Abstract
Peptides display many characteristics of efficient imaging agents such as rapid targeting, fast background clearance, and low non-specific cellular uptake. However, poor stability, low affinity, and loss of binding after labeling often preclude their use in vivo. Using the glucagon-like peptide-1 receptor (GLP-1R) ligands exendin and GLP-1 as a model system, we designed a novel alpha helix stabilizing linker to simultaneously address these limitations. The stabilized and labeled peptides showed an increase in helicity, improved protease resistance, negligible loss or an improvement in binding affinity, and excellent in vivo targeting. The ease of incorporating azidohomoalanine in peptides and efficient reaction with the dialkyne linker enables this technique to potentially be used as a general method for labeling alpha helices. This strategy should be useful for imaging beta cells in diabetes research and in developing and testing other peptide targeting agents.
Keywords: exendin, glucagon like peptide 1 receptor, stapled peptides
Introduction
The ability to image and observe cellular phenomena both in vivo and in vitro plays a crucial role in understanding disease progression and treatment response1, 2. Molecular imaging agents enable investigators to probe the dynamics of specific biologic interactions in normal and pathological conditions. In principle, peptides are ideal imaging agents due to their low molecular weight3, rapid clearance from background tissues, and ability to mimic protein-protein interactions for high binding specificity4. However, these agents suffer from several problems including poor protease stability, low affinity from lack of a stable conformation, and difficulty in labeling with fluorescent and/or radioactive probes without disrupting binding5. Due to recent advances in synthesis techniques, there is renewed interest in using stabilized alpha helices with improved protease resistance and binding affinity for targeting both intracellular6, 7 and extracellular8 proteins. Side chain cross-linking reactions to promote an alpha helix conformation include olefin metathesis9, copper catalyzed azide-alkyne reactions10, 11, lactam ring formation12, and disulfide bond formation13, 14 among others8.
In many cases, stabilizing the alpha helix increases helicity, improves binding affinity, and/or increases protease resistance15. However, this is not universally true since the introduction of the side chain cross linker necessarily introduces two mutations into the sequence and the linker addition itself, which can each change intra- and inter- molecular interactions16–18. The sequence mutations and cross linker can impart both positive and negative contributions to the free energy of helix formation and free energy of binding, making the net impact challenging to predict a priori from structural considerations alone. Typically biophysical characterization is necessary to test the overall effect, which we have done for the agents presented here.
In the context of imaging agent development, while these stabilizing structures have the potential to improve protease stability and binding affinity by locking molecules in a helical conformation, this exacerbates the problem of labeling the peptide since multiple modifications must now be done without disrupting the binding interface. To address this issue, we synthesized probes using a novel linker (Fig. 1A) to simultaneously label and stabilize an alpha helix to improve protease stability while maintaining in vitro and in vivo binding affinity. This was compared with two non-fluorescent stabilizing linkers (Fig. 1B) to separate the impact of the fluorophore from helix stabilization.
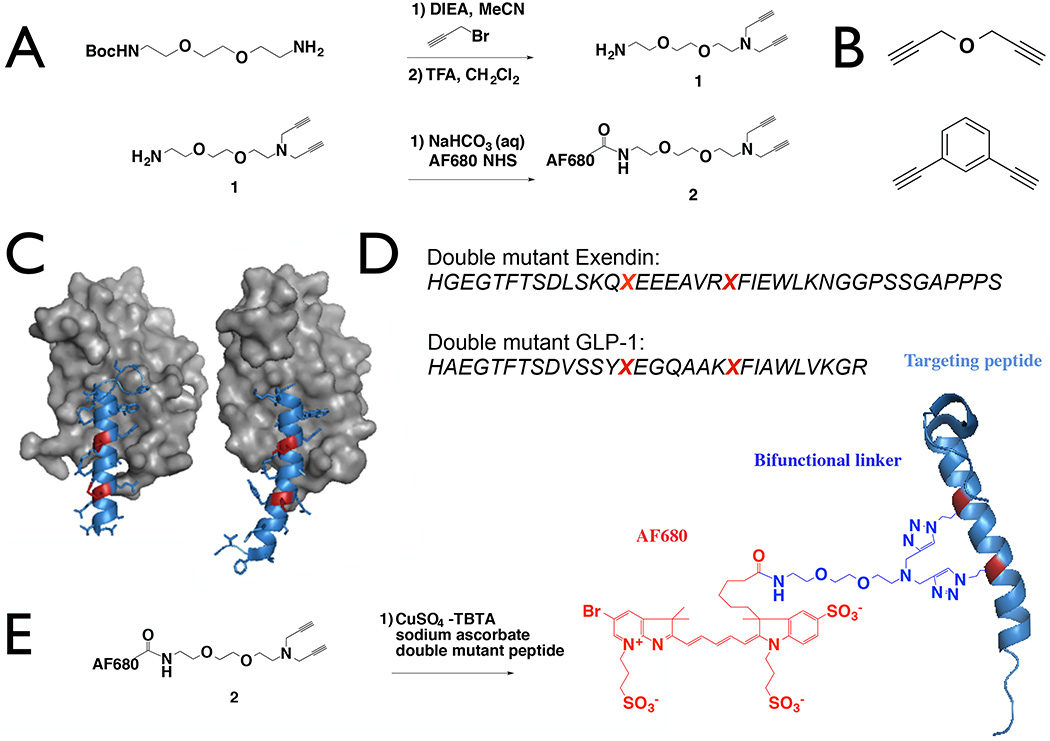
Figure 1.
