Abstract
Biofilms are organized multicellular communities encased in an extracellular polymeric substance (EPS). Biofilm-resident bacteria resist immunity and antimicrobials. The EPS provides structural stability and presents a barrier; however, a complete understanding of how EPS structure relates to biological function is lacking. This review focuses on the EPS of three Gram-negative pathogens: Pseudomonas aeruginosa, nontypeable Haemophilus influenzae, and Salmonella enterica serovar Typhi/Typhimurium. Although EPS proteins and polysaccharides are diverse, common constituents include extracellular DNA, DNABII (DNA binding and bending) proteins, pili, flagella, and outer membrane vesicles. The EPS biochemistry promotes recalcitrance and informs the design of therapies to reduce or eliminate biofilm burden.
Keywords: bacterial pathogenesis, biofilm, extracellular matrix, neutrophil, polysaccharide, eDNA, matrix, phagocyte, Pseudomonas aeruginosa, Haemophilus influenzae, Salmonella, pilin, DNABII, animal models
Introduction
Biofilms, which are defined as highly organized multicellular communities of bacteria encased in an extracellular polymeric substance (or EPS),3 contribute to most chronic infections in the body. These infections confer a significant socioeconomic burden with treatments costing billions of dollars annually. Biofilm formation is key to the establishment of most chronic bacterial infections, and a majority of the population will experience some form of chronic or recurrent bacterial disease in their lifetime. Bacteria resident within a biofilm are highly resistant to phagocytosis and other clearance mechanisms and demonstrate substantially increased resistance to immune effectors and antibiotics (see the accompanying review by van Acker and Coenye (1)). Much of this resistance can be attributed to the EPS, which presents a formidable physical barrier to cellular effectors of immunity and is highly recalcitrant to removal. Although the EPS can be diverse, depending on the microbe(s) that initiate its formation, most are composed of bacterial proteins, polysaccharides, and extracellular DNA (eDNA) (2). Moreover, as the EPS matrix encases the microbes, it is the first to interact with the human immune system. Although immune-mediated clearance of biofilms is often ineffective, the mechanistic basis is as yet undefined (3–5).
To develop new therapeutic strategies to limit chronic human infection, it is crucial to understand how the biofilm EPS provides protection against antimicrobials and the immune system. Addressing this requires a greater understanding of biofilm maturation in vivo with a focus on discerning what factors influence whether the immune response either augments biofilm formation, contributing to persistence, or results in effective biofilm disruption with clearance of the pathogen. This would include where, when, and why the immune system fails to mediate clearance and/or actually facilitates maintenance of long-term biofilms and carriage. The overall goal of this review is to describe the interface between host immunity and the biofilm EPS matrix of three important and well studied human pathogens (Pseudomonas aeruginosa (Pa), nontypeable Haemophilus influenzae (NTHI), and Salmonella enterica serovar Typhimurium/Typhi (St/Sty)). The human diseases under investigation in this proposal include airway and other chronic infections (wounds, gastrointestinal disease, and otitis media (OM)). Sophisticated, well established animal models to both study each of these infections and conduct pre-clinical evaluation of biofilm-focused therapeutic modalities will also be discussed.
EPS Components
Bacteria within biofilms are usually embedded in a matrix, which consists of protein, polysaccharide, and nucleic acid, and the matrix provides a critical role in the biofilm resistance phenotype (2, 6). In this section, a brief description of the major Pa, NTHI, and St/Sty EPS components will be described (Table 1) to allow readers a better understanding of their roles in biofilm biology, topics discussed later in this review.
TABLE 1.
EPS components of bacterial biofilms
| Bacteria and components |
References | ||
|---|---|---|---|
| Pa | NTHI | St/Sty | |
| Polysaccharide | |||
| Psl | LOS | O-antigen capsule | 7, 8, 70, 81, 101, 102 |
| Pel | Cellulose | ||
| Alginate | Colanic acida | ||
| Vi-Antigenb | |||
| Protein | |||
| CdrA | 18 biofilm-specific proteins including major OMPsc P1, P2, and P5 | Curli (amyloids) BapA | 19, 20, 26, 27, 103 |
| LecA/LecB | |||
| DNABII proteins (IHF and HU) | DNABII proteins (IHF and HU) | DNABII proteins (IHF and HU) | |
| eDNA | |||
| Yes | Yes | Yes | 6, 17 |
| Others | |||
| Outer membrane vesicles | Outer membrane vesicles | Flagella | 82, 12, 17, 25, 104 |
| Type IV pili | Type IV pili | ||
| Amyloid (Fap) | |||
a Produced in non-typhoidal serovars, but not in serovar Typhi.
b Produced by typhoidal strains (serovars Typhi, Paratyphi C) as well as serovar Dublin, but not other non-typhoidal serovars.
c OMPS, outer membrane proteins.
Exopolysaccharides
Exopolysaccharides contribute to the biofilm matrix of all three organisms discussed herein. Perhaps the best studied is Pa, which encodes at least three distinct polymers (Psl, Pel, and alginate). The composition, role, and regulation of each of these matrix components have been the subject of several recent reviews (7, 8). Expression of alginate confers a mucoid phenotype to the organism, seen most often in Pa isolated from later stages of cystic fibrosis (CF) lung infections. Psl and Pel are expressed in most non-mucoid Pa strains. Released Psl is a neutral charged polymeric pentasaccharide, yet the structure of the cell-associated Psl is not known (9, 10). Pel is a positively charged polymer (11). Each of these polysaccharides has distinct roles in biofilms formed by mucoid or non-mucoid Pa strains (7). In NTHI, there has yet to be an exopolysaccharide identified that clearly contributes to biofilms, yet the lipooligosaccharide (LOS) plays a prominent role in modulating biofilm structure. NTHI LOS can be modified by the addition of phosphorylcholine, although the role of this in biofilm function is not clear (12, 13). The composition of the Salmonella spp. biofilm matrix is complex and highly variable in response to altered environmental conditions and variable among serovars. The St/Sty polysaccharides identified to date include colanic acid, O-antigen capsule, and cellulose (14, 15). Sty produces a distinct polysaccharide, Vi-antigen, a primary marker for infection with this serovar and the basis of several current vaccine approaches.
eDNA
Another abundant biofilm matrix building block, eDNA, is a critical component of the NTHI, Pa, and St/Sty biofilm matrix (6, 16, 17). The source of eDNA appears to be random genomic sequences of varying lengths, with no apparent sequence selectivity. eDNA is apparently derived from stochastic lysis of a subpopulation of the bacteria within the biofilm. In most situations, eDNA is a contributing component of a more diverse biofilm matrix and typically interacts with proteins or polysaccharides to stabilize the matrix. In Pa and St, eDNA binds Pel polysaccharide or amyloid fibers, respectively (11, 18). Interactions of eDNA with other biofilm matrix components likely explain why DNase treatment of biofilms has variable effects on destabilizing the matrix.
