Abstract
Activation of death receptor-5 (DR5) leads to the formation of death inducing signaling complex (DISC) for apoptotic signaling. Targeting DR5 to induce breast cancer apoptosis is a promising strategy to circumvent drug resistance and present a target for breast cancer treatment. Calmodulin (CaM) has been shown to regulate DR5-mediated apoptotic signaling, however, its mechanism remains unknown. In this study, we characterized CaM and DR5 interactions in breast cancer cells with integrated experimental and computational approaches. Results show that CaM directly binds to DR5 in a calcium dependent manner in breast cancer cells. The direct interaction of CaM with DR5 is localized at DR5 death domain. We have predicted and verified the CaM-binding site in DR5 being 354WEPLMRKLGL363 that is located at the α2 helix and the loop between α2 helix and α3 helix of DR5 DD. The residues of Trp-354, Arg-359, Glu-355, Leu-363, and Glu-367 in DR5 death domain that are important for DR5 recruitment of FADD and caspase-8 for DISC formation to signal apoptosis also play an important role for CaM-DR5 binding. The changed electrostatic potential distribution in the CaM-binding site in DR5 DD by the point mutations of W354A, E355K, R359A, L363N, or E367K in DR5 DD could directly contribute to the experimentally observed decreased CaM-DR5 binding by the point mutations of the key residues in DR5 DD. Results from this study provide a key step for the further investigation of the role of CaM-DR5 binding in DR5-mediated DISC formation for apoptosis in breast cancer cells.
Keywords: breast cancer, calmodulin (CaM), death domain, electrostatics, protein-protein interaction, death receptor 5, integrated experimental and computational study
Introduction
Breast cancer is one of the most common malignancies among American women and affects an estimated 1.3 million women annually worldwide (1, 2). Chemotherapy has steadily increased breast cancer survival rate; however, drug resistance and systemic toxicity remain critical issues that hinder effective breast cancer treatment (3–7). Better understanding of the pathogenesis of breast cancer could facilitate the development of novel strategies for targeted therapy for effective breast cancer treatment. Classification of breast cancers into receptor-based subtypes, including estrogen receptor (ER)3-positive or triple-negative breast cancers, establish targets that guide individualized treatment for patients (8). Drug-resistant ER-positive breast cancers and triple-negative breast cancers are prolific breast cancers with poor clinical outcome (3). Death receptor-5 (DR5) is expressed in various breast cancer cell lines that represent the different breast cancer subtypes (9, 10), and DR5 expression is up-regulated in breast cancer cells compared with normal breast tissue (11).
DR5, also called tumor necrosis factor (TNF)-related apoptosis inducing ligand receptor 2 (TRAIL-R2), is a member of the TNF receptor superfamily and is one of five receptors bound by TNF receptor apoptosis-inducing ligand (TRAIL). TRAIL has been reported with strong antitumor activity (12–14), however, TRAIL can induce apoptosis of normal human hepatocytes (15). TRA-8 is a DR5-specific agonist antibody and TRA-8 activation of DR5 has shown tumoricidal activity in vitro and in vivo without inducing normal hepatocyte apoptosis (16). TRA-8 can induce DR5-mediated apoptosis in breast cancer cells (9, 17).
DR5 contains a cytoplasmic death domain (12) and transduces its apoptotic signal via death-inducing signaling complex (DISC) formation and activation of caspase signaling (18). Upon stimulation by binding of TRA-8 to its extracellular domains, DR5 oligomerizes into homotrimers (19). DR5 homotrimers are bound at their death domains (DD) by the adapter protein Fas-associated death domain protein (FADD). FADD recruits procaspase-8 forming DISC leading to caspase-8 activation (18, 20–22). Dysfunction of the DR5 DD or DISC components can lead to resistance of DR5-mediated apoptosis (18, 22). DR5 DD mutations have been identified in breast cancer (23). Studies show that some key residue mutations in DR5 DD result in DR5 unable to recruit FADD and caspase-8 for DISC formation to signal apoptosis (24–26). Leu-363 is the residue corresponding to the site of lpr mutation in the mouse Fas receptor, and L363N, L363A, and L363F (mutation related to non-small cell lung cancer (27)) mutations in DR5 DD result in the failure of inducing apoptosis. Arg-359 is the conserved residue among DR5, DR4, DR3, TNFR1, and Fas, and DR5 R359A mutant can completely lose the function of DR5. Glu-355 has been identified in human tumors, and DR5 E355K mutant (mutation related to non-small cell lung cancer and gastric cancer (27, 28)) results in the DR5 losing its ability to form DISC. Trp-354 has been identified as an important hydrophobic residue for the overall structure of DR5, and DR5 W354A mutant (i.e.W325A in Ref. 24) causes reduced ability of DR5 to induce apoptosis. Glu-367 has been identified in human tumor, and DR5 E367K mutant (i.e. E338K in Ref. 25) results in the DR5 losing its ability to form DISC.
Calmodulin (CaM) functions as an intracellular mediator of Ca2+ signals and regulates various cellular processes (29, 30). Breast tumor transformation to malignancy is associated with the increase in CaM expression (31, 32). CaM binding to Fas, a member of the TNF receptor superfamily, has been well characterized (33–37). CaM binds to Fas at the Fas death domain (34) and CaM is recruited into the Fas-mediated DISC after Fas activation in cholangiocarcinoma cells (33). A role of CaM in DR5-mediated apoptotic signaling regulation has been reported in human lung cancer cells (38) and hepatocellular carcinoma cells (39). The mechanism of CaM regulation of DR5-mediated apoptotic signaling has not been fully understood. The structure of CaM consists of 8 helices, forming four calcium-binding loops, which exhibit the characteristic EF hand structure (40). The dumbbell-shaped structure of CaM is composed of two globular terminal regions and two helices comprising the inter-domain region (41). CaM interacts with diverse proteins to regulate cellular activities in a Ca2+-dependent manner (34, 42, 43). Three main classes of recognition motifs exist for many of the known CaM-binding proteins (34, 45): [1] Ca2+-dependent binding 1–8-14 motif ([FILVW]1XXXXXX[FAILVW]8XXXXX[FILVW]14), [2] Ca2+-dependent binding 1–10 motif (XXX[FILVW]1XXXXXXXX[FILVW]10), in which the fifth residue is usually also a conserved residue, i.e. 1–5-10 motif (XXX[FILVW]1XXX[FAILVW]5XXXX[FILVW]10), and [3] Ca2+-independent binding IQ motif ([ILV]QXXXRXXXX[RK]XX[FILVWY]).
In this study, we characterized the Ca2+-dependent direct interactions between CaM and DR5 in estrogen receptor (ER)-positive ZR-75-1 and triple-negative MDA-MB-231 breast cancer cells. We identified the CaM-binding site in the DR5 death domain and the critical residues in the DR5 death domain for CaM-DR5 binding using integrated computational and experimental approaches.
Experimental Procedures
We characterized the direct interactions between CaM and DR5 in ER-positive ZR-75-1 and triple-negative MDA-MB-231 breast cancer cells using complex-immunoprecipitation (Co-IP) and CaM pull-down assay. We determined that CaM directly binds to the purified recombinant DR5 death domain using CaM pull-down assays. We identified the CaM-binding site in the DR5 death domain and the critical residues in the DR5 death domain for CaM-DR5 binding using integrated computational and experimental approaches.