Linker Design and Synthesis. A Boc-protected hydrophilic spacer was alkylated with propargyl bromide and deprotected to provide a free amine for fluorophore conjugation (A). Propargyl ether (top) and 1,3-diethynylbenzene (bottom) were used to test steric effects of the dye and rigidity of the linker (B). The crystal structure of exendin (left, PDB 3C59) and GLP-1 (right, PDB 3IOL) are shown with the modified residues highlighted in red (C) and the double mutant sequences (D). The peptides were reacted with the linker to form labeled and stabilized alpha helices (E).
One promising application of targeting peptides is to utilize modified glucagon-like peptide-1 receptor (GLP-1R) binding peptides for imaging beta cells in diabetes19, 20. Beta cells express moderate to high levels of GLP-1R resulting in excellent target specificity versus exocrine cells within the pancreas. These properties have led several investigators to explore the use of radiolabeled exendin molecules for tracking beta cell mass in diabetes19–25. However, the molecular properties of the radiolabeled peptide require further optimization before obtaining clinical utility23, 25. Using a novel dialkyne linker with a functional handle for attaching an imaging agent, we synthesized a dual-purpose linker to simultaneously stabilize and label alpha helices. Here we demonstrate improved protease resistance and in vivo targeting of these stabilized glucagon-like peptide-1 receptor (GLP-1R) ligands for visualizing the beta cell mass in the pancreas.
The ligands GLP-1 and exendin share significant homology and bind to the same pocket on GLP-1R, but they exhibit distinct differences. Exendin binds in a straight alpha helix conformation while GLP-1 has a small kink around glycine-2226 (Fig. 1C). Exendin also has significant helical structure in solution while GLP-1 is almost completely disordered27. This allowed us to investigate several aspects of the linker including the impact on helicity, flexibility, affinity, and stability.
One turn of an alpha helix constitutes approximately 3.6 amino acid residues. As a result, i, i+4 and i, i+7 are frequent candidates for crosslinking residues between either one or two adjacent loops12. To generate a staple across two loops, we postulated that non-natural azide-containing amino acids could be introduced and crosslinked at i, i+7 residues in exendin-4 and GLP-1 through incorporation of azidohomoalanine (AHA) during solid phase peptide synthesis (SPPS). A novel dialkyne linker is used to bridge the two AHA side chains and stabilize the helix while introducing a functional handle for labeling. The chemistry utilizes the well-studied copper catalyzed azide alkyne cycloaddition (CuAAC) for high-yielding and specific crosslinking of the desired residues.
The AHA substitution locations were selected using crystal structures26, 28 for the receptor-ligand interaction as well as alanine scan data of GLP-1 with sequence alignment for exendin-429. Previous work labeling exendin with fluorescent tags has shown variable tolerance (negligible impact to >30-fold reduction in binding) at several positions with different fluorophores along the peptide backbone30. The requirements for a stabilizing linker are even more stringent. First, the linker must be located within the alpha helix portion of the peptide, ruling out labeling of the c-terminal and n-terminal regions. Second, the cross-link necessitates a larger structure adjacent to the peptide compared to a single fluorophore. Ultimately, methionine at the 14th position (M-14) and leucine at the 21st (L-21) were substituted with AHA for exendin-4 (Fig. 1C and 1D) to generate double mutant exendin. Similarly, L-20 and E-27 substitutions were made to GLP-1 (7–36) for double mutant GLP-1. We present novel stabilized GLP-1R ligands crosslinked with dialkyne linkers (Fig. 1E) of varying rigidity and demonstrate that all conjugates maintain GLP-1R specific targeting. To abbreviate the structure names, the stabilized alpha helices of exendin generated with the AlexaFluor 680 linker, propargyl ether, and 1,3-diethynylbenzene are named AF680 sExendin, PropE sExendin, and DEB sExendin, respectively. AlexaFluor 680 linker, propargyl ether, and 1,3-diethynylbenzene stabilized GLP-1 are named AF680 sGLP-1, PropE sGLP-1, and DEB sGLP-1. The novel structures should prove useful for developing stabilized alpha helix imaging agents.
Results
Stabilized alpha helical imaging peptides were generated using a novel dialkyne linker as shown in Fig. 1A. To generate the dual-purpose linker, N-Boc- 2,2’-(ethylenedioxy)diethylamine was alkylated with excess propargyl bromide, followed by deprotection with trifluoroacetic acid and subsequently purified via flash chromatography. The resulting intermediate 1 (Fig. 1) was further purified on HPLC followed by conjugation to AF680 NHS ester to yield 2.
