Abstract
Fish remain nearly the same shape as they grow, but there are two different modes of bone growth. Bones in the tail fin (fin ray segments) are added distally at the tips of the fins and do not elongate once produced. On the other hand, vertebrae enlarge in proportion to body growth. To elucidate how bone growth is controlled, we investigated a zebrafish mutant, steopsel (stptl28d). Vertebrae of stptl28d/+ fish look normal in larvae (∼30 days) but are distinctly shorter (59–81%) than vertebrae of wild type fish in adults. In contrast, the lengths of fin rays are only slightly shorter (∼95%) than those of the wild type in both larvae and adults. Positional cloning revealed that stp encodes Connexin43 (Cx43), a connexin that functions as a gap junction and hemichannel. Interestingly, cx43 was also identified as the gene causing the short-of-fin (sof) phenotype, in which the fin ray segments are shorter but the vertebrae are normal. To identify the cause of this difference between the alleles, we expressed Cx43 exogenously in Xenopus oocytes and performed electrophysiological analysis of the mutant proteins. Gap junction coupling induced by Cx43stp or Cx43sof was reduced compared with Cx43-WT. On the other hand, only Cx43stp induced abnormally high (50× wild type) transmembrane currents through hemichannels. Our results suggest that Cx43 plays critical and diverse roles in zebrafish bone growth.
Keywords: bone, connexin, connexon (hemichannel), electrophysiology, gap junction, zebrafish, bone
Introduction
Vertebrate bones develop during embryonic stages and grow continuously as body size increases. Because bone shape and size determine body shape, the mechanisms that generate and maintain bone shape are of great interest to biologists. Recent studies have increased our knowledge of bone formation mechanisms. Interactions among three types of cells—osteoblasts, osteocytes, and osteoclasts—play major roles in bone formation and bone remodeling (1–8).
Zebrafish (Danio rerio) is a model organism for developmental and genetic studies, and several mutants related to bone formation have been identified and analyzed (9–13). Mutant bone formation phenotypes can be categorized into two types: bone development mutants and bone growth mutants. Genes corresponding to bone development mutants are usually expressed at early developmental stages of bone formation (type 1), and bone malformations are detected when the bones first appear in embryo (14–16). On the other hand there are several zebrafish bone growth mutants, in which bone development looks almost normal at early stages, but mutant phenotypes appear at late stages (type 2) (17–19).
In this study, we focused on the stoepsel (stp) mutant fish (20) to study bone morphogenesis in adult stages. The stp mutant was originally isolated by a large scale screen of zebrafish mutants induced by ENU2 (20). This mutant looks normal until ∼35 days after fertilization (dpf); however, the vertebrae of the mutant fish become shorter than those of wild type fish at later growth stages. The stp allele is dominant and homozygous lethal. We re-examined the stp phenotypes in detail using modern techniques, and we performed positional cloning to identify the stp gene. Surprisingly, we isolated a mutation in the connexin43 gene, which is known as the sof (short-of-fin) gene in zebrafish (21). sof is a well studied type 2 zebrafish mutant, in which fin segments are short in the caudal fin (21). Four sof mutant alleles were isolated by mutagenesis screening, and three single mutations that cause amino acid substitutions in Connexin43 protein were identified (22). In the sofb123 allele, no amino acid substitution was identified in the connexin43 gene, but cx43 expression was reduced in the mutant fish (21, 23, 24).
Connexin proteins are four-pass transmembrane proteins that are subunits of gap junctions (25). Approximately 20 connexin genes are known from the mammalian genome, and ∼36 connexin genes are predicted in the zebrafish genome (26, 27). Six connexin proteins make a hexamer called a connexon; docking of two connexons on adjacent cells creates a gap junction. Gap junctions allow movement of small molecules (<1000 Da; e.g. ATP, inositol trisphosphate, ions, etc.) between neighboring cells; a connexon acts as a hemichannel on the cytoplasmic membrane (25). connexin43 is one of the most studied connexin genes because it is linked to several human diseases (28–30). Oculodentodigital dysplasia is one Cx43-related human disease, which causes small eyes, underdeveloped teeth, and malformed fingers (31, 32). To identify the cause of the substantial difference in phenotype between the sof and stp alleles of cx43, we performed electrophysiological experiments and found that the growth-dependent malformation of stp vertebrae is likely caused by aberrant hemichannel activity of Cx43stp rather than reduced gap junction intercellular conductance observed in Cx43sof.
Experimental Procedures
Animal Care
All experiments in this study were conducted in accordance with the guidelines and approved protocols for animal care and use (approval number FBS-14-002-1) at Osaka University (Osaka, Japan). We used the Tu and AB strains as wild type zebrafish lines. stoepsel (stptl28d) mutant was obtained from the Tubingen Zebrafish Stock Center.
Clearing and Measuring of Vertebral Length and Height
Fish samples were fixed with 3.7% formaldehyde and treated with ethanol to remove lipids. Then they were transferred back to H2O. Next, samples were incubated in a saturated aqueous solution of sodium tetraborate for 1 day. Then they were incubated overnight in clearing solution (30% saturated aqueous solution of sodium tetraborate with 3 mg/ml trypsin) at 48 °C to clear the samples. Transparent samples were dyed with bone dye solution (90 mm KOH, 50 μg/ml alizarin red S) for 1 day and then incubated in 90 mm KOH for 1 day. Finally, samples were immersed in glycerol with thymol and preserved.
CT Scanning
A LaTheta LCT-100 (Hitachi Aloka Medical, Ltd.) x-ray micro-CT scanner at Osaka Bioscience Institute and an inspeXio SMX-100CT microfocus x-ray CT system at Shimadzu Corporation were used for taking CT images of bone structure. Fish samples were fixed with 3.7% formaldehyde and stored in PBS. Samples were scanned in air.
Positional Cloning and Sequence Analyses
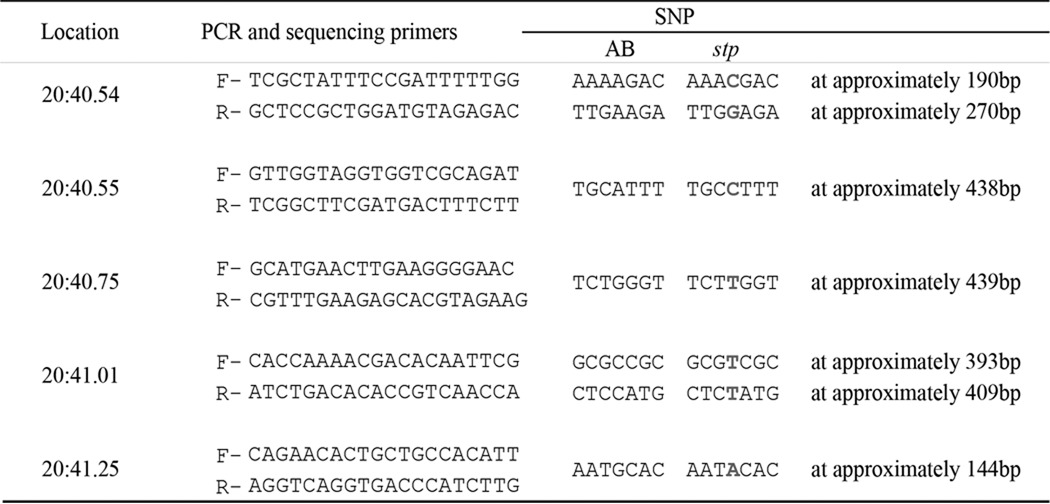
The stptl28d/+ (TE background) mutant was crossed with the wild type AB strain. TE is a mixture of Tu and TL lines. Here, we used Tu instead of TE. F1 heterozygous fish were backcrossed with wild type parents, and F2 fish were used for genetic mapping. stp was mapped to chromosome 20 by checking simple sequence length polymorphism markers. Then stp was mapped between single nucleotide polymorphisms listed in Table 1, which were identified within chromosome 20 (40.55–41.01 Mb). Gene predictions were derived from Ensemble (Sanger Institute). Candidate gene sequencing was performed using a 3130 Genetic Analyzer (Applied Biosystems).
TABLE 1.
SNP Markers used in positional cloning
SNP Markers were developed from comparison of sequences between AB and stp. Locations of SNP Markers are on the Zv9 assembly. Both forward (F) and reverse (R) primers were used for sequencing. All SNPs were assessed by sequencing reads.