Breast Cancer Cell Culture
ER-positive ZR-75-1 and triple-negative MDA-MB-231 breast cancer cells were kindly provided by Dr. Donald Buchsbaum or Dr. Tong Zhou at the University of Alabama at Birmingham. ZR-75-1 cells were cultured in HAM's K-12 medium with 20% FBS. Triple-negative MDA-MB-231 breast cancer cells were cultured in DMEM medium (Thermo Fisher Scientific, Waltham, MA) with 10% FBS and supplemented with MEM vitamins, nonessential amino acids, sodium pyruvate, and l-glutamine. Both cell lines were maintained in the medium with 10% FBS, supplemented with 1% penicillin, 1% streptomycin, and 1% amphotericin B antibiotic at 37 °C, 5% CO2, and 95% relative humidity atmosphere.
Prediction of CaM-binding Site on DR5 Death Domain (DR5 DD) using Computational Methods
The CaM-binding site in DR5 was predicted using the combined text pattern search method (46) that was based on the three CaM-binding motifs (34, 45), the hydropathic analysis method (47–53), and the experimentally known residues in DR5 DD critical for DR5-mediated death inducing signaling complex (DISC) formation for apoptosis (24, 25, 27, 54). CaM interacts with diverse proteins to regulate cellular activities in a Ca2+-dependent manner (34, 42, 43). Three main classes of recognition motifs for many of the known CaM-binding proteins (34, 45) include: 1) Ca2+-dependent binding 1–8-14 motif: ([FILVW]1XXXXXX[FAILVW]8XXXXX[FILVW]14), 2) Ca2+-dependent binding 1–10 motif (XXX[FILVW]1XXXXXXXX[FILVW]10), in which the fifth residue is usually a conserved residue, i.e. 1–5-10 motif (XXX[FILVW]1XXX[FAILVW]5XXXX[FILVW]10), and 3) Ca2+-independent binding IQ motif ([ILV]QXXXRXXXX[RK]XX[FILVWY]). The sequence of DR5 death domain (DD) was scanned to identify the CaM-binding motif(s) in DR5 DD using text pattern search method (46). To further identify the CaM-binding site in DR5 DD, we adopted complementary hydropathy index sign-matching strategy (55) that was based on the molecular recognition theory of Blalock (56) to predict protein-protein interaction sites. We used the hydropathy binary code (+ and −) of the predicted CaM-binding site(s) in DR5 DD from text pattern search method to match the binary code of hydropathy of CaM and rank the results by the number/percentage of the complementary pairs ((+)-(−)). Together with the consideration of the experimentally known residues in DR5 DD critical for DR5-mediated DISC formation for apoptosis (24, 25, 27, 54), the potential CaM-binding site in DR5 DD was identified and experimentally validated. The text pattern search and complementary hydropathy index sign-matching were performed using the in-house developed codes.
Generation of DNA Constructs of DR5 Fragments Using Polymerase Chain Reaction
Full-length human DR5 cDNA cloned into the pCMV-SPORT6 vector was kindly provided by Dr. John Mountz at the University of Alabama at Birmingham. DR5 cytoplasmic region (DR5 CR) was generated by PCR using pCMV-SPORT6 DR5 as the template. DR5 CR PCR products were cloned into the pET100 vector (Invitrogen Life Technologies, Grand Island, NY) for expression in Escherichia coli BL21 (DE3) strain cells. DR5 death domain (DR5 DD), DR5 cytoplasmic region with death domain deletion (DR5 ΔDD), DR5 cytoplasmic region with the predicted CaM-binding site (354WEPLMRKLGL363) deletion (DR5 BSD) were generated by deletion PCR. DR5 cytoplasmic region mutations: DR5 W354A, DR5 E355K, DR5 R359A, DR5 L363N, and DR5 E367K were generated by site mutation PCR using pET100 DR CR as the template. DR5 ΔDD, DR5 BSD, and DR5 cytoplasmic region mutation PCR products with overlapping ends were generated by using a forward primer (5′) together with the reverse primer (3′) coding for the mutation or deletion (supplemental Table S1).
Expression and Purification of Recombinant Proteins
pET100 6×His tag DR5 CR, DR5 DD, DR5 ΔDD, DR5 BSD, and DR5 CR mutations proteins were expressed in E. coli BL21 (DE3) strain cells (Invitrogen Life Technologies) as follows. Transformed E. coli cells were incubated in LB media with 100 μg/ml ampicillin (Fisher Scientific) on a shaking incubator at 37 °C for 3 h. 1 mm isopropyl β-d-thiogalactopyranoside (Promega) was added to cultures for induction of protein expression and then incubated for an additional 4 h at 37 °C. Recombinant protein expressions were evaluated by Western blot using an anti-6×His antibody (Sigma-Aldrich). After induction of protein expression, E. coli cultures were centrifuged and bacteria pellets were lysed by sonication, lysates were centrifuged and the supernatant collected. The 6×His-tagged recombinant proteins were purified by nickel ion affinity chromatography according to the manufacturer's direction for 6×His-tagged protein purification system (Qiagen). Recombinant proteins were further purified by size exclusion chromatography using a HiLoad 16/600 Superdex 75 prepgrade column on an AKTA purifier (GE Life Sciences).
DR5 Immunoprecipitation
To determine an interaction between CaM and DR5 in ER-positive ZR-75-1 and triple-negative MDA-MB-231 breast cancer cells, DR5 was immunoprecipitated (IP) from cell lysates using TRA-8-conjugated Sepharose beads (16), which were kindly provided by Dr. Tong Zhou at the University of Alabama at Birmingham. Iso-IgG-conjugated Sepharose beads (Cell Signaling Technologies, Beverly, MA, specificity: none, source: mouse, Cat. 3420S, Lot 3) were used for a control IP experiment. ER-positive or triple-negative breast cancer cells were lysed using a 50 mm Tris-HCL pH 7.5, 150 mm NaCl, 1% Triton X-100, and 1:100 Halt protease inhibitor (Thermo Fisher Scientific, Waltham, MA) buffer at 4 °C for 30 min. The total protein concentration was determined using the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). 5 mg/ml total protein of cell lysates was precleared with 40 μl of 4B-Sepharose beads (Sigma-Aldrich) at 4 °C for 1 h. After the clearance, the supernatants of each sample were incubated with TRA-8-conjugated Sepharose beads or mouse Iso-IgG-conjugated Sepharose beads, and rotated overnight at 4 °C. Following overnight incubation, TRA-8-conjugated Sepharose beads or mouse Iso-IgG conjugated Sepharose beads were washed using a 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Triton X-100 washing buffer five times. After washing, 40 μl of 2× Laemmli sample buffer with 2-mercaptoethanol was added to the beads to elute the proteins for Western blot analysis.
CaM Pull-down Assay
To determine the calcium-dependent interactions between CaM and DR5 death domain, ER-positive and triple-negative breast cancer cells were lysed with phosphate-buffered saline with 1% Triton X-100 (PBST), and 1:100 Halt protease inhibitor (Thermo Fisher Scientific) supplemented with 1 mm CaCl2 or 2 mm EGTA. Cell lysates were incubated with 100 μl of CaM-4B-Sepharose beads (GE Healthcare Bio-Science) or control 4B-Sepharose beads for 4 h at 4 °C. After CaM pull-down beads or control 4B-Sepharose beads were washed five times with the PBST buffer with 1 mm Ca2+ or 2 mm EGTA, and then 100 μl of 2× Laemmli buffer was added to elute proteins for Western blot analysis. To determine the direct interactions between CaM and DR5, purified recombinant DR5 cytoplasmic region (DR5 CR) and DR5 death domain (DR5 DD) proteins were generated. To determine the CaM-binding site in DR5, purified recombinant DR5 CR with death domain deletion (DR5 ΔDD) and DR5 CR with potential binding site deletion (DR5 BSD) proteins were generated. To identify the critical residues in DR5 death domain necessary for CaM-DR5 binding, purified recombinant DR5 CR mutants with the different point mutation were generated. All recombinant DR5 proteins and DR5 mutant proteins were dialyzed into a 50 mm Tris pH 7.6, 120 mm NaCl, 1 mm Ca2+, 1% Brij binding buffer. Recombinant proteins were incubated with 150 μl of CaM-4B-Sepharose beads or control Sepharose beads for 3 h at 4 °C. After pull-down, beads were washed with the 50 mm Tris, pH 7.6, 120 mm NaCl, 1 mm Ca 2+, 1% Brij binding buffer, and then 150 μl of 2× Laemmli buffer was added to elute recombinant proteins for Western blot.