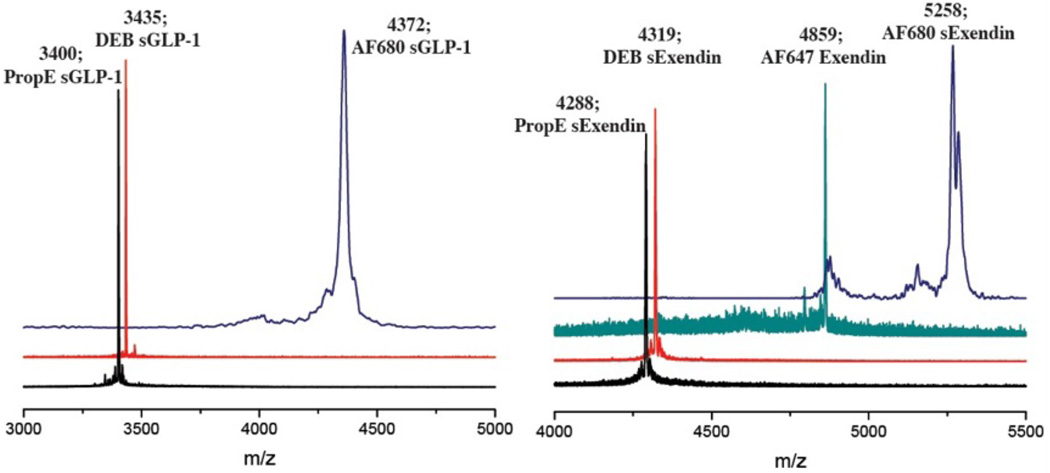
GLP-1R ligands exendin-4 and GLP-1 were chosen to demonstrate the ability of the dual-purpose linker to simultaneously stabilize and fluorescently tag targeting peptides under investigation for imaging in diabetes20, 21, 30. Based on crystallography data for the receptor-ligand interaction, the M14 and L21 residues in exendin and L20 and E27 residues of GLP-1 were substituted with azidohomoalanine (Fig. 1B, C). The i, i+7 residues on exendin-4 and GLP-1 double mutant peptides were then reacted with 2 to stabilize and label the agents (Fig. 1D). In addition to alpha helix stabilization with 2, we also investigated the effect of linker rigidity on the stabilized peptide with propargyl ether and the more rigid cross-linker 1,3-diethynylbenzene on the i, i+7 residues of the GLP-1R ligands. A non-stabilized single mutant exendin-4 labeled with AF647 was synthesized for quantifying the binding affinity of propargyl ether and 1,3-diethynylbenzene stabilized peptides in competition assays. Both AF647-exendin and stabilized peptides were purified using HPLC and characterized by MALDI-TOF (Fig. 2). MALDI for AF647 conjugated peptides, propargyl ether, and 1,3-diethynylbenzene stabilized peptides were collected using reflectron positive mode; fluorescently stabilized peptide MALDI spectra did not show up in reflectron positive mode and were collected using linear positive mode.
Figure 2.
MALDI-TOF mass spectrometry traces of purified peptides showing the successful synthesis of stabilized GLP-1 and exendin helices.
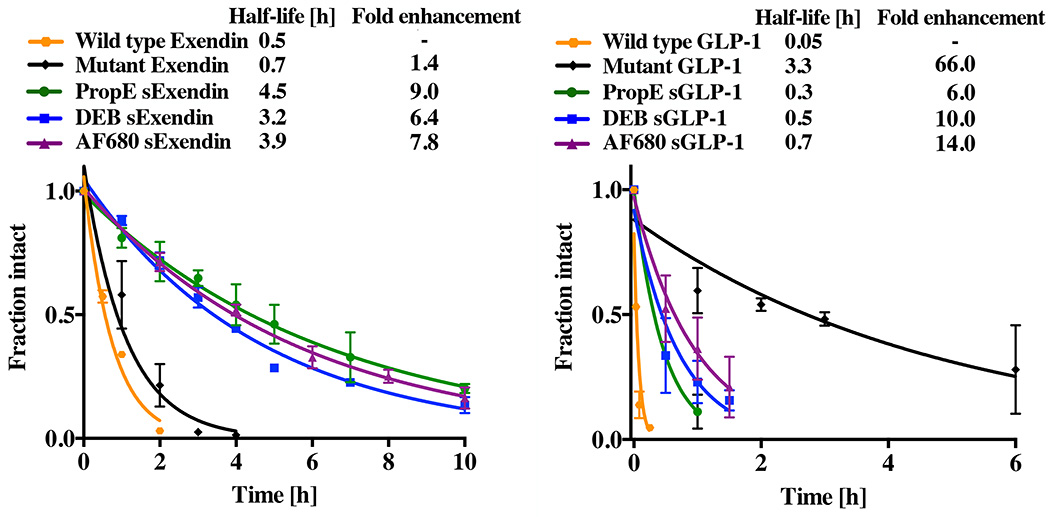
To determine the effect of the alpha helix stabilization on protease resistance, wild type GLP-1, wild type exendin, and both double mutant GLP-1R ligands with and without i, i+7 cross-links were subject to 500 ng/µl trypsin digests at room temperature. All the unstabilized (wild type and double mutant) peptides degraded rapidly except for double mutant GLP-1. Exendin is known to be more resistant to protease degradation, and the wild type peptide had a slower half-life than wild type GLP-1 (0.5 h versus 0.05 h) as anticipated. Unstabilized double mutant exendin with M14X and L21X modifications also digested rapidly in the presence of trypsin (half-life of 0.7 h, Fig. 3). Compared to wild type exendin, the stabilizing cross-linker improved protease resistance over nine-fold in the case of PropE sExendin (half-life of 4.5 h) and over six-fold for DEB sExendin (half-life of 3.2 h). AF680 sExendin demonstrated an almost eight-fold increase (half-life of 3.9 h). Stabilization of wild-type GLP-1 increased protease stability by 6, 10, and 14-fold for PropE sGLP-1, DEB sGLP-1, and AF680 sGLP-1, respectively. Surprisingly, the double mutant GLP-1 had a very long half-life that was 1 to 2 orders of magnitude slower than wild-type GLP-1.
Figure 3.
Protease stability of stabilized peptides. Digests were run on HPLC and monitored at 254 nm to separate intact versus degraded fragments and areas fit to an exponential decay.
