Abstract
5-Lipoxygenase activating protein (FLAP) plays a critical role in the metabolism of arachidonic acid to leukotriene A4, the precursor to the potent pro-inflammatory mediators leukotriene B4 and leukotriene C4. Studies with small molecule inhibitors of FLAP have led to the discovery of a drug binding pocket on the protein surface, and several pharmaceutical companies have developed compounds and performed clinical trials. Crystallographic studies and mutational analyses have contributed to a general understanding of compound binding modes. During our own efforts, we identified two unique chemical series. One series demonstrated strong inhibition of human FLAP but differential pharmacology across species and was completely inactive in assays with mouse or rat FLAP. The other series was active across rodent FLAP, as well as human and dog FLAP. Comparison of rodent and human FLAP amino acid sequences together with an analysis of a published crystal structure led to the identification of amino acid residue 24 in the floor of the putative binding pocket as a likely candidate for the observed speciation. On that basis, we tested compounds for binding to human G24A and mouse A24G FLAP mutant variants and compared the data to that generated for wild type human and mouse FLAP. These studies confirmed that a single amino acid mutation was sufficient to reverse the speciation observed in wild type FLAP. In addition, a PK/PD method was established in canines to enable preclinical profiling of mouse-inactive compounds.
Keywords: arachidonic acid (AA, ARA); computational biology; drug discovery; leukotriene; medicinal chemistry; membrane protein
Introduction
5-Lipoxygenase activating protein (FLAP)2 is a key accessory protein in the arachidonic acid metabolism pathway (1). Its function is to present arachidonic acid to 5-lipoxygenase for conversion to leukotriene A4 and subsequently present leukotriene A4 to leukotriene C4 synthase to generate the potent pro-inflammatory mediator leukotriene C4 (2–4). Because of this critical role in the biosynthesis of leukotrienes, FLAP has been the subject of multiple drug discovery efforts (5, 6), with several inhibitors reaching proof of concept in small clinical trials (7–9).
FLAP was originally identified via phenotypic screening for 5-lipoxygenase inhibitors and was described as an 18-kDa membrane protein (10). During the course of that effort, the compound MK-886 was found to bind to the FLAP polypeptide (11). Subsequently, the FLAP cDNA was cloned from multiple species, revealing high sequence homology (12, 13) (Fig. 1). X-ray crystallographic evidence indicated that FLAP exists as a homotrimer, similar to other members of the MAPEG family, with each monomer containing four transmembrane α-helices (14). A major compound binding site is embedded within the membrane, formed by the interface of α-helices 2 and 4 of one monomer and α-helix 1 of the adjacent monomer (14), resulting in three binding sites per trimer. Mutational analysis revealed several key interactions with the FLAP inhibitor MK-591, aiding in the understanding of its binding mode and SAR surrounding this series of indoles (15).
FIGURE 1.
Critical differences between human and mouse FLAP. A, graphic representation of human FLAP with non-orthologous residues of mouse FLAP shown as spheres and MK-591 in cyan to illustrate the proximity of the majority of non-orthologous residues to the MK-591 binding pocket. MK-591 is shown in one of the three binding pockets of the FLAP trimer. Actual distances of key residues from small molecule binding pocket are shown in the adjacent table. B, topology plot of human FLAP with differences from mouse shown in green. The key amino acid in mouse, Ala24 in transmembrane domain 1, is indicated with blue box (note, sequence truncated at Gly140 for presentation purposes). The topology plot was created with Protter. C, high sequence homology between species shown in alignment of amino acid sequences of human, dog, rat, mouse and pig FLAP. Sequences were aligned with Clustal Omega 1.2.1 multiple alignment tool (consensus symbols: *, fully conserved; :, strong similarity; ., weak similarity.
During our own high throughput screening efforts, we discovered benzimidazoles and a series of biaryl amino-heteroarenes (Fig. 2 and Table 2; Refs. 16–20) with distinct SAR relative to previously reported indole-containing FLAP inhibitors exemplified by MK-886, MK-591, and AM-803 (15, 21, 22). Unexpectedly, we found that the biaryl amino-heteroarenes lacked activity in rodent whole blood ex vivo and in vivo models. Here we propose that a single amino acid difference in the binding pocket that is conserved in murine, rat, and porcine FLAP is sufficient to render compounds of this series inactive in these species, based on ligand displacement analysis, whole blood activity assays, and computational studies. Because rodents are commonly used for pharmacokinetic and pharmacodynamics studies, we established an alternative path for the preclinical profiling of biaryl amino-heteroarenes and related compounds in canines.
FIGURE 2.

SAR of selected biaryl amino-heteroarenes in FLAP ligand displacement assay. Early SAR supported the importance of a lipophilic group one end of the molecule (t-butyl in blue circle, compound 3) and a single donor-acceptor pair at the other (compound 3, arrows). FLAP probe displacement HTRF pKi values are given in −log M.
