Abstract
Heat shock protein 47 (HSP47) is an endoplasmic reticulum (ER)-resident collagen-specific chaperone and essential for proper formation of the characteristic collagen triple helix. It preferentially binds to the folded conformation of its clients and accompanies them from the ER to the Golgi compartment, where it releases them and is recycled back to the ER. Unlike other chaperones, the binding and release cycles are not governed by nucleotide exchange and hydrolysis, but presumably the dissociation of the HSP47-procollagen complex is triggered by the lower pH in the Golgi (pH 6.3) compared with the ER (pH 7.4). Histidine residues have been suggested as triggers due to their approximate textbook pKa value of 6.1 for their side chains. We present here an extensive theoretical and experimental study of the 14 histidine residues present in canine HSP47, where we have mutated all histidine residues in the collagen binding interface and additionally all of those that were predicted to undergo a significant change in protonation state between pH 7 and 6. These mutants were characterized by biolayer interferometry for their pH-dependent binding to a collagen model. One mutant (H238N) loses binding, which can be explained by a rearrangement of the Arg222 and Asp385 residues, which are crucial for specific collagen recognition. Most of the other mutants were remarkably silent, but a double mutant with His273 and His274 exchanged for asparagines exhibits a much less pronounced pH dependence of collagen binding. This effect is mainly caused by a lower koff at the low pH values.
Keywords: collagen, heat shock protein (HSP), histidine, molecular chaperone, protein folding
Introduction
Collagen is the most abundant protein in mammals and constitutes a crucial component of the extracellular matrix. There are 28 different types of collagens known today. All of them have in common as a defining feature the so-called collagen triple helix, a structural element that is formed by three individual polypeptide chains (reviewed in Refs. 1–3). The three-dimensional structure of this extended, non-globular domain puts severe restrictions on the linear amino acid sequence of collagens, resulting in repeating triplets of the sequence Xaa-Yaa-Gly with proline and hydroxyproline as the preferred residues in the Xaa and Yaa positions, respectively. Biogenesis of collagen is a complicated process involving extensive post-translational modifications (4). After secretion into the endoplasmic reticulum (ER),4 a large network of modifying enzymes like proline hydroxylase or sugar transferases as well as molecular chaperones, such as protein-disulfide isomerase or binding immunoglobulin protein, are involved in the formation of a properly modified and folded trimeric procollagen molecule that is secreted via the Golgi compartment to the extracellular medium. HSP47 is an ER-resident procollagen-specific chaperone that binds to multiple sites on procollagen bearing an arginine in the Yaa positions and is essential for proper triple-helix formation (5, 6). HSP47 gene ablation results in impaired secretion, accompanied by overmodification and intracellular procollagen accumulation, as well as defective maturation of the procollagen chains (7). The resulting (pro)collagen from such HSP47-deficient cells exhibits a misfolded triple helix, as indicated by a proteolytic sensitivity of this otherwise very robust structure. As a consequence, the knock-out in mice is lethal at an early embryonic stage due to malformation of the basal membrane (8), and missense mutations in the HSP47/SERPINH1 gene are one cause of recessive osteogenesis imperfecta (9, 10).
Contrary to other chaperones, HSP47 binds preferentially to the folded (i.e. triple-helical) conformation of its client (11), and client binding and release events are not triggered by nucleotide binding and hydrolysis cycles but presumably rather by the pH shift the HSP47-collagen complex undergoes on its way from the ER (pH ∼7.4) to the cis-Golgi compartment (pH ∼6.3) (12, 13). After dissociation from collagen in the low pH compartments of the cis-Golgi or ER-Golgi intermediate compartment, HSP47 is recycled back to the ER due to its C-terminal RDEL sequence.
HSP47 is a member of the serpin family (14) but has lost all inhibitory function together with the characteristic serpin S → R transition that results upon proteolytic cleavage or latency transition in the insertion of a mobile element, the so-called reactive center loop, into the middle of a β-sheet. Based on CD and fluorescence spectra, conformational rearrangements of HSP47 at lower pH values have nevertheless been postulated (15, 16). Crystal structures of HSP47 in complex with collagen model peptides have been published, and the interface between the two proteins is known in atomic detail (17). However, a number of important questions, especially regarding the pH-triggered client release, are still open. Six histidine residues are found within this binding interface, thus opening an attractive explanation of client release at slightly acidic pH values. The involvement of imidazole side chains has also been previously postulated, due to their pKa values usually falling in the interval between 6 and 7, thus titrating in the pH interval relevant to the HSP47 client-binding and release cycle (15). Previous work has mainly assigned a pair of histidine residues in the so-called breach region of HSP47 as responsible triggers of collagen binding and release (18), together with another histidine located close by. The exact mechanism by which protonation of these histidines results in collagen dissociation from HSP47 is still unclear.
To gain a deeper insight into the function of HSP47 and the molecular nature of the interaction between chaperone and client, we have undertaken a systematic study involving the prediction of pKa values followed by an extensive mutagenesis approach. We show here that the previous assignment of histidine residues is in error and put forward a detailed model of how low pH triggers collagen release from HSP47.
Experimental Procedures
Molecular Dynamics Simulations
The crystal structure of HSP47 in complex with a collagen model peptide (Protein Data Bank entry 3ZHA (17), chains ABEFG) was used as a template for the starting structures. For closing the gap in the protein backbone between Ser-119 and Val-126 (chain A), a modeled loop was inserted using ModLoop (19, 20).
Titrating aspartate, glutamate, and histidine residues were renamed to prepare the pdb files for the constant pH molecular dynamics (CpHMD) simulation developed by Mongan et al. (21). Hydrogen atoms were added using the LEaP module of AMBER 12 (22). Subsequently, the topology and coordinate files of the systems were generated with LEaP. The CpHMD (and also all other MD) simulations were performed using Sander of AMBER 12 and the ff99SB force field (23) for proteins. At first, a minimization was carried out in three subsequent steps to optimize the geometry of the starting structures. In the first step of the minimization, the hydrogen atoms of the protein were minimized, whereas everything else was restrained with a constant force of 10 kcal/(mol·Å2) to the initial positions. In the second step, the modeled loop was minimized, whereas the remaining protein was restrained with 10 kcal/(mol·Å2). In the last step, no restraints were used, so that the whole system was minimized. All three minimization parts started with 2,500 steps using the steepest descent algorithm, followed by 2,500 steps of a conjugate gradient minimization.
The molecules were solvated with the implicit solvent developed by Onufriev et al. (control variable igb = 2) (24). The salt concentration was set to 0.1 m (based on Debye-Hückel). The cut-off distance for non-bonded interactions was set to 30.0 Å, and bonds involving hydrogen were constrained with SHAKE. The time step was 2 fs, and a Monte Carlo step was performed after every step. Thereafter, the systems were equilibrated in two successive steps. In the first step (0.1 ns long), the temperature was raised from 10 to 310 K, and the protein was restrained with a constant force of 5 kcal/(mol·Å2). In the second step (0.4 ns long), only the Cα atoms of the protein were restrained with a constant force of 5 kcal/(mol·Å2). Then the simulations ran for 25 ns (pH 4.0) or 100 ns (pH 5.0, 6.0, 7.0, and 8.0) without any restraints at physiological temperature (310 K) with a Langevin thermostat. For the analysis of the MD simulations, the fraction of doubly protonated histidines was measured over simulation.
The H238N mutant and, for comparison, also the wild-type HSP47 were simulated without the collagen molecule for 50 ns with the described CpHMD method at pH 8.0. For the wild-type HSP47 and the mutants (H273N, H274N, and H273N/H274N), a modified version of the CpHMD, namely a pH titrating MD (pHtMD) (25) simulation, was conducted. During these 201-ns-long simulations, the solution pH was decreased from 8.00 to 4.00 with a rate of −0.02 pH units/ns.
Steered molecular dynamics (SMD) simulations were performed for wild-type HSP47 and the H273N/H274N mutant, both complexed with the collagen model peptide, to investigate the work necessary for dissociation of the HSP47-collagen complex. Ten different structures from the pHtMD simulation at pH 6.0 were used as starting structures for the 10 independent SMD simulations. While the HSP47 molecule was restrained with a constant force of 5,000 kcal/(mol·Å2), the collagen was pulled away with a constant velocity of 0.5 Å/ps, thereby increasing the distance between the centers of mass of the collagen triple helix and the HSP47 molecule from 32 to 82 Å. Finally, the computed work, which is the integrated force over distances, was averaged over the 10 simulations.
Protein Constructs and Expression
A canine-derived synthetic HSP47 gene coding for residues 36LSP … RDEL418 was purchased as a codon-optimized cDNA for Escherichia coli from DNA 2.0 (Menlo Park, CA) with an additional C-terminal StrepII tag and cloned into the manufacturer's expression plasmid, pJExpress 411. The construct was transformed in E. coli BL21 (DE3) cells grown to A600 of 0.6–0.7, and protein expression was induced with 0.5 mm isopropyl-β-d-thiogalactoside. After a 4-h incubation at 37 °C/180 rpm, cells were harvested, and pellets were stored at −20 °C. Cells were resuspended in lysis buffer (50 mm Tris·HCl (pH 7.5), 150 mm NaCl, 100 μm PMSF, 4 mm DTT) and lysed by sonication. Cleared lysates were loaded onto 1 ml of Streptactin Superflow (IBA) and eluted with 2.5 mm d-desthiobiotin in lysis buffer excluding PMSF. The eluted proteins were further subjected to size exclusion chromatography (Superdex 200, GE Healthcare) in 50 mm Hepes (pH 7.5), 150 mm NaCl, 2 mm DTT, and the protein was frozen in liquid nitrogen and stored at −80 °C.
Mutations were introduced according to an optimized site-directed mutagenesis protocol (26) using overlapping primers. The mutants were expressed and purified following the protocol used for wild-type HSP47.