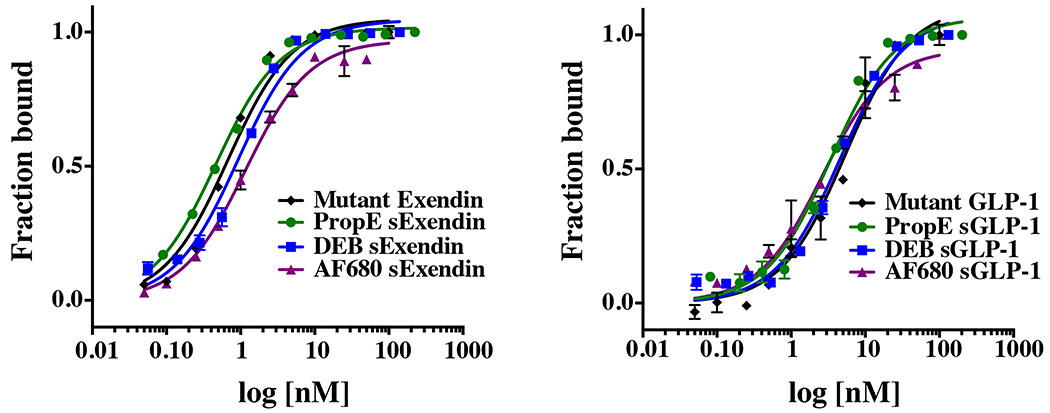
The stabilizing linker increased protease resistance relative to wild type exendin and GLP-1, but the impact on binding affinity from the stabilizing linker was unknown. To test the influence on target binding, the affinity was measured in a binding assay using NIT-1 cells (Fig. 4). Eleven or 12 point curves were done in triplicate for each experiment, and all experiments were repeated on 2–4 separate days. M-14, L-21 azidohomoalanine (AHA) substitutions in exendin-4 did not significantly lower the binding affinity as anticipated by their orientation away from the binding pocket (Table 1). Similarly, GLP-1 also maintained high affinity despite the two substitutions. Stabilization of PropE sExendin and DEB sExendin yielded a Kd = 0.9 ± 0.7 nM and Kd = 0.8 ± 0.2 nM, respectively. The affinity was not statistically significantly different than the double mutant exendin (p value of 0.6 compared to PropE sExendin and p = 0.3 compared to DEB sExendin), demonstrating that steric hindrance from the linker did not significantly increase the Kd. AF680 sExendin had a Kd slightly above the original peptide but still maintained high affinity. For GLP-1, PropE sGLP-1 resulted in increased affinity with a Kd = 3.5 ± 0.8 nM. In the case of the more rigid DEB sGLP-1, alpha helix stabilization resulted in similar affinity values. Stabilization with the new dual-purpose cross-linker in AF680 sGLP-1 also gave higher affinity for GLP-1R despite the presence of the fluorophore (Kd = 3.1 ± 0.7 nM). Both AF680 sGLP-1 and PropE sGLP-1 had statistically significantly higher affinity than the double mutant GLP-1 (p value of 0.04 for both), although DEB sGLP-1 did not (p = 0.08).
Figure 4.
Representative affinity curves show negligible loss of affinity for exendin and a small improvement in binding for GLP-1 stabilized peptides.
Table 1.
Binding affinity values for stabilized and non-stabilized exendin and GLP-1 peptides
| peptide | crosslinker | binding affinity (nM) |
|---|---|---|
| Exendin (M14X, L21X) | None | 0.5 (± 0.2) |
| Exendin (M14X, L21X) | Propargyl Ether | 0.9 (± 0.7) |
| Exendin (M14X, L21X) | 1,3-Diethynylbenzene! | 0.8 (± 0.2) |
| Exendin (M14X, L21X) | AF680 | 1.4 (± 0.3) |
| GLP-1 (L20, E27) | None | 5.6 (± 0.2) |
| GLP-1 (L20, E27) | Propargyl Ether | 3.5 (± 0.8) |
| GLP-1 (L20, E27) | 1,3-Diethynylbenzene! | 4.8 (± 0.3) |
| GLP-1 (L20, E27) | AF680 | 3.1 (± 0.7) |
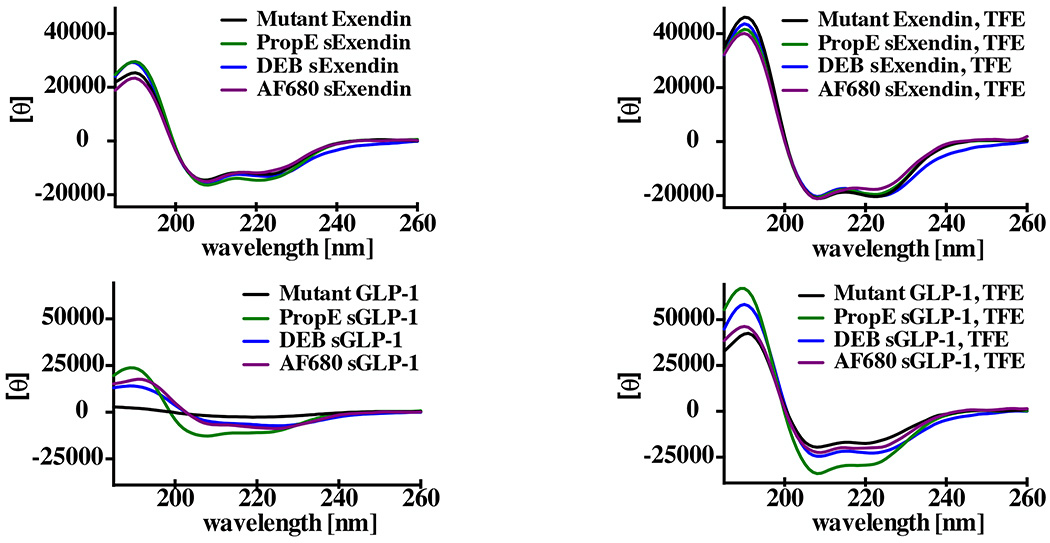
The increase in affinity for GLP-1 was unexpected given the typically negative impact of steric hindrance on Kd31. Helix stabilization is known to decrease the entropic penalty of binding by locking the peptide in an alpha helix and limiting the number of conformations8, although exceptions are known32. To investigate the effect of the dialkyne cross-linker on the helix structure, circular dichroism measurements were taken to determine the % helicity (Fig. 5). Non-stabilized double mutant exendin-4 displayed intermediate helicity (57%) in phosphate buffer (Table 2), consistent with the significant helicity of exendin27. The PropE sExendin and DEB sExendin had small increases in helicity (67 and 59%, respectively), and AF680 sExendin had 55% helicity. Calculations of the fractional helicity are based on estimates of the number of helical residues within the peptide sequence27. To experimentally validate these estimates, 50% trifluoroethanol (TFE) was used as a solvent to drive the peptides into a helical conformation. The highest values for exendin and GLP-1 derivatives were between 93–104% indicating agreement between the literature estimate and experimental values reported here. All the exendin peptides had elevated helicity in TFE (> 80%). On the contrary, GLP-1 had very little helicity in phosphate buffer (7%), but stabilization with propargyl ether, 1,3-diethynylbenzene, and the AF680 stabilizing linker resulted in a significant increase in % helicity (38, 23, and 30%). Higher helicity for stabilized peptides was maintained in 50% TFE buffer with 60% for the GLP-1 peptide and 104%, 82%, and 71% for PropE sGLP-1, DEB sGLP-1, and AF680 sGLP-1, respectively.