Western Blot Analysis
CaM binding to DR5 was assessed by Western blot analysis of eluted CaM or DR5 proteins from IP or CaM pull-downs. Samples from IP and CaM pull-down in Laemmli buffer were heated at 100 °C for 5 min. Protein samples were resolved by SDS-PAGE and transferred onto a 0.45 μm nitrocellulose membrane from Bio-Rad. For IP samples, membranes were incubated overnight at 4 °C with primary antibodies against DR5 (Abcam Cambridge, MA, specificity: DR5 isoform 1 and 2, source: rabbit, Cat. Ab47179, Lot# GR199335–1) and CaM (Millipore Billerica, MA, source mouse, Cat. 05173, Lot 2123697). For samples from CaM pull-down, membranes were incubated overnight at 4 °C with a primary antibody against the 6×His tag (Sigma-Aldrich, specificity: recognizes synthetic polyhistidine, as well as native or denatured, reduced forms of proteins tagged with 6× histidine, source: mouse, Cat. H1029-.2ML, Lot 121M4789) to detect 6×His-tagged DR5 cytoplasmic region or its mutants. Following overnight antibody incubation, membranes were washed three times with TBST, and then a 1-h incubation with horseradish peroxidase-conjugated secondary antibodies from Southern Biotechnologies (Birmingham, Alabama, specificity: anti-rabbit IgG (H+L) source: goat Cat. 405-05, Lot K0008-VJ213 or specificity: anti-mouse IgG (H+L), source: goat, Cat. 1031-05, Lot H2710-NA02) at room temperature. Proteins were evaluated using enhanced chemiluminescence detection reagents from Thermo Fisher Scientific according to the manufacturer's instructions.
DR5 Death Domain Structure Construction
The initial structure of DR5 death domain (DD) was constructed based on the structures of tumor necrosis factor receptor (TNFR)-1 DD (PDB ID: 1ICH) (57) and Fas DD (PDB ID: 1DDF) (58) with MODERLLER 9.2 (59). Fas, TNFR-1 and DR5 all belong to the TNFR superfamily and their death domains perform similar functions during the formation of signaling complex for apoptosis (54, 60) The T-Coffee software that is a multiple sequence alignment program provided by European Bioinformatics Institute (EBI) was used for the sequence alignment of the target protein DR5 DD and the templates of Fas DD and TNFR-1 DD. Transitive consistency score function (61) implemented in T-Coffee software was used to evaluate protein sequence alignment results and the transitive consistency score for Fas DD and TNFR-1 DD were separately 79 and 80 at a 0–100 scale. The stereochemical quality of the top five candidates for DR5 DD structure that are constructed based on the template of Fas DD and TNFR-1 DD was assessed with PROCHECK, (62) ERRAT (63), and WHATCHECK (64) using NIH structural analysis and verification server (SAVES) (65, 66). PROCHECK evaluates the residual ϕ/ψ angles in the Ramachandran plots (62). ERRAT analyze patterns of non-bonded interactions and gives an overall quality score for the entire model (63). WHATCHECK does extensive checking of many stereochemical parameters of the residues in the model (64).
Electrostatic Potential Calculations
The electrostatic potentials for DR5 DD and DR5 DD mutants were calculated with APBS software (67). To calculate the electrostatic potential, dielectric constants of 1 and 78.54 were used for the protein and solvent separately. The ion concentration of 150 mm was used in APBS calculation. The electrostatic potential was mapped onto the molecular surface of DR5 DD using the VMD program (68).
Results
CaM Interactions with DR5 in a Ca2+-dependent Manner in Triple-negative MDA-MB-231 and ER-positive ZR-75-1 Breast Cancer Cells
CaM may interact with target proteins via a calcium (Ca2+)-dependent or independent manner (45). We characterized the interaction of endogenous CaM and DR5 in a Ca2+-dependent manner in ER-positive ZR-75-1 and triple-negative MDA-MB-231 breast cancer cells that are sensitive to TRA-8 (9, 70, 71).
Co-immunoprecipitation (Co-IP) of DR5 and CaM from MDA-MB-231 and ZR-75-1 Cell Lysate
DR5 was immunoprecipitated (IP) from each cell lysate using TRA-8-conjugated Sepharose beads. Mouse Iso-IgG-conjugated Sepharose beads were used for the control IP experiments. Western blot analysis of Co-IP samples, using primary antibodies against DR5 and CaM, was performed to assess DR5 association with CaM. Results show that CaM was co-immunoprecipitated with DR5 from ER-positive ZR-75-1 and triple-negative MDA-MB-231 breast cancer cell lysates (IP: TRA-8 lane) (Fig. 1A). CaM or DR5 was not found in the control Co-IP using the mouse Iso-IgG-conjugated Sepharose beads, which indicated that CaM or DR5 did not interact with the Sepharose bead of mouse IgG portion of the TRA-8 antibody. Co-IP results using TRA-8-conjugated Sepharose beads demonstrated that CaM interacted with DR5 in both ER-positive ZR-75-1 and triple-negative MDA-MB-231 breast cancer cells.
FIGURE 1.
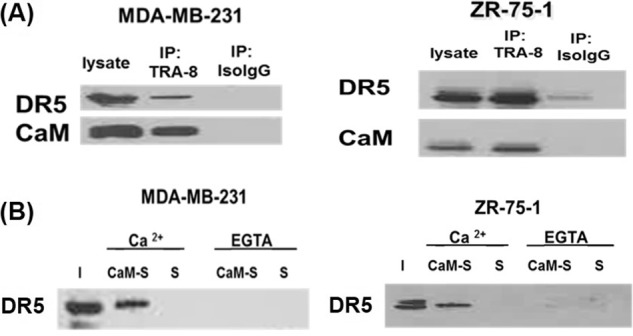
CaM interactions with DR5 in a Ca2+-dependent manner in triple-negative MDA-MB-231 and ER-positive ZR-75-1 breast cancer cells. A, co-immunoprecipitation of DR5 and CaM from MDA-MB-231 and ZR-75-1 cell lysates. Lysate lane: whole cell lysate of breast cancer cells; IP TRA-8 lane: IP using TRA-8-conjugated beads; IP Iso-IgG lane: IP using a mouse Iso-IgG-conjugated beads. B, CaM pull-down of DR5 from MDA-MB-231 and ZR-75-1 cell lysates supplemented with 1 mm Ca 2+ or 2 mm EGTA. I: input, CaM-S: pull-down using CaM-conjugated Sepharose beads. S: pull-down control using Sepharose beads. Representative results are shown from two independent experiments.