The plasmid coding for the GPR-foldon peptide (27) was a generous gift from Dr. M. Koch (University of Cologne). It encodes an N-terminal Twin-StrepII tag, a thrombin site, a collagen sequence ((GPP)5GPR(GPP)6), and a C-terminal phage T4 foldon domain for protein trimerization. The protein was expressed in BL21 (DE3), induced at A600 of 1.0 with 100 μm isopropyl-β-d-thio-galactoside, and incubated overnight at 20 °C/180 rpm. The foldon protein was purified analogously to HSP47 constructs but without DTT in the buffers.
Phylogenetic Analysis
Protein sequences of all currently annotated SerpinH1 orthologs in ENSEMBL (ensemble.org/GRCh38.p3) were retrieved and aligned using the ClustalO plugin of CLC Bio. Sequences containing too many gaps to be useful were deleted. The species were grouped together based on their phylogenetic class, and these classes were ordered by their phylogenetic relationship. Protein sequences of human SerpinF1 (PEDF), SerpinA1 (antitrypsin), and SerpinC1 (antithrombin) were included as an outgroup. The alignment was colored using BOXSHADE.
Kinetic Measurements Using Biolayer Interferometry
Biolayer interferometry was measured on a BLItz system (Pall ForteBIO LLC, Menlo Park, CA). For coupling on streptavidin biosensors, foldon peptides were biotinylated with NHS-Biotin following the manufacturer's protocols (Pierce) and loaded at a concentration of 10 μm to a response of ∼5.5 nm. Free biotin binding sites were blocked with 10 μm biocytin, and nonspecific interactions were blocked with two load/regeneration cycles with HSP47. The sensors were regenerated with McIlvaine buffer (28) at pH 6.0 for up to 40 s. Successful regeneration of the sensors was verified by comparison of HSP47 binding at pH 7.5 before and after regeneration. Kinetics were measured with a constant protein concentration of 5 μm HSP47 protein. The association time was fixed at 60 s, whereas the dissociation time was chosen differently at different pH to allow for sufficient dissociation (pH 7.5, 140 s; pH 7.0, 120 s; pH 6.5, 100 s; pH 6.0, 80 s). For each condition and sample, at least three sensorgrams were recorded, and all experiments were repeated at least twice on different days with different protein preparations. Kinetic parameters were extracted separately using the program Origin version 8.5 and a 1:1 Langmuir binding model. Sensorgrams are shown as representative results, and kinetic parameters (kon, koff, and KD) are presented with error bars indicating the S.D. in a single experiment.
ELISA Style Binding Assays
For ELISA style binding assays, rat tail collagen I (BD Biosciences) was diluted in TBS and coated at 10 μg/ml (500 ng/well) overnight at room temperature onto 96-well plates (MaxiSorp, Nunc). After washing with TBS-T (TBS containing 0.05% (v/v) Tween 20), plates were blocked for 1 h at room temperature with 5% (w/v) milk powder in TBS. Strep-tagged HSP47 proteins were added at a concentration of 3 μm in TBS-T containing 1% milk powder and incubated for 1 h. After washing away unbound protein with TBS-T, wells were incubated with McIlvaine buffers at the indicated pH levels for 5 min, shortly washed at the indicated pH, and then washed three times with TBS-T. Bound proteins were detected by Streptactin-HRP (IBA, Goettingen, Germany) and visualized with tetramethylbenzidine as substrate. The reaction was monitored at 370 nm. Data were analyzed using Origin version 8.5 and fitted using a four-parameter logistic model (29).
CD Spectra
Circular dichroism spectra were recorded using a Jasco J-715 instrument at 20 °C. All constructs were dialyzed against PBS, pH 7.0, and the indicated pH was adjusted shortly before the measurement by diluting the protein to a final concentration of 0.1 mg/ml in at least 10 volumes of target buffer. The samples were measured in a 1-mm cuvette with bandwidth at 2 nm, a response of 2 s, sensitivity set to standard (100 millidegrees), data pitch 0.2 nm, and a scan speed of 50 nm/min. Each curve was measured with 10 accumulations and plotted in Origin version 8.5. Data were analyzed between 200 and 240 nm with the CDPro package (30) using the CONTIN/LL algorithm (31) and the reference set 7 (30).
Thermal Shift Assays
Melting temperatures were measured using a total volume of 50 μl with 0.1 mg/ml protein concentration and a final dilution of 1× Sypro Orange (Molecular Probes). Samples were heated in a CFX96 real-time PCR system (Bio-Rad) at an approximate rate of 0.5 K/min, and the fluorescence intensity was measured every 0.5 K using the channel 6 filter set (excitation, 450–490 nm; emission, 560–580 nm). Curves were analyzed using the supplied software CFX Manager version 3.1, and melting temperatures were deduced from the minima of the first derivative.
Results
Theoretical Analysis of pKa Values
The crystal structure of HSP47 bound to trimeric collagen model peptides (17) (Fig. 1) enabled a structure-based analysis of the pH-dependent client release of HSP47 in the Golgi compartment. During the secretion process, the pH changes from 7.4 in the ER to 6.3 in the Golgi. It was already proposed earlier that histidine residues, whose imidazole side chains have a pKa of ∼6.1, might be responsible for the dissociation of the complex at low pH (15, 18). Canine HSP47 possesses 14 histidine residues (Fig. 1), with most of them being conserved in the HSP47 family. His108 and His353 appear to be general serpin features because they occur in all other serpin clades as well, such as antitrypsin (SerpinA1), antithrombin (SerpinC1), or PEDF (SerpinF1). Some histidines show a strict conservation in HSP47 (e.g. His238 or His315), whereas others are sometimes replaced (e.g. His216, His273, or His262) (Fig. 2). Of the 14 histidines in HSP47, six are directly involved in the collagen binding interface, namely His215, His216, His238, His273, His274, and His386 in the numbering of the protein from Canis lupus familiaris, UniProt ID C7C419 (Fig. 1). However, histidine residues outside of the interface could introduce changes in the overall structure upon protonation as well and might thereby also lead to a dissociation of the complex. Large conformational changes are very common for members of the serpin family, and pH-dependent changes in CD and fluorescence spectra have been reported (32).
FIGURE 1.
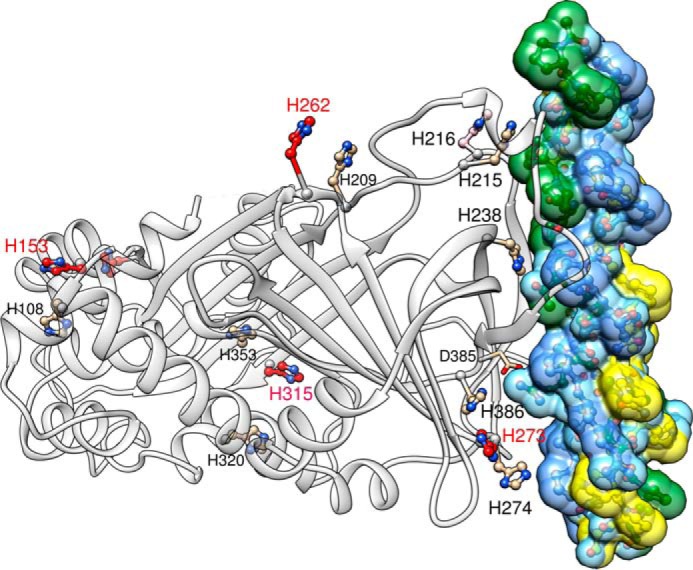
Positions of the histidine residues in the canine HSP47-collagen complex. Shown is a ribbon representation of HSP47 (left, gray ribbon) bound to a collagen model peptide (right). The trailing, middle, and leading strand of the collagen model peptide are colored in green, blue, and yellow, respectively. All 14 histidine residues are shown; those with a large change increase in double protonation between pH 7 and 6 are shown with red carbon atoms. The crucial Asp385, which interacts with an arginine in the Yaa position of the trailing strand, is shown as well. The second HSP47 protomer binding to this collagen model peptide has been omitted for the sake of clarity. Numbering is according to the full-length canine sequence, UniProt C7C419. This figure was prepared using Chimera (36).
FIGURE 2.
Conservation of titratable histidines in HSP47 orthologs. Protein sequences from all annotated SerpinH1 orthologs currently in ENSEMBL (ensemble.org/GRCh38.p3) were deduced and aligned using ClustalO. Sequence fragments containing histidines either in the HSP47-collagen interface or predicted to be of importance for pH dependence are shown, separated by white bars. The species were grouped together based on their phylogenetic class, and these classes were ordered by their phylogenetic relationship. Protein sequences of human SerpinF1 (PEDF), SerpinA1 (antitrypsin), and SerpinC1 (antithrombin) were included to differentiate between conservation within HSP47 orthologs and other members of the serpin protein family.
The pKa values of histidine side chains are known to depend on the chemical environment generated by the protein structure and can vary from about 4 to 9 with values between 5.0 and 8.0 being regularly observed (33). To narrow down possible key residues, we tried to calculate the pKa values of all histidine residues in HSP47 by CpHMD simulations.