Rescue of stp Homozygote Phenotype
A cx43 fragment was cloned from a bacterial artificial chromosome clone (DKEY-261A18) with a forward primer (5′-GCTCGAGTCTATGAATGGGATGAG-3′) and a reverse primer (5′-GGGCGGCCGCTAGACGTCCAGGTCATCAG-3′) and ligated into a Tol2 plasmid (pT2AL200R150G) (33). The cx43 sequence was injected into stptl28d/+ embryos to generate Tg(cx43pro-cx43)stptl28d/+ transgenic fish lines. Tg(cx43pro-cx43)stptl28d/tl28d transgenic fish were made by crossing stptl28d/+ fish and Tg(cx43pro-cx43)stptl28d/+ fish.
cx43 Reporter Line
The cx43 promoter sequence was cloned from a bacterial artificial chromosome clone (DKEY-261A18) with a forward primer (5′-CCGGCTCGAGGCTTAAAGGGTCACGAAACACC-3′) and a reverse primer (5′-GGCCGGATCCTTGAGGGAGTTCTAGCTGAAAATA-3′) and ligated into a Tol2 plasmid (pT2AL200R150G). A reporter construct of cx43 was made by inserting mCherry cDNA downstream of the cx43 promoter region.
TALEN-mediated cx43 Knock-out
Transcription activator-like effector nucleases (TALEN) pairs for cx43 knock-out were designed using the TALEN Targeter program (available at the Cornell University website). TAL repeats were assembled using a Golden Gate TALEN and TAL Effector kit 2.0 (34) with some modifications (35) and cloned into expression vectors pCS2TAL3DD or pCS2TAL3RR (36). The TALEN expression plasmids were linearized by NotI digestion, and then mRNAs were in vitro transcribed using an mMESSAGE mMACHINE SP6 transcription kit (Ambion). Synthetic TALEN mRNAs were injected into one-cell stage embryos (100 pg/embryo each). To test the efficacy of mutagenesis, TALEN mRNA-injected embryos were harvested 1 day after fertilization. The genomic DNA was extracted, and the loci containing the TALEN target sequence were amplified by PCR with a forward primer (5′-TTCAAGTGTCACCAAAGTGTCT-3′) and a reverse primer (5′-CTGCTGGGTATTGCACTTGA-3′). TALEN-induced mutations were detected by T7 endonuclease I assay. The most effective TALEN pair was used for cx43 knock-out. The target sequence is 5′-TCTGGCTCTCTGTGCTCTtcatcttccggatccTTGTTCTGGGAACAGCA-3′ (TALEN binding sequences are shown in uppercase, and spacer sequence is in lowercase).
Measurement of mRNA Expression for cx43 Alleles
To precisely compare the expression levels of the wild type and mutated alleles, which differ at only one nucleotide, we generated cDNA clones from the heterozygous fish and then sequenced them. mRNAs were collected from tail fins of stptl28d/+ fish and stptl28d/sofb123 fish by RNeasy Protect mini kit (Qiagen), respectively. Then mRNAs were reverse-transcribed using SuperScript III reverse transcriptase (Invitrogen). The cx43 sequence around the stp mutation site was amplified from the cDNAs, and the amplified fragments were cloned with plasmid vector. Then Escherichia coli colonies were picked up randomly, and DNA fragments in the plasmid were sequenced. The frequencies of DNA fragments derived from stp allele or wild type allele in stptl28d/+ fish and DNA fragments from the stp allele and sof allele in stptl28d/sofb123 were compared.
Preparation of Connexin mRNA and Injection into Xenopus Oocytes
Connexin cDNAs were amplified using PCR and cloned into pGEM-HeFx plasmid (37). The plasmids were linearized using restriction enzyme and then applied to the mRNA preparation kit (T7 mMessage mMachine; Ambion) according to the manufacturer's protocol. An adult Xenopus laevis female was anesthetized with MS-222, and the ovarian lobes were collected using a surgical knife and forceps. The eggs were treated with collagenase solution (2 mg/ml collagenase I (Sigma) and 2 mg/ml hyaluronidase (Sigma) in OR2 buffer: 82.5 mm NaCl, 2 mm KCl, 1 mm MgCl2, and 5 mm HEPES, adjusted to pH 7.5 using NaOH) at 18 °C for 2 h. Stage V and VI oocytes were collected manually and used for mRNA injection. Then mRNA (0.05 ng for gap junction recording; 5 ng for hemichannel recording) was co-injected with 10 ng of antisense oligonucleotide DNA for Xenopus cx38 into Xenopus oocytes. For the negative control, only antisense oligonucleotide was injected. Oocytes injected with mRNA were incubated overnight at 18 °C in 1/2× diluted L15 medium (Sigma) with 2 mm CaCl2 (HEPES-buffered and adjusted to pH 7.5 with NaOH).
Voltage Clamp Recording
Vitelline membranes of mRNA-injected oocytes were removed manually using forceps in a hypertonic solution (200 mm aspartic acid, 1 mm MgCl2, 10 mm EGTA, 20 mm KCl, and 10 mm HEPES, pH 7.5). The oocytes were manually paired, touching at the vegetal poles, while in ND96(+) solution (93.5 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 2 mm MgCl2, and 5 mm HEPES). The transjunctional current was measured using the dual whole cell voltage clamp technique. Current and voltage electrodes were prepared with a micropipette puller P-1000 (Sutter Instruments) to obtain a resistance of 0.5–1.0 MΩ. The pipette was filled with solution containing 3 m KCl, 10 mm EGTA, and 10 mm HEPES (pH 7.4). Voltage clamp experiments were performed using two Multi Electrode Clamp Amplifiers iTEV90 (HEKA). To measure the transjunctional current, both cells were initially clamped at −40 mV, and one cell was then subjected to alternating pulses of from −160 to +80 mV in 20-mV increments. Currents detected in the second oocyte were recorded, and junctional conductance was calculated using the current value at the end of each 3-s pulse (steady state). The conductance was obtained by Gj = Ij/(V1 − V0), and normalized Gj values were compared.
Hemichannel Recording
Hemichannel currents were recorded from a single oocyte using a Multi Electrode Clamp Amplifier iTEV90. Modified ND96(+) solution with/without 2 mm CaCl2 was used as a bath solution. To obtain the hemichannel current, the cells were initially clamped at −40 mV and then subjected to 5-s voltage steps from −30 to +60 mV in 10-mV increments.
Western Blotting and Immunofluorescence Staining
Xenopus oocytes, in which the antisense oligonucleotide for Xenopus cx38 and mRNA for cx43, cx43stp, or cx43sof were injected, were harvested after overnight incubation. Then each oocyte was used for Western blotting and immunofluorescence staining. Western blotting was performed using anti-Cx43 antibody (Santa Cruz Biotechnology). Anti-β-tubulin antibody (Cell Signaling) was used for the positive controls. Signals were detected using ECL Western blotting detection kit (GE Healthcare). Immunofluorescence staining was performed using the anti-Cx43 antibody and Alexa 488 conjugated antibody (Invitrogen). Pictures were taken using a BZ-X700 microscope (KEYENCE) with OP-66835 BZ filter GFP.