CaM Pull-down of DR5 from MDA-MB-231 and ZR-75-1 Cell Lysate
We characterized CaM and DR5 interactions in a Ca2+-dependent manner by using CaM-conjugated Sepharose beads to pull-down DR5 from ER-positive ZR-75-1 and triple-negative MDA-MB-231 breast cancer cell lysates supplemented with 1 mm Ca2+ or 2 mm EGTA. Sepharose beads were used for the control pull-down experiments. Western blot analysis of pull-down samples, using primary antibody against DR5, was performed to characterize DR5 association with CaM. Results show that DR5 was present in the CaM pull-down from both types of breast cancer cell lysates supplemented with calcium (Fig. 1B). However, DR5 was not present in the CaM pull-down experiments from breast cancer lysates supplemented with EGTA (Fig. 1B). DR5 was not present in the control experiments using Sepharose beads for pull-down control experiments. The results demonstrated that CaM and DR5 interaction in ER-positive ZR-75-1 and triple-negative MDA-MB-231 breast cancer cells was calcium dependent.
CaM Directly Interacts with DR5 at DR5 Death Domain in a Ca2+-dependent Manner
To determine a direct interaction between CaM and DR5, CaM pull-down assays were performed using purified recombinant 6×His-tagged DR5 cytoplasmic region (amino acids 232–440) (DR5 CR) and DR5 death domain (DD) proteins. DR5 CR or DR5 DD in 50 mm Tris, pH 7.6, 120 mm NaCl, 1% Brij buffer with 1 mm Ca2+, or 2 mm EGTA were incubated with CaM-Sepharose beads or Sepharose beads. The 6×His-tagged DR5 CR or DR5 DD proteins were detected using an anti-polyhistidine antibody by Western blot. Results show that both DR5 CR and DR5 DD bound to CaM but not to the control Sepharose beads in the presence of calcium (Fig. 2A). However, with the medium supplemented with EGTA, either DR5 CR or DR5 DD did not bind to CaM. The results show the direct interactions between CaM and DR5 in a Ca2+-dependent manner and CaM-DR5 interactions are not mediated by other DR5-binding proteins, such as FADD (20). To further confirm that CaM directly binds to DR5 at the DR5 death domain, we generated a DR5 cytoplasmic region mutant with the deletion of the DR5 death domain (DR5 CR ΔDD) and performed CaM pull-down assay of DR5 CR ΔDD with CaM-Sepharose beads or Sepharose beads. Results show that DR5 CR ΔDD did not bind to CaM (Fig. 2B). These results demonstrated that CaM directly binds to DR5 in a Ca2+-dependent manner, and the interaction of CaM with DR5 was localized at the DR5 death domain.
FIGURE 2.
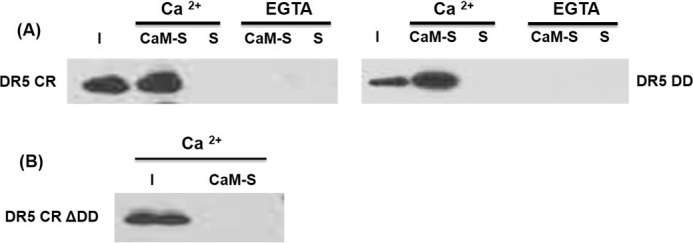
CaM directly interacts with DR5 in DR5 death domain in a Ca2+-dependent manner. A, CaM pull-down of purified DR5 cytoplasmic region (DR5 CR) and DR5 death domain (DR5 DD) in a 50 mm Tris, pH 7.6, 120 mm NaCl, 1% Brij buffer with 1 mm Ca 2+, or 2 mm EGTA. B, CaM pull-down of purified DR5 CR mutant with the deletion of DR5 death domain (DR5 CR ΔDD) in a 50 mm Tris pH 7.6, 120 mm NaCl, 1 mm Ca2+, 1% Brij buffer. I: input, CaM-S: pull-down using CaM-conjugated Sepharose beads. S: pull-down control using Sepharose beads. Representative results are shown from two independent experiments.
Prediction of the CaM-binding Site on DR5 Death Domain and the Residues in DR5 Death Domain Critical for CaM-DR5 Binding
The CaM-binding site on DR5 was firstly predicted with the pattern search method (46) by scanning the three CaM-binding motifs (34, 45) separately on the sequence of DR5 DD. Five potential CaM-binding sites on DR5 DD predicted from the pattern search method are: 1) 338LRQCFDDFADLVPF351; 2) 342FDDFADLVPFDSW354; 3) 390VNKTGRDASVHTLL403; 4) 406LETLGERLAKQKI418; and 5) 354WEPLMRKLGL363. To further narrow down the CaM-binding site in DR5 DD, we used complementary hydropathy index sign-matching strategy (55) that is based on the molecular recognition theory of Blalock (56) to predict protein-protein interaction sites. We used the hydropathy binary code (+ and −) of the predicted CaM-binding site(s) in DR5 DD from text pattern search method to match the binary code of hydropathy of the CaM and rank the results by the number/percentage of the complementary pairs ((+)-(−)). The sequence of 354WEPLMRKLGL363 on DR5 DD was ranked the highest as the potential CaM-binding site in DR5 DD. 354WEPLMRKLGL363 is located at the α2 helix and the loop between α2 helix and α3 helix of DR5 DD (Fig. 3A).
FIGURE 3.
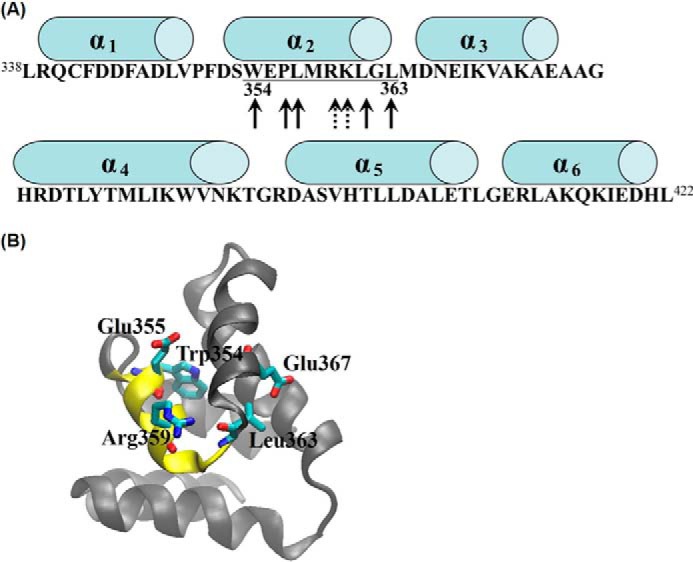
Predicted CaM-binding site in DR5 death domain (DR5 DD) and the key residues critical for CaM-DR5 binding. A, predicted CaM-binding site in DR5 DD (354WEPLMRKLGL363) is underlined. Arrows with solid lines indicate hydrophobic residues, and arrows with dotted lines indicate positively charged residues. Cylinders above the sequence represent α helical regions. B, structural view of DR5 DD with the predicted CaM-binding site (354WEPLMRKLGL363) shown in yellow color and the predicted key residues of Leu-363, Trp-354, Glu-355, Arg-359, and Glu-367 in DR5 DD shown as licorice.
Experimental studies have shown that the mutations of several key residues in DR5 DD including DR5 W354A, DR5 E355K, DR5 R359A, DR5 L363N, and DR5 E367K mutations result in DR5 unable to recruit FADD and caspase-8 for DISC formation to signal apoptosis (24–26, 32, 33, 44, 54). Residues of Trp-354, Arg-359, Glu-355, Leu-363, and Glu-367 are on or close to the predicted CaM-binding site in DR5 DD. Thus, we predicted that the mutations of these key residues could also affect CaM-DR5 binding, which could further affect DR5-mediated DISC formation for apoptosis. The structure of DR5 DD was constructed based on the structures of tumor necrosis factor receptor (TNFR)-1 DD (PDB ID: 1ICH) (57) and Fas DD (PDB ID: 1DDF) (58) with MODERLLER 9.2 (44, 59) and the constructed DR5 DD structure was evaluated with the NIH Structural Analysis and Verification Server as described under “Experimental Procedures.” The structure with the overall best score of stereochemical quality (supplemental Table S2) was used for this study. The predicted CaM-binding site in DR5 DD and the residues in DR5 DD critical for CaM-DR5 binding were shown in the three-dimensional structure of DR5 DD (Fig. 3B).