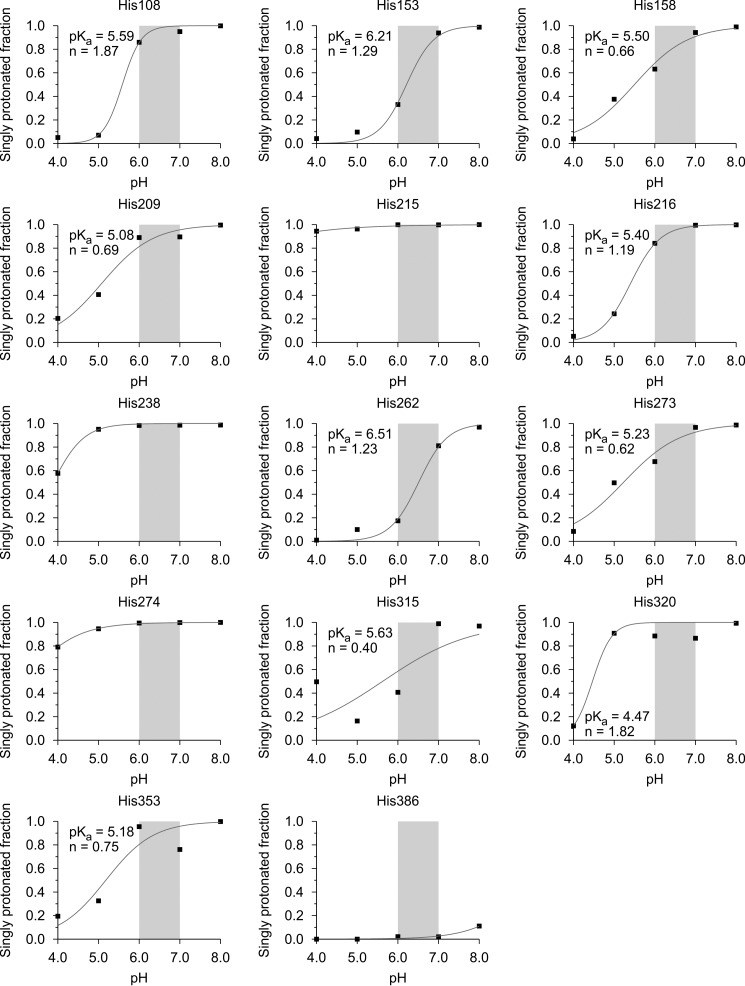
These simulations were performed for the HSP47 molecule in complex with the collagen triple helix at pH 8.0, 7.0, 6.0, 5.0, and 4.0, based on the assumption that these histidine residues should change their charge in a pH range between 6.0 and 7.0. Therefore, the doubly protonated (i.e. positively charged) fraction of each of the 14 histidine residues was calculated with an analysis tool of Amber 12 (Fig. 3 and Table 1).
FIGURE 3.
Protonation state of the HSP47 histidine residues versus pH. The protonation state (single/double) was measured as the fraction of monoprotonated histidines during constant pH molecular dynamics simulations that were performed for pH values 4.0, 5.0, 6.0, 7.0, and 8.0. The region shaded in gray corresponds to the physiologically interesting pH range from 7.0 to 6.0.
TABLE 1.
Protonation states of histidines in HSP47
Shown in boldface type are residues with a large change in protonation state between pH 7.0 and 6.0.
| Residue number | Interface | Fraction of doubly protonated histidine residues |
||||
|---|---|---|---|---|---|---|
| pH 4.0 | pH 5.0 | pH 6.0 | pH 7.0 | pH 8.0 | ||
| % | ||||||
| 108 | 94.9 | 92.9 | 14.1 | 5.0 | 0.1 | |
| 153 | 95.8 | 90.3 | 67.1 | 6.2 | 1.3 | |
| 158 | 96.1 | 62.4 | 36.9 | 5.8 | 1.0 | |
| 209 | 79.6 | 59.4 | 11.1 | 10.4 | 0.4 | |
| 215 | Yes | 5.6 | 3.7 | 0.1 | 0.2 | 0.0 |
| 216 | Yes | 94.8 | 75.7 | 15.8 | 0.5 | 0.0 |
| 238 | Yes | 42.3 | 4.9 | 1.7 | 1.4 | 1.3 |
| 262 | 99.0 | 90.0 | 82.6 | 18.9 | 3.1 | |
| 273 | Yes | 91.6 | 50.3 | 32.7 | 3.3 | 1.3 |
| 274 | Yes | 20.9 | 5.4 | 0.5 | 0.1 | 0.0 |
| 315 | 50.4 | 83.7 | 59.3 | 1.1 | 3.1 | |
| 320 | 87.8 | 9.2 | 11.7 | 13.4 | 0.7 | |
| 353 | 80.5 | 67.5 | 4.5 | 23.9 | 0.2 | |
| 386 | Yes | 100.0 | 100.0 | 97.9 | 98.0 | 88.9 |
Only 5 of the 14 histidine residues show a significant increase (>25%) in the doubly protonated (i.e. positively charged) state upon decreasing pH from 7.0 to 6.0 (Table 1 (boldface type) and Fig. 3). Only one of these five histidine residues (His273) is located close to the interface (Fig. 1, bottom right, red residue). Interestingly, His386, a direct neighbor of the important Asp385, is already doubly protonated at pH 8.0 and keeps this positive charge over the whole pH range, indicating a strong increase of its pKa value. This is presumably caused by the neighboring negative charge of Asp385, which stabilizes this doubly protonated form of His386.
Site-directed Mutagenesis and Preparation and Characterization of HSP47 Mutants
Assuming that the protonation of one or more histidine residues could trigger client release, we generated a set of mutants by changing histidines to asparagines with the rationale that the side chain of asparagine is structurally most similar to the imidazole moiety but cannot be protonated. Based on the simulations and our crystal structure, we replaced every histidine located within the binding interface as well as all of those predicted to change their protonation state by more than 25% between pH 6.0 and 7.0. With the exception of the mutants H386N and H315N, all proteins expressed as well as the wild type and generally exhibited a very similar thermostability, as judged by thermal shift assays (Table 2). The mutant H238N expressed well but had a significantly lower thermal stability, as will be discussed below.
TABLE 2.
Melting temperatures and dissociation rate constants (koff) of HSP47 variants
Melting temperatures were determined in McIlvain buffer, pH 7.5. koff values shown are means ± S.D. ND, not determined.
| Protein | Tm |
koff |
|||
|---|---|---|---|---|---|
| pH 6.0 | pH 6.5 | pH 7.0 | pH 7.5 | ||
| ºC | s−1 | ||||
| Wild typea | 57 | 0.192 ± 0.002 | 0.075 ± 0.001 | 0.042 ± 0.001 | 0.028 ± 0.000 |
| H153N | 56 | 0.179 ± 0.003 | 0.074 ± 0.006 | 0.045 ± 0.001 | 0.029 ± 0.001 |
| H158N | 56 | 0.187 ± 0.002 | 0.070 ± 0.002 | 0.045 ± 0.001 | 0.031 ± 0.003 |
| H215N | 58 | 0.313 ± 0.023 | 0.159 ± 0.015 | 0.126 ± 0.005 | 0.097 ± 0.005 |
| H216N | 57 | 0.184 ± 0.013 | 0.088 ± 0.004 | 0.055 ± 0.003 | 0.039 ± 0.001 |
| H215N/H216N | 58 | 0.273 ± 0.005 | 0.168 ± 0.019 | 0.124 ± 0.003 | 0.094 ± 0.002 |
| H238N | 45 | ND | ND | ND | ND |
| H262N | 57 | 0.236 ± 0.018 | 0.096 ± 0.002 | 0.047 ± 0.004 | 0.030 ± 0.001 |
| H273N | 55 | 0.148 ± 0.009 | 0.063 ± 0.003 | 0.038 ± 0.003 | 0.028 ± 0.001 |
| H274N | 56 | 0.170 ± 0.010 | 0.075 ± 0.004 | 0.038 ± 0.001 | 0.029 ± 0.001 |
| H273N/H274N | 56 | 0.071 ± 0.002 | 0.038 ± 0.002 | 0.026 ± 0.001 | 0.024 ± 0.001 |
| H273A | 57 | 0.111 ± 0.002 | 0.048 ± 0.004 | 0.030 ± 0.002 | 0.023 ± 0.002 |
| H274A | 57 | 0.169 ± 0.005 | 0.071 ± 0.008 | 0.036 ± 0.002 | 0.029 ± 0.001 |
| H273A/H274A | 57 | 0.080 ± 0.002 | 0.043 ± 0.002 | 0.031 ± 0.001 | 0.028 ± 0.002 |
| H273D | 57 | 0.110 ± 0.003 | 0.051 ± 0.002 | 0.028 ± 0.001 | 0.020 ± 0.001 |
| H273K | 58 | 0.134 ± 0.006 | 0.057 ± 0.005 | 0.034 ± 0.001 | 0.023 ± 0.001 |
| H274D | 58 | 0.100 ± 0.002 | 0.048 ± 0.004 | 0.032 ± 0.002 | 0.024 ± 0.001 |
| H274K | 57 | 0.285 ± 0.011 | 0.167 ± 0.006 | 0.124 ± 0.012 | 0.103 ± 0.010 |
| H273F | 57 | 0.139 ± 0.003 | 0.050 ± 0.002 | 0.029 ± 0.000 | 0.022 ± 0.000 |
| H273L | 57 | 0.112 ± 0.002 | 0.039 ± 0.001 | 0.024 ± 0.001 | 0.017 ± 0.000 |
| H273Y | 57 | 0.215 ± 0.001 | 0.076 ± 0.002 | 0.043 ± 0.003 | 0.029 ± 0.001 |
a Mean values for all experiments.