Results
Stoepsel (stptl28d) Phenotype
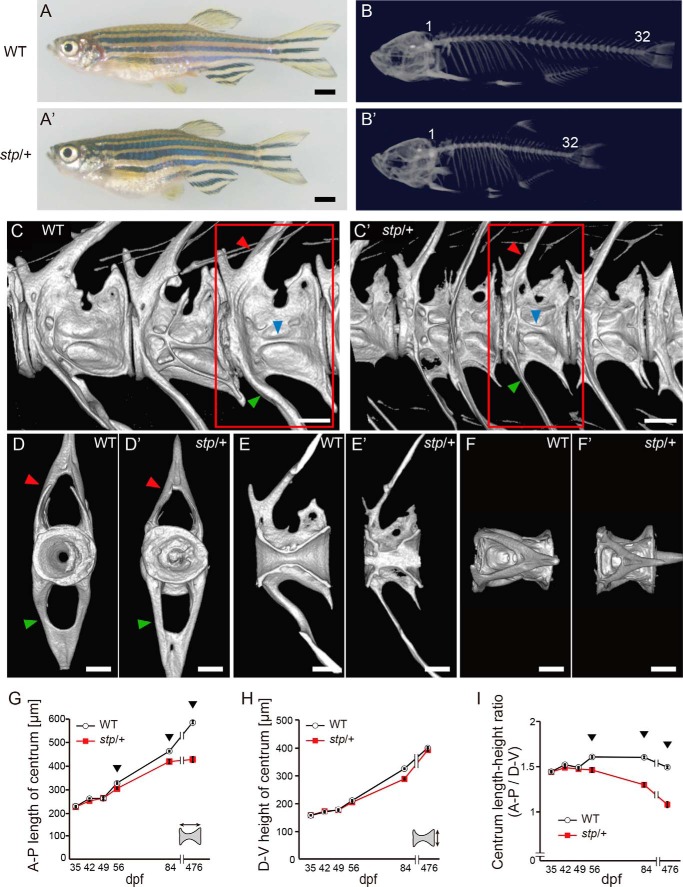
The stp mutation was isolated in a large scale screen of zebrafish mutants induced by ENU (20). In the original paper, stp was described as a mutant associated with abnormalities of the axial skeleton (Fig. 1, A and A′). stp homozygous (stptl28d/tl28d) fish die from edema of the eye and abdomen at ∼10 dpf (Fig. 2, A–C). Because wild type zebrafish start to form vertebrae along the notochord at ∼14 dpf, we could not observe the bone phenotype in stptl28d/tl28d fish. Heterozygous mutants (stptl28d/+) had similar body lengths to the wild type at early stages, but during later growth they gradually became shorter relative to the wild type along the anterior-posterior (AP) axis (Fig. 1, A and A′). The number of vertebrae appeared similar between the mutant and wild type fish. The body height along the dorsoventral (DV) axis was not affected by the stp mutation (Fig. 1, B and B′).
FIGURE 1.
stp vertebral phenotype. Panels show adult fish (A and A′), CT scans of their skeletons (B–F and B′–F′), and graphs of vertebral growth rate (G–I). B and B′, the numbers indicate vertebra identity along the AP axis. The stptl28d/+ fish had the same number of vertebrae as their wild type siblings (n = 6). C and C′, the red square indicates the third caudal vertebra. Arrowheads point to neural arches (red arrowheads), hemal arches (green arrowheads), and beam-like structures (blue arrowheads) on the centrums. D and D·, front views of the third vertebra. Abnormal calcification was seen in the inside of the centrum in stptl28d/+ fish. It was also seen in the wild type fish, but calcification was more severe in the stptl28d/+ fish. E and E′, sagittal images from the left side. F and F′, views from the top. G, centrum length at each time point. H, centrum DV height at each time point. I, centrum length to height ratio. G–I, the data shown are mean values from five vertebrae, the first through the fifth caudal vertebrae (n = 6). Arrowheads point to time points at which the stp vertebral phenotype was seen. Error bars, S.E. Scale bars, 1000 μm in A and A′ and 250 μm in C–F and C′–F′. stp/+, stptl28d/+.
FIGURE 2.
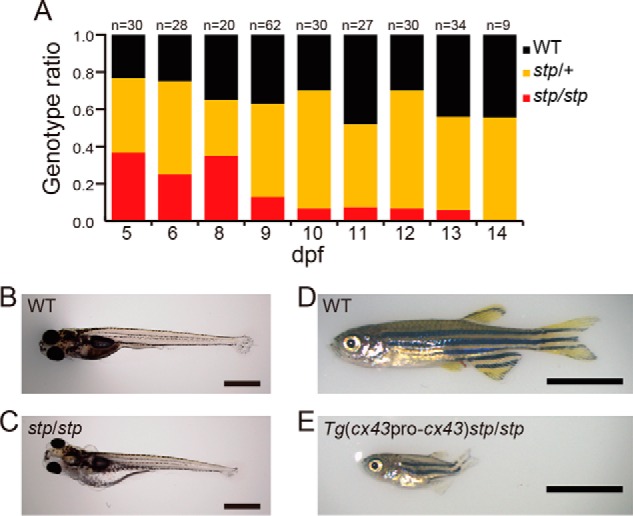
Phenotypes of stptl28d/tl28d and Tg(cx43pro-cx43)stptl28d/tl28d fish. A, genotype ratio of stptl28d/tl28d and siblings at each time point. Larvae derived from crossing stptl28d/+ and stptl28d/+ were harvested at indicated day and genotyped. B and C, wild type and stptl28d/tl28d larvae at 13 dpf. D and E, wild type and Tg(cx43pro-cx43)stptl28d/tl28d fish at 56 dpf. Scale bars, 1000 μm in B and C and 5000 μm in D and E. stp/+, stptl28d/+; stp/stp, stptl28d/tl28d.
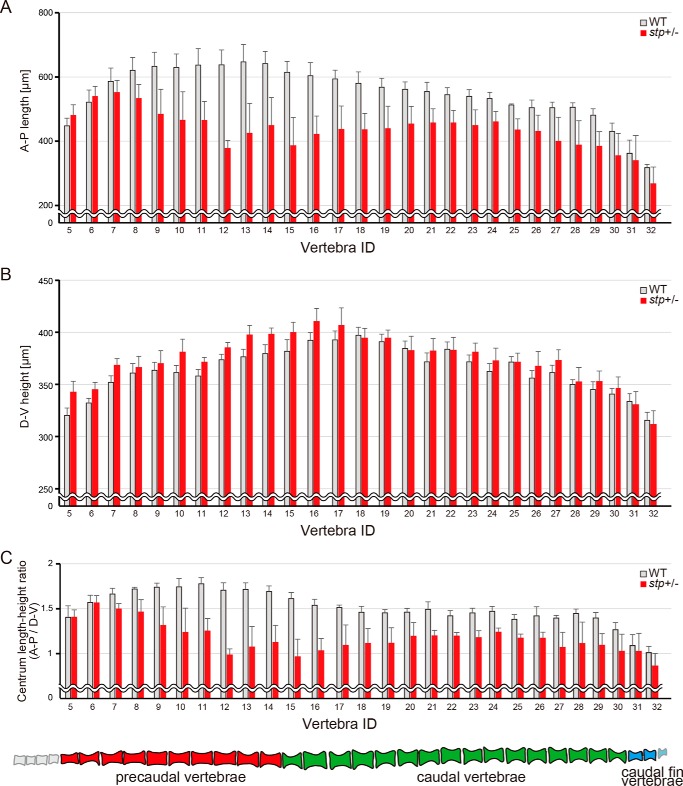
Next, we examined the bone phenotype in detail by microCT scanning (Figs. 1, B–F and B′–F′, and 3). AP length (Fig. 3A), DV height (Fig. 3B), and the proportions (Fig. 3C) of corresponding vertebrae (identification numbers from anterior to posterior) were compared between wild type and stp mutant fish. Vertebrae in stptl28d/+ fish were obviously shorter along the AP axis than those of wild type fish (Fig. 3A), whereas the head and fins did not differ notably (Fig. 1, B and B′).
FIGURE 3.
Graphs of centrum length and height. A–C, AP length (A), DV height (B), and the ratio (C) of centrum length/height of each vertebra at 476 dpf (n = 6). Six stptl28d/+ fish and their siblings were compared. Error bars, S.E. stp/+, stptl28d/+.
Each vertebra is composed of a centrum and appendixes, which include a ventral arch (Fig. 1, C, C′, D, and D′, red arrowheads), hemal arch (Fig. 1, C, C′, D, and D′, green arrowheads), and complex beam structures (Fig. 1, C and C′, blue arrowheads). The centrum appears at ∼14 dpf. The appendixes form after centrum growth. In a wild type centrum, an hourglass-like structure was seen along the AP axis (Fig. 1E). On the other hand, in stptl28d/+ mutants, the hourglass-like structure was distorted and sometimes filled with clots (Fig. 1, D′ and E′). Although the centrum was malformed in the stptl28d/+ mutant, the hemal arch (Fig. 1, C′ and D′, green arrowheads), neural arch (Fig. 1, C′ and D′, red arrowheads), and beam structures (Fig. 1C′, blue arrowhead) were approximately normal. These data indicate that the stp mutation mainly affected the formation of the centrum of the vertebrae.