Proteins
Proteins are increasingly recognized for their importance in biofilm structure and function. For many organisms, the structure and resistance properties of biofilms can be eliminated by protease treatment (19, 20). Matrix proteins include secreted proteins as well as components of adhesins or motility organelles. Large-scale proteomic studies have been performed on EPS recovered from both NTHI and Pa (20, 21). Consistent from these studies and others is the abundance of outer membrane proteins and type IV pili in these matrix preparations. The best studied matrix proteins produced by Pa include lectins LecA and LecB (22) and CdrA (23). Although both LecA and LecB are carbohydrate-binding proteins involved in biofilm formation, it is not clear how they contribute to matrix formation and neither appears to associate with Pel or Psl. CdrA, a large extracellular adhesin-like protein, associates with Psl and contributes to biofilm integrity (23). For NTHI, one of the best studied matrix proteins is type IV pilin protein, which has been shown to: 1) serve as a constituent of the EPS; 2) be necessary for twitching motility; and 3) contribute significantly to the architecture of an NTHI biofilm (17). The main proteinaceous component of St biofilms is curli pili, which structurally and biochemically are amyloid fibers. Curli appear to promote bacteria-surface and bacteria-bacteria interactions that enhance biofilm stability (24). Additionally, St surface flagella mediate attachment to cholesterol (primary constituent of gallstones) in the initial stages of biofilm development, whereas fimbriae do not appear to play a significant role in this process (25). Also, BapA is a large surface protein variably associated with the production of robust biofilm formation in Salmonella spp. (26). Finally, the DNABII family of proteins (e.g. HU and IHF) has been observed associated in a highly organized fashion with the eDNA outside of the bacterium (27) (see below).
Outer Membrane Vesicles (OMVs)
Most Gram-negative bacteria produce OMVs that contain a diverse array of molecules, which contribute to a variety of biological processes. Bacterial OMVs can allow trafficking of biomolecules to other cells in their environment. Studies showed that OMVs are definitive components of the Pa and NTHI biofilm EPS (28, 29), and they contain cargo that may contribute to the matrix EPS (Table 1). Although mutants that regulate both Escherichia coli and NTHI OMV biogenesis have recently been identified (30), their precise role in modulating biofilm/EPS structure has not been evaluated.
Role of EPS in Promoting Recalcitrance to Host Immunity
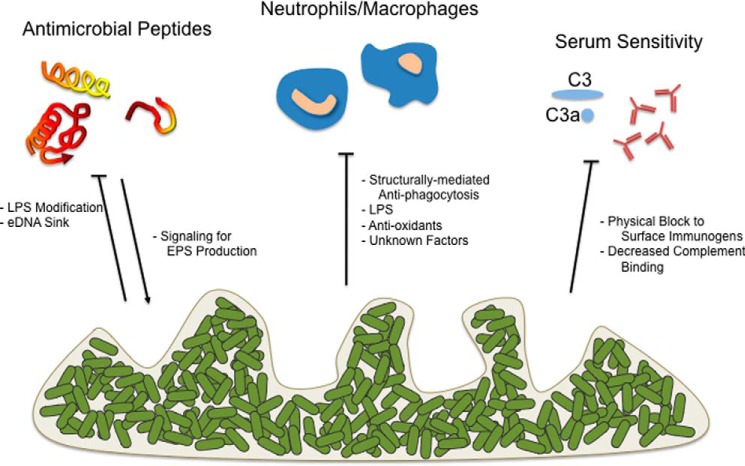
The biofilm EPS of Pa, NTHI, and St/Sty promote resistance to killing by innate immune constituents (Fig. 1) including antimicrobial peptides (AMPs), professional phagocytes, and serum factors. Part of this resistance is mediated by the biofilm community structure, which likely sterically limits engulfment by host cells, as well as the penetration of immune components. The eDNA and polysaccharides can bind and sequester immune components, particularly those with a charge differential such as AMPs and matrix eDNA. Opsonization by complement and by immunoglobulins is also negatively affected by EPS. The bacteria within the biofilm respond by producing factors that limit the oxidative and non-oxidative capabilities of phagocytic cells, aiding bacterial survival. Additionally, as exemplified by Pa, host immune components can promote Pa diversification with variants having enhanced EPS production providing further recalcitrance (31, 32).
FIGURE 1.
Role of EPS in promoting recalcitrance to host immunity. The biofilm architecture, as well as products released by biofilms, can help mediate resistance to immune clearance. Categories of factors to which biofilms provide recalcitrance include, in addition to traditional antibiotics, antimicrobial peptides, engulfment/killing by professional phagocytes, and opsonization (antibodies/serum complement). Additionally, in some instances, host inflammation can signal biofilm EPS production (arrow). Horizontal line termini denote biofilm-mediated resistance to the immune effector.
It is well accepted that the biofilm mode of growth affords Pa protection from host immune effectors (33). This appears to be independent of where Pa biofilms form in its host. One of the first studies to address this revealed that human neutrophils are capable of penetrating biofilms and carry out phagocytosis and granule secretion. However, the neutrophils exhibit a distinct non-reactive morphology and increase oxygen consumption, yet fail to kill bacteria (4). The precise mechanisms underlying this mitigation of neutrophil defense function is currently unknown. Perhaps the best recognized recalcitrance mechanism is the production of quorum sensing-dependent rhamnolipid by biofilm-grown Pa. Rhamnolipid has potent cytotoxicity toward polymorphonuclear leukocytes (3). As with several other exopolysaccharides, the presence of Psl or alginate on the Pa surface inhibits phagocytosis by limiting opsonin deposition (34, 35). Alginate also provides Pa protection from IFN-γ-mediated macrophage killing (36). Expression of both Pel and Psl in Pa affords protection against clearance in murine models of acute infection (35, 37). Likewise, eDNA provides resistance to AMPs and aminoglycosides by chelating cations, which otherwise could lead to perturbation of Pa membrane structure/function and induction of the PhoPQ and PmrAB regulon. Positively charged AMPs can bind eDNA, sequestering them from the bacterial cell surface (38). Of note, Pa variants that emerge during chronic infection have a hyper-biofilm phenotype and express elevated levels of EPS, which provides these bacteria further protection from host defenses. Often, products of the robust inflammatory response (e.g. H2O2 or antimicrobial peptides) enhance the frequency of variants that emerge (31, 32, 39).
The NTHI EPS also confers immune resistance to the resident microbes via a variety of mechanisms. One of the first examples was that LOS provided resistance to phagocytosis by polymorphonuclear leukocytes in vitro as well as to extracellular killing mediated by the action of histones and elastase within neutrophil extracellular traps (40). Izano et al. (41) utilized both proteinase K and DNase treatment to measure resistance of NTHI biofilms to a variety of biocides. They concluded that the cohesive properties conferred by proteinaceous intercellular adhesins and the eDNA within the EPS contributed to resistance, perhaps by providing a barrier to penetration and/or by sequestration of these agents within the matrix. A recent study (42) demonstrated that NTHI-produced peroxiredoxin-glutaredoxin and catalase promote resistance to oxidants and survival within neutrophil extracellular traps, whereas another study showed that production of the DNA-binding protein Dps also confers resistance of NTHI to oxidative stress in vitro and to clearance in vivo (43). Finally, in addition to providing structure to NTHI biofilms, eDNA present within the biofilm matrix binds the AMP human β-defensin 3, thus limiting its access to bacteria resident within the biofilm and reducing its antimicrobial activity (44).