TABLE 2.
Structure-activity relationship of select FLAP inhibitors
HTRF values are average pKa (in m) with standard deviations in parentheses. Whole blood values are average IC50 μ values (in μm) with standard deviations in parentheses. HWB, human whole blood; MWB, mouse whole blood; DWB, dog whole blood.

Experimental Procedures
Preparation of FITC-labeled MK-591
MK-591 (30 mg) was suspended in a 2:1 mixture of N,N-dimethylformamide and water to a total volume of 1.5 ml, followed by addition of 34 mg (5 equivalents) of hydroxybenzotriazole, 10 mg (1 equivalent) of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, 5 μl (1 equivalent) of N-methylmorpholine, and 25 mg (1 equivalent) of 5-(((2-(carbohydrazino) methyl)thio)acetyl)aminofluorescein (FITC; catalog no. C356; Invitrogen). The reaction was stirred overnight in the dark at room temperature, after which the crude mixture was filtered and purified by reverse phase chromatography to afford the final product as a yellow powder (25 mg). Mass spectroscopy analysis (electrospray ionization) was performed on the final product. The mass calculated for C58H52ClN5O9S2 is 1061.29; the m/z found was 1062.2 [M+H]+, which is consistent with the desired product.
FLAP Expression and Membrane Preparation
FLAP cDNA was amplified by PCR and cloned into pFASTBac1 (Invitrogen) with an N-terminal His6 tag according to standard techniques. After virus production and amplification, Sf9 cells were infected for 48 h and harvested by centrifugation, washed once with ice-cold PBS, and frozen at −80 °C. Subsequently, the cells were suspended at 2 × 107 cells/ml in ice-cold TE (10 mm Tris, 1 mm EDTA, pH 8.0) containing 1 mm DTT and Complete protease inhibitor tablets (Sigma). The cells were lysed by sonication (Branson) on ice with a large probe for 20 s at 50% duty cycle, setting 5, until the cells reached quantitative lysis (as monitored, intermittently, by phase contrast microscopy). Lysates were centrifuged at 9,000 × g for 10 min, and supernatants were harvested and centrifuged for 1 h at 100,000 × g in a Ti70 rotor. The pellets were resuspended in TE with sonication, as above, and maintained at a protein concentration of >5 mg/ml. Aliquots were frozen in liquid nitrogen after the addition of glycerol to 20% and stored at −80 °C.
FLAP Homogeneous Time-resolved Fluorescence (HTRF) Assay
The compounds were diluted to 4× final concentration in assay buffer (PBS, 2 mm EDTA, 0.5 mm DTT, 0.01% Triton X-100) such that the final DMSO concentration was not greater than 1.25%. Then a 4× HTRF mixture was prepared by diluting FITC-labeled MK-591 first to 10 μm in DMSO, from a 10 mm DMSO stock, and then to 100 nm in cold assay buffer in combination with 25 μg/ml terbium-labeled anti-His6 (catalog no. 61HISTLA; Cisbio). The membranes were diluted to 0.4 mg/ml (2× final concentration) in cold assay buffer. The following were added to a black 384-well, non-binding, plate (catalog no. 784900; Greiner): 5 μl of compound or buffer, 5 μl of HTRF mixture, and 10 μl of membrane preparation. After sealing, the plate was incubated with shaking for 2 h and read on a laser-equipped Envision plate reader (PerkinElmer Life Sciences). The data are presented as an HTRF ratio of FITC fluorescence (HTRF signal) divided by terbium fluorescence × 10,000, and specific signals were typically 10-fold greater than background. IC50 values were calculated with a non-linear single site competition model (Y = bottom + (top − bottom)/(1 + 10 ∧ (X-LogEC50))), and inhibition constant (Ki) with the standard Cheng-Prusoff transformation (23) (Ki = IC50/1+[L]/Kd), where the equilibrium dissociation constant, Kd, of FITC-591 was determined previously by standard one-site non-linear regression analysis (Y = Bmax * X/(Kd + X)) of a FITC-labeled MK-591 dose response versus the different FLAP isoforms (not shown).