Figure 5.
The stabilizing linker dramatically improves the helicity of GLP-1 with a smaller increase in helicity for the more highly structured exendin peptide.
Table 2.
Fractional helicity for stabilized and non-stabilized exendin and GLP-1 peptides
| peptide | crosslinker | χhelix | χhelix |
|---|---|---|---|
| Exendin (M14X, L21X) | None | 0.57 | 0.93 |
| Exendin (M14X, L21X) | Propargyl Ether | 0.67 | 0.9 |
| Exendin (M14X, L21X) | 1,3-Diethynylbenzene! | 0.59 | 0.93 |
| Exendin (M14X, L21X) | AF680 | 0.55 | 0.82 |
| GLP-1 (L20, E27) | None | 0.07 | 0.6 |
| GLP-1 (L20, E27) | Propargyl Ether | 0.38 | 1.04 |
| GLP-1 (L20, E27) | 1,3-Diethynylbenzene! | 0.23 | 0.81 |
| GLP-1 (L20, E27) | AF680 | 0.3 | 0.71 |
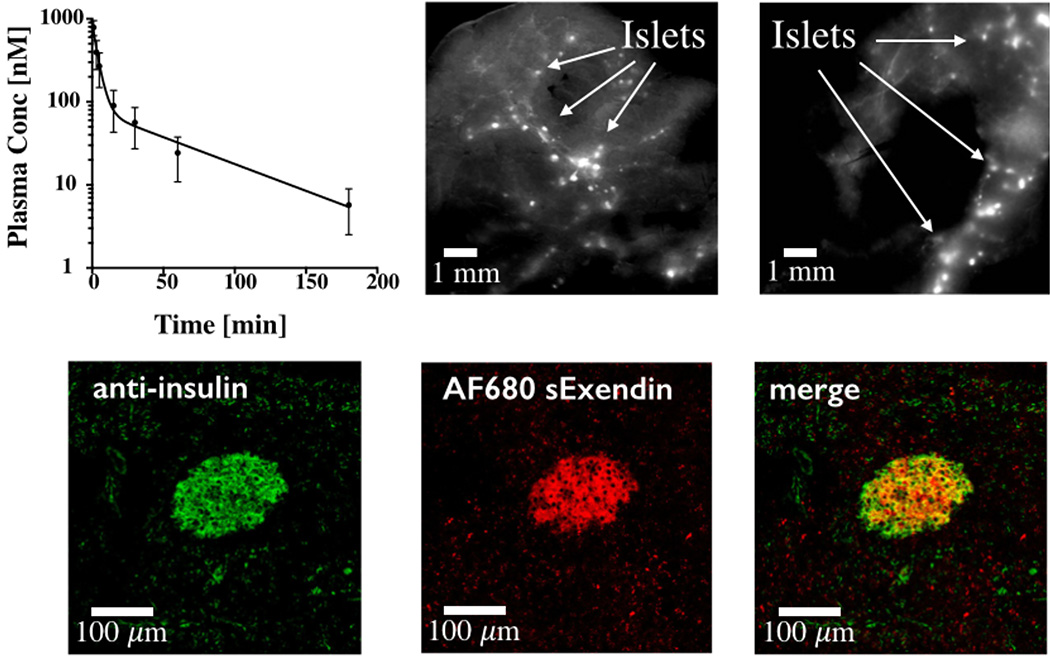
Owing to its higher stability, AF680 sExendin was used to demonstrate the in vivo imaging potential of the fluorescent helix stabilizing cross-linker. To show the specificity of the imaging agent for beta cells in the islets of Langerhans, AF680 sExendin was injected in C57BL/6 mice and allowed to circulate for 3 h. Blood samples were taken and the half-life of AF680 sExendin in plasma was determined by fitting to a biexponential decay. We observed a fast decay t1/2 of 2.7 min (91%) and a slow decay t1/2 of 67 min (Fig. 6). To confirm specific beta cell targeting, the animal was sacrificed after 3 h and the pancreas removed. Macroscopic scans of the organ indicate AF680 sExendin signal in distinct, punctate spots spread along the pancreatic vasculature indicating efficient targeting of the islets of Langerhans (Fig. 6). A validated K12 labeled exendin peptide21, 33 is shown for comparison. The pancreas was then processed for histology, and islets were stained with an anti-insulin antibody (Fig. 6). Colocalization between in vivo delivered AF680 sExendin and anti-insulin demonstrate specific targeting of islets in vivo.
Figure 6.