Salmonella species produce numerous EPS components in vitro; however, comparatively little is known about the in vivo condition. These EPS components include cellulose, colanic acid, O-antigen capsule, Vi antigen, curli pili, eDNA, and other protein components. Some of these EPS members are species-specific: for example, colanic acid (produced only in non-typhoidal species) and Vi-antigen (produced in typhoidal species). Winter and colleagues (45, 46) showed that the Sty Vi-antigen prevents complement receptor 3 (CR3)-mediated clearance and neutrophil chemotaxis, and results in reduced IL-8 production. O-antigen capsule and Vi capsular polysaccharide prevent complement-mediated clearance of Salmonella (45, 47). Deletion of colanic acid genes enhances antibody production to vaccine strains, likely by unmasking surface antigens (47). Curli pili, on the other hand, are sensed by Toll-like receptor (TLR) 1/2 and NLRP3 (48, 49). Additionally, the two-component regulatory systems PhoPQ and PmrAB mediate AMP resistance in Salmonella in part by divalent cation sensing via a periplasmic acidic patch of PhoQ and induction of LPS modification. The production of eDNA is a key component of Salmonella biofilms, and the chelation of Mg2+ in biofilm eDNA mediates AMP resistance in a PmrAB/PhoPQ-dependent manner (16).
Animal Models to Interrogate EPS Biofilm Attributes
Experiments in animals are often essential for defining the virulence potential of microbes and the host response to infection, and validating treatment modalities. The development of models that faithfully mimic human chronic biofilm diseases has been challenging. Striking an appropriate balance between modeling the persistent disease state (biofilm) yet limiting overt systemic spread typically requires careful attention to the pathogen burden, providing localized, confined delivery, and monitoring/manipulation of the immune system. Such methods have been applied to the development of chronic infections involving Pa, NTHI, or Sty. The advantages and potential limitations of each method are discussed.
Pa causes an array of both chronic and acute infections (50). As such, there are several models that have been developed to mimic such infections (for a review, see Ref. 51). Although most models provide insights into the pathogenesis of acute disease, there are often issues with the chronic models. In fact, progress in our understanding of the role of biofilms in persistence and the development/testing of therapeutics in many Pa chronic infections is hampered by the absence of suitable models. Nonetheless, murine models do exist for the study of CF airway diseases. Perhaps the most widely used is the intratracheal instillation in rodents of Pa embedded in either agar or alginate beads (52). The agar or alginate matrix provides protection against clearance. Although variations of this have also been adapted to cftr-defective mice, these models do not exhibit characteristic CF ion transporter defects in the lungs and fail to faithfully reproduce the chronic infections that occur in humans. More promising CF models include the epithelial sodium channel (ENaC)-overexpressing mouse (53) and the porcine CF model (54). Perhaps the best systems to study chronic Pa infections lie in the various wound models that have been developed. These incorporate murine systems, including those with chronic diabetes syndrome (55, 56). As wound healing dynamics and pathogenesis in murine systems differ from those in humans, others have implemented porcine wound models of persistent infections with Pa growing as single or mixed species biofilms (57, 58). Other biofilm-associated models, including those that mimic device related/implant or corneal infections, have also been applied with success (50, 51, 59).
NTHI induces multiple diseases of the upper and lower respiratory tract including OM, sinusitis, bronchitis, and exacerbations of both chronic obstructive pulmonary disease (COPD) and CF. The majority of these include a biofilm component, which contributes greatly to the chronicity and/or recurrence of disease. To model these diseases and investigate the role of biofilms in pathogenesis, two rodent hosts have been relied upon. For OM, the chinchilla has served as the predominant mammalian host (60). This has become a robust and highly reproducible model of this prevalent pediatric disease (61) with the formation of large biofilms that remain in the middle ear for many weeks. In the majority of these studies, NTHI was inoculated directly into the middle ear space wherein biofilms form rapidly; however, in chinchillas that have been co-challenged with respiratory syncytial virus (RSV), NTHI, and Moraxella catarrhalis, both M. catarrhalis and NTHI ascend the Eustachian tube to induce mixed species biofilm formation in the middle ear (62). The murine host has served as the predominant rodent model for demonstration of biofilm formation by NTHI in the lower airway and investigation of its role in chronic obstructive pulmonary disease and other pulmonary infections (43, 63); however, the significantly smaller size of this host has limited the biomass of samples available for evaluation.
Salmonella colonize humans and animals to cause a spectrum of diseases, with the primary clinical manifestations of gastroenteritis and typhoid fever. Sty is the primary etiologic agent of typhoid fever, which is an acute illness, but can result in a chronic, asymptomatic infection primarily localized to the gallbladder (14). Gallbladder colonization by Salmonella during chronic infection has been known for more than a century. A high percentage of human carriers harbor gallstones, and biofilms form on gallstone surfaces during chronic carriage (64, 65). A mouse model of carriage has been developed based on the documented long-term survival of St (the mouse model for the human-specific Sty) in NRAMP1+/+ (SLC11A1) mice (129X1/SvJ) and the ability to induce gallstone formation in the mouse with a lithogenic diet (66). Such gallstone-containing mice harbor a 5000-fold increase in bacteria in the gallbladder and demonstrate a 500-fold increased shedding in feces, while bacterial biofilms can be observed on gallstone surfaces. Studies have been carried out to 1 year after infection in the 129X1/SvJ model, showing colonization of the gallbladder, as well as distal sites (mesenteric lymph nodes and bone marrow) (67, 68). Biofilm formation on the gallbladder epithelium and epithelial cell invasion have also been observed in the chronic mouse model and may account for persistence in the absence of gallstones (67). In human carriers and in acute mouse models, gallbladder tissues possess Salmonella within and/or on the epithelium (69). Explanted tissue from the chicken intestinal epithelium has also been used to study Salmonella biofilms and intestinal colonization (70).