Ex Vivo Assays (Human, Mouse, and Dog)
Freshly drawn whole blood collected into sodium heparin tubes was diluted 1:1 with RPMI 1620 medium (catalog no. SH30096.01; HyClone). 200 μl of the diluted blood were dispensed to each well of a 96-well round-bottomed plate. Compounds were diluted in RPMI to 11× final concentration, 20 μl of this working solution were added to the diluted blood, followed by incubation for 15 min at 37 °C (5% CO2). Calcium ionophore A23187 (catalog no. C7522-10MG; Sigma-Aldrich) was dissolved in 100% DMSO to generate a 1.67 mg/ml stock solution, which was further diluted 1–10 times dropwise with vortexing into deionized water to generate a working solution. At the end of the compound preincubation, 9 μl of the ionophore working solution was added to each well, and the plate was returned to 37 °C for 30 min. The final DMSO concentration was 0.68%. Following incubation, the plate was centrifuged at 300 × g for 10 min, supernatants were removed, and LTB4 was quantitated by ELISA (catalog no. ADI-900-068; Enzo Life Sciences).
For in vivo studies, compounds may be formulated in 20% hydroxypropyl-β-cyclodextrin and dosed to animals. Blood was sampled by jugular venipuncture into collection tubes containing sodium heparin prior to and at various time points following oral or intravenous dosing in dogs and then diluted 1:1 in RPMI before plating. Calcium ionophore stimulation and quantification were performed as described above.
Structural Biology and Computational Modeling
To enable computational studies, we evaluated the available crystal structures. Two public crystal structures of FLAP were known and registered as 2Q7M and 2Q7R in the Protein Data Bank (14). Both have a relatively low resolution of ∼4 Å. In each asymmetric unit there are two protein assemblies, and a molecule of MK591 (in 2Q7M) or an analog of MK591 (in 2Q7R) is bound to the interface of each pair of monomers (three in each protein assembly). Thus, there were 12 different conformations of the MK-591 binding site available for docking.
Molecular Operating Environment (2010.10; Chemical Computing Group Inc., Montreal, Canada) was used to align the human, mouse, rat, dog, and pig sequences of FLAP; to identify residues near to the MK-591 binding site; and to dock novel compounds. Novel compounds were manually docked into the MK-591 structure, and the MMFF94 force field was used to refine the resulting ligand poses, holding the protein fixed.
Results
Comparative Modeling of the MK-591 Binding Pocket in Human and Mouse FLAP Orthologs and Functional Assessment of Binding Mode
The SAR shown in Fig. 2 yielded a simple pharmacophore needed for activity of the biaryl amino-heteroarenes. For example, compound 7 (see Table 2), which has a methyl group instead of an amino group, was inactive, whereas compound 9, which is N-methylated, shows activity comparable with that of compound 3, suggesting that only a single donor is required. Compound 5, in which the pyrazine of compound 3 is replaced with a pyridine, is active, but the alternate pyridine replacement, in compound 6, is inactive, suggesting that only a single acceptor is required. At the other end of the molecule, reducing the lipophilicity of the t-butyl group to a methyl, as in compound 4, also resulted in a loss of activity. We therefore hypothesized that a single donor acceptor pair on the biaryl amino-heteroarenes was important, as well as a hydrophobic group on the opposite end of the molecule.
We examined the 12 different crystallographic conformations of the FLAP binding pocket. There was significant variation of the loop in the back of the pocket among the 12 structures, but we were able to identify a conformation that could plausibly provide the complementary hydrogen bond donor/acceptor pair needed to explain the pharmacophore.
Compound 3 was docked into the pocket occupied by MK-591 using this pharmacophore as a guide in Molecular Operating Environment. The t-butyl group of compound 3 was superposed onto the t-butyl of MK-591, and the critical donor-acceptor pair of the amino-pyrazine were positioned in the interior of the pocket, where they were able to make hydrogen-bonding pairs with the backbone amides of residues Tyr112 and Phe114 (Fig. 3, C and D). In support of this idea, compounds 1 and 3 were tested in FLAP ligand displacement HTRF assays with membranes containing human FLAP carrying an Y112A mutation. Compound 1 had a pKi versus human wild type FLAP of 8.2 ± 0.4 and 7.2 ± 0.1 m at Y112A FLAP, whereas compound 3 had a pKi versus human wild type of 6.9 ± 0.1 and 5.7 ± 0.26 m at Y112A FLAP (samples run in duplicate, n = 4 for compound 1; n = 2 for compound 3). Therefore, the substantial loss in binding affinity observed for these compounds between wild type FLAP and Y112A FLAP supports the proposed hydrogen bond requirement at Tyr112 and the subsequent docking model.
FIGURE 3.
Alanine 24 of mouse FLAP blocks access of biaryl amino-heteroarenes to small molecule pocket. Compound 1 (MK-591, green) and compound 3 (yellow) modeled in the binding pocket formed between helices 2 and 4 of one monomer and helix 1 of the adjacent monomer of human wild type and A24G mutant FLAP. Binding pocket surface is shown as cyan mesh, whereas glycine 24 is shown with brown surface (A and C), and alanine 24 is shown as pink surface (B and D). Dotted lines depict compound interaction with back bone amides Tyr112 and Phe114. E, compounds 1 (gray) and 3 (green) docked in wild type human FLAP showing distance in angstrom to nearest atom in each compound to nearest atom in Gly24. F, compounds docked as in E but in G24A FLAP, with distances to nearest atom in Ala24.