Stabilized fluorescent exendin clears rapidly from the plasma (top left) and efficiently targets islets, providing high tissue contrast (top middle). Beta cells form ~1–2% of the pancreas and are located in 100–300 µm Islets of Langerhans that are labeled intensely by the fluorescent peptides (arrows). K12C labeled fluorescent exendin is shown for comparison (top right). Histology slide of pancreas showing ex vivo anti-insulin staining of islets (left), in vivo delivered AF680 sExendin (middle) and merge (right).
Discussion
Peptides possess many properties that are ideal for imaging agents34, 35. Their low molecular weight allows rapid uptake in target tissue, fast clearance from background regions, and low nonspecific uptake. Despite these advantages, significant challenges exist including poor stability, low affinity, and difficulty in labeling without lowering affinity due to steric hindrance of binding. To help address some of these issues, we designed a dual-purpose linker to simultaneously label an alpha helix peptide while stabilizing the secondary structure. Because not all stabilized helices exhibit improved properties, we analyzed the protease stability, helicity, and binding affinity of several helix stabilized derivatives.
For the linker design, propargyl bromide was used to react with the N-Boc- 2,2′-(ethylenedioxy)diethylamine spacer for optimal cross-linker length. Previous reports of i, i+7 linkers indicated that a 13-atom linker stabilizes an alpha helix conformation8. Linker design was critical to avoid the stabilization of other types of helices36 and the instability of longer linkers 37. The facile incorporation of azidohomoalanine, either by synthetic techniques (SPPS) or biological systems38 makes this crosslinking chemistry advantageous for synthesizing stabilized helices. The 7-carbon span of the dialkyne generates a 13-atom cross-link for efficient stabilization of an alpha helix.
Stabilization of alpha helices reduces solvent interaction of the hydrophilic backbone and reportedly increases cell permeability for some peptides 6, 39. While membrane permeability is desirable for intracellular targets, this would increase the background signal of an imaging agent. A diethylene glycol spacer was placed between the amine functional handle and the dialkyne to impart additional hydrophilicity to the linker. Although the fluorescent dyes used in this work are charged and hydrophilic, the linker could aid in radiolabelling applications where the tag may be more lipophilic21.
As a model system for testing the dual-purpose linker, we chose exendin and GLP-1 peptides. We and others19–21, 23,25, 30, 33 have used exendin and stabilized GLP-122 for imaging beta cells and increasing therapeutic efficacy, and the peptides are well characterized. Several GLP-1R agonists are in the clinic for treating type 2 diabetes40, and the crystal structures of both peptides bound to their target have been published.
The stabilized peptides were synthesized with moderate yield (9–63%) with the expected molecular weight shown by MALDI-TOF. Despite the homobifunctional crosslinking chemistry, little to no oligomerization was detected. This is likely due to templating effects from the peptide backbone11 and is in agreement with the higher yield for the more helical exendin-4 peptide compared to GLP-1. An additional linker, 1,4-diethynylbenzene, was tested to see if the increased strain from a more rigid linear crosslinking agent would stabilize or destabilize the helix. However, yields of this reaction were extremely low likely due to inefficient crosslinking (data not shown). The stabilized helices showed up to a 14-fold increase in trypsin protease stability. For exendin, the three predicted cleavage sites include one site within the cross-link, one just outside the cross-link, and a third several residues away. Proteases recognize an extended beta strand conformation, so either direct steric hindrance by non-natural amino acids and cross links or a helical conformation can prevent degradation41. A combination of these factors likely contributes to the lack of protease digestion in our system.
Unexpectedly, the double mutant GLP-1 had significantly increased protease stability. CD measurements indicated that the azidohomoalanine substitutions did not result in high helicity. One of the non-natural amino acids is located adjacent to a lysine cleavage site potentially slowing the rate of proteolysis. However, LC-MS data indicated that this site was still cleaved (data not shown). While no aggregation was detected at lower concentrations during experiments, the substitution of a glutamic acid residue for azidohomoalanine reduced the charge of the peptide, and aggregation of double mutant GLP-1 at high concentrations during synthesis and protease digestion could have contributed to the low GLP-1 stabilization reaction yields and high protease stability. The data also did not fit an exponential decay very well, indicating that aggregation may play a role in its protease resistance. However, mutant GLP-1 protease resistance could be an example where the sequence mutations impart unique properties independent of the linker16. Notably, the linker was required for the large increase in exendin protease resistance.
Despite the larger crosslinking label, the Kd for AF680 sExendin only increased by 0.9 nM compared to double mutant exendin. Locating the linker opposite the binding interface and including a hydrophilic spacer allowed the peptide to maintain high binding affinity in addition to increased protease stability. The other crosslinking agents maintained high affinity of the exendin to GLP-1R but did not increase in affinity. This is likely due to the high helicity of the peptide even without a stabilizing side chain cross-link. To corroborate this argument, a three state thermodynamic model of binding for helical peptides (supplementary data) was used to show a negligible predicted increase in the binding affinity (0.42 to 0.53 nM) for exendin. The larger increase in helicity for GLP-1 derivatives results in a small but significant predicted increase in binding affinity (Kd ~ 1.5 nM) in the absence of steric effects and changes in the enthalpy of binding. Larger decreases in Kd are predicted for lower affinity peptides (supplementary data).
The trade-off between a more rigid alpha helix (lowering the entropic penalty but reducing the enthalpy of binding) and a more flexible linker has been established for stabilized helices8. The stabilized GLP-1 peptides demonstrated a small but statistically significant increase in affinity and a large increase in helicity, consistent with the thermodynamic model. GLP-1 is disordered in solution, so stabilizing the alpha helix increases the affinity by lowering the entropic penalty upon binding. In principle, locking the GLP-1 peptide in a straight alpha helix could reduce affinity by removing the kink present in the crystal structure, and this level of detail is not captured in the simple three state model. For GLP-1 however, it appears that the i,i+7 cross-link is flexible enough to allow for high affinity interactions, or at the very least, the reduction in the entropic penalty dominates the free energy of binding.