Therapies Targeting EPS to Reduce or Eliminate Biofilm Burden
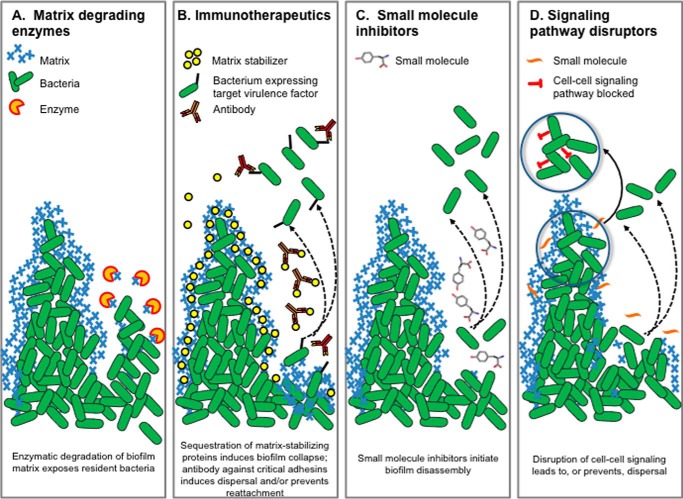
Diseases wherein a biofilm contributes to the chronic and recurrent nature of the disease course require novel methods for diagnosis, treatment, and prevention. Given the recalcitrant nature of biofilm-resident bacteria to the action of antibiotics, a variety of non-antibiotic approaches are being investigated (Fig. 2), including those that focus on physical disruption, surgical removal, and even ex vivo thermal mitigation to eradicate biofilms present on implanted medical devices (71). This area of research is beyond the scope of this article; however, there have been several recent excellent reviews (72, 73). The following strategies have been explored for their ability to disrupt established Pa, NTHI, and St/Sty biofilms, either in vitro or in vivo.
FIGURE 2.
Therapies that target the EPS to reduce or eliminate biofilm burden. Four therapeutic strategies targeting EPS (A–D) are indicated and discussed further in the text. Symbols depicting methods and targets are indicated along the top, and proposed mechanisms of action are indicated at the bottom of each panel.
Matrix-degrading Enzymes
Given that eDNA is a common EPS constituent, treatment of biofilms with DNase has been explored as a mechanism for biofilm disruption for many microbes including NTHI (74) (Fig. 2A). In certain diseases, disruption of the heavy DNA strands contributed by neutrophil netting (e.g. as in the viscous middle effusion recovered from children with chronic OM) by DNase provides an additional desired clinical outcome. Given the abundance of alginate in biofilms produced by many mucoid Pa isolates, the use of alginate lyase to disperse biofilms has been investigated (75). A recent study (76) reinforced the conclusions about the biofilm-disruptive properties of this approach as well as its synergistic interaction with antibiotics. However, they present data that question the mechanisms that underlie alginate lyase enzyme-based therapies. Another approach for disruption of biofilms formed by Pa has involved the use of glycoside hydrolases to target exopolysaccharides present within the EPS, as the enzymes PelAh, PslGh, and Sph3h can disrupt existing Pa biofilms in vitro (77, 78). The accompanying review by Sheppard and Howell discusses this class of enzymes in more detail (79). Likewise, the enzyme Dispersin B in combination with an AMP showed synergistic antibiofilm/antibacterial activity in a chronic wound model of Pa infection (80). For St, cellulase has been used in vitro to target cellulose, which often (depending on growth conditions) has a dramatic negative effect on biofilm formation (81). DNase is also highly effective at disrupting eDNA-rich biofilms formed by St (18).
Immunotherapeutics
Constituents of the biofilm EPS can also serve as targets for immune intervention as a result of either a natural immune response or one directed by immunization (Fig. 2B). Biofilms already established in the middle ears of chinchillas were resolved following transcutaneous immunization with a chimeric immunogen that incorporated epitopes of type IV pili and outer membrane protein P5, delivered with the adjuvant dmLT, a double mutant of the E. coli heat labile enterotoxin (83). Clearance of these established biofilms was attributed to the action of immunogen-specific IgG and IFNγ- and IL-17-producing CD4+ T-cells and secretion of host defense peptides within the middle ear (84). Within this same line of investigation, by targeting a lynchpin protein that is positioned at the vertices of crossed strands of eDNA present within the biofilm matrix (e.g. either IHF or HU of the DNABII family of DNA-binding proteins), biofilms formed by NTHI, St/Sty, and Pa can be significantly disrupted when exposed to antiserum directed against a DNABII protein. This treatment is proposed to induce an equilibrium shift, which removes these proteins from the biofilm matrix, thereby mediating catastrophic structural collapse (27).
Small Molecule Inhibitors
In an early study, the ability of a mixture of d-amino acids to prevent Pa biofilm formation (85) suggested that they might similarly be useful to disrupt these biofilms (Fig. 2C). Others found that specific d-amino acids disrupted Pa biofilms and were particularly effective when combined with antibiotics (86). When used alone, however, treatment of Pa biofilms with a mixture of d-amino acids induced ∼30% increase in matrix production, thereby suggesting the potential to inadvertently provide protection for any remaining viable or persister cells (86). Recently, Leiman et al. (87) indicated that d-tyrosine actually inhibited bacterial growth, and others have not found d-amino acids to be effective (88). Additional studies are clearly needed to resolve the exact mechanism of action. Much of the effort with St biofilm disruption has focused on chemical disruption to mediate sterilization of food processing surfaces (not reviewed here), but a broad-spectrum anti-biofilm peptide (peptide 1018) was utilized to induce a disruptive cellular stress response in St, Pa, and other bacteria (89). This peptide eradicated mature biofilms when used at low concentrations by targeting the enzymes RelA and SpoT, which mediate the synthesis of two small signaling nucleotides (collectively referred to as guanosine tetra- or pentaphosphate guanosine ((p)ppGpp)) involved in the stringent response. Although little additional literature exists regarding St/Sty dispersal agents, an ATP-mimetic biofilm inhibitory compound that reduced initial St binding but did not disrupt existing biofilms has been identified (90). This compound also showed activity against Acinetobacter baumannii. Furthermore, several different classes of potent St and Pa biofilm inhibitors have been identified, centered on brominated furanones, 2-aminoimidazoles, and 2-aminoimidazoline-based compounds (91, 92).
Signaling Pathway Targets
Many signaling pathways have been implicated in biofilm development, including two-component regulators, cyclic nucleotides, nitric oxide, phenazines, small peptides, and quorum sensing (QS) (Fig. 2D). Due to its pivotal importance in biofilm development, much of the work in this area has focused on the action of the second messenger cyclic di-GMP (see the accompanying background in the Valentini and Filloux review (93)). A recent study showed that the diguanylate cyclase GcbA mediated Pa biofilm dispersal via activation of the chemosensory protein BdlA (94). In a related line of investigation, a substituted fatty acid messenger, cis-2-decenoic acid (CDA), produced by Pa can disperse biofilms formed by a range of bacteria and even Candida albicans (95). A recent study (96) using a microarray-based approach to better define the CDA-mediated signaling pathways and mechanisms involved identified enhanced motility, altered metabolic activity, virulence, and persistence at varied temperatures. Use of CDA in combination with antimicrobials mediated the best results. Although characterization of the QS pathways utilized by NTHI, as well as defining their important role in pathobiology, is an area of active investigation, to date this approach has not been extensively explored as a means to disrupt existing biofilms despite recognition of its potential (97). One recent in vivo study, however, showed that the ability to disrupt pre-existing NTHI biofilms in the middle ears of chinchillas following immunization with the majority subunit of the type IV pilus was dependent upon the production of AI-2 quorum signaling molecules via the activity of the 4,5-dihydroxy-2,3-pentanedione (DPD) synthase, LuxS (98). This suggests that immune pressure could induce a dispersal response. Moreover, the addition of DPD to type IV pili+ NTHI biofilms mediated their dispersal. Although comparatively less is appreciated regarding QS signaling and biofilm development in St, several investigators have considered the feasibility of this approach, and although not tested for their ability to disrupt an existing biofilm, an alkyl-DPD panel of AI-2 inhibitors potently inhibits St QS (99). The accompanying review by Kavanaugh and Horswill (100) provides an excellent discussion of the Staphylococcus aureus agr QS pathway, which controls the production of exotoxins and exoenzymes required for infection and biofilm production and dispersal.