Next, to reveal when vertebral malformation occurred, we investigated the time course of centrum growth (Fig. 1, G–I). We measured the AP length (Fig. 1G) and DV height (Fig. 1H) of the centrums of caudal (No. 1–5) vertebrae (Fig. 3). Graphs in Fig. 1 (G–I) show that early vertebral development (35 dpf) was not affected by the stp mutation; however, after 35 dpf, centrum growth was abnormally low along the AP axis (Fig. 1G), but not along the DV axis (Fig. 1H). This suggests that the stp mutation affects the mechanism that maintains appropriate vertebral shape during growth.
Cloning the Gene Affected by the stp Mutation
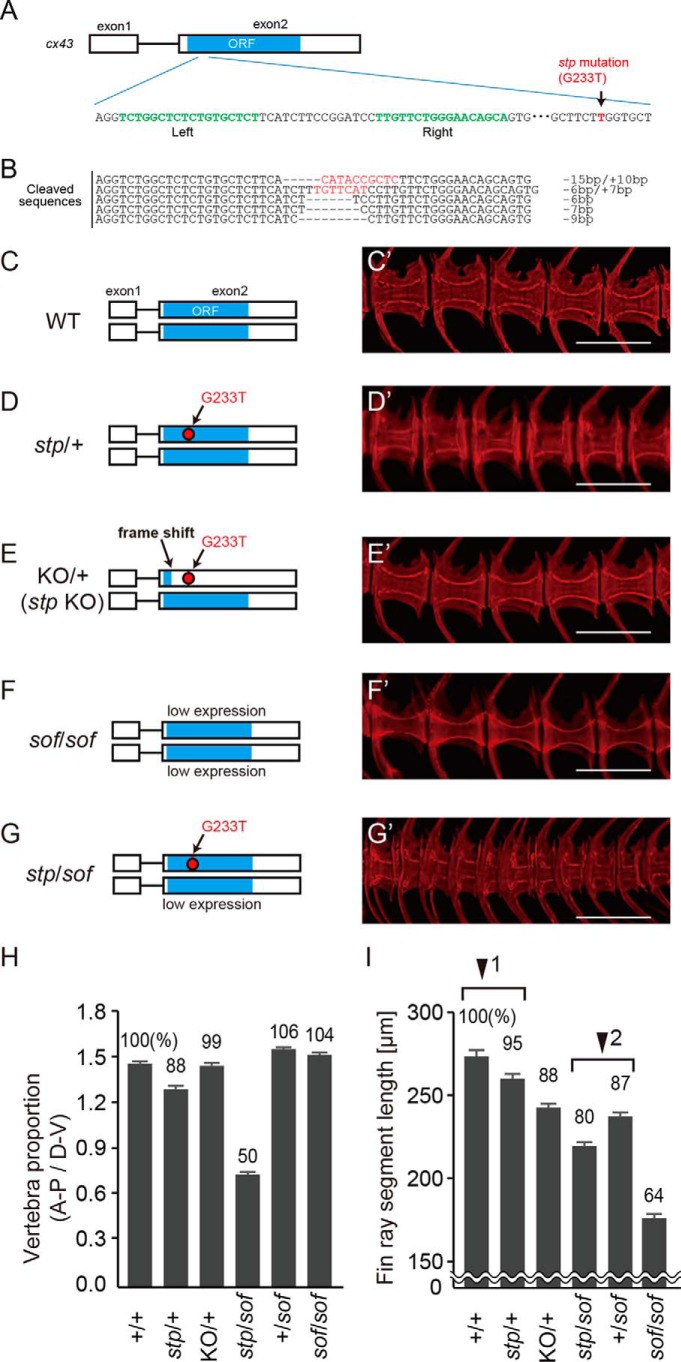
To identify the gene affected by the stp mutation, we next performed positional cloning and identified a critical region of 460 kb in chromosome 20 (Fig. 4A). This region contains eight genes, of which six genes code for connexins (cx). The coding sequences of the stp allele were sequenced, and their amino acid sequences were compared with the wild type. Among the eight genes, only the cx43 gene had a point mutation (G233T) specific to the stp allele, which altered the amino acid sequence (tryptophan → leucine: W78L) (Fig. 4B). The amino acid substitution occurs in the second transmembrane domain of the Cx43 protein (Fig. 4C). A BLAST search revealed that the region around the mutation was highly conserved in vertebrates (Fig. 4D). To confirm cx43 expression in vertebrae, we generated transgenic fish that expressed mCherry under the control of the cx43 promoter (Fig. 4, E, E′, F, and F′). This transgenic fish showed mCherry expression at the growing edges of the centrum, where the defect resulting in the stp phenotype was expected to occur (Fig. 4, E and E′), and at the junctions between fin bones (Fig. 4, F and F′).
FIGURE 4.
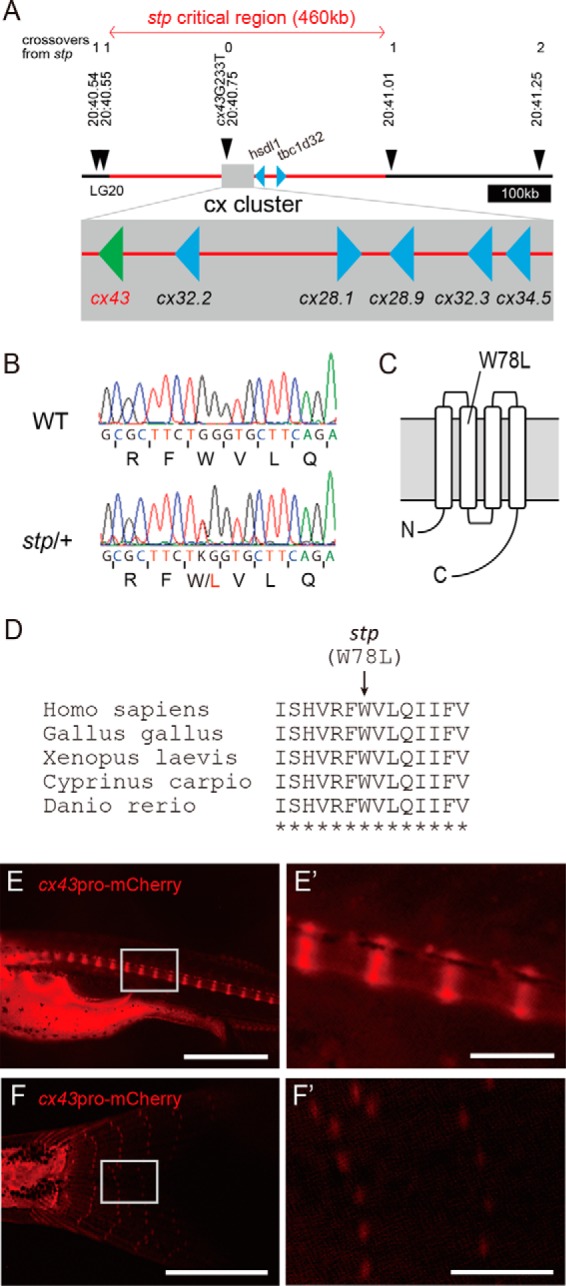
The stp allele exhibits a lesion in cx43. A, meiotic mapping of the stp allele revealed ∼460 kb of contiguous sequence of the stp region. Only cx43 exhibits an ENU-induced lesion. Red lines represent the region spanning the stp critical interval. B, electrophoretograms showing the mutation identified in the coding sequence of the stp allele. C, cartoon of Cx43 protein in a double-lipid bilayer indicating the location of the ENU-induced lesion. D, BLAST search results showing the highly conserved amino acid sequence of Cx43. E, E′, F, and F′, expression of mCherry fluorescent protein in the vertebrae and caudal fin, driven by the cx43 promoter. E and E′, mCherry was expressed at the growing edges of vertebral centrums of 21 dpf fish. F and F′, mCherry was expressed at junctions between fin ray segments of 36 dpf fish. Scale bars, 1000 μm in E and F and 200 μm in E′ and F′.
stp Gene Knock-out by TALEN
To confirm that the mutation in cx43 is responsible for the stp phenotype, we knocked out the stp allele of cx43 using TALEN. The TALEN construct was designed to target the middle region of the first extracellular loop of Cx43, just at the 5′ side of the stp mutation (Fig. 5A). TALEN mRNA was injected into embryos of stptl28d/+ fish, and we isolated the F1 fish that had a frameshift in the stp allele (Fig. 5B). Because stp (Fig. 5, D and D′) is a dominant mutation, knock-out of the stp allele in stptl28d/+ fish should rescue the phenotype. The heterozygous fish, which had the frameshifted stp allele and the wild type cx43 allele (KO/+; Fig. 5, E and E′), did not show any difference in centrum structure compared with wild type siblings (Fig. 5, C, C′, and H), indicating that knock-out of the stp allele rescued vertebral structure. Vertebrae in stp mutants were 12% shorter than in the wild type, but vertebral lengths were similar in KO/+ fish and the wild type (Fig. 5H). This result confirmed that the mutation in cx43 (Cx43W78L) is responsible for the stp phenotype.