Next, we examined the human, mouse, dog, rat, and pig sequences of FLAP, which show high amino acid sequence homology between these species (Fig. 1). Despite this high degree of conservation, there is a single residue difference between human and mouse that occurs near the MK-591 binding site: Gly24 (human and dog) or Ala24 (mouse, rat, and pig; see Fig. 1). We then used the proposed docking model to compare the binding mode of various compounds in wild type and G24A human FLAP. A close examination of compounds bound to human and G24A FLAP using this docking model suggested that compounds from the biaryl amino-heteroarene series were likely to be in closer proximity to Ala24 than was MK-591. The computational mutation of G24A predicts a steric clash that would be introduced in the predicted binding mode of the biaryl amino-heteroarenes, but not of MK-591. As shown in Fig. 3, MK-591 (green) avoids any steric hindrance by sitting higher in the binding pocket (Fig. 3B). Conversely, when compound 3 is modeled in the binding pocket of WT and G24A FLAP, its phenyl ring appears to clash with the surface of the protein at Ala24. In addition, the closest atom of compound 1 to Ala24 is predicted to be 3.12 Å, whereas that of compound 3 is predicted to be 2.65 Å (Fig. 3, E and F). Thus, these computational results offered plausible rationale for the lack of activity of the biaryl amino-heteroarene series against mouse FLAP.
Functional Characterization of Human and Mouse FLAP in Ligand Displacement Assays
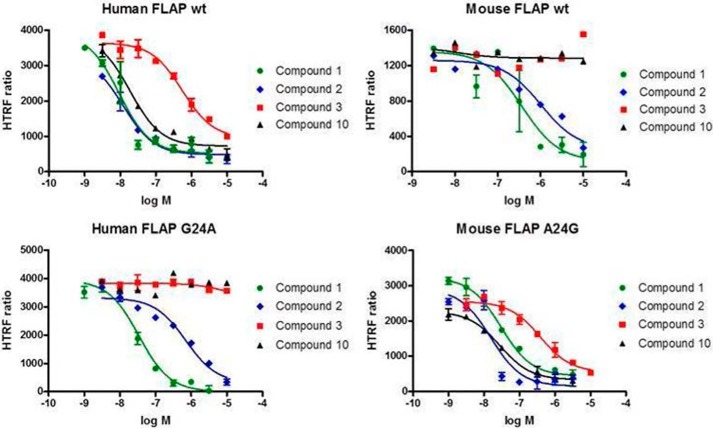
To functionally characterize compound-FLAP interactions, we adopted an HTRF probe displacement assay, using a modified version of established methods (Ref. 24; see “Experimental Procedures”). With this system, we first determined the Kd of FITC-labeled MK591 (FITC-591) for binding to wild type human and mouse FLAP, as well as the variants hG24A and mA24G (see “Experimental Procedures” and Table 1). The FITC-591 ligand had a very similar Kd for human wild type and G24A FLAP isoforms at 30 ± 14 and 21 ± 18 nm, respectively. FITC-591 had somewhat lower affinity for the mouse isoforms of FLAP with Kd of 80 ± 41 and 55 nm for wild type and A24G, respectively. Next, we evaluated the Ki of a number of compounds from the literature, as well as our molecules. A known substituted indole, exemplified by MK-591 (compound 1; Table 2), did show some loss of potency when tested in the mouse FLAP binding assay (human pKi = 8.2 versus mouse pKi = 6.5), as did a distinct substituted benzimidazole, such has compound 2, pKi 8.3 versus 6.7 (Fig. 4 and Table 2). In contrast, biaryl amino-heteroarenes identified in our screening efforts (for example, compound 3), and an optimized ter-aryl amino-heteroarene (Table 2, compound 10) (20), were inactive against mouse FLAP (Fig. 4 and Table 2).
TABLE 1.
Kd values of FITC-labeled MK 591 at human, mouse, and FLAP variants
Kd values are presented in nm with standard deviations in parentheses. Individual experiment samples run in duplicate.
| Human | Mouse | Human G24A | Mouse A24G | |
|---|---|---|---|---|
| Kd | 30.3 (14.1; n = 5) | 80.2 (41; n = 3) | 20.7 (18.2; n = 4) | 55.4 (n = 1)a |
a Insufficient membrane preparations for additional runs.
FIGURE 4.