Wild-type HSP47 Releases Its Client Due to a Larger koff at Low pH
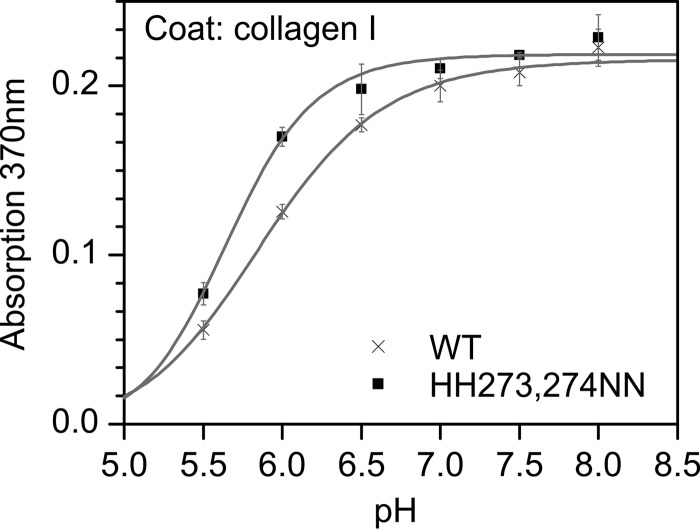
We first measured the interaction after washing with buffers of different pH values in an ELISA style binding assay utilizing coated rat tail collagen I. As expected, a decrease in the amount of bound HSP47 can be observed below a pH of 7.0, with a midpoint at a pH of ∼5.9 (Fig. 4). To examine this more closely, we investigated the kinetic parameters of the HSP47 interaction with collagen using biolayer interferometry (Fig. 5). When incubated at constant HSP47 concentrations but under different pH values, there was a reduced binding at lower pH to the foldon collagen model peptide (Fig. 5, first panel, WT). Interestingly, the koff rate constants were more strongly affected than the kon values. For the wild type, the koff increased from 0.028 s−1 at pH 7.5 to 0.192 s−1 at pH 6.0 (Table 2), similarly to the KD change from about 0.74 to 6.23 μm (Fig. 6 (WT) and Table 3). This change is mainly driven by the increase in koff because the kon stays relatively constant at about 40,000 (m·s)−1 and 33,000 (m·s)−1 (Fig. 6). Interestingly, the kinetic parameters of all of the mutants not located within the binding interface (H153N, H158N, and H262N) were similar to those of the wild-type protein (Figs. 3 and 6 and Tables 2 and 3), thus excluding a long range conformational rearrangement upon protonation.
FIGURE 4.
The pH dependence of client binding agrees with the titration curves of histidine residues. There is a clear pH dependence in an ELISA style HSP47-collagen binding assay employing rat tail collagen I as coat and recombinant canine HSP47 as analyte, indicating an inflection point around pH 5.88 ± 0.03 for the WT protein. The double mutant H273N/H274N shows a decreased inflection point at pH 5.66 ± 0.02. Data points were measured in triplicates, and the results were confirmed using at least two independent biological samples. Error bars, S.D.
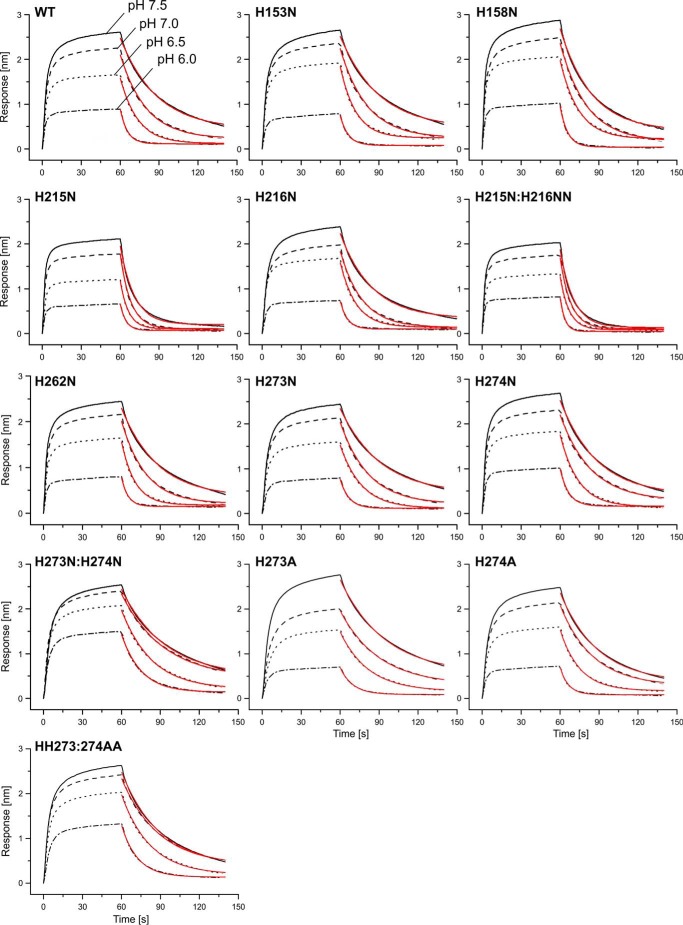
FIGURE 5.
The interaction of wild-type HSP47 and various histidine mutants with collagen as measured by biolayer interferometry. A foldon-stabilized collagen mimetic peptide was loaded on a streptavidin chip and incubated at constant concentration (5 μm) at the indicated pH values. Association was measured for 60 s, and dissociation time varied between 80 and 140 s (only the first 80 s are shown). koff values were fitted by a 1:1 Langmuir model. All dissociation curves show a clear dependence of the koff with the pH. Most mutants behave similarly to the wild-type protein. However, H215N and H215N/H216N have an overall faster dissociation rate, whereas the dissociation rates of H273N/H274N, H273A/H274A, and to some degree H273A change less with pH. Data show representative curves from at least three technical replicates. Results for wild type, H273N, H274N, and H273N/H274N were confirmed using at least three independent biological samples.
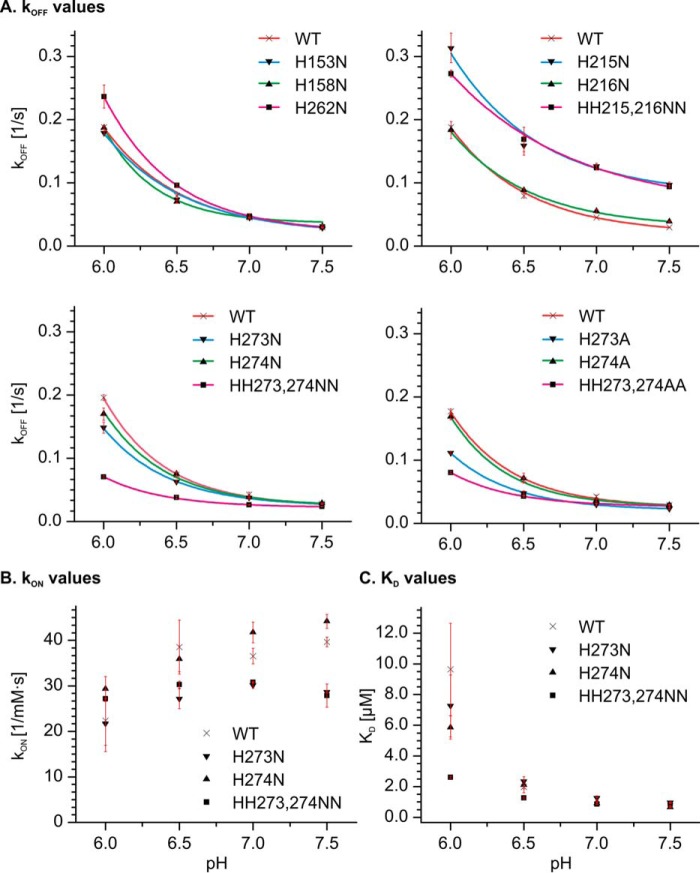
FIGURE 6.
Kinetic analysis of the biolayer interferometry data. A foldon-stabilized collagen mimetic peptide was loaded on a streptavidin chip and incubated at constant concentration (5 μm) with HSP47 variants at the indicated pH values. Kinetic parameters were extracted using a 1:1 Langmuir binding model. For wild type and most variants, there is a clear increase in koff with decreasing pH observable, which is typically about 5-fold, as seen in A (top left). The histidine pair previously assigned as being responsible for the pH switch, His215 and His216, shows minor changes in pH dependence upon double mutation to asparagine (A, top right). However, the binding of the H215N/H216N double mutant is weaker than the wild type. The H273N/H274N double mutant has a very much decreased dependence of koff (A, bottom left) and KD (C) on the pH value, and the H273A/H274A variant behaves very similarly (A, bottom right). The kon values are less sensitive to pH changes (B). Error bars, S.D.
TABLE 3.
Dissociation constants (KD) of HSP47 variants
Values shown are means ± S.D. ND, not determined.