Synopsis and Perspectives
The ability of a bacterium to form a biofilm aids the establishment and development of recurrent and chronic infection. Moving forward, challenges include the development of animal models that accurately model human infection, the definition of EPS components produced in vivo and their relative contributions to the establishment and maintenance of chronic infection, anti-biofilm discovery with in vivo efficacy, and a more complete understanding of immune modulation by biofilm components. Much remains to be learned regarding persistent bacterial infections and immune system interactions, but the study of Pa, NTHI, and St/Sty biofilms in vitro and in vivo has helped to advance this work.
Acknowledgments
We thank Drs. Paul Stoodley, Kevin Mason, and Chris Jones for critiquing this review.
This work was supported by the Public Health Preparedness for Infectious Diseases (PHPID) and Biofilms in Human Medicine. This is the second article in the Thematic Minireview series “Biofilms.” The authors declare that they have no conflicts of interest with the contents of this article.
- EPS
- extracellular polymeric substance
- DNABII
- DNA binding and bending
- eDNA
- extracellular DNA
- HU
- histone-like protein
- Pa
- Pseudomonas aeruginosa
- NTHI
- nontypeable Haemophilus influenzae
- St/Sty
- Salmonella enterica serovar Typhimurium/Typhi
- OM
- otitis media
- CF
- cystic fibrosis
- LOS
- lipooligosaccharide
- AMP
- antimicrobial peptide
- OMV
- outer membrane vesicle
- IHF
- integration host factor
- CDA
- cis-2-decenoic acid
- QS
- quorum sensing
- DPD
- 4,5-dihydroxy-2,3-pentanedione.
References
- 1.van Acker H., and Coenye T. (2016) The role of efflux and physiological adaptation in biofilm tolerance and resistance. J. Biol. Chem. 291, 12565–12572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karatan E., and Watnick P. (2009) Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alhede M., Bjarnsholt T., Jensen P. Ø., Phipps R. K., Moser C., Christophersen L., Christensen L. D., van Gennip M., Parsek M., Høiby N., Rasmussen T. B., and Givskov M. (2009) Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155, 3500–3508 [DOI] [PubMed] [Google Scholar]
- 4.Jesaitis A. J., Franklin M. J., Berglund D., Sasaki M., Lord C. I., Bleazard J. B., Duffy J. E., Beyenal H., and Lewandowski Z. (2003) Compromised host defense on Pseudomonas aeruginosa bofilms: characterization of neutrophil and biofilm interactions. J. Immunol. 171, 4329–4339 [DOI] [PubMed] [Google Scholar]
- 5.Leid J. G., Shirtliff M. E., Costerton J. W., and Stoodley P. (2002) Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect. Immun. 70, 6339–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitchurch C. B., Tolker-Nielsen T., Ragas P. C., and Mattick J. S. (2002) Extracellular DNA required for bacterial biofilm formation. Science 295, 1487. [DOI] [PubMed] [Google Scholar]
- 7.Franklin M. J., Nivens D. E., Weadge J. T., and Howell P. L. (2011) Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, Alginate, Pel, and Psl. Front. Microbiol. 2, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limoli D. H., Jones C. J., and Wozniak D. J. (2015) Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 3, 10.1128/microbiolspec.MB-0011-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrd M. S., Sadovskaya I., Vinogradov E., Lu H., Sprinkle A. B., Richardson S. H., Ma L., Ralston B., Parsek M. R., Anderson E. M., Lam J. S., and Wozniak D. J. (2009) Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73, 622–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kocharova N. A., Hatano K., Shaskov A. S., Knirel Y. A., Kochetkov N. K., and Pier G. B. (1989) The structure and serologic distribution of an extracellular neutral polysaccharide from Pseudomonas aeruginosa immunotype 3. J. Biol. Chem. 264, 15569–15573 [PubMed] [Google Scholar]
- 11.Jennings L. K., Storek K. M., Ledvina H. E., Coulon C., Marmont L. S., Sadovskaya I., Secor P. R., Tseng B. S., Scian M., Filloux A., Wozniak D. J., Howell P. L., and Parsek M. R. (2015) Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Nat.l Acad. Sci. U.S.A. 112, 11353–11358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong W., Mason K., Jurcisek J., Novotny L., Bakaletz L. O., and Swords W. E. (2007) Phosphorylcholine decreases early inflammation and promotes the establishment of stable biofilm communities of nontypeable Haemophilus influenzae strain 86–028NP in a chinchilla model of otitis media. Infect. Immun. 75, 958–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puig C., Marti S., Hermans P. W., de Jonge M. I., Ardanuy C., Liñares J., and Langereis J. D. (2014) Incorporation of phosphorylcholine into the lipooligosaccharide of nontypeable Haemophilus influenzae does not correlate with the level of biofilm formation in vitro. Infect. Immun. 82, 1591–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Escobedo G., Marshall J., and Gunn J. (2011) Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat. Rev. Micro. 9, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simm R., Ahmad I., Rhen M., Le Guyon S., and Römling U. (2014) Regulation of biofilm formation in Salmonella enterica serovar Typhimurium. Future Microbiol. 9, 1261–1282 [DOI] [PubMed] [Google Scholar]
- 16.Johnson L., Horsman S. R., Charron-Mazenod L., Turnbull A. L., Mulcahy H., Surette M. G., and Lewenza S. (2013) Extracellular DNA-induced antimicrobial peptide resistance in Salmonella enterica serovar Typhimurium. BMC Microbiol. 13, 115–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurcisek J. A., and Bakaletz L. O. (2007) Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J. Bacteriol. 189, 3868–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallo P. M., Rapsinski G. J., Wilson R. P., Oppong G. O., Sriram U., Goulian M., Buttaro B., Caricchio R., Gallucci S., and Tükel Ç. (2015) Amyloid-DNA composites of bacterial biofilms stimulate autoimmunity. Immunity 42, 1171–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong J., and Yildiz F. (2015) Biofilm matrix proteins. Microbiol. Spectr. 3, 10.1128/microbiolspec.MB-0004-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S., Baum M. M., Kerwin J., Guerrero D., Webster S., Schaudinn C., VanderVelde D., and Webster P. (2014) Biofilm-specific extracellular matrix proteins of nontypeable Haemophilus influenzae. Pathog. Dis. 72, 143–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toyofuku M., Roschitzki B., Riedel K., and Eberl L. (2012) Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J. Proteome Res. 11, 4906–4915 [DOI] [PubMed] [Google Scholar]
- 22.Imberty A., Wimmerová M., Mitchell E. P., and Gilboa-Garber N. (2004) Structures of the lectins from Pseudomonas aeruginosa: insights into the molecular basis for host glycan recognition. Microbes Infect. 6, 221–228 [DOI] [PubMed] [Google Scholar]