Biaryl amino-heteroarenes are inactive in ligand displacement assays with membranes from mouse wild type or human G24A FLAP. Activity of select FLAP inhibitors versus human wild type (wt), human G24A, mouse wild type, and mouse A24G FLAP in the FITC-591 HTRF probe displacement assay (see “Experimental Procedures” for description). Biaryl amino-heteroarenes (compounds 3 and 10) show consistent lack of activity in mouse wild type, as well as human G24A mutant. A representative experiment of three is shown with samples run in duplicate.
Mutational Analysis in MK-591 Binding Pocket of Human and Mouse
To test the hypothesis that mouse Ala24 prevents biaryl amino-heteroarenes compounds from accessing the binding pocket in FLAP, human G24A and mouse A24G variants were prepared for comparison with wild type in the HTRF probe displacement assay. Benzimidazoles (Table 2, compound 2), as well as the literature compound MK-591 (compound 1), did show some loss of activity when tested in mouse and the human G24A variant. Importantly, however, biaryl amino-heteroarenes inactive in wild type mouse were also inactive in the mutant human G24A FLAP (compounds 3 and 10, Fig. 4; and compounds 3–10, Table 2). Likewise, compounds had similar potency between mouse A24G and wild type human (Fig. 4 and Table 2).
Establishment of Alternative Compound Advancement Paradigm for Mouse-inactive Compounds
Because compounds from the biaryl amino-heteroarene series were inactive in mouse binding and whole blood assays, an alternative path was required to assess their pharmacokinetics and pharmacodynamics (PK/PD). Thus, we evaluated FLAP orthologs and found that canine FLAP met the criteria for choosing an appropriate species for preclinical profiling because it also contains a Gly residue at position 24 and is, overall, highly homologous to human FLAP (96% sequence identity). In addition, canines are a commonly utilized species for preclinical development studies (25). Therefore, we established a canine whole blood assay and confirmed that representative compounds from the different series, including mouse inactive biaryl amino-heteroarenes, had similar potency in blocking LTB4 production in human and dog whole blood (Table 2). Next, we developed a canine PK/PD model to enable the advanced profiling of lead compounds. To that end, dogs were dosed with compound 10 (as described under “Experimental Procedures”), and blood samples were drawn at various time points to quantify plasma drug levels and perform ex vivo whole blood activity assays. In this model, compound 10 demonstrated dose-dependent inhibition of A23187-induced LTB4 production after administration, which correlated closely with the human and canine ex vivo whole blood assays (Fig. 5 and Table 2). Thus, these data support the utilization of canines for whole blood assays and PK/PD models, as well as the choice of this species as an adequate substitute for rodents in the establishment of pharmacodynamics parameters.
FIGURE 5.

Pharmacokinetic/pharmacodynamic analysis of compound 10 in canines. Compound 10 was formulated and dosed as described under “Experimental Procedures.” Subsequently, blood samples were assessed for compound concentration and LTB4 levels after stimulation with the calcium ionophore, A23187.
Discussion
FLAP is an essential component of the eicosanoid biosynthetic pathway that leads to the generation of the pro-inflammatory leukotrienes LTB4, the cysteinyl leukotriene C4, and its derivatives leukotrienes D4 and E4. Those are potent bioactive lipids that mediate inflammation, increase in vascular permeability, and bronchoconstriction (26). A number of pharmaceutical companies have placed significant drug discovery resources in search of FLAP inhibitors for asthma and atherosclerosis. Through screening and SAR studies, we found potent and selective inhibitors of FLAP, including benzimidazoles and biaryl amino-heteroarenes (16–20). Profiling of the latter compounds revealed differential pharmacology in rodents. We present evidence indicating that this speciation stems from a single amino acid difference between human and mouse FLAP. Through homology modeling and mutational analysis, we confirmed that amino acid residue 24 in the base of the small molecule binding pocket is likely to be this key residue.
FLAP exhibits very high cross-species homology. However, there are subtle, but impactful, differences that can affect compound affinities. Remarkably, an alanine mutational scan of key residues around the MK-591 binding pocket identified several residues important for molecular interactions. For example, mutating Tyr66 and Tyr112 to alanine resulted in significant losses in MK-591 binding activity (14). Similarly, the G24A mutation that we describe here did have a slight effect on the activity of indole- and benzimidazole-based inhibitors (Table 2). Using the published crystallographic model of human FLAP, our computational analysis suggested that the benzimidazole (compound 2) maintains activity in mouse by avoiding a clash with Ala24 of murine FLAP, whereas compounds from the biaryl amino-heteroarene family, because of a different binding mode, had no activity in mouse or human G24A FLAP. Together with subsequent mutational analysis, this enabled the discovery that mouse Ala24 is likely the causative residue for a complete loss of activity of this series. Thus, a common theme emerges for the MAPEG family in that the previously described compound binding site that sits at the interface between two monomers is well conserved and that species that lack homology with human, in or around the pocket, have the possibility of being resistant to even the most potent inhibitors.
