Abstract
Background
Patients with low-grade glioma (LGG) have a relatively long survival, and a balance is often struck between treating the tumor and impacting quality of life. While lesions may remain stable for many years, they may also undergo malignant transformation (MT) at the time of recurrence and require more aggressive intervention. Here we report on a state-of-the-art multiparametric MRI study of patients with recurrent LGG.
Methods
One hundred and eleven patients previously diagnosed with LGG were scanned at either 1.5 T or 3 T MR at the time of recurrence. Volumetric and intensity parameters were estimated from anatomic, diffusion, perfusion, and metabolic MR data. Direct comparisons of histopathological markers from image-guided tissue samples with metrics derived from the corresponding locations on the in vivo images were made. A bioinformatics approach was applied to visualize and interpret these results, which included imaging heatmaps and network analysis. Multivariate linear-regression modeling was utilized for predicting transformation.
Results
Many advanced imaging parameters were found to be significantly different for patients with tumors that had undergone MT versus those that had not. Imaging metrics calculated at the tissue sample locations highlighted the distinct biological significance of the imaging and the heterogeneity present in recurrent LGG, while multivariate modeling yielded a 76.04% accuracy in predicting MT.
Conclusions
The acquisition and quantitative analysis of such multiparametric MR data may ultimately allow for improved clinical assessment and treatment stratification for patients with recurrent LGG.
Keywords: cancer imaging, low-grade glioma, malignant transformation, magnetic resonance, spectroscopy
Infiltrating low-grade gliomas (LGGs) are a class of terminal central nervous system tumors that comprise malignant neuroglia. Histopathological diagnosis of tumor grade is performed using criteria set by the World Health Organization (WHO) and is based on factors that include nuclear atypia, proliferative capacity, tumor neovascularization, and necrosis.1 The clinical outcome for patients with LGG is variable, with some lesions following a more indolent disease course, while others recur more rapidly and often after undergoing malignant transformation (MT) to a higher grade.2 Lesions that have thus transformed to a grade III anaplastic glioma or grade IV secondary glioblastoma multiforme (GBM) are managed with additional, more aggressive treatments. To date, little is known regarding the nature of recurrent disease, and the most significant prognostic factors for patients diagnosed with LGG are the presence of somatic driver mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2) oncogenes and the codeletion of the 1p and 19q chromosomal arms, which have been associated with increased survival and sensitivity to the treatment given.2–5
IDH mutations have been implicated as an initiating event in gliomagenesis and are key to reprogramming the tumor epigenome and metabolome, largely through neomorphic production and accumulation of 2-hydroxyglutarate.6 Given uncertainties regarding the effectiveness of options available for treating LGGs, IDH mutations have garnered significant attention as a targetable therapeutic pathway, and there are several novel therapies on the horizon.7–9 Until these therapeutics become available, the clinical mainstay of treatment for patients with LGG consists of surgical resection, with radiation therapy and alkylating chemotherapy being typically reserved for recurrences that were either subtotally resected or have undergone MT.
MRI is an integral component of brain tumor diagnosis and monitoring. Recent advances in state-of-the-art techniques have enabled the noninvasive assessment of tumor morphology and physiology, with primary GBM having been studied most extensively. Although the ability to noninvasively detect MT would be of significant clinical interest for diagnosis and treatment planning, few studies have focused on addressing this question. Multiparametric MRI holds significant promise for comprehensively characterizing the structural and physiological properties of the tumor. We hypothesize that these advanced imaging techniques may also provide improved characterization of MT in patients with recurrent LGG.
The objective of this study was to establish multiparametric MRI profiles of patients with tumors prior to image-guided surgery in order to relate metrics obtained from these methodologies to histopathological grade. Volumetric regions with abnormal imaging features were calculated to provide a robust assessment of the entire recurrent tumor lesion. Regions of viable tumor were targeted for image-guided tissue sampling in order to associate in vivo parameters with histopathological features and to strengthen our understanding of the link between glioma tumor biology and parameters from noninvasive imaging.
Materials and Methods
Patient Accrual
Institutional review board approval was obtained to study patients who had an original pathological diagnosis of WHO grade II glioma. Patients were recruited immediately prior to surgical resection for a suspected recurrence, when MT to a higher grade is often observed.