FIGURE 5.
Vertebral and fin ray segment phenotypes of zebrafish mutants. A and B, knock out of cx43stp by TALEN. A, schematic of cx43 gene structure. The green letters are the DNA binding sites by TALEN. The red letter is the stp mutation (G233T). B, Mutated sequences produced by TALEN. Three F1 lines had frame-shifting mutations, which were (−15 bp/+10 bp), (−6 bp/+7 bp), and (−7 bp). The (−15 bp/+10 bp) line was used for the stp gene knock-out investigation. C–G, vertebral structure of wild type (C), stp/+ (D), KO/+ (stp allele knock-out) (E), sofb123/b123 (F), and stp/sofb123 (G) fish. Cleared, alizarin red S-stained specimens were made from 70 dpf fishes. KO/+ fish had a wild type allele and a knock-out stp allele. sofb123/b123 fish showed normal vertebra structure, but stp/sofb123 fish had markedly shorter vertebrae. H and I, six mutant fish were randomly chosen and compared with six of their wild type siblings at 70 dpf. H, length to height ratio of vertebral centrums. I, length of fin ray segment. The segments second from the tail bud were measured, and average length was calculated. Arrowheads 1 and 2 indicate pairs of fish genotypes that differ in the presence or absence of the stp allele. Error bars, S.E. Scale bars, 500 μm in C′–G′. stp/+, stptl28d/+; sof/sof, sofb123/b123; stp/sof, stp/sofb123.
We also examined the lethality of the stp homozygote. If the lethality was caused by the loss of normal Cx43 protein, survival of stptl28d/tl28d larvae should be rescued by introducing exogenous wild type cx43. We generated Tg(cx43pro-cx43)stptl28d/tl28d fish by introducing a wild type cx43 gene. Tg(cx43pro-cx43)stptl28d/tl28d fish did not die at larval stages and grew into adults (Fig. 2, D and E). Interestingly the rescued fish showed a unique phenotype (Fig. 2E): they had very short body lengths compared with the wild type fish (Fig. 2D) and stp/+ heterozygous fish (Fig. 1A′). These results suggest that the lethality of stptl28d/tl28d results from the loss of cx43 function. In addition this transgenic fish also suggested that Cx43W78L has a dominant negative effect on wild type Cx43 because the transgene, cx43pro-cx43, did not rescue the stp phenotype perfectly (Fig. 2E).
Another cx43 Mutant, Short Fin
The zebrafish mutant, sof, which exhibits short fin bone segments, also involves cx43 (21). There are four reported sof alleles. All of these alleles show similar recessive phenotypes. Three alleles have amino acid substitutions in cx43, and one allele causes low expression of cx43. It is very puzzling that these alleles of the same gene do not cause shortening of vertebrae. We first examined the amount of cx43 mRNA between stp and sofb123 (low expression allele) and found that the expression of mRNA from the stp allele was comparable with that from the wild type cx43 allele (Fig. 6A); however, expression of mRNA from the sof123 allele was less than one-tenth that of the stp allele (Fig. 6B).
FIGURE 6.
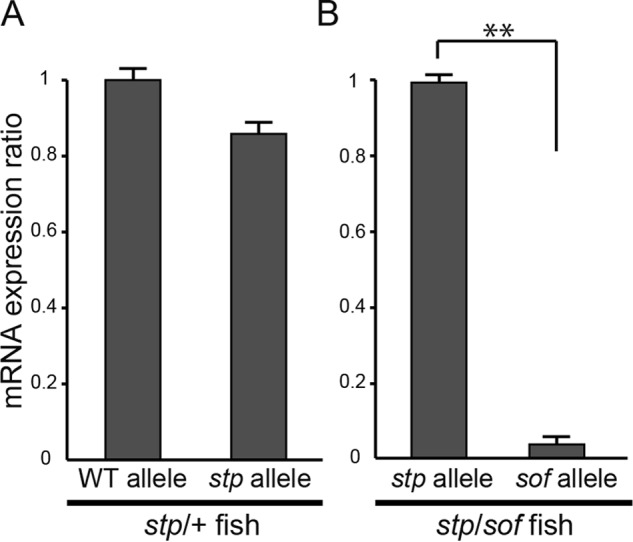
cx43 mRNA expression ratios in mutant fish. A, cx43 mRNA expression ratio in stptl28d/+ fish, normalized to WT fish. 39–48 colonies were sequenced for each fish (n = 6). B, cx43 mRNA expression ratio in stp/sofb123 fish, normalized to stp fish. 28–48 colonies were sequenced for each fish (n = 5). Error bars, S.E. **, p < 0.01. stp/+ = stptl28d/+; stp/sof = stp/sofb123.
We then investigated the shape of sofb123 mutant fish to determine whether they had morphologically normal vertebrae. As shown in Fig. 5 (F, F′, and H), vertebrae of both sofb123/b123and sofb123/+ fish are normal in size and shape. These data indicated that a simple decrease of cx43 mRNA level did not affect vertebral growth. However, the vertebrae of stp/sofb123 fish are drastically shorter than those of stptl28d/+ fish (Fig. 5, G, G′, and H), suggesting that the wild type Cx43 protein compensates for the dominant effect of the stp allele. When the amount of normal Cx43 protein was decreased, the stp phenotype (short vertebrae) became more evident.
Next, we measured the length of fin ray segments for fish carrying the different cx43 alleles (Fig. 5I) to see whether the stp allele causes defects in fin bones. Segment lengths of sofb123/b123 and sofb123/+ fish were 64 and 87% of wild type, respectively. This showed that fin ray length is sensitive to the amount of Cx43 protein. stptl28d/+ fish also had shorter fin ray segments than wild type fish, but the difference was only 5% (Fig. 5I, arrowhead 1). The difference of the fin ray segment length between stp/sofb123 and sofb123/+ fish was 7% (Fig. 5I, arrowhead 2). Therefore, the stp allele also causes the short fin bone phenotype, but the severity is less than that of the sof alleles.
Functional Analysis of Cx43stp
As described above, the influence of the stp allele is severe in vertebrae but mild in fin bones. On the other hand, the influence of the sofb123 allele is undetectable in vertebrae (Fig. 5H) and severe in fin bones. This apparent discrepancy suggests that the Cx43 protein has more than one distinct function. It is known that there are two distinct functions of connexins: formation of gap junctions or hemichannels. We examined both functions of Cx43sof and Cx43stp electrophysiologically. There are three known sof missense mutations: Cx43F30V, Cx43P191S, and Cx43F209I. A previous paper reported that all of them have similar hemichannel and gap junction activity (22). Here, we used cx43j7e2 (which codes for Cx43P191S) as a representative sof mutant.
To examine gap junction activity of these Cx43 mutants, we performed voltage clamp current recording using Xenopus oocytes. In this study, we injected only 0.05 ng of mRNAs for Connexin proteins into each oocyte because we detected very large transjunctional currents when we injected 5 ng of mRNA for wild type Cx43 (22). Fig. 7A shows examples of junctional currents induced by wild type Cx43 (Cx43-WT) and Cx43W78L (Cx43stp). Cx43W78L (Cx43stp) showed reduced junctional currents between the oocytes compared with Cx43-WT (Fig. 7B). On the other hand Cx43P191S (Cx43sof) did not show significant junctional current in this experimental condition (0.05 ng of mRNA/oocyte).
FIGURE 7.