Differential pharmacology between species, or speciation, is a common problem encountered during the course of drug discovery that can severely hamper efficient advancement of clinical candidates. Many examples of speciation that have impacted drug discovery efforts exist in the literature and cover a broad range of protein families including G protein-coupled receptors (27), cytochrome P450 isoforms (28), and the MAPEG member microsomal prostaglandin E synthase-1 (29), for example. In the simplest of cases, research teams have identified an appropriate preclinical species, with homologous target protein sequence to human, to enable translational models. In more challenging settings, where this is not possible because of the lack of an acceptable preclinical species, the use of humanized mice may be required (30). In our own efforts to combat the issue of speciation, we identified an acceptable species for PK/PD studies, in which the FLAP protein sequence is highly homologous to human, enabling an alternative preclinical development pathway to enable the advancement of an unprecedented series of FLAP inhibitors. Thus, we established a canine whole blood assay, the results from which correlated well with the human whole blood assay, thereby providing a workable translational component to our compound advancement paradigm (Table 2 and Fig. 5). In a particularly compelling example for comparison to our findings, microsomal prostaglandin E synthase-1 inhibitors also demonstrated a dramatic loss in activity when tested in rodents (29). With this case, it was shown that in microsomal prostaglandin E synthase-1 of rats and mice, residues 131, 135, and 138 of TM4 block inhibitor access to the enzyme active site, rendering compounds identified by their activity against the human protein, inactive in these two rodent species (31). To establish translational models in which to test compounds, the authors generated a human microsomal prostaglandin E synthase-1 transgenic mouse strain. An interesting point arises from this comparison in that, for well validated targets for which there is clinical experience, more simplistic translational studies such as whole blood assays coupled with PK/PD can satisfy the institutional requirements for compound advancement. However, for new targets with poorly understood mechanisms of action and no clinical experience, more involved methods are required, such as the use of knock-in mice or exotic species with appropriate sequence homology.
In conclusion, we have identified a key residue in mouse FLAP that appears to prevent access of novel biaryl amino-heteroarene-based inhibitors to the previously described small molecule binding pocket of the protein. Computational studies coupled with mutational analyses have demonstrated that Ala24 in mouse FLAP is likely the residue responsible for this speciation effect because of a significant alteration in the floor of the binding pocket. Because both the mouse and rat orthologs contain this amino acid substitution, we adopted alternative methods for compound advancement and successfully implemented a canine whole blood assay and canine PK/PD procedures that enabled the advanced profiling of lead compounds.
Author Contributions
J. M. B. and M. D. H. wrote the manuscript; L. C. and K. H. performed biological studies; J. M. K. and A. D. L. synthesized compounds; M. D. H. performed computational studies; L. C., K. H., N. L. R., A. D. L., M. E. M., J. M. B., and M. D. H. analyzed the data; and N. L. R., A. D. L., J. M. K., M. E. M., T. M., J. M. B., and M. D. H. conceived the work.
Acknowledgments
We thank Annie Fourie, Tadimeti Rao, and members of Immunology Discovery and Discovery Sciences Teams, Janssen Research and Development, for contributions and support of this work.
The authors declare that they have no conflicts of interest with the contents of this article.
- FLAP
- 5-lipoxygenase activating protein
- HTRF
- homogeneous time resolved fluorescence
- MAPEG
- membrane-associated proteins in eicosanoid and glutathione metabolism
- PK
- pharmacokinetics
- PD
- pharmacodynamics
- LTB4
- leukotriene B4
- SAR
- structure and activity relationship.
