FSHβ and PRLR genes with litter size traits in Yunan black pig and three foreign pig breeds [J]. Acta Agriculturae Boreali-Sinica 5, 015 [Google Scholar]
- 57.Kmieć M., and Terman A. (2006) Associations between the prolactin receptor gene polymorphism and reproductive traits of boars. J. Appl. Genet. 47, 139–141 [DOI] [PubMed] [Google Scholar]
- 58.Tempfli K., Farkas G., Simon Z., and Bali Papp Á. (2011) Effects of prolactin receptor genotype on the litter size of Mangalica. Acta Vet. Hung. 59, 269–277 [DOI] [PubMed] [Google Scholar]
- 59.Wang X. P., Wang L. X., Luo Reng Z. M., and Sun S. D. (2008) Analysis of PRLR and BF genotypes associated with litter size in Beijing black pig population. Agricultural Sci. China 7, 1374–1378 [Google Scholar]
- 60.Kavarthapu R., Tsai Morris C. H., and Dufau M. L. (2014) Prolactin induces up-regulation of its cognate receptor in breast cancer cells via transcriptional activation of its generic promoter by cross-talk between ERα and STAT5. Oncotarget 5, 9079–9091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peck A. R., Witkiewicz A. K., Liu C., Stringer G. A., Klimowicz A. C., Pequignot E., Freydin B., Tran T. H., Yang N., Rosenberg A. L., Hooke J. A., Kovatich A. J., Nevalainen M. T., Shriver C. D., Hyslop T., Sauter G., Rimm D. L., Magliocco A. M., and Rui H. (2011) Loss of nuclear localized and tyrosine phosphorylated Stat5 in breast cancer predicts poor clinical outcome and increased risk of antiestrogen therapy failure. J. Clin. Oncol. 29, 2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamashita H., Nishio M., Ando Y., Zhang Z., Hamaguchi M., Mita K., Kobayashi S., Fujii Y., and Iwase H. (2006) Stat5 expression predicts response to endocrine therapy and improves survival in estrogen receptor-positive breast cancer. Endocr. Relat Cancer 13, 885–893 [DOI] [PubMed] [Google Scholar]
- 63.Nevalainen M. T., Xie J., Torhorst J., Bubendorf L., Haas P., Kononen J., Sauter G., and Rui H. (2004) Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J. Clin. Oncol. 22, 2053–2060 [DOI] [PubMed] [Google Scholar]
- 64.Peck A. R., Witkiewicz A. K., Liu C., Klimowicz A. C., Stringer G. A., Pequignot E., Freydin B., Yang N., Ertel A., Tran T. H., Girondo M. A., Rosenberg A. L., Hooke J. A., Kovatich A. J., Shriver C. D., Rimm D. L., Magliocco A. M., Hyslop T., and Rui H. (2012) Low levels of Stat5a protein in breast cancer are associated with tumor progression and unfavorable clinical outcomes. Breast Cancer Res. 14, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.May M., Mosto J., Vazquez P. M., Gonzalez P., Rojas P., Gass H., Lanari C., and Molinolo A. A. (2016) Nuclear staining of fgfr-2/stat-5 and runx-2 in mucinous breast cancer. Exp. Mol. Pathol. 100, 39–44 [DOI] [PubMed] [Google Scholar]
- 66.Nouhi Z., Chughtai N., Hartley S., Cocolakis E., Lebrun J. J., and Ali S. (2006) Defining the role of prolactin as an invasion suppressor hormone in breast cancer cells. Cancer Res. 66, 1824–1832 [DOI] [PubMed] [Google Scholar]
- 67.Hachim I. Y., Hachim M. Y., Lopez V. M., Lebrun J.-J., and Ali S. (2016) Prolactin receptor expression is an independent favorable prognostic marker in human breast cancer. Appl. Immunohistochem. Mol. Morphol. 24, 238–245 [DOI] [PubMed] [Google Scholar]
- 68.Zhang Z., Yamashita H., Toyama T., Sugiura H., Ando Y., Mita K., Hamaguchi M., Kawaguchi M., Miura Y., and Iwase H. (2005) ATBF1-a messenger RNA expression is correlated with better prognosis in breast cancer. Clin. Cancer Res. 11, 193–198 [PubMed] [Google Scholar]
- 69.Roylance R., Gorman P., Papior T., Wan Y. L., Ives M., Watson J. E., Collins C., Wortham N., Langford C., Fiegler H., Carter N., Gillett C., Sasieni P., Pinder S., Hanby A., and Tomlinson I. (2006) A comprehensive study of chromosome 16q in invasive ductal and lobular breast carcinoma using array CGH. Oncogene 25, 6544–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun X., Frierson H. F., Chen C., Li C., Ran Q., Otto K. B., Cantarel B. L., Vessella R. L., Gao A. C., Petros J., Miura Y., Simons J. W., and Dong J. T. (2005) Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nat. Genet. 37, 407–412 [DOI] [PubMed] [Google Scholar]