In vivo Multimodal MRI and Spectroscopy
MR examinations were performed on either a 1.5 T or 3 T EXCITE GE Signa Echospeed scanner (GE Healthcare Technologies) using an 8-channel phased-array headcoil (MRI Devices). Standard anatomic imaging included T2-weighted (fluid attenuated inversion recovery [FLAIR] and fast spin echo) as well as T1-weighted pre- and post-gadolinium (Gd) contrast images. Diffusion weighted imaging (DWI) was obtained in the axial plane with 6 gradient directions and twofold acceleration with sensitivity encoding parallel imaging (repetition time [TR]/echo time [TE] = 1000/108 ms, voxel size = 1.7 × 1.7 × 3 mm3, b = 1000 s/mm2). Dynamic susceptibility contrast perfusion weighted imaging (PWI) was obtained with a 5 mL/s bolus injection of 0.1 mmol/kg body weight Gd-DTPA (diethyltriamine pentaacetic acid) acquired using a series of T2*-weighted echo-planar images (TR/TE/flip angle = 1250–1500/35–54 ms/30–35 degrees, 128 × 128 matrix, slice thickness = 3–5 mm, 7–15 slices with 60–80 timepoints) before, during, and after injection. Lactate-edited 3D proton MR spectroscopic imaging (MRSI) was applied using point-resolved spectroscopic selection for volume localization and very selective saturation pulses for lipid signal suppression (approximate excited volume = 80 × 80 × 40 mm3, TR/TE = 1104/144 ms, overpress factor = 1.5, field of view = 16 × 16 × 16 cm3, nominal voxel size = 1 × 1 × 1 cm3, flyback echo-planar readout gradient in the superior-to-inferior direction, 988 Hz sweep width, and 712 dwell points).10
Postprocessing of MR Exam
The in vivo data were de-identified and transferred to a local Linux workstation. Software developed in house was applied to estimate relevant DWI, PWI, and MRSI parameters and normalize between field strengths using estimates from normal appearing brain tissue (NABT). Normalization factors were defined for each study using the mode of the histogram from the whole brain minus the region of hyperintensity on T2-weighted FLAIR images, which was previously shown to be representative of values in NABT. Maps of the normalized apparent diffusion coefficient (ADC) were generated on a voxel-by-voxel basis according to a published algorithm.11 Perfusion datasets were nonrigidly aligned using the VTK CISG software package.12 Normalized cerebral blood volume (CBV), percent ΔR2* signal recovery (%-REC), ΔR2* peak height (PH), and recirculation factor were calculated for each voxel using software developed by our lab. CBV intensities and PH parameters were obtained by fitting the dynamic perfusion data by a modified gamma-variate function with a recirculation parameter.13 Peak height and percent recovery values were also estimated using a simplified nonparametric procedure.14
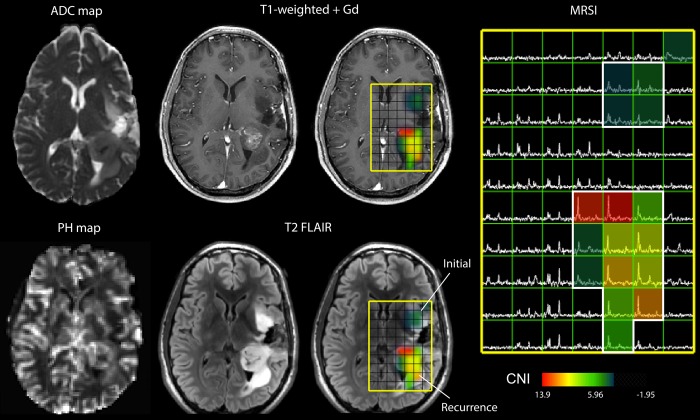
Lactate-edited MRSI data were reconstructed and the signal from the individual channels combined to quantify total choline (tCho), N-acetyl-aspartate (NAA), creatine (Cr), lactate (Lac), and lipid (Lip) levels. The values of the Cho-to-NAA index (CNI) and the Cho-to-Cr index (CCRI) were generated from a linear regression–based algorithm, which represents changes in Cho and NAA or Cr levels compared with voxels in NABT. Excess choline (exCho) and excess creatine (exCr) values were calculated using the formulas (tCho-[tCho/NAA]NABT * NAA)/tCho NABT and (Cr-[Cr/NAA] NABT * NAA)/Cr NABT,15 respectively. Imaging data were aligned to the post-Gd scan using the Linear Image Registration Tool of Functional Magnetic Resonance Imaging of the Brain. Anatomic imaging was resampled for overlay with DWI, PWI, and MRSI, and the software package by SIVIC (Spectroscopic Image Visualization and Computing) was used to select target locations for intraoperative tissue sampling.16 An example of the multimodal MRI data is shown in Fig. 1.
Fig. 1.
Multimodality MRI exam of a subject with a recurrent LGG that had undergone MT. Neurosurgical tissue targets were planned based on regions of suspected tumor using additional functional MR techniques. The imaging revealed a heterogeneously enhancing region of recurrent tumor situated in the left posterior temporal and parietal white matter. An additional, masslike non–contrast enhancing region of residual tumor was also seen in the left insular white matter. This lesion was consistent with residual low-grade neoplasm with marked differences in ADC, PH, and CNI (abnormal regions highlighted in white).