Validation of the Predicted CaM-binding Site in DR5 DD and the Key Residues in DR5 DD Critical for CaM-DR5 Binding
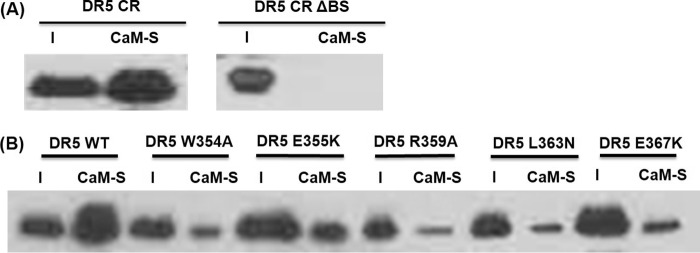
We performed biochemical experiments to verify the predicted CaM-binding site in DR5 DD (354WEPLMRKLGL363) and the key residues in DR5 DD critical for CaM-DR5 binding. We generated the constructs for DR5 cytoplasmic region (DR5 CR), DR5 CR mutant with the deletion of the predicted CaM-binding site in DR5 DD (DR5 CR ΔBS), and DR5 CR mutants with the point mutation of W354A, E355K, R359A, L363N, or E367K. Residues of Trp-354, Arg-359, Glu-355, Leu-363, and Glu-367 are on or close to the predicted CaM-binding site in DR5 DD. We expressed and purified DR5 CR protein and DR5 CR mutants as described under “Experimental Procedures.” We performed pull-down experiments for the purified recombinant DR5 CR, DR5 CR ΔBS mutant, and the mutants of DR5 CR W354A, DR5 CR E355K, DR5 CR R359A, DR5 CR L363N, and DR5 CR E367K separately using CaM-Sepharose beads. Results show that the deletion of the predicted CaM-binding site (354WEPLMRKLGL363) in DR5 cytoplasmic region resulted in the loss of the CaM-DR5 binding (Fig. 4A). The point mutation of W354A, E355K, R359A, L363N, or E367K in DR5 CR resulted in the decreased CaM-DR5 binding compared with CaM binding to wild type DR5 CR (Fig. 4B). These results show that the predicted CaM-binding site in DR5 DD is required for CaM-DR5 binding, validating the predicted CaM-binding site in DR5: 354WEPLMRKLGL363. The results also demonstrate that the point mutations of the residues of Trp-354, Arg-359, Glu-355, Leu-363, and Glu-367 that are on or close to the predicted CaM-binding site in DR5 death domain directly affect CaM-DR5 binding. These residues have been shown to be important for DR5 recruitment of FADD and caspase-8 for DISC formation to signal apoptosis (24–26). These results suggest the important role of CaM-DR5 binding in DR5-mediated DISC formation for apoptosis.
FIGURE 4.
Validation of the predicted CaM-binding site in DR5 DD and the effect of critical residues in DR5 DD on CaM-DR5 binding. A, CaM pull-down of DR5 cytoplasmic region (DR5 CR) or DR5 CR mutant with the deletion of the predicted CaM-binding site (354WEPLMRKLGL363) (DR5 CR ΔBSD). B, CaM pull-down of wild type DR5 CR (WT) or DR5 CR mutants: DR5 W354A, DR5 E355K, DR5 R359A, DR5 L363N, and DR5 E367K. I: input, CaM-S: pull-down using CaM-conjugated Sepharose beads. Representative results are shown from two independent experiments.
Electrostatic Potential Changes in DR5 DD Resulted from the Point Mutation of the Key Residues in CaM-binding Site in DR5 DD
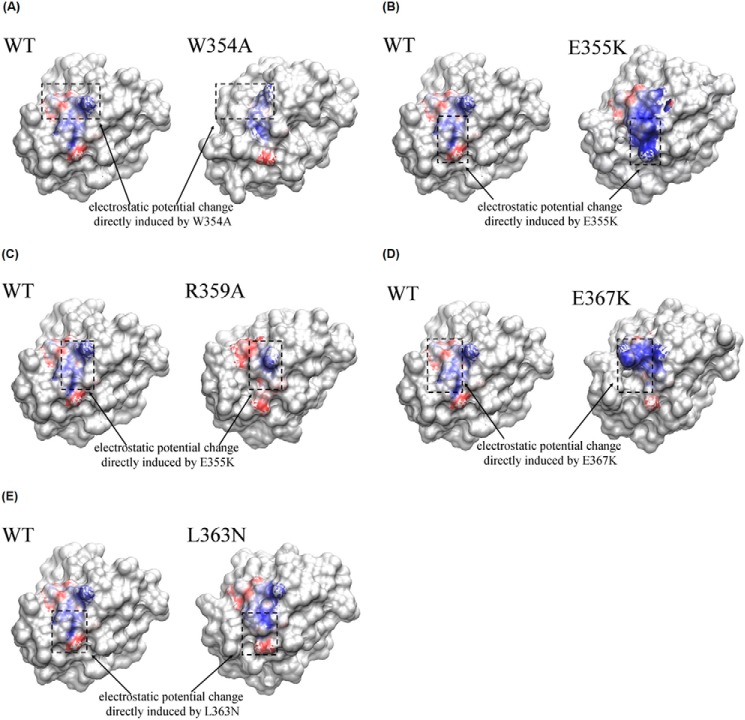
The electrostatic potential changes of the CaM-binding site in DR5 DD could directly affect CaM-DR5 binding. To understand the effect of the point mutations of the key residues in DR5 DD, including W354A, E355K, R359A, L363N, or E367K mutation on CaM-DR5 binding, we performed electrostatic potential analyses for DR5 DR5 DD WT and DR5 DD mutants using APBS software (67). The comparisons of the electrostatic potential of the CaM-binding site in DR5 DD WT and in DR5 DD mutants are shown in Fig. 5, in which the blue color represents the positive electrostatic potential whereas the red color represents the negative electrostatic potential. The results show that for DR5 DD WT, the positive electrostatic potential was distributed in the middle region of the CaM-binding site, whereas the negative electrostatic potential was distributed in the upper and lower regions of the CaM-binding site (Fig. 5). For DR5 DD W354A mutant, the positive electrostatic potential distribution in the middle region of the CaM-binding site in DR5 DD was much smaller compared with that in DR5 DD WT, and the negative electrostatic potential was only distributed in the lower region (Fig. 5A). For the DR5 DD E355K mutant, the positive electrostatic potential in the middle region of the CaM-binding site in DR5 DD was significantly increased compared with that in DR5 DD WT and the negative electrostatic potential was only distributed in the left upper region of the CaM-binding site in DR5 DD (Fig. 5B). For the DR5 DD R359A mutant, the positive electrostatic potential only appeared in a small region of the middle of the CaM-binding site in DR5 DD and the negative electrostatic potential was significantly increased in the left top and bottom region (Fig. 5C). For the DR5 DD E367K mutant, although Glu367 is not on the CaM-binding site in DR5 DD, the E367K mutation also affects electrostatic potential distributions in the CaM-binding site in DR5 DD, resulting in the increased positive electrostatic potential distribution in the top half region of the CaM-binding site in DR5 DD (Fig. 5D). For the DR5 DD L363N mutant, although the electrostatic potential pattern was similar to that in DR5 DD WT, however, the degree/magnitude of electrostatic potential distribution resulting from the L363N mutation was different from that for DR5 DD WT (Fig. 5E). The electrostatic potential changes resulting from the key residue mutations in DR5 DD could directly affect CaM-DR5 interactions, further affecting the degree of CaM-DR5 binding.