- 71.Grasso C. S., Wu Y.-M., Robinson D. R., Cao X., Dhanasekaran S. M., Khan A. P., Quist M. J., Jing X., Lonigro R. J., Brenner J. C., Asangani I. A., Ateeq B., Chun S. Y., Siddiqui J., Sam L., et al. (2012) The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim N. S., Kim H. J., Koo B. K., Kwon M. C., Kim Y. W., Cho Y., Yokota Y., Penninger J. M., and Kong Y. Y. (2006) Receptor activator of NF-κB ligand regulates the proliferation of mammary epithelial cells via Id2. Mol. Cell Biol. 26, 1002–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim N. S., Kim H. T., Kwon M. C., Choi S. W., Kim Y. Y., Yoon K. J., Koo B. K., Kong M. P., Shin J., Cho Y., and Kong Y. Y. (2011) Survival and differentiation of mammary epithelial cells in mammary gland development require nuclear retention of Id2 due to RANK signaling. Mol. Cell Biol. 31, 4775–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mori S., Nishikawa S. I., and Yokota Y. (2000) Lactation defect in mice lacking the helix-loop-helix inhibitor Id2. EMBO J. 19, 5772–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi Y. S., Chakrabarti R., Escamilla-Hernandez R., and Sinha S. (2009) Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of Stat5 activation and functional differentiation in the absence of Elf5. Dev. Biol. 329, 227–241 [DOI] [PubMed] [Google Scholar]
- 76.Chakrabarti R., Wei Y., Romano R. A., DeCoste C., Kang Y., and Sinha S. (2012) Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells 30, 1496–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parsons M. J., Brancaccio M., Sethi S., Maywood E. S., Satija R., Edwards J. K., Jagannath A., Couch Y., Finelli M. J., Smyllie N. J., Esapa C., Butler R., Barnard A. R., Chesham J. E., Saito S., Joynson G., Wells S., Foster R. G., Oliver P. L., Simon M. M., Mallon A. M., Hastings M. H., and Nolan P. M. (2015) The regulatory factor ZFHX3 modifies circadian function in SCN via an AT motif-driven Axis. Cell 162, 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Casey T. M., Crodian J., Erickson E., Kuropatwinski K. K., Gleiberman A. S., and Antoch M. P. (2014) Tissue-specific changes in molecular clocks during the transition from pregnancy to lactation in mice. Biol. Reprod. 90, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Metz R. P., Qu X., Laffin B., Earnest D., and Porter W. W. (2006) Circadian clock and cell cycle gene expression in mouse mammary epithelial cells and in the developing mouse mammary gland. Dev. Dyn. 235, 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hesling C., Lopez J., Fattet L., Gonzalo P., Treilleux I., Blanchard D., Losson R., Goffin V., Pigat N., Puisieux A., Mikaelian I., Gillet G., and Rimokh R. (2013) Tif1γ is essential for the terminal differentiation of mammary alveolar epithelial cells and for lactation through SMAD4 inhibition. Development 140, 167–175 [DOI] [PubMed] [Google Scholar]
- 81.Zhou J., Chehab R., Tkalcevic J., Naylor M. J., Harris J., Wilson T. J., Tsao S., Tellis I., Zavarsek S., Xu D., Lapinskas E. J., Visvader J., Lindeman G. J., Thomas R., Ormandy C. J., Hertzog P. J., Kola I., and Pritchard M. A. (2005) Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. EMBO J. 24, 635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pal R., Janz M., Galson D. L., Gries M., Li S., Jöhrens K., Anagnostopoulos I., Dörken B., Mapara M. Y., Borghesi L., Kardava L., Roodman G. D., Milcarek C., and Lentzsch S. (2009) C/EBPbeta regulates transcription factors critical for proliferation and survival of multiple myeloma cells. Blood 114, 3890–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]