Tissue Sample Acquisition
Tissue sample targets were planned for each patient based on surgically accessible regions of abnormally decreased ADC, decreased %-REC, and increased PH and CBV, as well as elevated CNI, which are expected to represent viable, cellular regions of tumor with elevated proliferation and neovascularization. These locations were designated as 5-mm-diameter spherical targets on coregistered MRIs using Brainlab surgical navigation software. Intraoperative navigation guided the neurosurgeons to these locations, and tissue samples were excised if possible to do so safely. A research assistant was present in the operating room to obtain 3D screenshots and coordinates of the precise location where the tissue sample was removed. Samples were immediately bisected, with half being fixed in 10% zinc formalin, dehydrated by graded ethanols, and embedded in Paraplast Plus wax (McCormick Scientific) using standardized techniques for tissue processing and immunohistochemistry, and the other half being snap-frozen in liquid nitrogen and stored at 80°C for 1H high-resolution magic angle spinning spectroscopy. The results of the ex vivo metabolic analysis of a subset of these patients have been published by our group in previous manuscripts.17–19
Histopathology and IDH Analysis
Tissue samples were reviewed and scored for standard WHO II criteria by a board-certified neuropathologist. Antibodies used in the assessment of the samples and histological scoring are described in the Supplementary material.20
Analysis of MR Parameters
Volumetric regions of interest (ROIs) were defined manually for the T2 hyperintensity (T2ALL), contrast enhancement, necrosis, and nonenhancement throughout the entire tumor lesion. In-house software was applied to quantify normalized intensity values at a patient level within each T2 lesion and to evaluate the 10th percentile, median, and 90th percentile values of parameters estimated from DWI, PWI, and MRSI. T1 difference subtraction images were obtained through registration and subtraction of the T1 precontrast images from the T1 postcontrast images after normalization to make their intensities consistent.
To assess variations in the spatial extent of the anatomic lesion, the volumes of regions with intensities in the T1 post-Gd image from the contrast enhancing lesion ROI >1.2× NABT (T1c12) or >1.2× NABT in the image obtained after T1 difference subtraction (T1s12) were evaluated. Additionally, we analyzed volumes of regions with abnormal ADC, the normalized ADC <1.5 or <1.25 times NABT in the T2ALL (ADC15, ADC125); the perfusion volumes of normalized CBV >2 or >3 (CBV2, CBV3), nonparametric PH >2 or >3 (PH2, PH3); spectroscopy volumes of CNI >2 or >3 within the T2ALL (CNI2t, CNI3t), and the region with CNI >2 or >3 that overlapped with T2ALL (CNI2p, CNI3p).
To compare imaging parameters with histological findings, normalized intensity values were calculated at the individual tissue target locations using 5 mm spherical ROIs. Additional perfusion analysis is presented in the Supplementary material.21
Heatmap Generation
Histopathology data were categorized by histology and grade and imported into Gitools version 2.2.1 (www.gitools.org),22 and a linear, hierarchical clustering algorithm was performed within grade as well as within grade and histological subtype. Heatmaps of abnormal imaging volumes were similarly generated based on the difference from the median values within each parameter and normalized by the 90th percentile for relative visualization.
Statistical Analysis
A Wilcoxon rank sum test was used to assess the statistical significance of imaging parameters between lesions that had remained grade II and those that had undergone MT. Kaplan–Meier curves were generated using Stata version 11.23 If the curves did not cross, the association of parameter and outcome was assessed via the log-rank test, otherwise the Tarone–Ware test was employed. Statistical significance was assessed at P < .05.
In order to identify variables that could play a role in predicting MT, we built logistic regression models implementing a forward/backward selection process using the minimum Akaike information criterion in JMP Pro 11.0. Among anatomic, diffusion, perfusion, and spectroscopic modalities there were a total of 80 variables included in this selection process, which included both volumetric and intensity measurements. This process indicated around a dozen variables for a logistic model, which decreased the smallest outcome group to 40–50 patients due to missingness in the variables. To maximize the number of patients included, we constrained the final predictive models to at most 4 variables. We chose models for which (i) variables came from multiple different imaging modalities and (ii) prediction error rate estimations via leave-one-out cross-validation on our patient dataset were minimized. A custom R script (version 3.1.2)24 was used to iterate over the different variable combinations to determine several models that satisfied these criteria. The model presented in the results had the lowest overall error rate of those generated.
The Kendell's-tau rank correlation test was used to assess pairwise correlations between tissue sample level histopathology and imaging parameters. The Holmes step-down algorithm was used to determine a conservative cutoff of P < .00008 to account for multiple testing in our correlation analysis. Network analysis and visualization were performed using Cytoscape version 3.2.0 (www.cytoscape.org).25 Due to the exploratory nature of this type of network analysis, a cutoff value of P < .05 was applied for that component.
Results
Characterization of Patient Population with Recurrent Low-Grade Glioma
A total of 120 patients with recurrent LGG were initially recruited into the study. Nine patients were excluded due to diagnosis inconsistent with LGG, leaving 111 patients available for analysis. Ten patients received scans at 2 distinct recurrence timepoints, giving a total of 121 scans for analysis. Ninety percent of scans (109/121) were performed using a 3T scanner, while 10% (12/121) were scanned at 1.5 T. The population included 54 females and 57 males, with a median age of 36 years (range 15–70) at their original diagnosis and 43 years (range 18–70) at the current surgery. Age was not found to be associated with tumor grade. The median time from diagnosis to the surgery being considered was 2244 days (6.1 y); and for the patients who had records available, 26% had received prior radiation therapy, while 43% had been treated with temozolomide and 25% with other experimental agents. For the majority of cases, there had been one or more prior recurrences, but the surgery considered was the first since their original diagnoses. It was observed that 59% of scans were of patients who had undergone MT to a higher-grade lesion: 50 patients (41%, 92 tissue samples) remained grade II, while 55 patients (46%, 107 tissue samples) had upgraded to anaplastic glioma (grade II → III) and 16 patients (13%, 37 tissue samples) to secondary GBM (grade II → IV). There were 55 subjects with astrocytoma, 45 with oligodendroglioma, and 21 with mixed oligoastrocytoma histological subtypes (Table 1). IDH mutation status was assessed in 100 patients with sufficient size tissue, with 19 patients (19%) having lesions that were IDH wild-type and 81 patients (81%) having lesions that were mutated. Deletion status for 1p-19q had been evaluated for 61 patients, of whom 33 (54%) were codeleted and 28 (46%) were not. A total of 52 patients had p53 mutation status assessed, among whom 37 (71%) were mutated.