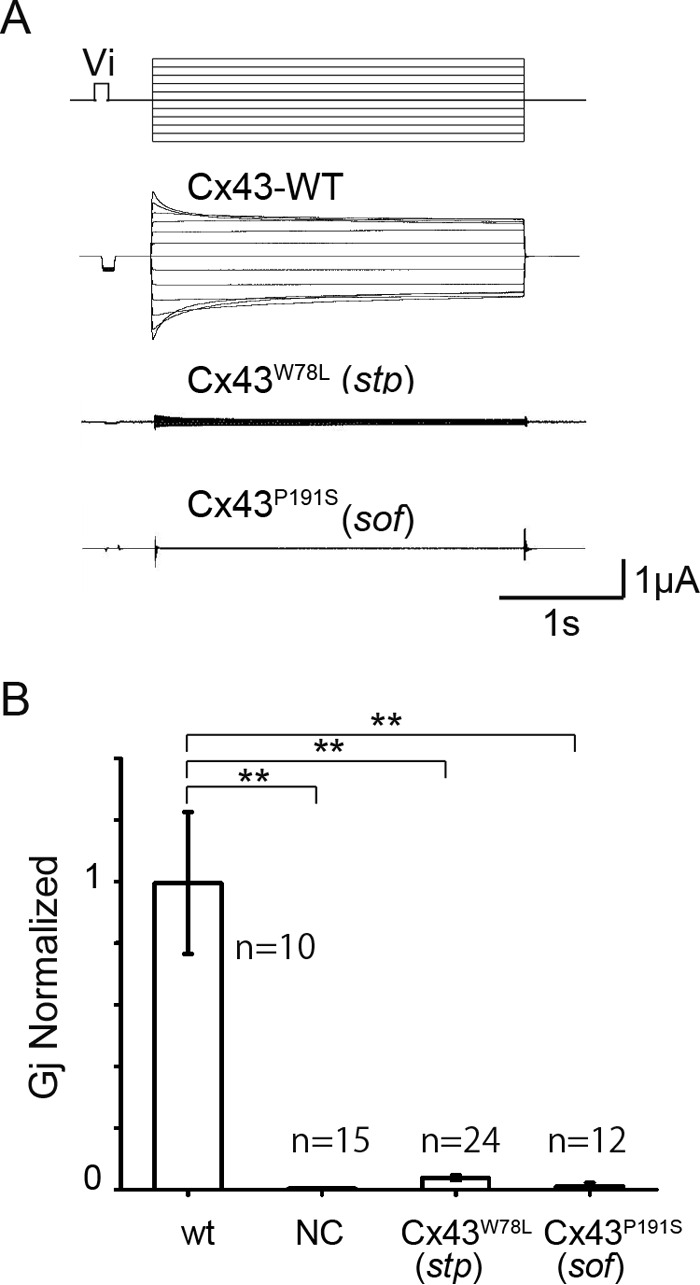
Electrophysiological analysis of Cx43 gap junctions. A, examples of junctional current traces for Cx43 (Cx43-WT), Cx43W78L (Cx43stp), and Cx43P191S (Cx43sof) shown below voltage protocol (top). B, normalized junctional conductance of mutants compared with Cx43-WT. NC, negative control (only antisense oligonucleotide for Xenopus cx38 was injected). Error bars, S.E. **, p < 0.01.
We next examined the hemichannel activities of wild type and mutant Cx43s. Panels A and B of Fig. 8 show current traces of Cx43-WT and Cx43W78L (Cx43stp), and panels C–G summarizes the current-voltage relationship of these hemichannels. As reported previously, Cx43-WT and Cx43P191S (Cx43sof) did not increase current in comparison with the negative control (Fig. 8, C, E, and G). On the other hand Cx43W78L (Cx43stp) induced extraordinarily high hemichannel activity (Fig. 8D), which was more than 50-fold greater than that of Cx43-WT. We examined the hemichannel activity of Cx43W78L (Cx43stp) co-expressed with Cx43-WT, mimicking the situation in stptl28d/+ fish in vivo. The hemichannel activity of Cx43W78L (Cx43stp) co-expressed with Cx43-WT was also very high (Fig. 8F). Assuming that wild type and mutant connexins interact heteromerically, these results suggest that heteromeric hemichannels of Cx43-WT and Cx43W78L (Cx43stp) in stptl28d/+ fish would have increased activity. These results suggest that the hemichannel activity of Cx43P191S (Cx43sof) is similar to that of Cx43-WT, whereas that of Cx43W78L (Cx43stp) is abnormally increased.
FIGURE 8.
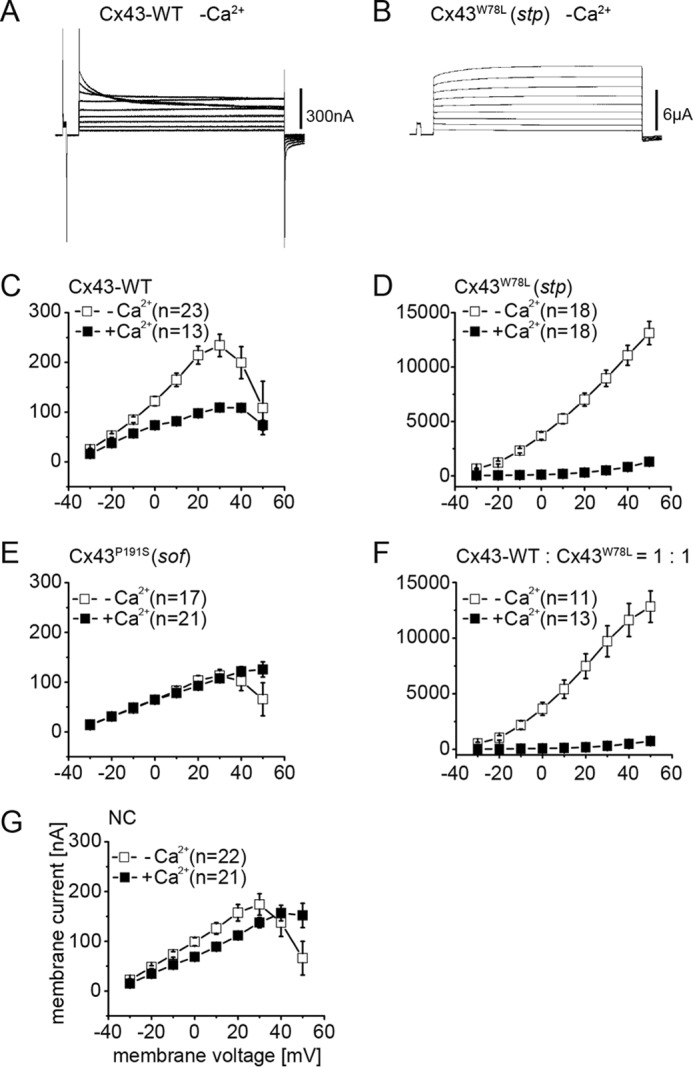
Non-junctional current recording. A and B, examples of hemichannel current traces for Cx43-WT (A) and Cx43W78L (Cx43stp) (B). C–G, plots of current versus membrane voltage in single oocytes expressing Cx43-WT (C), Cx43W78L (Cx43stp) (D), Cx43P191S (Cx43sof) (E), Cx43-WT and Cx43W78L (Cx43stp) (F), and negative control (only antisense oligonucleotide for Xenopus cx38 was injected) (G). Oocytes voltage was pulsed from −30 to +60 mV in 10-mV steps in the presence (■) or absence (□) of 2 mm Ca+2 ion. Error bars, S.E. NC, negative control.
To determine the protein expression and functional changes of connexin proteins in Xenopus oocytes, we performed Western blotting and immunofluorescence staining. The bracket in Fig. 9A indicated that Cx43 and its mutants were expressed in Xenopus oocyte, although several bands were detected in the Cx43W78L (Cx43stp) lane. The bands detected in Cx43W78L imply that the W78L mutation causes a protein modification defect in Cx43. Further study is needed to determine whether this modification of Cx43 is related to the stp phenotype. 50-kDa bands in Fig. 9A are due to nonspecific binding of the anti-Cx43 antibody used in this study. The arrow in Fig. 9A′ indicates the expression of β-tubulin (positive control). Protein localization of Cx43 and its mutants were also examined, and membrane localization was detected by immunofluorescence staining (Fig. 9, B—E and B′–E′). Compared with the wild type Cx43 (Fig. 9B), the signals of Cx43W78L (Cx43stp) (Fig. 9D) and Cx43P191S (Cx43sof) (Fig. 9E) detected at the cell membranes looked weak (22). For the negative controls (NC), only antisense oligonucleotide for Xenopus cx38 was injected into Xenopus oocytes (Fig. 9, A, A′, and C).
FIGURE 9.