FIGURE 5.
Electrostatic potential comparison between the CaM-binding site in wild type DR5 DD and DR5 DD mutants. The blue regions show the positive electrostatic potential, whereas the red regions show the negative electrostatic potential. The black dot boxes show the electrostatic potential comparison between the CaM-binding site in wild type DR5 DD and DR5 DD mutants (A) W354A mutant, (B) E355K mutant, (C) R359A mutant, (D) E367K mutant, and (E) L363N mutant.
Discussion
DR5 is one of the well characterized death receptors, and DR5 contains a cytoplasmic death domain (12) and similar to the Fas receptor transduces its apoptotic signal via DISC formation and activation of caspase signaling for apoptosis (18, 20). Moreover, not only mediating apoptosis, DR5 also mediate anti-apoptotic signaling in breast cancer cells (69). These suggest that DR5 engage a variety of biological effect. So far, significant studies about the downstream molecules related to DR5-mediated apoptotic and anti-apoptotic functions have been reported, including FADD, glycogen synthase kinase-3 DDX3 and cellular inhibitor of apoptosis protein-1 (18, 69). DR5 is expressed in various breast cancer cell lines that represent the different breast cancer subtypes (9, 10), and DR5 expression is up-regulated in breast cancer cells compared with normal breast tissue (11). Although the studies about DR5-mediated apoptotic signaling and anti-apoptotic signaling in breast cancer cells has been reported (9, 69). However, many aspects of DR5-mediated biological effects remain unknown, which could be mediated by the interactions of DR5 with various other molecules.
In this study, we characterized the interactions between CaM and DR5 in ER-positive ZR-75-1 and triple-negative MDA-MB-231 breast cancer cells with integrated experimental and computational approaches. The results show that CaM directly binds to DR5 in a calcium-dependent manner in both ER-positive ZR-75-1 and triple-negative MDA-MB-231 breast cancer cells. Results also show that the direct interaction of CaM with DR5 was localized at the DR5 death domain. We have predicted and verified the CaM-binding site in DR5 being 354WEPLMRKLGL363 that is located at the α2 helix and the loop between α2 helix and α3 helix of DR5 DD. These findings are important not only for understanding the DR5-mediated apoptosis but also important for expanding the knowledge of CaM. CaM functions as an intracellular mediator of Ca2+ signals and regulates various cellular processes (29, 30). Breast tumor transformation to malignancy is associated with the increase in CaM expression (31, 32). CaM binding to Fas has been well characterized (33–37). Results from this provide the potential mechanism for CaM regulation of DR5-mediated apoptotic signaling in breast cancer.
The residues of Trp-354, Arg-359, Glu-355, Leu-363, and Glu-367 in the DR5 death domain have been shown important for DR5 recruitment of FADD and caspase-8 for DISC formation to signal apoptosis (24–26). Results from this study show that the point mutations of W354A, E355K, R359A, L363N, or E367K in the DR5 death domain directly affect CaM-DR5 binding, which suggest the important role of CaM-DR5 binding in DR5-mediated DISC formation for apoptosis. Our electrostatic potential analysis results show that the mutations of W354A, E355K, R359A, L363N, or E367K in DR5 DD directly affect the electrostatic potential distribution in the CaM-binding site in DR5 DD, which help to interpret our experimentally observed decreased CaM-DR5 binding by the point mutation of these key residues in DR5 DD. The results from this study provide the basis for the further investigation of the role of CaM-DR5 binding in DR5-mediated DISC formation for apoptosis in ER-positive and triple-negative breast cancer cells.
Author Contributions
R. M. F., Y. S., and T. Z. designed the research. R. M. F., Q. Z., and H. W. conducted the biochemical experiments and analyzed experimental data. L. W. conducted the computational studies. L. W. and Y. S. analyzed computational data. T. Z. and D. J. B. provided guidance for biochemical experiments. R. M. F., L. W., and Y. S. wrote the manuscript. Q. Z., H. W., T. Z. D. J. B. provide inputs and assistance for the manuscript writing. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Jun Li for technical assistance with generation of the pET 100 DR5 and DR5 mutation constructs.
This work was supported in part by an National Institutes of Health K25 award (5K25 CA140791) (to Y. S.). The authors declare that they have no conflicts of interest with the contents of this article except that Dr. Tong Zhou and Dr. Donald J. Buchsbaum have intellectual property right in TRA-8. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1 and S2
- ER
- estrogen receptor
- CaM
- calmodulin
- DISC
- death-inducing signaling complex
- DR5
- death receptor 5
- DD
- death domain
- CR
- cytoplasmic region
- BSD
- binding site deletion
- FADD
- Fas-associated death domain protein
- TRAIL
- TNF receptor apoptosis-inducing ligand.
References
- 1.DeSantis C., Siegel R., Bandi P., and Jemal A. (2011) Breast cancer statistics, 2011. CA Cancer J. Clin. 61, 409–418 [DOI] [PubMed] [Google Scholar]
- 2.Patterson R., Saquib N., Natarajan L., Rock C., Parker B., Thomson C., and Pierce J. (2011) Improvement in self-reported physical health predicts longer survival among women with a history of breast cancer. Breast Cancer Res. Treat. 127, 541–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guarneri V., and Conte P. F. (2004) The curability of breast cancer and the treatment of advanced disease. Eur. J. Nucl. Med. Mol. Imaging 31, S149–161 [DOI] [PubMed] [Google Scholar]
- 4.Siegel R., Desantis C., Virgo K., Stein K., Mariotto A., Smith T., Cooper D., Gansler T., Lerro C., Fedewa S., Lin C., Leach C., Cannady R. S., Cho H., Scoppa S., Hachey M., Kirch R., Jemal A., and Ward E. (2012) Cancer treatment and survivorship statistics, 2012. CA Cancer J. Clin. 62, 220–241. [DOI] [PubMed] [Google Scholar]
- 5.Plasschaert S. L., Van Der Kolk D. M., De Bont E. S., Vellenga E., Kamps W. A., and De Vries E. G. (2004) Breast cancer resistance protein (BCRP) in acute leukemia. Leuk. Lymphoma 45, 649–654 [DOI] [PubMed] [Google Scholar]
- 6.Shou J., Massarweh S., Osborne C. K., Wakeling A. E., Ali S., Weiss H., and Schiff R. (2004) Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J. Natl. Cancer Inst. 96, 926–935 [DOI] [PubMed] [Google Scholar]
- 7.Osborne C. K., Bardou V., Hopp T. A., Chamness G. C., Hilsenbeck S. G., Fuqua S. A., Wong J., Allred D. C., Clark G. M., and Schiff R. (2003) Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J. Natl. Cancer Inst. 95, 353–361 [DOI] [PubMed] [Google Scholar]
- 8.Sørlie T. (2009) Introducing molecular subtyping of breast cancer into the clinic? J. Clin. Oncol. 27, 1153–1154 [DOI] [PubMed] [Google Scholar]