Table 1.
Recurrent LGG presurgical scans by grade and histopathological subtype
| Grade | Total Patient Scans (samples) | Astrocytoma (samples) | Oligodendroglioma (samples) | Oligoastrocytoma (samples) | |
|---|---|---|---|---|---|
| Acquired | All | 130 (251) | – | – | – |
| Excluded | All | 9 (15) | – | – | – |
| Analyzed | All | 121 (236) | 55 (105) | 45 (85) | 21 (46) |
| II | 50 (92) | 9 (16) | 29 (52) | 12 (24) | |
| II→III | 55 (107) | 30 (52) | 16 (33) | 9 (22) | |
| II→IV | 16 (37) | 16 (37) | – | – |
The population comprised astrocytoma, oligodendroglioma, and mixed oligoastrocytoma histological subtypes. Nine patients and 15 tissue samples were excluded due to their having diagnoses other than LGG. Ten of the 121 patient scans were done at 2 distinct recurrences.
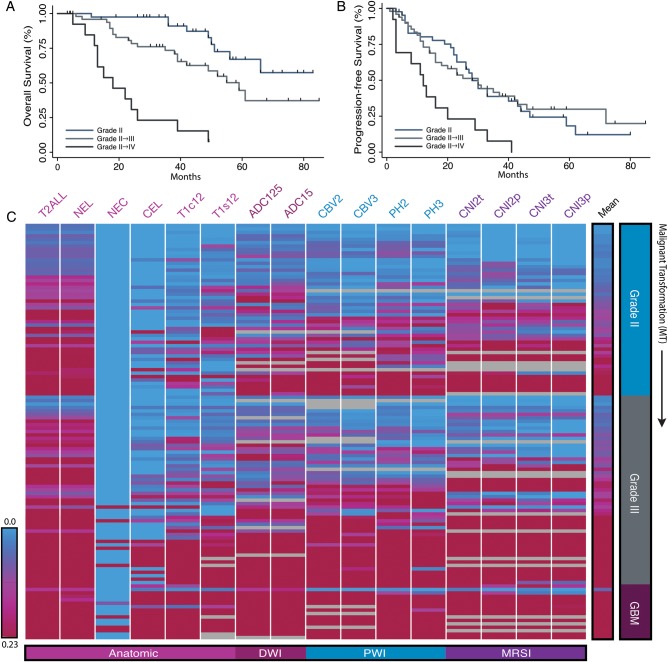
For patients who had clinical follow-up available (109/113), the median progression-free survival (PFS) from the time of the current surgery was 27 months (95% CI: 21–32 mo), with the time for subjects who remained grade II being 28 months (95% CI: 21–40 mo), those in the grade II → III cohort being 31 months (95% CI: 19–46 mo), and those in the grade II → IV cohort being 13 months (95% CI: 3–19 mo). PFS and overall survival (OS) curves are presented in Fig. 2. Although there was insufficient follow-up to assess the median OS for the cohort that remained grade II, the median OS for the grade II → III cohort was estimated to be 52 months (95% CI: 38 mo, with 25 subjects dead and 26 subjects censored) and for the grade II → IV cohort 22 months (95% CI: 12–26 mo and all 18 subjects having died).
Fig. 2.
Heatmaps of volumetric MR parameters and clinical outcome of recurrent LGG. Kaplan–Meier curves generated from (A) OS and (B) PFS demonstrated statistically significant differences among WHO grades for OS between grade II and grade II → III (P = .006), grades II/II → III and GBM (P < .001), while PFS only distinguished GBM from grade II and grade II → III lesions (P < .001). (C) The heatmap generated from patient scans (rows) and hierarchical clustering revealed subgroups of grade II and grade III lesions that displayed abnormal imaging features similar to GBM. Zero volume is shown in blue and approximately twice the median normalized volume is shown in red. Header column denotes the mean parameter value. Gray cells denote cases where there were no data available. NEL, nonenhancing lesion; CEL, contrast enhancing lesion; NEC, necrosis.
Findings from Presurgery Anatomic MRI
The right-hand rows in the heatmap shown in Fig. 2C give a visual interpretation of the volumetric imaging data from individual subjects, and Fig. 3 shows summary parameters separated by grade and histological subtype. Recurrence with larger T2 lesions was associated with a higher probability of MT (see Supplementary Table 1). The median volume of the T2 lesions for the 47 subjects who remained grade II was 15.6 cc; for the 47 subjects in the grade II → III cohort, 30.9 cc; and for the 18 subjects in the grade II → IV cohort, 69.7 cc. Only 8 of the lesions had visually identifiable regions of necrosis, which corresponded to 4 lesions with a diagnosis of grade III oligodendroglioma and 4 lesions with a diagnosis of GBM.