References
- 1.Vickers P. J. (1995) 5-Lipoxygenase-activating protein (FLAP). J. Lipid Mediat. Cell Signal. 12, 185–194 [DOI] [PubMed] [Google Scholar]
- 2.Haeggstrom J. Z., Wetterholm A., Medina J. F., and Samuelsson B. (1993) Leukotriene A4 hydrolase: structural and functional properties of the active center. J. Lipid Mediat. 1–3, 1–13 [PubMed] [Google Scholar]
- 3.Lam B. K., Penrose J. F., Freeman G. J., and Austen K. F. (1994) Expression cloning of a cDNA for human leukotriene C4 synthase, an integral membrane protein conjugating reduced glutathione to leukotriene A4. Proc. Natl. Acad. Sci. U.S.A. 91, 7663–7667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welsch D. J., Creely D. P., Hauser S. D., Mathis K. J., Krivi G. G., and Isakson P. C. (1994) Molecular cloning and expression of human leukotriene-C4 synthase. Proc. Natl. Acad. Sci. U.S.A. 91, 9745–9749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sampson A. P. (2009) FLAP inhibitors for the treatment of inflammatory diseases. Curr. Opin. Investig. Drugs 10, 1163–1172 [PubMed] [Google Scholar]
- 6.Lemurell M., Ulander J., Winiwarter S., Dahlén A., Davidsson Ö., Emtenäs H., Broddefalk J., Swanson M., Hovdal D., Plowright A. T., Pettersen A., Rydén-Landergren M., Barlind J., Llinas A., Herslöf M., et al. (2015) Discovery of AZD6642, an inhibitor of 5-lipoxygenase activating protein (FLAP) for the treatment of inflammatory diseases. J. Med. Chem. 58, 897–911 [DOI] [PubMed] [Google Scholar]
- 7.Dahlén B., Kumlin M., Ihre E., Zetterström O., and Dahlén S. E. (1997) Inhibition of allergen-induced airway obstruction and leukotriene generation in atopic asthmatic subjects by the leukotriene biosynthesis inhibitor BAY X1005. Thorax 52, 342–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamant Z., Timmers M. C., van der Veen H., Friedman B. S., De Smet M., Depré M., Hilliard D., Bel E. H., and Sterk P. J. (1995) The effect of MK-0591, a novel 5-lipoxygenase-activating protein inhibitor, on leukotriene biosynthesis and allergen-induced airway responses in asthmatic subjects in vivo. J. Allergy Clin. Immunol. 95, 42–51 [DOI] [PubMed] [Google Scholar]
- 9.Singh D., Boyce M., Norris V., Kent S. E., and Bentley J. H. (2013) Inhibition of the early asthmatic response to inhaled allergen by the 5-lipoxygenase activating protein inhibitor GSK2190915: a dose-response study. Int. J. Gen. Med. 6, 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rouzer C. A., Ford-Hutchinson A. W., Morton H. E., and Gillard J. W. (1990) MK886, a potent and specific leukotriene biosynthesis inhibitor blocks and reverses the membrane association of 5-lipoxygenase in ionophore-challenged leukocytes. J. Biol. Chem. 265, 1436–1442 [PubMed] [Google Scholar]
- 11.Mancini J. A., Prasit P., Coppolino M. G., Charleson P., Leger S., Evans J. F., Gillard J. W., and Vickers P. J. (1992) 5-lipoxygenase-activating protein is the target of a novel hybrid of two classes of leukotriene biosynthesis inhibitors. Mol. Pharmacol. 41, 267–272 [PubMed] [Google Scholar]
- 12.Dixon R. A., Diehl R. E., Opas E., Rands E., Vickers P. J., Evans J. F., Gillard J. W., and Miller D. K. (1990) Requirement of a 5-lipoxygenase-activating protein for leukotriene synthesis. Nature 343, 282–284 [DOI] [PubMed] [Google Scholar]
- 13.Vickers P. J., O'Neill G. P., Mancini J. A., Charleson S., and Abramovitz M. (1992) Cross-species comparison of 5-lipoxygenase-activating protein. Mol. Pharmacol. 42, 1014–1019 [PubMed] [Google Scholar]
- 14.Ferguson A. D., McKeever B. M., Xu S., Wisniewski D., Miller D. K., Yamin T. T., Spencer R. H., Chu L., Ujjainwalla F., Cunningham B. R., Evans J. F., and Becker J. W. (2007) Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Science 317, 510–512 [DOI] [PubMed] [Google Scholar]
- 15.Evans J. F., Ferguson A. D., Mosley R. T., and Hutchinson J. H. (2008) What's all the FLAP about: 5-lipoxygenase-activating protein inhibitors for inflammatory diseases. Trends. Pharm. Sci. 29, 72–78 [DOI] [PubMed] [Google Scholar]
- 16.Bacani G. M., Eccles W., Fitzgerald A. E., Goldberg S. D., Hack M. D., Hawryluk N. A., Jones W. M., Keith J. M., Krawczuk P., Lebsack A. D. Lee-Dutra A., Liu J., McClure K. J., Meduna S. P., Pippel D. J., et al. (2014) Flap modulators. PCT Int. Appl. WO 2014121055 A2 20140807 [Google Scholar]