- 9.Buchsbaum D. J., Zhou T., Grizzle W. E., Oliver P. G., Hammond C. J., Zhang S., Carpenter M., and LoBuglio A. F. (2003) Antitumor efficacy of TRA-8 anti-DR5 monoclonal antibody alone or in combination with chemotherapy and/or radiation therapy in a human breast cancer model. Clin. Cancer Res. 9, 3731–3741 [PubMed] [Google Scholar]
- 10.Dong H. P., Kleinberg L., Silins I., Flørenes V. A., Tropé C. G., Risberg B., Nesland J. M., and Davidson B. (2008) Death receptor expression is associated with poor response to chemotherapy and shorter survival in metastatic ovarian carcinoma. Cancer 112, 84–93 [DOI] [PubMed] [Google Scholar]
- 11.McCarthy M. M., Sznol M., DiVito K. A., Camp R. L., Rimm D. L., and Kluger H. M. (2005) Evaluating the Expression and Prognostic Value of TRAIL-R1 and TRAIL-R2 in Breast Cancer. Clinical Cancer Res. 11, 5188–5194 [DOI] [PubMed] [Google Scholar]
- 12.Pitti R. M., Marsters S. A., Ruppert S., Donahue C. J., Moore A., and Ashkenazi A. (1996) Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J. Biol. Chem. 271, 12687–12690 [DOI] [PubMed] [Google Scholar]
- 13.Walczak H., Miller R. E., Ariail K., Gliniak B., Griffith T. S., Kubin M., Chin W., Jones J., Woodward A., Le T., Smith C., Smolak P., Goodwin R. G., Rauch C. T., Schuh J. C., and Lynch D. H. (1999) Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 5, 157–163 [DOI] [PubMed] [Google Scholar]
- 14.Ashkenazi A., Pai R. C., Fong S., Leung S., Lawrence D. A., Marsters S. A., Blackie C., Chang L., McMurtrey A. E., Hebert A., DeForge L., Koumenis I. L., Lewis D., Harris L., Bussiere J., Koeppen H., Shahrokh Z., and Schwall R. H. (1999) Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Invest. 104, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jo M., Kim T. H., Seol D. W., Esplen J. E., Dorko K., Billiar T. R., and Strom S. C. (2000) Apoptosis induced in normal human hepatocytes by tumor necrosis factor-related apoptosis-inducing ligand. Nat. Med. 6, 564–567 [DOI] [PubMed] [Google Scholar]
- 16.Ichikawa K., Liu W., Zhao L., Wang Z., Liu D., Ohtsuka T., Zhang H., Mountz J. D., Koopman W. J., Kimberly R. P., and Zhou T. (2001) Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat. Med. 7, 954–960 [DOI] [PubMed] [Google Scholar]
- 17.Amm H. M., Zhou T., Steg A. D., Kuo H., Li Y., and Buchsbaum D. J. (2011) Mechanisms of drug sensitization to TRA-8, an agonistic death receptor 5 antibody, involve modulation of the intrinsic apoptotic pathway in human breast cancer cells. Mol. Cancer Res. 9, 403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sprick M. R., Weigand M. A., Rieser E., Rauch C. T., Juo P., Blenis J., Krammer P. H., and Walczak H. (2000) FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 12, 599–609 [DOI] [PubMed] [Google Scholar]
- 19.Hymowitz S. G., Christinger H. W., Fuh G., Ultsch M., O'Connell M., Kelley R. F., Ashkenazi A., and de Vos A. M. (1999) Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol. Cell 4, 563–571 [DOI] [PubMed] [Google Scholar]
- 20.Kischkel F. C., Lawrence D. A., Chuntharapai A., Schow P., Kim K. J., and Ashkenazi A. (2000) Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity 12, 611–620 [DOI] [PubMed] [Google Scholar]
- 21.Kischkel F., Hellbardt S., Behrmann I., Germer M., Pawlita M., Krammer P., and Peter M. (1995) Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 14, 5579–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniel P. T., Wieder T., Sturm I., and Schulze-Osthoff K. (2001) The kiss of death: promises and failures of death receptors and ligands in cancer therapy. Leukemia 15, 1022–1032 [DOI] [PubMed] [Google Scholar]
- 23.Shin M. S., Kim H. S., Lee S. H., Park W. S., Kim S. Y., Park J. Y., Lee J. H., Lee S. K., Lee S. N., Jung S. S., Han J. Y., Kim H., Lee J. Y., and Yoo N. J. (2001) Mutations of tumor necrosis factor-related apoptosis-inducing ligand receptor 1 (TRAIL-R1) and receptor 2 (TRAIL-R2) genes in metastatic breast cancers. Cancer Res. 61, 4942–4946 [PubMed] [Google Scholar]
- 24.McDonald E. R. 3rd., Chui P. C., Martelli P. F., Dicker D. T., and El-Deiry W. S. (2001) Death domain mutagenesis of KILLER/DR5 reveals residues critical for apoptotic signaling. J. Biol. Chem. 276, 14939–14945 [DOI] [PubMed] [Google Scholar]
- 25.Bin L., Thorburn J., Thomas L. R., Clark P. E., Humphreys R., and Thorburn A. (2007) Tumor-derived mutations in the TRAIL receptor DR5 inhibit TRAIL signaling through the DR4 receptor by competing for ligand binding. J. Biol. Chem. 282, 28189–28194 [DOI] [PubMed] [Google Scholar]
- 26.Özören N., and El-Deiry W. S. (2003) Cell surface death receptor signaling in normal and cancer cells. Immunity 13, 135–147 [DOI] [PubMed] [Google Scholar]
- 27.Lee S. H., Shin M. S., Kim H. S., Lee H. K., Park W. S., Kim S. Y., Lee J. H., Han S. Y., Park J. Y., Oh R. R., Jang J. J., Han J. Y., Lee J. Y., and Yoo N. J. (1999) Alterations of the DR5/TRAIL receptor 2 gene in non-small cell lung cancers. Cancer Res. 59, 5683–5686 [PubMed] [Google Scholar]
- 28.Park W. S., Lee J. H., Shin M. S., Park J. Y., Kim H. S., Kim Y. S., Park C. H., Lee S. K., Lee S. H., Lee S. N., Kim H., Yoo N. J., and Lee J. Y. (2001) Inactivating mutations of KILLER/DR5 gene in gastric cancers. Gastroenterology 121, 1219–1225 [DOI] [PubMed] [Google Scholar]
- 29.Cohen P., and Klee C. B. (1988) in Calmodulin, Elsevier Science Publishing Co. Inc., New York [Google Scholar]
- 30.Eldik L. V., and Watterson D. (1988) in Calmodulin and Signal Transduction, Academic Press, San Diego, CA [Google Scholar]
- 31.Hait W. N., and Lazo J. S. (1986) Calmodulin: a potential target for cancer chemotherapeutic agents. J. Clin. Oncol. 4, 994–1012 [DOI] [PubMed] [Google Scholar]
- 32.Wei J., Morris H., and Hickie R. (1982) Positive correlation between calmodulin content and hepatoma growth rates. Cancer Res. 42, 2571–2574 [PubMed] [Google Scholar]
- 33.Chen Y., Pawar P., Pan G., Ma L., Liu H., and McDonald J. (2008) Calmodulin binding to the Fas-mediated death-inducing signaling complex in cholangiocarcinoma cells. J. Cell Biochem. 103, 788–799 [DOI] [PubMed] [Google Scholar]
- 34.Ahn E. Y., Lim S. T., Cook W. J., and McDonald J. M. (2004) Calmodulin binding to the Fas death domain. Regulation by Fas activation. J. Biol. Chem. 279, 5661–5666 [DOI] [PubMed] [Google Scholar]
- 35.Wu X., Ahn E., McKenna M., Yeo H., and McDonald J. (2005) Fas binding to calmodulin regulates apoptosis in osteoclasts. J. Biol. Chem. 280, 29964–29970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fancy R. M., Wang L., Napier T., Lin J., Jing G., Lucius A. L., McDonald J. M., Zhou T., and Song Y. (2014) Characterization of calmodulin-Fas death domain interaction: an integrated experimental and computational study. Biochemistry 53, 2680–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suever J. D., Chen Y., McDonald J. M., and Song Y. (2008) Conformation and free energy analyses of the complex of calcium-bound calmodulin and the Fas death domain. Biophys. J. 95, 5913–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang M. K., Min Y. K., and Kim S. H. (2009) Calmodulin inhibition contributes to sensitize TRAIL-induced apoptosis in human lung cancer H1299 cells. Biochem. Cell Biol. 87, 919–926 [DOI] [PubMed] [Google Scholar]