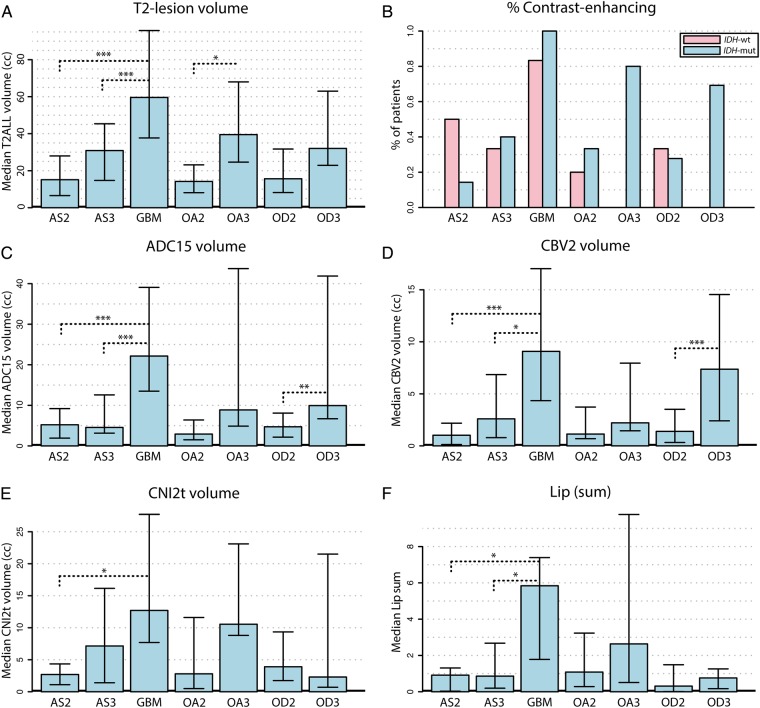
Fig. 3.
Differences in lesions that had undergone MT. The bar plots represent median differences of volumetric imaging parameter values among the various histological grades and subtypes. Error bars are representative of the 25th and 75th percentiles. Statistical significance was assessed using a Wilcoxon rank sum test at P < .05 (*), P < .01 (**), and P < .005 (***).
Of the cohort that remained grade II, only 13/47 (28%) were characterized as having small regions of enhancement on postcontrast T1-weighted images, while 23/47 (49%) of the grade II → III cohort were enhancing, and all of the lesions for the grade II → IV cohort were enhancing. Visual assessment of the volume of contrast enhancement, as well as analysis of intensities in the postcontrast T1-weighted image and in T1 subtraction images indicated that these regions were relatively small compared with the T2 lesion but were larger for cases that had undergone MT. Oligodendrogliomas tended to have modest T2 lesion volumes but were more likely to have regions of contrast enhancement (69% of the grade II → III oligodendrogliomas compared with 37% of the grade II → III astrocytomas; see Fig. 3A and B). Univariate statistical analysis indicated that several parameters derived from the histograms of intensities within the T2 lesion were also significantly different between lesions that remained grade II and those that had undergone MT. These included the median and 90th percentile values from T1 subtraction and T1 postcontrast images (as presented in Supplementary Table 1).
Findings from Presurgery Diffusion Weighted MRI
Analysis of diffusion parameters within the T2 lesion indicated that the 10th percentile of the ADC and radial diffusion (λ-rad) were significantly lower for lesions that had undergone MT. While this trend held for all 3 histological subtypes, it was noted that oligodendroglioma lesions had lower median ADC values than astrocytomas. As can be seen in Supplementary Table 1, the larger overall T2 lesion volumes and lower ADC values associated with MT translated to the diffusion volume metrics (ADC15, ADC125) for these lesions being significantly larger relative to lesions that had remained grade II. When separated based on histological subtype, it was noted that astrocytomas transforming to grade III had median ADC15 volumes that were similar to those remaining grade II (Fig. 2C). The heatmap in Fig. 2 also reveals that a subset of the lesions that remained grade II had similar diffusion volumes as their higher-grade counterparts.
Findings from Presurgery Perfusion Weighted MRI
As shown in Supplementary Table 1, the 90th percentile CBV values and 75th percentile nonparametric PH values were significantly higher for lesions that had undergone MT. When considering the volumes of regions with elevated CBV and PH within the T2 lesion, it was clear that lesions transforming to GBM and to grade III oligodendroglioma also had larger abnormal perfusion volumes (CBV2, CBV3, PH2, and PH3; Fig. 2D). Interestingly, as can be seen in the heatmap, the same group of patients who had remained grade II and who had larger diffusion volumes also had larger abnormal perfusion volumes.