| Protein |
KD |
|||
|---|---|---|---|---|
| pH 6.0 | pH 6.5 | pH 7.0 | pH 7.5 | |
| μm | ||||
| Wild type | 6.23 ± 0.88 | 2.25 ± 0.24 | 1.14 ± 0.04 | 0.74 ± 0.02 |
| H153N | 6.00 ± 0.21 | 1.93 ± 0.36 | 1.20 ± 0.04 | 0.76 ± 0.02 |
| H158N | 3.85 ± 0.52 | 1.38 ± 0.13 | 1.00 ± 0.08 | 0.55 ± 0.05 |
| H215N | 8.93 ± 1.20 | 3.70 ± 0.96 | 2.21 ± 0.27 | 1.60 ± 0.19 |
| H216N | 7.65 ± 1.93 | 2.76 ± 0.04 | 1.55 ± 0.14 | 1.18 ± 0.08 |
| H215N/H216N | 4.58 ± 0.92 | 4.70 ± 1.96 | 2.15 ± 0.12 | 1.63 ± 0.12 |
| H238N | ND | ND | ND | ND |
| H262N | 11.39 ± 5.18 | 2.55 ± 0.17 | 1.13 ± 0.20 | 0.67 ± 0.06 |
| H273N | 7.27 ± 2.01 | 2.34 ± 0.29 | 1.27 ± 0.13 | 0.97 ± 0.04 |
| H274N | 5.86 ± 0.75 | 2.12 ± 0.26 | 0.92 ± 0.05 | 0.65 ± 0.03 |
| H273N/H274N | 2.60 ± 0.08 | 1.26 ± 0.09 | 0.86 ± 0.02 | 0.85 ± 0.09 |
| H273A | 6.21 ± 1.20 | 2.58 ± 0.16 | 1.48 ± 0.21 | 0.82 ± 0.09 |
| H274A | 6.85 ± 1.93 | 2.37 ± 0.19 | 1.12 ± 0.05 | 1.23 ± 0.12 |
| H273A/H274A | 3.17 ± 0.92 | 1.69 ± 0.22 | 1.00 ± 0.06 | 1.07 ± 0.07 |
| H273D | 4.04 ± 0.58 | 1.74 ± 0.07 | 0.89 ± 0.02 | 0.64 ± 0.04 |
| H273K | 5.08 ± 0.67 | 2.11 ± 0.56 | 1.09 ± 0.06 | 0.69 ± 0.02 |
| H274D | 4.02 ± 0.44 | 1.40 ± 0.07 | 1.11 ± 0.18 | 0.73 ± 0.04 |
| H274K | 16.29 ± 7.56 | 5.17 ± 0.81 | 3.40 ± 0.78 | 2.98 ± 0.28 |
| H273F | 5.39 ± 0.20 | 2.20 ± 0.32 | 1.01 ± 0.20 | 0.61 ± 0.04 |
| H273L | 3.84 ± 0.26 | 1.28 ± 0.06 | 0.72 ± 0.06 | 0.47 ± 0.06 |
| H273Y | 10.37 ± 1.90 | 3.16 ± 0.20 | 1.53 ± 0.24 | 0.83 ± 0.02 |
His238 Is Essential for Collagen Binding by Organizing an Extended Hydrogen Bond Network but Does Not Govern the pH Switch
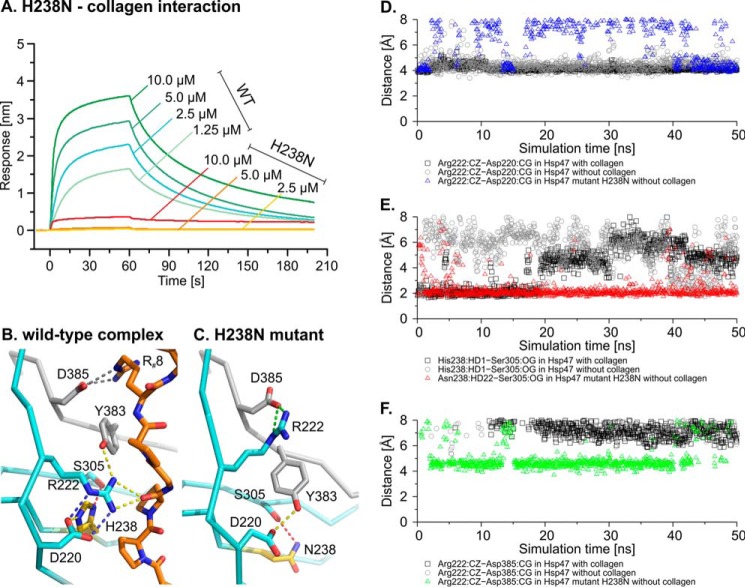
His238 is located in the center of the interaction surface between collagen and HSP47. The single mutant H238N has no detectable binding to collagen anymore (Fig. 7A). The side chain of His238 forms few and rather weak contacts with the collagen chains. To understand the destabilization of the HSP47-collagen complex by the H238N mutation, we examined this mutant more closely by molecular dynamics simulations.
FIGURE 7.
Analysis of the interaction of HSP47 H238N with collagen. A, interaction of HSP47 WT (blue, cyan, and green sensorgrams) and H238N (yellow, orange, and red) was measured in biolayer interferometry. A foldon-stabilized collagen mimetic peptide was loaded on a streptavidin chip and incubated at the indicated concentrations. For H238N, no significant signal even at the highest concentration could be detected. B, in the crystal structure, His238 can interact via a hydrogen bond with Asp220, which stabilizes Arg222 (blue dashed lines). Arg222 and Asp385 make important contacts to the collagen helix (green and yellow dashed lines). C, representative snapshot of the uncomplexed H238N mutant obtained from an constant pH MD simulation, revealing a rearrangement of the hydrogen bonding network compared with the wild type; Asn238 now interacts with Ser305 (red dashed line), and the Asp220-Arg222 salt bridge is lost. Arg222 forms a stable salt bridge with Asp385 (green dashed lines), both important residues for collagen recognition. HSP47 is shown in ribbon (cyan/gray) and collagen as sticks in orange, and all numbers are according to full-length canine protein. R#8, arginine in the collagen model peptide crystallized (Protein Data Bank entry 4AU2). D–F, comparison of the conformational stability of several side-chain interactions in MD simulations between wild type and H238N HSP47. For wild-type HSP47, two plots are shown in each panel corresponding to the unliganded and collagen-bound form. Black and gray, wild-type protein with and without collagen, respectively. Blue, red, and green triangles, distances between the indicated atoms in the H238N mutant. Biolayer interferometry data show representative curves from three technical replicates. The general result was confirmed in three independent biological replicates.
In the wild-type situation, His238 is able to form hydrogen bonds with the side chains of Asp220 and Ser305 (Fig. 7B, dashed lines). Asp220 forms an additional stable salt bridge to the Nϵ and Nη atoms of Arg222 (Fig. 7B, blue dashed lines), thus, together with an additional hydrogen bond from Tyr383, stabilizing this side chain in a conformation capable of binding to the collagen backbone (Fig. 7B, dashed lines). Calculation of the protonation states (Fig. 3) indicates that His238 starts to titrate only at pH <5; thus, it should not govern the pH switch. Its shifted pKa value can be attributed to the adjacent positive charge of the Arg222 side chain, which hampers protonation at the Nϵ nitrogen.
To understand the effect of the H238N mutation, we modeled it in collagen-unbound HSP47 and simulated the conformational changes by constant pH molecular dynamics simulations. For comparison, additional simulations of collagen-bound and unliganded wild-type HSP47 were performed. Monitoring the Asp220-Arg222 side-chain interaction over the simulation time reveals that this interaction is both present in the collagen-bound and in the unbound wild-type HSP47 (Fig. 7D, gray and black). This implies that the respective interaction plays an important role in prepositioning Arg222 for a proper interaction with collagen. In contrast, this interaction is significantly less stable in the unliganded H238N mutant (Fig. 7D, blue), indicating that conformational changes in the collagen binding site occurred. A closer inspection of the H238N mutant shows a considerable rearrangement of the hydrogen bonding network that includes the formation of several interactions not present in the wild-type (Fig. 7C). One prominent difference is the hydrogen bond between the oxygen atom of the Ser305 side chain and the side chain of Asn238 (Fig. 7C, red dashed line), which is more stable than observed for His238 in the simulation of wild-type structures (Fig. 7E, compare gray with red). The strong hydrogen bond between Asn238 and Ser305 together with the differences in the side-chain geometry between Asn and His also lead to a loss of the Asn238-Asp220 interaction. As an additional consequence of these rearrangements, the Asp220-Arg222 interaction becomes less stable in the mutant (Fig. 7D), and Arg222 forms a stable interaction with Asp385 instead (Fig. 7, C (green dashed lines) and F (green and gray lines)). Both residues, Arg222 and Asp385, are highly important for the collagen interaction of the wild-type HSP47 (17). The reorganization of the hydrogen bond network and the lock of the two residues, which made direct electrostatic interaction with collagen in the wild-type situation provide an explanation of the significantly weakened binding of the H238N mutant.
The disruption of the hydrogen-bonding network centered on His238 has an influence on the stability of the three-dimensional structure, as revealed by a significantly lower melting temperature in a thermal shift assay experiment, where the H238N mutant melts at 45 °C and the wild-type at 57 °C (Table 2).
The Double Mutant H273N/H274N Shows an Altered pH Dependence of Collagen Binding
Of all of the mutants that could be studied, only H238N, which was discussed above, and the tandem histidine pair variants H215N/H216N and H273N/H274N showed a significant change in their kinetic behavior. These two tandem histidine pairs are located at opposite ends of the binding interface (Fig. 1).
The H215N/H216N double mutant has been reported to show a loss of the pH switch in binding to collagen when the pH-dependent elution profiles of wild type and mutant in collagen affinity chromatography are compared (18). Therefore, we investigated these residues in more detail by biolayer interferometry. Whereas the H216N variant behaved similar to the wild type in our experiments, the H215N mutant showed an increased koff over the whole pH range (0.097–0.313 s−1) (Figs. 5 and 6A and Table 2). Consequently, the double mutant H215N/H216N also showed an overall increase in koff.
In contrast, the single mutations H273N and H274N individually did not show a significant change compared with the wild-type, whereas the double mutant H273N/H274N showed a markedly reduced pH dependence of the koff rate constant (Figs. 5 and 6A). In other words, the koff of the double mutant does not increase as much at lower pH values as the koff of the wild-type protein or the other histidine mutants that we examined in this study. This is also reflected in the KD values of this mutant at the different pH values, which only increase from 0.85 μm at pH 7.5 to 2.60 μm at pH 6.0. This is a significant reduction compared with the wild-type situation (0.74–6.23 μm) (Fig. 6C and Table 3). The tighter binding of the H273N/H274N mutant at pH 6.0 could also be observed in steered molecular dynamics simulations of the dissociation process. Here, more energy was needed to displace the collagen helix from the H273N/H274N double mutant than from the wild-type protein (Fig. 8).
FIGURE 8.
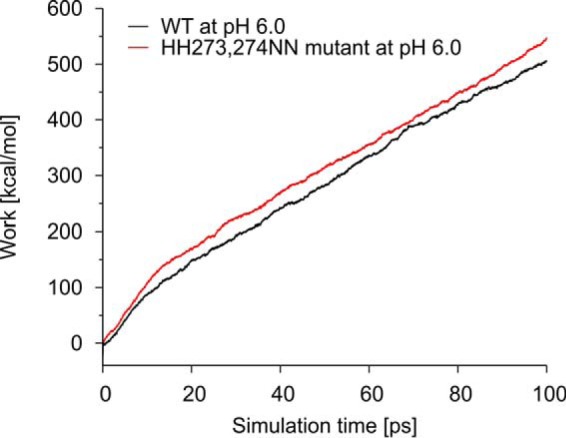
Steered molecular dynamics (SMD) simulation of wild-type (red) and H273N/H274N (black) HSP47 performed with snapshots from pHtMD simulations at pH 6. The SMD simulation measures the work that is required to remove collagen from HSP47. The work required in the double mutant is higher compared with the wild type, which is consistent with the tighter binding observed in experiment.