- 23.Borlee B. R., Goldman A. D., Murakami K., Samudrala R., Wozniak D. J., and Parsek M. R. (2010) Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75, 827–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnhart M. M., and Chapman M. R. (2006) Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crawford R. W., Reeve K. E., and Gunn J. S. (2010) Flagellated but not hyperfimbriated Salmonella enterica serovar Typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J. Bacteriol. 192, 2981–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Latasa C., Roux A., Toledo-Arana A., Ghigo J.-M., Gamazo C., Penadés J. R., and Lasa I. (2005) BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58, 1322–1339 [DOI] [PubMed] [Google Scholar]
- 27.Goodman S. D., Obergfell K. P., Jurcisek J. A., Novotny L. A., Downey J. S., Ayala E. A., Tjokro N., Li B., Justice S. S., and Bakaletz L. O. (2011) Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 4, 625–637 [DOI] [PubMed] [Google Scholar]
- 28.Hong W., Pang B., West-Barnette S., and Swords W. E. (2007) Phosphorylcholine expression by nontypeable Haemophilus influenzae correlates with maturation of biofilm communities in vitro and in vivo. J. Bacteriol. 189, 8300–8307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schooling S. R., and Beveridge T. J. (2006) Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188, 5945–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roier S., Zingl F. G., Cakar F., Durakovic S., Kohl P., Eichmann T. O., Klug L., Gadermaier B., Weinzerl K., Prassl R., Lass A., Daum G., Reidl J., Feldman M. F., and Schild S. (2016) A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 7, 10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Limoli D. H., Rockel A. B., Host K. M., Jha A., Kopp B. T., Hollis T., and Wozniak D. J. (2014) Cationic antimicrobial peptides promote microbial mutagenesis and pathoadaptation in chronic infections. PLoS Pathog. 10, e1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathee K., Ciofu O., Sternberg C., Lindum P. W., Campbell J. I., Jensen P., Johnsen A. H., Givskov M., Ohman D. E., Molin S., Høiby N., and Kharazmi A. (1999) Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145, 1349–1357 [DOI] [PubMed] [Google Scholar]
- 33.Alhede M., Bjarnsholt T., Givskov M., and Alhede M. (2014) Chapter One: Pseudomonas aeruginosa biofilms: Mechanisms of immune evasion. in Advances in Applied Microbiology (Sima S., and Geoffrey M. G., eds), pp. 1–40, Academic Press, Orlando, FL: [DOI] [PubMed] [Google Scholar]
- 34.Krieg D. P., Helmke R. J., German V. F., and Mangos J. A. (1988) Resistance of mucoid Pseudomonas aeruginosa to nonopsonic phagocytosis by alveolar macrophages in vitro. Infect. Immun. 56, 3173–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra M., Byrd M. S., Sergeant S., Azad A. K., Parsek M. R., McPhail L., Schlesinger L. S., and Wozniak D. J. (2012) Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell. Microbiol. 14, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leid J. G., Willson C. J., Shirtliff M. E., Hassett D. J., Parsek M. R., and Jeffers A. K. (2005) The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-γ-mediated macrophage killing. J. Immunol. 175, 7512–7518 [DOI] [PubMed] [Google Scholar]
- 37.Yang L., Hengzhuang W., Wu H., Damkiær S., Jochumsen N., Song Z., Givskov M., Høiby N., and Molin S. (2012) Polysaccharides serve as scaffold of biofilms formed by mucoid Pseudomonas aeruginosa. FEMS Immunol. Med. Microbiol. 65, 366–376 [DOI] [PubMed] [Google Scholar]
- 38.Mulcahy H., Charron-Mazenod L., and Lewenza S. (2008) Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 4, e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodríguez-Rojas A., Makarova O., Müller U., and Rolff J. (2015) Cationic peptides facilitate iron-induced mutagenesis in bacteria. PLoS Genet. 11, e1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong W., Juneau R. A., Pang B., and Swords W. E. (2009) Survival of bacterial biofilms within neutrophil extracellular traps promotes nontypeable Haemophilus influenzae persistence in the chinchilla model for otitis media. J. Innate Immun. 1, 215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izano E. A., Shah S. M., and Kaplan J. B. (2009) Intercellular adhesion and biocide resistance in nontypeable Haemophilus influenzae biofilms. Microb. Pathog. 46, 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juneau R. A., Pang B., Armbruster C. E., Murrah K. A., Perez A. C., and Swords W. E. (2015) Peroxiredoxin-glutaredoxin and catalase promote resistance of nontypeable Haemophilus influenzae 86–028NP to oxidants and survival within neutrophil extracellular traps. Infect. Immun. 83, 239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pang B., Hong W., Kock N. D., and Swords W. E. (2012) Dps promotes survival of nontypeable Haemophilus influenzae in biofilm communities in vitro and resistance to clearance in vivo. Front. Cell. Infect. Microbiol. 2, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones E. A., McGillivary G., and Bakaletz L. O. (2013) Extracellular DNA within a nontypeable Haemophilus influenzae-induced biofilm binds human β defensin-3 and reduces its antimicrobial activity. J. Innate Immun. 5, 24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson R. P., Winter S. E., Spees A. M., Winter M. G., Nishimori J. H., Sanchez J. F., Nuccio S.-P., Crawford R. W., Tükel Ç., and Bäumler A. J. (2011) The Vi capsular polysaccharide prevents complement receptor 3-mediated clearance of Salmonella enterica Serotype Typhi. Infect. Immun. 79, 830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winter S. E., Raffatellu M., Wilson R. P., Rüssmann H., and Bäumler A. J. (2008) The Salmonella enterica serotype Typhi regulator TviA reduces interleukin-8 production in intestinal epithelial cells by repressing flagellin secretion. Cell. Microbiol. 10, 247–261 [DOI] [PubMed] [Google Scholar]
- 47.Marshall J. M., and Gunn J. S. (2015) The O-antigen capsule of Salmonella enterica serovar Typhimurium facilitates serum resistance and surface expression of FliC. Infect. Immun. 83, 3946–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rapsinski G. J., Wynosky-Dolfi M. A., Oppong G. O., Tursi S. A., Wilson R. P., Brodsky I. E., and Tükel Ç. (2015) Toll-Like receptor 2 and NLRP3 cooperate to recognize a functional bacterial amyloid, curli. Infect. Immun. 83, 693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tükel Ç., Nishimori J. H., Wilson R. P., Winter M. G., Keestra A. M., Van Putten J. P. M., and Bäumler A. J. (2010) Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell. Microbiol. 12, 1495–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mulcahy L. R., Isabella V. M., and Lewis K. (2014) Pseudomonas aeruginosa biofilms in disease. Microb. Ecol. 68, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lebeaux D., Chauhan A., Rendueles O., and Beloin C. (2013) From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2, 288–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cash H. A., Woods D. E., McCullough B., Johanson W. G. Jr., and Bass J. A. (1979) A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am. Rev. Respir. Dis. 119, 453–459 [DOI] [PubMed] [Google Scholar]