Findings from Presurgery Spectroscopic Imaging Data
Although there were fewer patients for whom presurgery spectroscopic imaging data were acquired (83/113), there were a number of metabolic intensity parameters significantly different for lesions that had undergone MT. These included the median, 90th percentile, and maximum values of exCho; 90th percentile and maximum values of CCRI; maximum values of CNI and tCho levels; and the minimum and 10th percentile values of Cr (Supplementary Table 1). The volumes of abnormal metabolism (CNI2t, CNI2p, CNI3t, and CNI3p) were all assessed as significantly larger for cases that had undergone MT. As was the case with abnormal perfusion volumes, the same group of subjects who had larger abnormal diffusion volumes also had larger metabolic lesions (Fig. 3). When evaluated by histological subtype, oligodendrogliomas tended to have lower CNI, Lac, and Lip than astrocytomas and oligoastrocytomas. The subjects with lesions that had transformed to GBM had the highest integrated CNI, Lac, and Lip values in the CNI2t region (Fig. 2F).
Multivariate Analysis of Imaging Parameters
In an effort to provide predictive modeling that utilized the strongest statistical differentiators of MT, we developed several multivariate logistical regression models utilizing parameters that spanned multiple MR modalities and both volumetric and intensity parameter measurements. Although there were several models that provided comparable results, the one with the highest overall accuracy included estimates of the 10th percentile of Cr in regions with CNI >2; the volume of ADC >1.5× that of NABT divided by the T2ALL volume; the median ADC in the T2ALL region; and the 10th percentile fast spin echo in the T2ALL. This model had a predictive accuracy of 76.04%, with a 10.42% false-negative and 13.54% false-positive rate (Supplementary Fig. 1). Lesions that were misclassified corresponded to 17% of the astrocytomas, 20% of the oligoastrocytomas, and 35% of the oligodendrogliomas, suggesting that there may be differences among the histological subtypes which influence the accuracy of the model.
Differences in Histopathological Parameters with Grade and Subtype
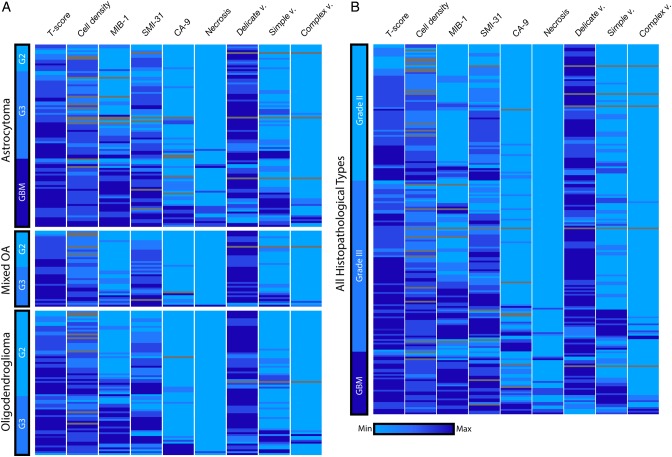
To examine the histopathological features associated with recurrent LGG, we produced a heatmap based on the neuropathology assessment of each of the image-guided tissue samples (Fig. 4). As expected, tissue samples that had undergone MT were characterized by increased measures of relative tumor content (T-score), proliferation (MIB-1), and axonal disruption (SMI-31). Interestingly, several image-guided tissue samples from WHO grade II oligodendrogliomas contained very elevated MIB-1 scores of 10%–20%, two of which were >20%. Thus, image-guided sampling identified tumor regions with MT that clinical sampling did not. Anaplastic glioma and secondary GBM samples were characterized by increased hypoxic conditions (carbonic anhydrase 9), the presence of neovascularization, and necrosis, which was found primarily in GBM samples.
Fig. 4.
Heatmap of image-guided tissue samples. (A) To assess the biological features within each distinct histology, the neuropathology data were separated by glioma subtype and used hierarchical clustering within each grade. Based on image-guided targeting criteria, several grade II samples were identified that had an unusually high MIB-1 score, and 2 grade III astrocytomas or mixed oligoastrocytomas (OA) had necrosis present. Although few in number, these examples highlight the utility of image guidance to improve tumor sampling. A portion of grade III oligodendroglioma patients had elevated carbonic anhydrase 9 (CA-9) scores of hypoxia. (B) When evaluating the entire mixed population of glioma histologies, we observed a decrease in normal, delicate vasculature (delicate v.), and increases in the presence of simple and complex neovascularization (simple v. and complex v.) were noted within a subset of tumors that had undergone MT.
Correlation of Histopathology and In vivo Imaging Parameters
Analysis of imaging intensities that were calculated at the spatial location of the tissue sample excision with histopathology scores provided insight into the biological underpinnings of specific MR-based imaging measurements (Supplementary Table 2). ADC values from DWI were inversely correlated with cell proliferation. Nonlinear CBV and nonparametric PH were correlated with cell density, mitotic figures, and hypoxia. Metrics of T1 imaging (T1c, T1s) were also correlated with mitotic figures and simple vascular hyperplasia, while T1 contrast enhancement was correlated with necrosis.