To test whether this effect is specific for the histidine to asparagine exchange, we also replaced these histidine residues with alanines. Biolayer interferometry analysis of the double mutant H273A/H274A showed a behavior similar to that of the asparagine double mutant (Figs. 5 and 6A). However, a small decrease of the koff at lower pH could already be observed with the single mutant H273A, indicating that this type of exchange might be slightly more important for the pH dependence of the koff.
The H273N/H274N Double Mutant Is Structurally Similar to the Wild-type Protein
Based on CD and fluorescence spectra, it was reported earlier that HSP47 undergoes structural changes at low pH (15, 16). To investigate whether the H273N/H274N double mutant has any influence on the overall structure of HSP47, we measured far-UV CD spectra of the mutant and the wild-type protein at pH 7.5 and pH 6.0 (Fig. 9). For the wild-type protein, the CD spectra are indistinguishable between pH 7.5 and pH 6.0 down to a wavelength of 200 nm. For the H273N/H274N mutant, the shape in this region remains constant, but the amplitude changes slightly (Fig. 9). Analysis of the spectra for the amount of secondary structure using the CONTIN/LL algorithm gave very similar results for wild-type and double mutant proteins at neutral and low pH (wild type, pH 6.0: 21% α-helices, 26% β-sheets, 21% turns, and 33% unordered; wild type, pH 7.5: 21, 26, 20, and 33%; H273N/H274N, pH 6.0: 26, 22, 21, and 31%; H273N/H274N, pH 7.5: 23, 24, 21, and 32%). It should be noted that the amount of secondary structure is slightly underestimated compared with data from our crystal structure (28% α-helices, 31% β-sheets). This is most likely the case because the data below 200 nm are not of sufficient quality and probably the reason why other algorithms failed to give good results.
FIGURE 9.
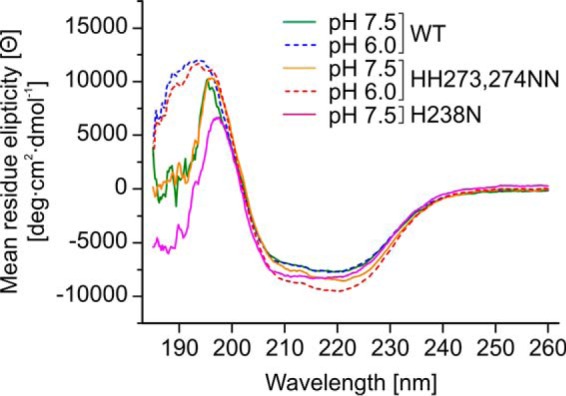
Far-UV CD spectra indicating structural integrity of the mutants H273N/H274N and H238N. The CD spectra indicate no significant difference in conformation of wild-type HSP47 and the H273N/H274N double mutant at pH 7.5 and 6.0. HSP47 H238N at pH 7.5 has a slightly more pronounced minimum at 208 nm but is overall very similar to HSP47 wild type. The data were measured with 10 accumulations, and the general trend was confirmed on two independent biological samples.
The H273N Mutation Significantly Affects the Titration Behavior of His274
To understand the effects of the mutation of residues 273 and 274 on collagen binding, computational simulations were performed. For that purpose, in silico titration experiments were calculated for WT HSP47 and for the single point mutations H273N and H274N. Whereas the titration behavior of His273 in the H274N mutant is almost unaffected compared with the wild type (Fig. 10A), the H273N mutation has a significant effect on the titration properties of His274 (Fig. 10B). Compared with the wild type, titration of His274 starts at significantly higher pH values in the H273N mutant (Fig. 10B).
FIGURE 10.
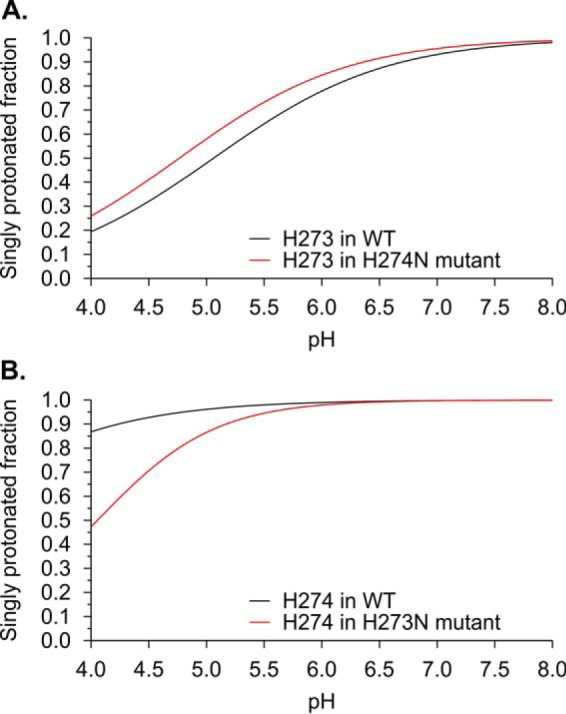
Dependence of the protonation state of His273 and His274 on the identity of the adjacent residue calculated from molecular dynamics simulations. A, fraction of singly protonated His273 in the wild type (black) and in H274N (red) HSP47 as a function of the pH value. Note that the titration behavior of His273 is almost unaffected by the exchange at position 274. B, fraction of singly protonated His274 in the wild type (black) and in H273N (red) HSP47 as a function of the pH value. Note that the titration behavior of His274 is significantly affected by the exchange at position 273.
We noted that the titration of His274 in the H273N mutant still starts at lower values than one would expect for a residue that participates in pH-dependent collagen release. This finding can most likely be attributed to the limited accuracy of the modeled mutant structure used for pKa value calculations, which are highly sensitive to the local geometry. Nevertheless, the differences between the wild type and H273N mutant are so large that they at least allow the qualitative statement that the identity of the residue at position 273 significantly affects His274 titration behavior.
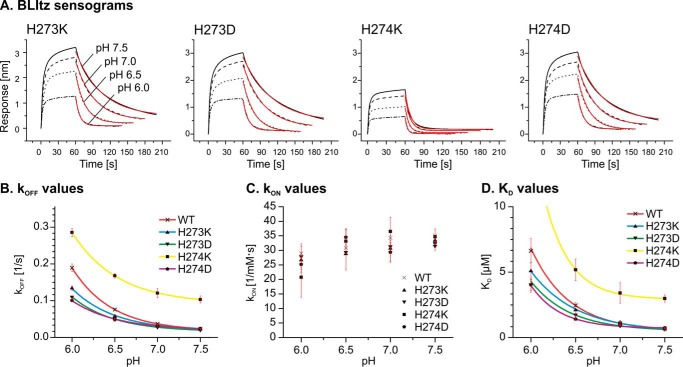
Introducing Charged Residues at Positions 273 and 274
To further test whether charges at the positions of the tandem histidine pair 273/274 impair collagen binding, we mutated these residues to lysine and aspartic acid. Mutation of either histidine to aspartic acid resulted in a slower dissociation at lower pH of the HSP47-client complex (Fig. 11), thus emphasizing again the importance of a positive charge at these positions for client release. As expected, the H274K mutant showed a larger koff over the whole pH range observed (i.e. also at neutral pH (pH 7.0–7.5), where the wild type has presumably not acquired yet a significant positive charge at this position). However, this mutant still shows a very pronounced further increase of koff at a lower pH. Unexpectedly, the H273K mutant exhibited a decreased dependence of its koff on the pH value (Fig 11B, blue curve). A possible explanation is the influence of residue 273 on the pKa of residue 274; a positive charge could potentially impede protonation of His274, whereas the flexibility of the lysine side chain and its position further away from the interface as compared with residue 273 could avoid interference with client binding.
FIGURE 11.
The influence of charged residues at positions 273 and 274 on collagen binding. A foldon-stabilized collagen mimetic peptide was loaded on a streptavidin chip and incubated at constant concentration (5 μm) with HSP47 variants at the indicated pH values (A). Kinetic parameters were extracted using a 1:1 Langmuir binding model. KD for H274K at pH 6.0 is not visible at the used scale (B–D). Introduction of the negatively charged Asp at either position, 273 or 274, reduces the pH dependence of koff. Interestingly, also the H273K mutant shows a slightly tighter binding to collagen at low pH. Data show representative curves from at least three technical replicates. Results were confirmed using at least two independent biological samples.
Phylogenetic Analysis Confirms the Importance of His215, His238, and His274 and Implies Certain Restrictions on His273 Replacements
To investigate the conservation of the identified histidines on HSP47, we analyzed all currently listed orthologs of human SerpinH1 in the Ensembl database (ensemble.org/Ch38.p3). As described for many other genes, SerpinH1 exists in two copies in most fish (class Actinopterygii) due to a genome-wide duplication in their common ancestor (34, 35). For medaka and fugu, however, no second gene is currently annotated. Genes originating from whole genome duplication are called ohnologs and may differ in function. Interestingly, many residues in HSP47 are strictly conserved in both fish ohnologs (e.g. MMHRT cluster around His238; Fig. 2), indicating that many of these genes still fulfill a functional role. Most of the histidine residues are relatively well conserved. However, some ohnologs have accumulated exchanges of otherwise well conserved histidines (compare cod genes; Fig. 2, blue boxes).
We have identified four histidine residues that have an effect on binding of HSP47 to collagen. His238 (Fig. 2, blue residues), which is essential for binding to collagen, is strictly conserved throughout all species. The tandem pair His215/His216 is only conserved in tetrapods (mammals, amphibia, reptiles, and birds) with the exception of the platypus (Fig. 2, green residues), where the correct protein sequence has not yet been confirmed, though. In fish (Actinopterygii and coelacanth) His215 is mostly conserved with the exception of one ohnolog in the stickleback. His216, however, is replaced by various residues in a number of genes and also in two ohnologs of one species simultaneously (e.g. platyfish, cod, and tilapia), indicating that this residue is not important for proper HSP47 function at least in these species.