Comparing the more rigid 1,3-diethynylbenzene cross-linker versus the propargyl ether, it appears the latter has more helix inducing propensity in these two peptides. While both linkers increased the protease stability and helicity of exendin and GLP-1, along with the affinity of GLP-1, the propargyl ether did so to a larger extent in each case. The additional flexibility likely makes the alpha helix more energetically favorable, thereby increasing protease stability, affinity, and helicity.
The stabilized fluorescent peptide maintained efficient targeting of beta cells in vivo. The rapid clearance from the plasma is beneficial for lowering background fluorescence but could potentially reduce targeting if insufficient amounts reach the beta cells. For tumors, an analysis of molecular weight versus targeting efficiency concluded that for small targeting agents, the lower the molecular weight, the more efficient the uptake provided high affinity is maintained3. Low molecular weight agents were all filtered rapidly by the kidneys, but the smaller agents extravasated into the tumor more quickly. This has also driven protein engineers to find smaller scaffolds for rapid targeting42–44. It remains to be seen whether the same quantitative conclusions apply to the pancreas. Intravital microscopy experiments of exendin show rapid uptake in islets within several minutes after injection33. Islets are highly vascularized (500 cm2/cm3 blood vessel surface area to volume21) with fenestrated endothelium which appears to allow efficient access to the target cells even over short circulation times.
We anticipate this dual-purpose linker will have applications with other peptides in imaging and therapeutic development. Recently, Lau et al. used a poly-arginine conjugated 1,3-diethynylbenzene cross-linker to increase cellular uptake of a p53 alpha helix45. In their MDM2 targeting peptide, they found a similar trend where a more flexible aliphatic dialkyne produced better stabilization than a more rigid 1,3-diethynylbenzene46. This is analogous to our findings where flexible propargyl ether resulted in more efficient binding and helicity than 1,3-diethynylbenzene. Importantly, the propargyl ether and fluorescent linker maintain hydrophilicity for this extracellular target compared to the intracellular MDM2-targeting peptides. The ability to both stabilize the alpha helix and impart additional functionality through the tertiary amine in our linker minimizes the impact of adding multiple sterically bulky groups. Successful alpha helix stabilization in both the GLP-1R targeting peptides and MDM2 peptides provides evidence that this method may work as a general strategy. As seen with lactam bridges and hydrocarbon stapling, results can be sequence dependent, and individual cases need to be tested16, 18. Nevertheless, these functional linkers may have applications in several stabilized alpha helix applications including imaging agents, drug design, surface modification, and affinity separations47, 48.
In conclusion, we have reported a novel dual-purpose linker that is capable of labeling GLP-1 receptor ligands with an imaging agent modality, stabilizing the alpha helix structure, and increasing protease stability while having minimal impact or an improvement on binding affinity.
Experimental Procedures
Materials
Double mutant exendin-4 (HGEGTFTSDLSKQXEEEAVRXFIEWLKNGGPSSGAPPPS), double mutant GLP-1 (7-36, HAEGTFTSDVSSYXEGQAAKXFIAWLVKGR), and single mutant exendin-4 (HGEGTFTSDLSKQXEEEAVRLFIEWLKNGGPSSGAPPPS), where X is the non-natural amino acid AHA, were purchased from Innopep (San Diego, CA). Fluorochromes AF647 alkyne and AF680 N-hydroxysuccinimidyl (NHS) ester were purchased from Life Technologies (Carlsbad, CA). NBoc-2,2’-(ethylenedioxy)diethylamine, rabbit anti-insulin, and goat anti-rabbit IgG FITC antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX). All other reagents, unless specified, were purchased from Sigma-Aldrich (Milwaukee, WI) and were used as received. Reverse phase high performance liquid chromatography (RP-HPLC) was performed on a Shimadzu LC unit using analytical and preparative reversed phase Phenomenex Luna C18(2) columns. MALDI-MS spectra were collected using a Bruker Autoflex mass spectrometer, and ESI-MS analysis was performed on an Agilent Q-TOF 1200 series. NMR spectra were collected using a Varian MR400 spectrometer. Fluorescence microscopy images were collected using an Olympus FV 1200 confocal microscope.
Preparation of (1)
N-Boc- 2,2′-(ethylenedioxy)diethylamine (805 µmol) was added to diisopropylethylamine (2.40 mmol) in 3.8 mL MeCN. Propargyl bromide in toluene was added dropwise (2.62 mmol). The reaction mixture was stirred overnight at room temperature before being concentrated under reduced pressure and then subjected to flash chromatography (80:7:1 CHCl3: MeOH: NH4OH). The desired fraction was concentrated, deprotected using 50% trifluoroacetic acid in dichloromethane, and purified using preparative RP-HPLC using a linear gradient of MeCN in H2O to yield 1 (475 µmol, 59%).1H NMR (400 MHz, CD3OD) δ 3.89 (4H, d), 3.77 (2H, t), 3.73 (2H, t), 3.70 (4H, s), 3.18 (2H, t), 3.15 (2H, t), 3.04 (2H, t). 13C NMR(400 MHz, CD3OD) δ 76.8, 74.6, 69.9, 69.8, 66.5, 51.9, 42.2, 39.2. HRMS: m/z calculated for C12H20N2O2: 225.1603, found: 225.1600.