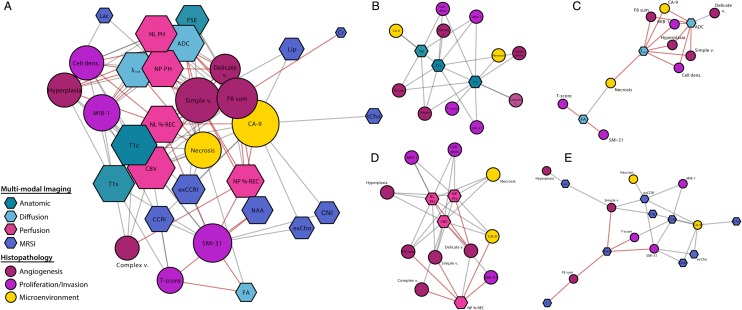
Visual network maps that were generated based on Kendell's-tau correlations are presented in Fig. 5. High levels of connectivity and decreased average edge length within diffusion (λ-rad, ADC), perfusion (PH, CBV, %-REC), and anatomic T1 imaging metrics (T1c, T1s) were observed, as well as MRSI measures of excess Cho-to-Cr indices (exCCRIs) in the complete multimodal imaging and histopathology network (Fig. 5A). Due to increased centrality and connectivity in these parameters, these metrics may provide an increasingly objective assessment of tumor histopathology.
Fig. 5.
Network linkage map of histopathology and advanced imaging of image-guided tissue samples. Nodes were color coded using meta-labels of angiogenesis, proliferation/invasion, and microenvironment for histopathology parameters as well as by modality for imaging parameters. Node size corresponds with average shortest path length. Imaging nodes are presented as hexagons and histopathology nodes as circles. Although imaging and histopathology parameters were intracorrelated, only edges between histopathology and imaging were generated for clarity of network visualization. Red connections denote negative correlation.
To better assess the specific nature of the various imaging modalities, we generated modality-specific networks (Fig. 5B–E). The anatomic imaging metrics were highly central and shared edges with several pathological measures of tumor; however, the T1 imaging correlated more specifically with necrosis and vascularization and the fast spin echo with hypoxia. The diffusion network consisted of 2 major clusters, with the larger containing both ADC and λ-rad and being associated with several measures of proliferative tumor. The presence of normal, delicate vasculature was correlated with ADC values, while necrosis shared a node with λ-rad and the second, smaller cluster around fractional anisotropy, T-score, and SMI-31. In the perfusion network, nonlinear CBV held common edges with all histopathology parameters, while PH and %-REC were each correlated with several distinct nodes. In the metabolic imaging correlation network, measures of CCRI and exCCRI had the highest levels of connectivity with histopathology, and increased dispersion and specificity were observed among the other metabolites.
Discussion
This study demonstrated the feasibility of utilizing multiparametric MRI to identify patients with recurrent LGG whose lesions had undergone MT. Quantitative analysis of anatomic, diffusion, perfusion, and metabolic images provided parameters that described lesion volumes and the magnitude of abnormal intensities relative to normal appearing brain within the same individual. A bioinformatics approach was applied for data visualization, with heatmaps and networks proving helpful in summarizing and interpreting variations in the relatively large number of parameters and complex information contained within the data. The tools that were developed and the metrics that they produce may be helpful for evaluating temporal changes within the lesion during routine patient follow-up or when surgical resection is not feasible.
When considering all patients as a single group, we found many multivariate models with similar accuracy for predicting grade II versus transformation of grade II → III or of grade II → IV. The model with the lowest error rate assessed by cross-validation had an accuracy of 76%. Given that there were differences in imaging and histological parameters among different subtypes and that transformation to grade IV had the biggest impact upon survival, a second cohort of patients are now being accumulated to allow further tests of these models and exploratory analyses that include partitioning patients into more specific subgroups.
The imaging heterogeneity found within this patient population illustrates the challenges that exist for clinicians in interpreting imaging findings from a subject with suspected recurrence, where the results obtained are reflective of both inherent and therapy-driven features of the tumor.26 Based on the heterogeneity of the histological findings for the image-guided tissue samples, it is clear that surrogate noninvasive markers of proliferation, angiogenesis, and invasion are likely to be important for making decisions about both upgrade status and the effectiveness of different treatments. The correlations between imaging and histological parameters observed in our study provide strong evidence that integrating metrics derived from multiparametric imaging can be helpful for assessing spatial heterogeneity within the lesion. Of particular interest is the ability to use maps of CBV, ADC, and CNI from the presurgery imaging examination to target tissue sampling to regions that are likely to be the most malignant. The criteria that were used in our study selected regions of CBV >2, ADC <1.5, and CNI >3. While it was not always possible to obtain tissue from these exact locations, the relatively high yield with positive tumor scores (201/212) indicates that this was a successful strategy.