- 53.Zhou Z., Duerr J., Johannesson B., Schubert S. C., Treis D., Harm M., Graeber S. Y., Dalpke A., Schultz C., and Mall M. A. (2011) The ENaC-overexpressing mouse as a model of cystic fibrosis lung disease. J. Cyst. Fibros. 10, Suppl. 2, S172–S182 [DOI] [PubMed] [Google Scholar]
- 54.Rogers C. S., Stoltz D. A., Meyerholz D. K., Ostedgaard L. S., Rokhlina T., Taft P. J., Rogan M. P., Pezzulo A. A., Karp P. H., Itani O. A., Kabel A. C., Wohlford-Lenane C. L., Davis G. J., Hanfland R. A., Smith T. L., et al. (2008) Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science 321, 1837–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sullivan S. R., Underwood R. A., Gibran N. S., Sigle R. O., Usui M. L., Carter W. G., and Olerud J. E. (2004) Validation of a model for the study of multiple wounds in the diabetic mouse (db/db). Plast. Reconst. Surg. 113, 953–960 [DOI] [PubMed] [Google Scholar]
- 56.Watters C., DeLeon K., Trivedi U., Griswold J. A., Lyte M., Hampel K. J., Wargo M. J., and Rumbaugh K. P. (2013) Pseudomonas aeruginosa biofilms perturb wound resolution and antibiotic tolerance in diabetic mice. Med. Microbiol. Immunol. 202, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pastar I., Nusbaum A. G., Gil J., Patel S. B., Chen J., Valdes J., Stojadinovic O., Plano L. R., Tomic-Canic M., and Davis S. C. (2013) Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS ONE 8, e56846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy S., Elgharably H., Sinha M., Ganesh K., Chaney S., Mann E., Miller C., Khanna S., Bergdall V. K., Powell H. M., Cook C. H., Gordillo G. M., Wozniak D. J., and Sen C. K. (2014) Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. J. Pathol. 233, 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fleiszig S. M., and Evans D. J. (2002) The pathogenesis of bacterial keratitis: studies with Pseudomonas aeruginosa. Clin. Exp. Optom. 85, 271–278 [DOI] [PubMed] [Google Scholar]
- 60.Ehrlich G. D., Veeh R., Wang X., Costerton J. W., Hayes J. D., Hu F. Z., Daigle B. J., Ehrlich M. D., and Post J. C. (2002) Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 287, 1710–1715 [DOI] [PubMed] [Google Scholar]
- 61.Bakaletz L. O. (2009) Chinchilla as a robust, reproducible and polymicrobial model of otitis media and its prevention. Expert. Rev. Vaccines 8, 1063–1082 [DOI] [PubMed] [Google Scholar]
- 62.Brockson M. E., Novotny L. A., Jurcisek J. A., McGillivary G., Bowers M. R., and Bakaletz L. O. (2012) Respiratory syncytial virus promotes Moraxella catarrhalis-induced ascending experimental otitis media. PLoS ONE 7, e40088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morey P., Viadas C., Euba B., Hood D. W., Barberán M., Gil C., Grilló M. J., Bengoechea J. A., and Garmendia J. (2013) Relative contributions of lipooligosaccharide inner and outer core modifications to nontypeable Haemophilus influenzae pathogenesis. Infect. Immun. 81, 4100–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dongol S., Thompson C. N., Clare S., Nga T. V. T., Duy P. T., Karkey A., Arjyal A., Koirala S., Khatri N. S., Maskey P., Poudel S., Jaiswal V. K., Vaidya S., Dougan G., Farrar J. J., et al. (2012) The microbiological and clinical characteristics of invasive Salmonella in gallbladders from cholecystectomy patients in Kathmandu, Nepal. PLoS ONE 7, e47342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schiøler H., Christiansen E. D., Høybye G., Rasmussen S. N., and Greibe J. (1983) Biliary calculi in chronic Salmonella carriers and healthy controls: a controlled study. Scand. J. Infect. Dis. 15, 17–19 [DOI] [PubMed] [Google Scholar]
- 66.Crawford R. W., Rosales-Reyes R., Ramírez-Aguilar Mde L., Chapa-Azuela O., Alpuche-Aranda C., and Gunn J. S. (2010) Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc. Natl. Acad. Sci. U.S.A. 107, 4353–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gonzalez-Escobedo G., and Gunn J. S. (2013) Gallbladder epithelium as a niche for chronic Salmonella carriage. Infect. Immun. 81, 2920–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monack D. M., Bouley D. M., and Falkow S. (2004) Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNγ neutralization. J. Exp. Med. 199, 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menendez A., Arena E. T., Guttman J. A., Thorson L., Vallance B. A., Vogl W., and Finlay B. B. (2009) Salmonella infection of gallbladder epithelial cells drives local inflammation and injury in a model of acute typhoid fever. J. Infect. Dis. 200, 1703–1713 [DOI] [PubMed] [Google Scholar]
- 70.Ledeboer N. A., and Jones B. D. (2005) Exopolysaccharide sugars contribute to biofilm formation by Salmonella enterica serovar Typhimurium on HEp-2 cells and chicken intestinal epithelium. J. Bacteriol. 187, 3214–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O'Toole A., Ricker E. B., and Nuxoll E. (2015) Thermal mitigation of Pseudomonas aeruginosa biofilms. Biofouling 31, 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gupta P., Sarkar S., Das B., Bhattacharjee S., and Tribedi P. (2016) Biofilm, pathogenesis and prevention—a journey to break the wall: a review. Arch. Microbiol. 198, 1–15 [DOI] [PubMed] [Google Scholar]
- 73.Kostakioti M., Hadjifrangiskou M., and Hultgren S. J. (2013) Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 3, a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cavaliere R., Ball J. L., Turnbull L., and Whitchurch C. B. (2014) The biofilm matrix destabilizers, EDTA and DNaseI, enhance the susceptibility of nontypeable Hemophilus influenzae biofilms to treatment with ampicillin and ciprofloxacin. MicrobiologyOpen 3, 557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Høiby N., Bjarnsholt T., Givskov M., Molin S., and Ciofu O. (2010) Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332 [DOI] [PubMed] [Google Scholar]