- 39.Wang C., Chen T., Zhang N., Yang M., Li B., Lü X., Cao X., and Ling C. (2009) Melittin, a major component of bee venom, sensitizes human hepatocellular carcinoma cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis by activating CaMKII-TAK1-JNK/p38 and inhibiting IκBα kinase-NFκB. J. Biol. Chem. 284, 3804–3813 [DOI] [PubMed] [Google Scholar]
- 40.Babu Y. S., Sack J. S., Greenhough T. J., Bugg C. E., Means A. R., and Cook W. J. (1985) Three-dimensional structure of calmodulin. Nature 315, 37–40 [DOI] [PubMed] [Google Scholar]
- 41.Ganoth A., Friedman R., Nachliel E., and Gutman M. (2006) A molecular dynamics study and free energy analysis of complexes between the Mlc1p protein and two IQ motif peptides. Biophys. J. 91, 2436–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eldik L., and Watterson D. (1998) Calmodulin and Calcium Signal Transduction. Academic Press, San Diego [Google Scholar]
- 43.Moser M. J., Lee S. Y., Klevit R. E., and Davis T. N. (1995) Ca2+ binding to calmodulin and its role in Schizosaccharomyces pombe as revealed by mutagenesis and NMR spectroscopy. J. Biol. Chem. 270, 20643–20652 [DOI] [PubMed] [Google Scholar]
- 44.Sali A., and Blundell T. L. (1993) Comparative Protein Modeling by Satisfaction of Spatial Restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 45.Rhoads A. R., and Friedberg F. (1997) Sequence motifs for calmodulin recognition. Faseb J. 11, 331–340 [DOI] [PubMed] [Google Scholar]
- 46.Friedl J. E. F. (1997) Mastering Regular Expressions: Powerful Techniques for Perl and Other Tools, Softcover Ed., Oreilly & Associates Inc [Google Scholar]
- 47.Goicoechea S., Pallero M. A., Eggleton P., Michalak M., and Murphy-Ullrich J. E. (2002) The anti-adhesive activity of thrombospondin is mediated by the N-terminal domain of cell surface calreticulin. J. Biol. Chem. 277, 37219–37228 [DOI] [PubMed] [Google Scholar]
- 48.Houtman R., Ten Broeke R., Blalock J. E., Villain M., Koster A. S., and Nijkamp F. P. (2001) Attenuation of very late antigen-5-mediated adhesion of bone marrow-derived mast cells to fibronectin by peptides with inverted hydropathy to EF-hands. J. Immunol. 166, 861–867 [DOI] [PubMed] [Google Scholar]
- 49.Villain M., Jackson P. L., Manion M. K., Dong W. J., Su Z., Fassina G., Johnson T. M., Sakai T. T., Krishna N. R., and Blalock J. E. (2000) De novo design of peptides targeted to the EF hands of calmodulin. J. Biol. Chem. 275, 2676–2685 [DOI] [PubMed] [Google Scholar]
- 50.Manion M. K., Villain M., Pan Z. G., McDonald J. M., and Blalock J. E. (2000) Cellular uptake and in situ binding of a peptide agonist for calmodulin. Biochem. Biophys. Res. Commun. 277, 462–469 [DOI] [PubMed] [Google Scholar]
- 51.Weigent D. A., Clarke B. L., and Blalock J. E. (1994) Peptide design using a genetically patterned binary code: growth hormone-releasing hormone as a model. ImmunoMethods 5, 91–97 [DOI] [PubMed] [Google Scholar]
- 52.Maier C. C., Moseley H. N., Zhou S. R., Whitaker J. N., and Blalock J. E. (1994) Identification of interactive determinants on idiotypic-anti-idiotypic antibodies through comparison of their hydropathic profiles. ImmunoMethods 5, 107–113 [DOI] [PubMed] [Google Scholar]
- 53.Dillon J., Woods W. T., Guarcello V., LeBoeuf R. D., and Blalock J. E. (1991) A peptide mimetic of calcium. Proc. Natl. Acad. Sci. U.S.A. 88, 9726–9729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaudhary P. M., Eby M., Jasmin A., Bookwalter A., Murray J., and Hood L. (1997) Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-κB pathway. Immunity 7, 821–830 [DOI] [PubMed] [Google Scholar]
- 55.Jacchieri S. G., Torquato R., and Brentani R. R. (2003) Structural study of binding of flagellin by Toll-like receptor 5. J. Bacteriol. 185, 4243–4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blalock J. E. (1995) Genetic origins of protein shape and interaction rules. Nat. Med. 1, 876–878 [DOI] [PubMed] [Google Scholar]
- 57.Sukits S. F., Lin L. L., Hsu S., Malakian K., Powers R., and Xu G. Y. (2001) Solution structure of the tumor necrosis factor receptor-1 death domain. J. Mol. Biol. 310, 895–906 [DOI] [PubMed] [Google Scholar]
- 58.Huang B., Eberstadt M., Olejniczak E. T., Meadows R. P., and Fesik S. W. (1996) NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature 384, 638–641 [DOI] [PubMed] [Google Scholar]
- 59.Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., and Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophysi. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 60.Schneider P., Thome M., Burns K., Bodmer J., Hofmann K., Kataoka T., Holler N., and Tschopp J. (1997) TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-κB. Immunity 7, 831–836 [DOI] [PubMed] [Google Scholar]
- 61.Chang J. M., Di Tommaso P., and Notredame C. (2014) TCS: a new multiple sequence alignment reliability measure to estimate alignment accuracy and improve phylogenetic tree reconstruction. Mol. Biol. Evol. 31, 1625–1637 [DOI] [PubMed] [Google Scholar]
- 62.Laskowski R. A., MacArthur M. W., Moss D. S., and Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 63.Colovos C., and Yeates T. O. (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2, 1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8, 52-56, 29. [DOI] [PubMed] [Google Scholar]
- 65.Eisenberg D., Lüthy R., and Bowie J. U. (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 277, 396–404 [DOI] [PubMed] [Google Scholar]
- 66.Wiederstein M., and Sippl M. J. (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, W407–W410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 69.Sun M., Song L., Li Y., Zhou T., and Jope R. S. (2008) Identification of an antiapoptotic protein complex at death receptors. Cell Death Differ. 15, 1887–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oliver P., LoBuglio A., Zhou T., Forero A., Kim H., Zinn K., Zhai G., Li Y., Lee C., and Buchsbaum D. (2012) Effect of anti-DR5 and chemotherapy on basal-like breast cancer. Breast Cancer Res. Treat 133, 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chinnaiyan A. M., Prasad U., Shankar S., Hamstra D. A., Shanaiah M., Chenevert T. L., Ross B. D., and Rehemtulla A. (2000) Combined effect of tumor necrosis factor-related apoptosis-inducing ligand and ionizing radiation in breast cancer therapy. Proc. Natl. Acad. Sci. 97, 1754–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.