The tandem His273/His274 shows a similar behavior. In mammals, both residues are completely conserved (Fig. 2, red residues). In reptiles and birds (sauropsida), however, these residues are exchanged to an asparagine/histidine motif (NH). Because the amphibia still retain an HH motif, the NH of reptiles and birds is very likely to be evolutionarily derived, whereas the two histidines (HH) appear to be more ancestral. In ray-finned fish (Actinopterygii), YH is the most common sequence motif (14 of 18 species) and therefore is likely to be ancestral to ray-finned fish. It is difficult to judge which of the two motifs, HH or YH, is ancestral to Actinopterygii and Sarcopterygii. There are appearances of both motifs in both clades (e.g. YH in coelacanth and HH in platyfish). The more ancestral lamprey carries an LH motif at this position. LH and HH are interchangeable by a single base substitution on the DNA level (changing Leu for His), whereas the mutation of LH to YH requires two bases to be exchanged. This slightly favors an HH motif either as ancestral to all vertebrates (and the lamprey sequence would be a derivative) or as a derived motif from an ancestral LH motif. In summary, substitution of His273 by an asparagine or a tyrosine seems not to interfere with HSP47 function. In contrast, His274 is evolutionarily more strictly conserved.
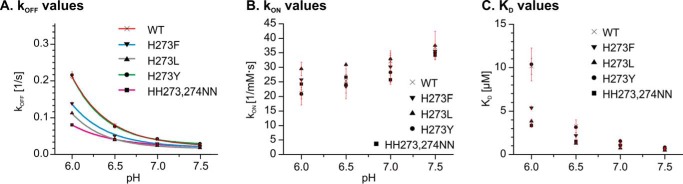
A Tyrosine Residue Is Equally Tolerated at the Position of His273
To confirm our theory based on the phylogenetic analysis, we simulated the situation in fish by changing in the canine HSP47 the position His273 to tyrosine (most fish), leucine (lamprey and one ohnolog in cod), and phenylalanine (one ohnolog in spotted gar) (Fig. 12). Interestingly, H273Y behaved exactly as the wild-type protein, indicating that the tyrosine at the position 273 is equally able to shift the pKa value of His274 to higher values. In contrast, H273F showed only a small increase in the dissociation rate constants from 0.022 s−1 at pH 7.5 to 0.139 s−1 at pH 6.0 (Fig. 12 (blue line) and Table 2). This is similar to the effect of the H273A variant. The H273L showed a significantly reduced pH dependence similar to the H273N/H274N double mutant (Fig. 12, gray line), although at pH 6, the koff is slightly larger.
FIGURE 12.
The effect of single amino acid exchanges present in various fish species on collagen binding. A foldon-stabilized collagen mimetic peptide was loaded on a streptavidin chip and incubated at constant concentration (5 μm) with HSP47 variants at the indicated pH values. Kinetic parameters were extracted using a 1:1 Langmuir binding model. The pH dependence of HSP47 WT and H273Y are indistinguishable. H273L and H273F, however, have an intermediate pH dependence similar to that of H273A (Fig. 6). Data show representative fits from at least three technical replicates. Results were confirmed using at least two independent biological samples. A, dissociation rate constant (kOFF). B, association rate constant (kON). C, dissociation constant (KD) calculated as kOFF/kON.
Discussion
Our recent crystal structure allowed for the first time the analysis of all histidine residues in HSP47 in their spatial context. For a histidine residue to play a major role in the pH-dependent release, it should change its protonation state from single to doubly protonated between pH 7.0 and 6.0. A total of 14 histidines are present in C. lupus familiaris HSP47, of which only 5 are predicted to undergo significant changes in protonation (His153, His158, His262, His273, and His315). Other, especially buried, histidines are not predicted to get significantly protonated even at pH 6.0.
We reasoned that an exchange of histidines to a non-protonatable analogue should prevent the pH-dependent release. From the 20 canonical amino acids, asparagine is considered to be structurally the closest; it carries a nitrogen in the form of an NH at a similar position as the Nδ1 atom of histidine but lacks a further proton acceptor similar to the Nϵ2 atom. From our analysis of the wild-type interaction, we know that the pH dependence is mostly conveyed by an increase in koff between pH 7 and pH 6. We therefore exchanged all histidines that are predicted to undergo a large change in protonation state as well as all the histidines in the interface to asparagine residues and measured their kinetic parameters.
Two of these HSP47 mutants (H315N and H386N) did not express in a soluble form. This can be explained by the fact that both residues are buried in the structure and are probably important for the integrity of the folded conformation.
H238N did not show any binding to collagen. This residue is located in a catalytic triad-like arrangement with Ser305 and Asp220. Analysis of this mutant in molecular dynamics simulations showed that His238 is critical for the stabilization of a hydrogen network locking Arg222 in a binding-capable conformation, thus preventing an intramolecular salt bridge between Arg222 and Asp385, two residues of pivotal importance for the HSP47 collagen interface.
In an earlier study employing recombinant mouse HSP47, His191, His197, and His198 (numbering according to the mature mouse protein), which correspond to His209, His215, and His216 (numbering according to the full-length canine HSP47 as used here) were reported to be important for the pH-dependent client release. It was observed that a double mutant corresponding to H215N/H216N and a single mutant corresponding to H209A were released early in a pH elution from a collagen affinity column. H215N/H216N showed a particularly broad elution profile, leading the authors to conclude that this mutant is unable to undergo the typical pH switch of HSP47 (18). In the study presented here, we do see that His215 and His216 are important for the general HSP47-collagen affinity, but we cannot detect any effect on the pH dependence of these residues; the pH dependence of the koff and KD of the H215N/H216N double mutant shows a similar behavior as the wild type, with koff increasing from 0.094 s−1 at pH 7.5 to 0.273 s−1 at pH 6.0, but compared with the wild type, the binding is weakened by a factor of 2.3. We believe that the weaker binding explains the observed early elution from the collagen affinity column and does not imply that those residues play a role in the pH sensitivity of the interaction. For a mutant unable to undergo the pH switch, a delayed elution (i.e. at even lower pH) from a collagen affinity column would be expected.
If protonation of certain histidine residues triggers collagen release, removal of these protonation sites by mutagenesis should result in a dissociation rate constant (koff) that is not (or at least is less) pH-dependent in comparison with the wild type. Only the double mutant H273N/H274N showed a decrease in the pH dependence of the koff and the KD values. This means that protonation at either site will decrease the affinity of HSP47 with collagen because both of the single mutants (H273N and H274N) show a similar pH-dependent behavior as the wild-type protein. In other words, protonation of either His273 or His274 is sufficient to induce collagen release. Interestingly, the double mutant H273A/H274A shows a behavior very similar to that of H273N/H274N, indicating that this is not a particular effect of the newly introduced asparagine side chains but rather of the missing imidazole moieties (i.e. the proton acceptor sites). However, in contrast to H273N, the single mutant H273A alone exhibits a small but significant reduction in pH dependence. This fits well with our observation from molecular dynamics analyses of wild-type HSP47, where His273 but not His274 was predicted to change its protonation state between ER and Golgi compartments, but opened the question of how His274 is able to compensate for His273 in the H273N mutant. Furthermore, His274 but not His273 is conserved in evolution.
Our computer simulations of the protonation states of His274 in the H273N protein indicate that the H273N mutation has a significant effect on the titration properties of His274. In the H273N mutant, His274 starts to be protonated at significantly higher pH values than in the wild-type protein (Fig. 10B), allowing His274 to compensate for His273 as a pH-dependent trigger. The finding that an asparagine is actually present at position 273 in several HSP47 orthologs supports the idea that this shift in the pKa of His274 might indeed be sufficient to allow His274 to exert a function in pH-dependent collagen release. In addition to an asparagine in birds and reptiles, in “fish” (clades Actinopterygii, Sarcopterygii, and Hyperoartia), tyrosine, leucine, and phenylalanine can be found at position 273. Our experiments using single amino acid substitutions in a canine protein background suggest that a tyrosine at position 273 can induce a shift in the pKa of His274 in a similar fashion as observed previously for H273N (Fig. 12), whereas phenylalanine and leucine cannot. For the H273F variant, this failure in pH-dependent release might easily be compensated for by the second ohnolog in spotted gar, which has the canonical tyrosine at position 273. Additionally, the phenylalanine carrying ohnolog has accumulated many additional amino acid exchanges. Compared with other fish sequences, the Tyr273 ohnolog is ∼70% identical; the Phe273 variant, however, is only about 50% identical. This huge discrepancy might indicate that the Phe273 variant has lost its function as a collagen chaperon and might play a different role in the spotted gar. In cod and lamprey, there are no canonical ohnologs present, which could compensate for the lack of pH dependence of the H273L HSP47 variant. However, because pKa changes are highly dependent on the local environment, it still might be possible that Leu273 in the background of a fish protein is able to introduce the necessary pKa shifts to His274. These data suggest that only a limited set of amino acids is capable of conferring a suitable environment for His274 that allows this residue to act in pH-dependent collagen release in those species.
In this context, our earlier finding on the charged mutants at position 273 is also understandable (Fig. 11). Introduction of charged amino acids (H273D or H273K) at position 273 as well as significantly more hydrophobic side chains (Phe, Leu, and Ala) is less able to induce pH-triggered client release. In all of these cases, a reduced pH dependence is observed. This indicates that the H273A, H273K, and H273D mutations are unable to induce a shift of His274 pKa to higher values.