Our previous work in patients with high-grade gliomas showed that several of the imaging metrics used in the current study were associated with PFS and OS.15 It is too early to say whether the subset of lesions that remained grade II but had similar imaging characteristics as those that upgraded had worse OS. Future studies will investigate this possibility. Although the current definition of tumor grade based upon WHO II criteria has been driving decisions about when to use more aggressive therapy, recent studies have suggested that molecular markers such as IDH and TERT mutations provide a better assessment of prognosis.27,28 Because the majority (79%) of the patients in our study were IDH mutated, it was not possible to determine whether IDH status was an important factor in driving outcome. Future studies will be designed to include a full analysis of genomic and epigenetic markers that have been shown to be relevant to the natural history and response to treatment for LGG. This will shed light on the nature of the clonal outgrowths present in recurrent lesions and how they influence imaging. The long-term goal of this and future studies is to improve our understanding of the evolution of malignant glioma and the clinical management of such patients.
Supplementary Material
Funding
This work was supported as a project in the NCI Brain Tumor SPORE grant, number P50CA097257.
Supplementary Material
Acknowledgments
We would like to acknowledge support from the Brain Tumor Research Center at UCSF in collecting and analyzing the tissue samples, as well as from staff in the Margaret Hart Surbeck Laboratory for Advanced Imaging. We would particularly like to express our gratitude to J. Kurhanewicz, S. Ronen, D. Vigneron, J. Costello, J. Crane, and N. Strauli for their technical assistance during this project.
Conflict of interest statement. None declared.
References
- 1.Riemenschneider MJ, Reifenberger G. Molecular neuropathology of low-grade gliomas and its clinical impact. Adv Tech Stand Neurosurg. 2010;35(1):35–64. [DOI] [PubMed] [Google Scholar]
- 2.Grier JT, Batchelor T. Low-grade gliomas in adults. Oncologist. 2006;11(6):681–693. [DOI] [PubMed] [Google Scholar]
- 3.Yan H, Parsons DW, Jin G et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houillier C, Wang X, Kaloshi G et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75(17):1560–1566. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Chou AP, Chen W et al. Overexpression of isocitrate dehydrogenase mutant proteins renders glioma cells more sensitive to radiation. Neuro Oncol. 2013;15(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang L, White DW, Gross S et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kernytsky A, Wang F, Hansen E et al. IDH2 mutation-induced histone and DNA hypermethylation is progressively reversed by small-molecule inhibition. Blood. 2015;125(2):296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turcan S, Fabius AW, Borodovsky A et al. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT inhibitor decitabine. Oncotarget. 2013;4(10):1729–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yen KE, Bittinger MA, Su SM et al. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29(49):6409–6417. [DOI] [PubMed] [Google Scholar]
- 10.Park I, Chen AP, Zierhut ML et al. Implementation of 3T lactate-edited 3D 1H MR spectroscopic imaging with flyback echo-planar readout for gliomas patients. Ann Biomed Eng. 2011;39(1):193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson. 1996;111(3):209–219. [DOI] [PubMed] [Google Scholar]
- 12.Hartkens T, Rueckert D, Schnabel JA et al. VTK CISG registration toolkit: an open source software package for affine and non-rigid registration of single- and multimodal 3D images. Leipzig: Springer-Verlag; 2002. http://www.bvm-workshop.org/BVM2002. [Google Scholar]
- 13.Lee MC, Cha S, Chang SM et al. Dynamic susceptibility contrast perfusion imaging of radiation effects in normal-appearing brain tissue: changes in the first-pass and recirculation phases. J Magn Reson Imaging. 2005;21(6):683–693. [DOI] [PubMed] [Google Scholar]
- 14.Lupo JM, Cha S, Chang SM et al. Dynamic susceptibility weighted perfusion imaging of high-grade gliomas: characterization of spatial heterogeneity. Am J Neuroradiol. 2005;26(6):1446–1454. [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Lupo JM, Parvataneni R et al. Survival analysis in patients with newly diagnosed glioblastoma using pre- and postradiotherapy MR spectroscopic imaging. J Neurooncol. 2013;15(5):607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crane JC, Olson MP, Nelson SJ. SIVIC: open-source, standards-based software for DICOM MR spectroscopy workflows. Int J Biomed Imaging. 2013;2013:169526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkhaled A, Jalbert LE, Phillips JJ et al. Magnetic resonance of 2- hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4(116):116ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elkhaled A, Jalbert L, Constantin A et al. Characterization of metabolites in infiltrating gliomas using ex vivo ¹H high-resolution magic angle spinning spectroscopy. NMR Biomed. 2014;27(5):578–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Constantin A, Elkhaled A, Jalbert L et al. Identifying malignant transformations in recurrent low grade gliomas using high resolution magic angle spinning spectroscopy. Artif Intell Med. 2012;55(1):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capper D, Zentgraf H, Balss J et al. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009;118(5):599–601. [DOI] [PubMed] [Google Scholar]
- 21.Lupo JM, Wen Q, Phillips JJ et al. Weighted-average model curve preprocessing strategy for quantification of DSC perfusion imaging metrics from image-guided tissue samples in patients with brain tumors. Proc Intl Soc Mag Reson Med. 2015;4392. [Google Scholar]
- 22.Perez-Llamas C, Lopez-Bigas N. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS ONE. 2011;6(5):e19541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.StataCorp. Stata Statistical Software: Release 11. College Station, TX: StataCorp LP; 2009. [Google Scholar]
- 24.R Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson BE, Mazor T, Hong C et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckel-Passow JE, Lachance DH, Molinaro AM et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.