- 76.Lamppa J. W., and Griswold K. E. (2013) Alginate lyase exhibits catalysis-independent biofilm dispersion and antibiotic synergy. Antimicrob. Agents Chemother. 57, 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baker P., Whitfield G. B., Hill P. J., Little D. J., Pestrak M. J., Robinson H., Wozniak D. J., and Howell P. L. (2015) Characterization of the Pseudomonas aeruginosa glycoside hydrolase PslG reveals that Its levels are critical for Psl polysaccharide biosynthesis and biofilm formation. J. Biol. Chem. 290, 28374–28387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu S., Su T., Wu H., Liu S., Wang D., Zhao T., Jin Z., Du W., Zhu M.-J., Chua S. L., Yang L., Zhu D., Gu L., and Ma L. Z. (2015) PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 25, 1352–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sheppard D. C., and Howell P. L. (2016) Biofilm exopolysaccharides of pathogenic fungi: lessons from bacteria. J. Biol. Chem. 291, 12529–12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gawande P. V., Leung K. P., and Madhyastha S. (2014) Antibiofilm and antimicrobial efficacy of DispersinB®-KSL-W peptide-based wound gel against chronic wound infection associated bacteria. Curr. Microbiol. 68, 635–641 [DOI] [PubMed] [Google Scholar]
- 81.Solano C., García B., Valle J., Berasain C., Ghigo J.-M., Gamazo C., and Lasa I. (2002) Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43, 793–808 [DOI] [PubMed] [Google Scholar]
- 82.Seviour T., Hansen S. H., Yang L., Yau Y. H., Wang V. B., Stenvang M. R., Christiansen G., Marsili E., Givskov M., Chen Y., Otzen D. E., Nielsen P. H., Geifman-Shochat S., Kjelleberg S., and Dueholm M. S. (2015) Functional amyloids keep quorum-sensing molecules in check. J. Biol. Chem. 290, 6457–6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Novotny L. A., Clements J. D., and Bakaletz L. O. (2011) Transcutaneous immunization as preventative and therapeutic regimens to protect against experimental otitis media due to nontypeable Haemophilus influenzae. Mucosal Immunol. 4, 456–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Novotny L. A., Clements J. D., and Bakaletz L. O. (2013) Kinetic analysis and evaluation of the mechanisms involved in the resolution of experimental nontypeable Haemophilus influenzae-induced otitis media after transcutaneous immunization. Vaccine 31, 3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kolodkin-Gal I., Romero D., Cao S., Clardy J., Kolter R., and Losick R. (2010) d-Amino acids trigger biofilm disassembly. Science 328, 627–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sanchez Z., Tani A., and Kimbara K. (2013) Extensive reduction of cell viability and enhanced matrix production in Pseudomonas aeruginosa PAO1 flow biofilms treated with a d-amino acid mixture. Appl. Environ. Microbiol. 79, 1396–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leiman S. A., Richardson C., Foulston L., Elsholz A. K., First E. A., and Losick R. (2015) Identification and characterization of mutations conferring resistance to d-amino acids in Bacillus subtilis. J. Bacteriol. 197, 1632–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sarkar S., and Pires M. M. (2015) d-Amino acids do not inhibit biofilm formation in Staphylococcus aureus. PLoS ONE 10, e0117613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de la Fuente-Núñez C., Reffuveille F., Haney E. F., Straus S. K., and Hancock R. E. (2014) Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10, e1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koopman J. A., Marshall J. M., Bhatiya A., Eguale T., Kwiek J. J., and Gunn J. S. (2015) Inhibition of Salmonella enterica biofilm formation using small-molecule adenosine mimetics. Antimicrob. Agents Chemother. 59, 76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Janssens J. C. A., Steenackers H., Robijns S., Gellens E., Levin J., Zhao H., Hermans K., De Coster D., Verhoeven T. L., Marchal K., Vanderleyden J., De Vos D. E., and De Keersmaecker S. C. J. (2008) Brominated furanones inhibit biofilm formation by Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 74, 6639–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Steenackers H. P. L., Ermolat'ev D. S., Savaliya B., Weerdt A. D., Coster D. D., Shah A., Van der Eycken E. V., De Vos D. E., Vanderleyden J., and De Keersmaecker S. C. J. (2011) Structure-activity relationship of 2-hydroxy-2-aryl-2,3-dihydro-imidazo[1,2-a]pyrimidinium salts and 2N-substituted 4(5)-aryl-2-amino-1H-imidazoles as inhibitors of biofilm formation by Salmonella Typhimurium and Pseudomonas aeruginosa. Bioorg. Med. Chem. 19, 3462–3473 [DOI] [PubMed] [Google Scholar]
- 93.Valentini M., and Filloux A. (2016) Biofilms and c-di-GMP signaling: lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 291, 12547–12555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Petrova O. E., Cherny K. E., and Sauer K. (2015) The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. J. Bacteriol. 197, 174–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Davies D. G., and Marques C. N. (2009) A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191, 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rahmani-Badi A., Sepehr S., Fallahi H., and Heidari-Keshel S. (2015) Dissection of the cis-2-decenoic acid signaling network in Pseudomonas aeruginosa using microarray technique. Front. Microbiol. 6, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Armbruster C. E., and Swords W. E. (2010) Interspecies bacterial communication as a target for therapy in otitis media. Expert Rev. Anti. Infect. Ther. 8, 1067–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Novotny L. A., Jurcisek J. A., Ward M. O. Jr., Jordan Z. B., Goodman S. D., and Bakaletz L. O. (2015) Antibodies against the majority subunit of type IV pili disperse nontypeable Haemophilus influenzae biofilms in a LuxS-dependent manner and confer therapeutic resolution of experimental otitis media. Mol. Microbiol. 96, 276–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lowery C. A., Abe T., Park J., Eubanks L. M., Sawada D., Kaufmann G. F., and Janda K. D. (2009) Revisiting AI-2 quorum sensing inhibitors: direct comparison of alkyl-DPD analogues and a natural product fimbrolide. J. Am. Chem. Soc. 131, 15584–15585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kavanaugh J. S., and Horswill A. R. (2016) Impact of environmental cues on staphylococcal quorum-sensing and biofilm development. J. Biol. Chem. 291, 12556–12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gibson D. L., White A. P., Snyder S. D., Martin S., Heiss C., Azadi P., Surette M., and Kay W. W. (2006) Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J. Bacteriol. 188, 7722–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murphy T. F., and Kirkham C. (2002) Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol. 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Römling U., Bokranz W., Rabsch W., Zogaj X., Nimtz M., and Tschäpe H. (2003) Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol. 293, 273–285 [DOI] [PubMed] [Google Scholar]
- 104.Couto N., Schooling S. R., Dutcher J. R., and Barber J. (2015) Proteome profiles of outer membrane vesicles and extracellular matrix of Pseudomonas aeruginosa biofilms. J. Proteome Res. 14, 4207–4222 [DOI] [PubMed] [Google Scholar]