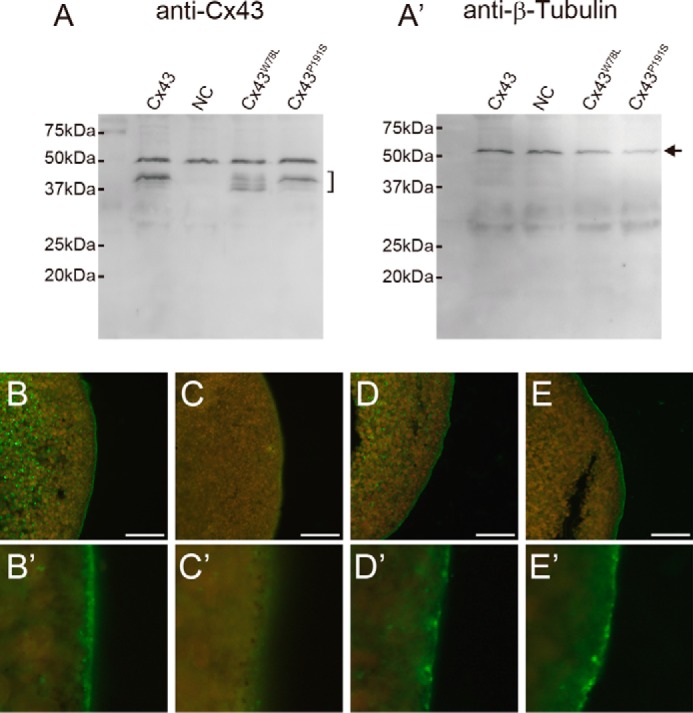
Protein expression. Protein expression and membrane localization in Xenopus oocytes were analyzed by Western blotting (A and A′) and immunofluorescence staining (B–E and B′–E′). A, bracket indicates Cx43 bands detected by anti-Cx43 antibody. Bands at ∼50 kDa are nonspecific labeling. A′, arrow indicates β-tubulin bands (positive control). mRNA for each connexin was injected into Xenopus oocytes along with an antisense oligonucleotide for Xenopus cx38. NC, negative control (only antisense oligonucleotide for Xenopus cx38 was injected). B–E, membrane localization of Cx43 or its mutants was detected. B, wild type Cx43. C, negative control. D, Cx43W78L (Cx43stp). E, Cx43P191S (Cx43sof). B′–E′, magnified images of B–E, respectively. Scale bars, 50 μm.
Discussion
In this paper we analyzed a zebrafish mutation that disrupts normal vertebra formation. Homozygous (stptl28d/tl28d) fish die; heterozygous (stptl28d/+) fish grow to adults but show a specific phenotype of shorter vertebrae along only the AP axis. Because this phenotype is not seen in young fish (∼35 dpf) and becomes evident only after ∼60 dpf, we deduced that the mutation caused some defect in the control of bone growth. To identify the gene altered in the stp mutant, we performed positional cloning and TALEN knock-out of the genomic sequence and found that a single base mutation in cx43 causes the phenotype.
Curiously, a previous paper reported that another mutation in cx43 is responsible for the sof phenotype, in which fins are short but vertebrae are normal. To determine why these alleles of the same gene differed in their effects, we investigated two different functions of connexins: as gap junctions and hemichannels. Both Cx43stp and Cx43sof showed reduced gap junction currents relative to wild type, which confirmed that the sof phenotype was caused by the defect in gap junction activity, as suggested by Hoptak-Solga et al. (22). On the other hand, Cx43stp hemichannels showed abnormally high (50-fold higher than wild type) conductance, whereas Cx43sof hemichannel conductance was almost the same as Cx43-WT. These results are consistent with the hypothesis that extremely high hemichannel activity causes the vertebral phenotype of the stp allele.
Although the mechanism by which gap junctions and hemichannels affect different bones is unknown, we assume that it is related to the difference in the bone forming process between vertebrae and fin bones. Fins are mechanically supported by fin rays arrayed in parallel. Each fin ray is composed of short segments of bone called lepidotrichia. Each segment is almost the same size along the AP axis (for tail fins). When the fin grows, a new fin ray segment is made at the tip of each fin ray. The newly made segments are nearly as long as the old segments along the AP axis and do not grow longer with time or with increasing fin length. On the other hand, vertebrae initially appear at ∼14 days post fertilization and continuously enlarge during the growth of the fish. Therefore, we infer that Cx43 gap junctions are involved in initial bone formation, and Cx43 hemichannels are involved in later bone growth.
One proposed mechanism of mechanical stress sensing by osteocytes is as follows. When bones are exposed to mechanical stress, they deform slightly, causing movement of the fluid surrounding the osteocytes. This movement of fluid causes shear stress that induces effusion of prostaglandin E2 through hemichannels (38). Prostaglandin E2 promotes osteoclast differentiation by down-regulating Osteoprotegerin (39), a protein that blocks RANKL-RANK interaction and promotes osteoclast differentiation.
We hypothesize that the stp allele reduces vertebral elongation by mimicking high mechanical stress. We found that cx43 expression in zebrafish vertebrae is highest in the growing edges (Fig. 4, E and E′). Because these parts of the vertebra contact the adjacent vertebra, we expect that mechanical stress is highest there. Therefore, one possible explanation for the effect of the stp mutation on vertebral shape is that it enhances hemichannel activity, allowing more prostaglandin E2 effusion. This would mimic the effects of high mechanical stress and lead to increased osteoclast differentiation and, hence, increased rates of bone breakdown at the edges of the centrum.
The major finding of our study is that there are two distinct functions of Cx43 in bone formation, one of which affects early development only and one of which affects subsequent growth only. These two functions correlate with differences in the electrophysiological properties of gap junctions and hemichannels among cx43 alleles. We hypothesize that the differences in gap junction and hemichannel function among alleles cause their differing effects on bone development and growth. Future studies are needed to test this hypothesis in vivo.
Author Contributions
A. M., H. Y., S. K., and M. W. designed the experimental strategy. A. M. performed positional cloning experiments. A. M. and T. A. performed transgenic experiments. A. M., H. Y., S. K., and M. K. I analyzed bone development. M. W. and I. M. S. performed electrophysiological experiments and analyzed data. A. M. prepared the manuscript, and all authors commented on the manuscript.
Acknowledgments
We thank the Shimadzu Corporation and the Osaka Bioscience Institute for lending CT scanners. We also thank H. Shirota and C. Sato at Osaka University for technical assistance in the positional cloning experiment, and Dr. T. W. White at SUNY Stony Brook for technical advice on the oocyte clamp experiments.
This work was supported by Ministry of Education, Culture, Sports, Science, and Technology in Japan KAKENHI Grant 22127003 (to S. K.); by the Core Research for Evolutional Science and Technology (CREST), Japan Science and Technology Agency (to S. K.); and by Japanese Society for the Promotion of Science KAKENHI Grant 26291049 (to M. W.). The authors declare that they have no conflicts of interest with the contents of this article.
- ENU
- N-ethyl-N-nitrosourea
- AP
- anterior-posterior
- dpf
- days after fertilization
- DV
- dorsoventral
- TALEN
- transcription activator-like effector nucleases.