- 17.Fitzgerald A. E., Hack M. D., Hawryluk N. A., Jones W. M., Keith J. M., Krawczuk P., Lebsack A. D., Liu J. M., Mani N. S., McClure K. J., Meduna S. P., and Rosen M. D. (2014) Flap modulators. PCT Int. Appl. WO 2014121040 A1 20140807 [Google Scholar]
- 18.Chai W., Deckhut C., Dvorak C. A., Eccles W., Edwards J. P., Goldberg S. D., Krawczuk P. J., Lebsack D., Liu J., Tanis V. M., and Tarantino K. T. (September 18, 2014) 1,2,5-Substituted benzimidazoles as FLAP modulators. U. S. Patent 20140275028 A1
- 19.Chai W., Dvorak C. A., Eccles W., Edwards J. P., Goldberg S. D., Krawczuk P. J., Lebsack A. D., Liu J., Pippel D. J., Sales Z. S., Tanis V. M., Tichenor M. S., and Wiener J. J. (September 18, 2014) 1,2,6-Substituted benzimidazoles as FLAP modulators. U. S. Patent 20140275029 A1 20140918
- 20.Song J., Liu X., Zhu J., Tootoonchi M., Keith J. M., Meduna S. P., Dvorak C. A., Eccles W., Krawczuk P. J., Blevitt J. M., Wu J., Rao N. L., Lebsack A. D., and Milla M. E. (2016) Polypharmacology of small-molecule modulators of the 5-liopoxygenase activating protein (FLAP) observed via a high-throughput lipidomics platform. J. Biomol. Screen. 21, 127–135 [DOI] [PubMed] [Google Scholar]
- 21.Brideau C., Chan C., Charleson S., Denis D., Evans J. F., Ford-Hutchinson A. W., Fortin R., Gillard J. W., Guay J., and Guévremont D. (1992) Pharmacology of MK-0591 (3-[1-(4-chlorobenzyl)-3-(t-butylthio)-5-(quinolin-2-yl-methoxy)- indol-2-yl]-2,2-dimethyl propanoic acid), a potent, orally active leukotriene biosynthesis inhibitor. Can. J. Physiol. Pharmacol. 70, 799–807 [DOI] [PubMed] [Google Scholar]
- 22.Lorrain D. S., Bain G., Correa L. D., Chapman C., Broadhead A. R., Santini A. M., Prodanovich P. P., Darlington J. V., Stock N. S., Zunic J., King C. D., Lee C., Baccei C. S., Stearns B., Roppe J., et al. (2010) Pharmacology of AM803, a novel selective five-lipoxygenase-activating protein (FLAP) inhibitor in rodent models of acute inflammation. Eur. J. Pharmacol. 640, 211–218 [DOI] [PubMed] [Google Scholar]
- 23.Cheng Y., and Prusoff W. H. (1973) Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 24.Miller D. K., Yamin T. T., Cummings R. T., Zhao A., and Wisniewski D. (2004) Development of a high capacity HTRF assay for measurement of antagonists of 5-lipoxygenase activation protein (FLAP). Proceedings of the Inflammation Research Association: 12th International Conference of the IRA, Bolton Landing, NY, October 3–7, 2014 [Google Scholar]
- 25.Lin J. H. (1995) Species similarities and differences in pharmacokinetics. Drug Metab. Dispos. 23, 1008–1021 [PubMed] [Google Scholar]
- 26.Song J., Liu X., Rao T. S., Chang L., Meehan M. J., Blevitt J. M., Wu J., Dorrestein P. C., and Milla M. E. (2015) Phenotyping drug polypharmacology via eicosanoid profiling of blood. J. Lipid Res. 56, 1492–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tichenor M. S., Thurmond R. L., Venable J. D., and Savall B. M. (2015) Functional profiling of 2-aminopyrimidine histamine H4 receptor modulators. J. Med. Chem. 58, 7119–7127 [DOI] [PubMed] [Google Scholar]
- 28.Lewis D. F., Ito Y., and Lake B. G. (2009) Molecular modelling of CYP2F substrates: comparison of naphthalene metabolism by human, rat and mouse CYP2F subfamily enzymes. Drug Metabol. Drug Interact. 24, 229–257 [DOI] [PubMed] [Google Scholar]
- 29.Xu D., Rowland S. E., Clark P., Giroux A., Côté B., Guiral S., Salem M., Ducharme Y., Friesen R. W., Méthot N., Mancini J., Audoly L., and Riendeau D. (2008) MF63 [2-(6-chloro-1H-phenanthro[9,10-d]imidazol-2-yl)-isophthalonitrile], a selective microsomal prostaglandin E synthase-1 inhibitor, relieves pyresis and pain in preclinical models of inflammation. J. Pharmacol. Exp. Ther. 326, 754–763 [DOI] [PubMed] [Google Scholar]
- 30.Shultz L. D., Ishikawa F., and Greiner D. L. (2007) Humanized mice in translational biomedical research. Nat. Rev. Immunol. 7, 118–130 [DOI] [PubMed] [Google Scholar]
- 31.Pawelzik S. C., Uda N. R., Spahiu L., Jegerschöld C., Stenberg P., Hebert H., Morgenstern R., and Jakobsson P. J. (2010) Identification of key residues determining species differences in inhibitor binding of microsomal prostaglandin E synthase-1. J. Biol. Chem. 285, 29254–29261 [DOI] [PMC free article] [PubMed] [Google Scholar]