In other studies, conformational changes were often suspected of being important for the pH-dependent release. This was based on the differences observable in CD spectra under neutral and acidic conditions for recombinant mouse protein (15). However, we could only find minor differences in the overall CD spectra, thus indicating only minor structural changes over the pH range tested here (Fig. 9). We believe that the structural changes described previously (15) only occur at pH values below 6.0, an observation that has also been reported earlier (16). Most importantly, the less pH-dependent double mutant H273N/H274N shows a behavior in the CD spectrum similar to that of the wild type, hence excluding large structural rearrangements as the major driving force for pH-dependent client release. This is also confirmed by our steered molecular dynamics simulation (Fig. 8), in which dissociation of the HSP47 complex was easier for protonated HSP47 wild-type protein than for the non-protonatable H273N/H274N mutant. Here again, no major structural rearrangements were necessary to explain the effect.
Therefore, the mechanism of HSP47 dissociation in the Golgi apparatus seems to be driven by two factors. First, the protonation of HSP47 locally disrupts the collagen binding interface, most likely due to electrostatic repulsions. This leads to an increase in the koff and allows a rapid dissociation of the collagen client. In our experiments, we find a difference of ∼7-fold in binding affinity between pH 7.0 and 6.0, translating into a small difference in the Gibbs free energy of about 5 kJ/mol. High resolution crystal structures will be needed to explain this in terms of an atomic model. Second, the released HSP47 is transported out of the Golgi in the ER via the KDEL receptor pathway, thus withdrawing the protein from the equilibrium. Our in-depth analysis of the protonation behavior of the HSP47 histidine residues and their influence on collagen binding and release identified positions 273 and 274 for the first part of this mechanism. Compared with the wild type, these two residues alone explain half of the increase in koff upon shifting the surrounding pH from 7 to 6. Most interestingly, His273 is not strictly conserved in all HSP47 orthologs; however, the residue at this position influences the pKa of His274.
Author Contributions
J. M. G., H. S., and U. B. designed the research; J. M. G, S. O., M. U., C. E., and E. S. performed the research; J. M. G, S. O., E. S., H. S., F. Z., and U. B. analyzed the data; and J. M. G., S. O., E. S., H. S., and U. B. wrote the paper.
Acknowledgments
We thank Daniela Zwolanek and Manuel Koch for the kind gift of expression constructs for foldon-stabilized collagen peptides.
This work was supported by German Research Council Grants SFB829/B11, SFB796/A2, and BA 1264/2-1 and by the University of Cologne. The authors declare that they have no conflicts of interest with the contents of this article.
- ER
- endoplasmic reticulum
- MD
- molecular dynamics
- CpHMD
- constant pH molecular dynamics
- SMD
- steered molecular dynamics
- pHtMD
- pH titrating molecular dynamics.
References
- 1.Kadler K. E., Baldock C., Bella J., and Boot-Handford R. P. (2007) Collagens at a glance. J. Cell Sci. 120, 1955–1958 [DOI] [PubMed] [Google Scholar]
- 2.Ricard-Blum S. (2011) The collagen family. Cold Spring Harb. Perspect. Biol. 3, a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoulders M. D., and Raines R. T. (2009) Collagen structure and stability. Annu. Rev. Biochem. 78, 929–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koide T., Nagata K., Asada S., Takahara Y., Nishikawa Y., and Kitagawa K. (2005) in Collagen: Topics in Current Chemistry (Brinckmann J., Notbohm H., and Müller P. K., eds) pp. 85–114, Springer, Berlin [Google Scholar]
- 5.Hirayoshi K., Kudo H., Takechi H., Nakai A., Iwamatsu A., Yamada K. M., and Nagata K. (1991) HSP47: a tissue-specific, transformation-sensitive, collagen-binding heat shock protein of chicken embryo fibroblasts. Mol. Cell. Biol. 11, 4036–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishida Y., and Nagata K. (2011) Hsp47 as a collagen-specific molecular chaperone. Methods Enzymol. 499, 167–182 [DOI] [PubMed] [Google Scholar]
- 7.Ishida Y., Kubota H., Yamamoto A., Kitamura A., Bächinger H. P., and Nagata K. (2006) Type I collagen in Hsp47-null cells is aggregated in endoplasmic reticulum and deficient in N-propeptide processing and fibrillogenesis. Mol. Biol. Cell 17, 2346–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagai N., Hosokawa M., Itohara S., Adachi E., Matsushita T., Hosokawa N., and Nagata K. (2000) Embryonic lethality of molecular chaperone hsp47 knockout mice is associated with defects in collagen biosynthesis. J. Cell Biol. 150, 1499–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christiansen H. E., Schwarze U., Pyott S. M., AlSwaid A., Al Balwi M., Alrasheed S., Pepin M. G., Weis M. A., Eyre D. R., and Byers P. H. (2010) Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am. J. Hum. Genet. 86, 389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drögemüller C., Becker D., Brunner A., Haase B., Kircher P., Seeliger F., Fehr M., Baumann U., Lindblad-Toh K., and Leeb T. (2009) A missense mutation in the SERPINH1 gene in Dachshunds with osteogenesis imperfecta. PLoS Genet. 5, e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono T., Miyazaki T., Ishida Y., Uehata M., and Nagata K. (2012) Direct in vitro and in vivo evidence for interaction between Hsp47 protein and collagen triple helix. J. Biol. Chem. 287, 6810–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paroutis P., Touret N., and Grinstein S. (2004) The pH of the secretory pathway: measurement, determinants, and regulation. Physiology 19, 207–215 [DOI] [PubMed] [Google Scholar]
- 13.Wu M. M., Llopis J., Adams S., McCaffery J. M., Kulomaa M. S., Machen T. E., Moore H. P., and Tsien R. Y. (2000) Organelle pH studies using targeted avidin and fluorescein-biotin. Chem. Biol. 7, 197–209 [DOI] [PubMed] [Google Scholar]
- 14.Whisstock J. C., Silverman G. A., Bird P. I., Bottomley S. P., Kaiserman D., Luke C. J., Pak S. C., Reichhart J.-M., and Huntington J. A. (2010) Serpins flex their muscle: II. structural insights into target peptidase recognition, polymerization, and transport functions. J. Biol. Chem. 285, 24307–24312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Thaher T. S. H., Drake A. F., Yokota S., Nakai A., Nagata K., and Miller A. D. (1996) The pH-dependent, ATP-independent interaction of collagen specific serpin/stress protein HSP47. Protein Pept. Lett. 3, 1–8 [Google Scholar]
- 16.Thomson C. A., and Ananthanarayanan V. S. (2000) Structure-function studies on hsp47: pH-dependent inhibition of collagen fibril formation in vitro. Biochem. J. 349, 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Widmer C., Gebauer J. M., Brunstein E., Rosenbaum S., Zaucke F., Drögemüller C., Leeb T., and Baumann U. (2012) Molecular basis for the action of the collagen-specific chaperone Hsp47/SERPINH1 and its structure-specific client recognition. Proc. Natl. Acad. Sci. U.S.A. 109, 13243–13247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdul-Wahab M. F., Homma T., Wright M., Olerenshaw D., Dafforn T. R., Nagata K., and Miller A. D. (2013) The pH sensitivity of murine heat shock protein 47 (HSP47) binding to collagen is affected by mutations in the breach histidine cluster. J. Biol. Chem. 288, 4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiser A., Do R. K., and Sali A. (2000) Modeling of loops in protein structures. Protein Sci. 9, 1753–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiser A., and Sali A. (2003) ModLoop: automated modeling of loops in protein structures. Bioinformatics 19, 2500–2501 [DOI] [PubMed] [Google Scholar]
- 21.Mongan J., Case D. A., and McCammon J. A. (2004) Constant pH molecular dynamics in generalized Born implicit solvent. J. Comput. Chem. 25, 2038–2048 [DOI] [PubMed] [Google Scholar]
- 22.Case D. A., Darden T. A., Cheatham T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Hayik S., Roitberg A., Seabra G., Swails J., et al. (2012) AMBER 12, University of California, San Francisco [Google Scholar]
- 23.Wang J., Cieplak P., and Kollman P. A. (2000) How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules. J. Comput. Chem. 21, 1049–1074 [Google Scholar]
- 24.Onufriev A., Bashford D., and Case D. A. (2004) Exploring protein native states and large-scale conformational changes with a modified generalized Born model. Proteins 55, 383–394 [DOI] [PubMed] [Google Scholar]
- 25.Socher E., and Sticht H. (2016) Mimicking titration experiments with MD simulations: a protocol for the investigation of pH-dependent effects on proteins. Sci. Rep. 6, 22523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng L., Baumann U., and Reymond J. L. (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du C., Wang M., Liu J., Pan M., Cai Y., and Yao J. (2008) Improvement of thermostability of recombinant collagen-like protein by incorporating a foldon sequence. Appl. Microbiol. Biotechnol. 79, 195–202 [DOI] [PubMed] [Google Scholar]
- 28.McIlvaine T. C. (1921) A buffer solution for colorimetric comparison. J. Biol. Chem. 49, 183–186 [Google Scholar]
- 29.Findlay J. W., and Dillard R. F. (2007) Appropriate calibration curve fitting in ligand binding assays. AAPS J. 9, E260-E267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sreerama N., and Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 31.Provencher S. W., and Glöckner J. (1981) Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 32.Macdonald J. R., and Bächinger H. P. (2001) HSP47 binds cooperatively to triple helical type I collagen but has little effect on the thermal stability or rate of refolding. J. Biol. Chem. 276, 25399–25403 [DOI] [PubMed] [Google Scholar]
- 33.Edgcomb S. P., and Murphy K. P. (2002) Variability in the pKa of histidine side-chains correlates with burial within proteins. Proteins 49, 1–6 [DOI] [PubMed] [Google Scholar]
- 34.Gebauer J. M., Kobbe B., Paulsson M., and Wagener R. (2016) Structure, evolution and expression of collagen XXVIII: lessons from the zebrafish. Matrix Biol. 49, 106–119 [DOI] [PubMed] [Google Scholar]
- 35.Meyer A., and Schartl M. (1999) Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 11, 699–704 [DOI] [PubMed] [Google Scholar]
- 36.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]