References
- 1.Attanasio C., Nord A. S., Zhu Y., Blow M. J., Li Z., Liberton D. K., Morrison H., Plajzer-Frick I., Holt A., Hosseini R., Phouanenavong S., Akiyama J. A., Shoukry M., Afzal V., Rubin E. M., et al. (2013) Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342, 1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gluhak-Heinrich J., Ye L., Bonewald L. F., Feng J. Q., MacDougall M., Harris S. E., and Pavlin D. (2003) Mechanical loading stimulates dentin matrix protein 1 (DMP1) expression in osteocytes in vivo. J. Bone Miner. Res. 18, 807–817 [DOI] [PubMed] [Google Scholar]
- 3.Schoenebeck J. J., Hutchinson S. A., Byers A., Beale H. C., Carrington B., Faden D. L., Rimbault M., Decker B., Kidd J. M., Sood R., Boyko A. R., Fondon J. W. 3rd, Wayne R. K., Bustamante C. D., Ciruna B., et al. (2012) Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet. 8, e1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonewald L. F. (2011) The amazing osteocyte. J. Bone Miner. Res. 26, 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caetano-Lopes J., Canhão H., and Fonseca J. E. (2007) Osteoblasts and bone formation. Acta Reumatol. Port. 32, 103–110 [PubMed] [Google Scholar]
- 6.Duplomb L., Dagouassat M., Jourdon P., and Heymann D. (2007) Concise review: embryonic stem cells: a new tool to study osteoblast and osteoclast differentiation. Stem Cells 25, 544–552 [DOI] [PubMed] [Google Scholar]
- 7.Harada S., and Rodan G. A. (2003) Control of osteoblast function and regulation of bone mass. Nature 423, 349–355 [DOI] [PubMed] [Google Scholar]
- 8.Komori T. (2006) Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 99, 1233–1239 [DOI] [PubMed] [Google Scholar]
- 9.Haffter P., Granato M., Brand M., Mullins M. C., Hammerschmidt M., Kane D. A., Odenthal J., van Eeden F. J., Jiang Y. J., Heisenberg C. P., Kelsh R. N., Furutani-Seiki M., Vogelsang E., Beuchle D., Schach U., et al. (1996) The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123, 1–36 [DOI] [PubMed] [Google Scholar]
- 10.Kimmel C. B., Miller C. T., and Moens C. B. (2001) Specification and morphogenesis of the zebrafish larval head skeleton. Dev. Biol. 233, 239–257 [DOI] [PubMed] [Google Scholar]
- 11.Neuhauss S. C., Solnica-Krezel L., Schier A. F., Zwartkruis F., Stemple D. L., Malicki J., Abdelilah S., Stainier D. Y., and Driever W. (1996) Mutations affecting craniofacial development in zebrafish. Development 123, 357–367 [DOI] [PubMed] [Google Scholar]
- 12.Piotrowski T., Schilling T. F., Brand M., Jiang Y. J., Heisenberg C. P., Beuchle D., Grandel H., van Eeden F. J., Furutani-Seiki M., Granato M., Haffter P., Hammerschmidt M., Kane D. A., Kelsh R. N., Mullins M. C., et al. (1996) Jaw and branchial arch mutants in zebrafish II: anterior arches and cartilage differentiation. Development 123, 345–356 [DOI] [PubMed] [Google Scholar]
- 13.Schilling T. F., Walker C., and Kimmel C. B. (1996) The chinless mutation and neural crest cell interactions in zebrafish jaw development. Development 122, 1417–1426 [DOI] [PubMed] [Google Scholar]
- 14.Clément A., Wiweger M., von der Hardt S., Rusch M. A., Selleck S. B., Chien C. B., and Roehl H. H. (2008) Regulation of zebrafish skeletogenesis by ext2/dackel and papst1/pinscher. PLoS Genet. 4, e1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper K. L., Oh S., Sung Y., Dasari R. R., Kirschner M. W., and Tabin C. J. (2013) Multiple phases of chondrocyte enlargement underlie differences in skeletal proportions. Nature 495, 375–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray R. S., Wilm T. P., Smith J., Bagnat M., Dale R. M., Topczewski J., Johnson S. L., and Solnica-Krezel L. (2014) Loss of col8a1a function during zebrafish embryogenesis results in congenital vertebral malformations. Dev. Biol. 386, 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huycke T. R., Eames B. F., and Kimmel C. B. (2012) Hedgehog-dependent proliferation drives modular growth during morphogenesis of a dermal bone. Development 139, 2371–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perathoner S., Daane J. M., Henrion U., Seebohm G., Higdon C. W., Johnson S. L., Nüsslein-Volhard C., and Harris M. P. (2014) Bioelectric signaling regulates size in zebrafish fins. PLoS Genet. 10, e1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iovine M. K., and Johnson S. L. (2000) Genetic analysis of isometric growth control mechanisms in the zebrafish caudal Fin. Genetics 155, 1321–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haffter P., Odenthal J., Mullins M. C., Lin S., Farrell M. J., Vogelsang E., Haas F., Brand M., van Eeden F. J., Furutani-Seiki M., Granato M., Hammerschmidt M., Heisenberg C. P., Jiang Y. J., Kane D. A., et al. (1996) Mutations affecting pigmentation and shape of the adult zebrafish. Dev. Genes Evol. 206, 260–276 [DOI] [PubMed] [Google Scholar]
- 21.Iovine M. K., Higgins E. P., Hindes A., Coblitz B., and Johnson S. L. (2005) Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev. Biol. 278, 208–219 [DOI] [PubMed] [Google Scholar]
- 22.Hoptak-Solga A. D., Klein K. A., DeRosa A. M., White T. W., and Iovine M. K. (2007) Zebrafish short fin mutations in connexin43 lead to aberrant gap junctional intercellular communication. FEBS Lett. 581, 3297–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sims K. Jr., Eble D. M., and Iovine M. K. (2009) Connexin43 regulates joint location in zebrafish fins. Dev. Biol. 327, 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoptak-Solga A. D., Nielsen S., Jain I., Thummel R., Hyde D. R., and Iovine M. K. (2008) Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration. Dev. Biol. 317, 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar N. M., and Gilula N. B. (1996) The gap junction communication channel. Cell 84, 381–388 [DOI] [PubMed] [Google Scholar]
- 26.Abascal F., and Zardoya R. (2013) Evolutionary analyses of gap junction protein families. Biochim. Biophys. Acta 1828, 4–14 [DOI] [PubMed] [Google Scholar]
- 27.Eastman S. D., Chen T. H., Falk M. M., Mendelson T. C., and Iovine M. K. (2006) Phylogenetic analysis of three complete gap junction gene families reveals lineage-specific duplications and highly supported gene classes. Genomics 87, 265–274 [DOI] [PubMed] [Google Scholar]
- 28.Becker D. L., Phillips A. R., Duft B. J., Kim Y., and Green C. R. (2016) Translating connexin biology into therapeutics. Semin. Cell Dev. Biol. 50, 49–58 [DOI] [PubMed] [Google Scholar]
- 29.Merrifield P. A., and Laird D. W. (2016) Connexins in skeletal muscle development and disease. Semin. Cell Dev. Biol. 50, 67–73 [DOI] [PubMed] [Google Scholar]
- 30.Bai D. (2016) Structural analysis of key gap junction domains-Lessons from genome data and disease-linked mutants. Semin. Cell Dev. Biol. 50, 74–82 [DOI] [PubMed] [Google Scholar]
- 31.Laird D. W. (2014) Syndromic and non-syndromic disease-linked Cx43 mutations. FEBS Lett. 588, 1339–1348 [DOI] [PubMed] [Google Scholar]
- 32.Pizzuti A., Flex E., Mingarelli R., Salpietro C., Zelante L., and Dallapiccola B. (2004) A homozygous GJA1 gene mutation causes a Hallermann-Streiff/ODDD spectrum phenotype. Hum. Mutat. 23, 286. [DOI] [PubMed] [Google Scholar]
- 33.Urasaki A., Morvan G., and Kawakami K. (2006) Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J. A., Somia N. V., Bogdanove A. J., and Voytas D. F. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakuma T., Hosoi S., Woltjen K., Suzuki K., Kashiwagi K., Wada H., Ochiai H., Miyamoto T., Kawai N., Sasakura Y., Matsuura S., Okada Y., Kawahara A., Hayashi S., and Yamamoto T. (2013) Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells 18, 315–326 [DOI] [PubMed] [Google Scholar]
- 36.Dahlem T. J., Hoshijima K., Jurynec M. J., Gunther D., Starker C. G., Locke A. S., Weis A. M., Voytas D. F., and Grunwald D. J. (2012) Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8, e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furutani K., Ohno Y., Inanobe A., Hibino H., and Kurachi Y. (2009) Mutational and in silico analyses for antidepressant block of astroglial inward-rectifier Kir4.1 channel. Mol. Pharmacol. 75, 1287–1295 [DOI] [PubMed] [Google Scholar]
- 38.Cherian P. P., Siller-Jackson A. J., Gu S., Wang X., Bonewald L. F., Sprague E., and Jiang J. X. (2005) Mechanical strain opens connexin 43 hemichannels in osteocytes: a novel mechanism for the release of prostaglandin. Mol. Biol. Cell 16, 3100–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suda K., Udagawa N., Sato N., Takami M., Itoh K., Woo J. T., Takahashi N., and Nagai K. (2004) Suppression of osteoprotegerin expression by prostaglandin E2 is crucially involved in lipopolysaccharide-induced osteoclast formation. J. immunol. 172, 2504–2510 [DOI] [PubMed] [Google Scholar]