Preparation of (2)
AF680 NHS ester (1 µmol in DMSO) was added to an aqueous solution containing 1 (10 µmol) buffered with 7.5% sodium bicarbonate. The reaction was stirred at room temperature for 30 min followed by purification on preparative RP-HPLC (12 mL/min; A: 0.1% trifluoroacetic acid in water, B: 0.1% trifluoroacetic acid in acetonitrile; 20% B 0.1–3 min, 20–40% B 3–11 min ) to give 2: tR = 10.2 min. MALDI-TOF: m/z calculated: 1063.11, found: 1064.38. All MALDI-TOF and ESI-Mass Spectrometry data were collected at the University of Michigan Department of Chemistry’s Core Facility.
Preparation of Stabilized Peptides
Propargyl ether (0.3 µmol), 1,3 diethynylbenzene (0.3 µmol), 2 (0.3 µmol), or AF647 alkyne (0.3 µmol) was first added to 200 µL of 1:1 water:tert-butanol, followed by Cu-TBTA (30 nmol) and sodium ascorbate (0.3 µmol). Lastly, double mutant peptide (0.3 µmol) was added and the solution was gently stirred at room temperature for 12 h followed by purification on preparative RP-HPLC (propargyl ether stabilized exendin (PropE sExendin) and 1,3-diethynylbenzene stabilized exendin (DEB sExendin): 28% B 0.1–7 min, 28–70% B 7–23 min (PropE sExendin: 97% purity; DEB sExendin: 98% purity); AF680 linker stabilized exendin (AF680 sExendin), AF680 linker stabilized GLP-1 (AF680 sGLP-1), AF647 labeled exendin: 25% B 0.1–7 min, 25–70% B 7–20 min (AF680 sExendin: 92% purity; AF680 sGLP-1: 89% purity; AF647 exendin: 94% purity); propargyl ether stabilized GLP-1 (PropE sGLP-1): 37% B 0.1–7 min (PropE sGLP-1: 93% purity); 1,3-diethynylbenzene stabilized GLP-1 (DEB sGLP-1): 23–38% B 0.1–12 min, 38–50% B 12–20 min (DEB sGLP-1: 92% purity)) at a flow rate of 12 mL/min. MALDI-TOF: PropE Exendin: m/z calculated 4288, found: 4288; DEB sExendin: m/z calculated 4320, found: 4319; AF680 sExendin: m/z calculated 5257, found: 5258; AF680 sGLP-1: m/z calculated 4370, found: 4372; AF647 sExendin: m/z calculated: MW of dye unpublished, found: 4859; PropE sGLP-1: m/z calculated 3401, found: 3400; DEB sGLP-1: m/z calculated 3433, found: 3435. For quantifying peptide concentrations to determine yield and circular dichroism measurements, amino acid analysis was carried out by the University of Michigan Proteomics and Peptide Synthesis Core.
Cell Culture
NIT-1 cells, a GLP-1R positive mouse beta cell line, were generously provided by Dr. Ralph Weissleder’s Laboratory and used for receptor binding studies. Cells were grown in F12K containing 10% (v/v) heat-inactivated FBS, 50U/mL penicillin, 50 µg/mL streptomycin, and 1.5 g/L sodium bicarbonate. The passage number for NIT-1 cells used in affinity measurements for all peptides was between 4 and 16
In Vitro Receptor Binding Assay
NIT-1 cells were grown for 48 hours before being harvested with trypsin-EDTA. The cells were then washed with PBS, centrifuged, and resuspended in PBS with 1.0% BSA. Cells were aliquoted and suspended in binding buffer containing stabilized GLP-1 or exendin on ice (0.05–250 nM). After 3 h, cells were centrifuged and washed (for fluorescent constructs) or the buffer was replaced with a second binding buffer containing 20 nM of AF647 exendin. After 1 h, cells were washed with PBS with 1.0% BSA and immediately analyzed using an Attune Acoustic Focusing Cytometer (Applied Biosystems). Binding affinity curves and statistical analysis were carried out using Prism 6.0 software.
Animals
All animal experiments were conducted in compliance with the University of Michigan University Committee on Use and Care of Animals (UCUCA). For measuring plasma clearance, AF680 sExendin (1.2 nmol) or a validated K12C fluorescent exendin control peptide21, 33 was injected in the lateral tail vein of C57BL/6 mice (6 mice total). At predetermined time points (1, 3, 5, 15, 30, 60, 180 min), retro orbital blood samples were collected. A LICOR Odyssey CLx scanner (Lincoln, NE) was used to measure the fluorescence intensity for each sample and the intensities were converted to concentration using a dilution series of AF680 sExendin in mouse plasma. After 3 h, the mice were sacrificed and the pancreas resected. Islets were visualized by a near-infrared scan on a LICOR Odyssey CLx scanner.
Histology and Microscopy
Pancreata resected from C57BL/6 mice injected with AF680 sExendin were submerged in OCT and frozen in chilled 2-methylbutane. The organ was then sectioned into 6 µm slices, fixed with 4% paraformaldehyde for 10 min, and incubated overnight at 4°C with a rabbit anti-insulin primary antibody (dilution 1:150 in PBS with 0.1% BSA supplemented with 1% goat serum), followed by a 30 min room temperature incubation with goat anti-rabbit IgG FITC secondary antibody (dilution 1:50 in PBS with 0.1% BSA).
Trypsin Digest
To assess the proteolytic resistance of stabilized peptides, both stapled and unstapled peptides (75 µM) were subject to a trypsin digest (500 ng/µL, pH 7.4, room temperature). Enzymatic digest was monitored at 254 nm using RP-HPLC to determine the degradation half-life.
Circular Dichroism (CD) Measurements
To quantify peptide secondary structure, CD spectra were collected on a Jasco-815 CD spectrometer with a 1 mm Hellma quartz cuvette at 23 °C. Five scans from 185 nm to 260 nm at 20 nm/min were averaged for peptide samples prepared in 5 mM potassium phosphate buffer, pH 7.0 at concentrations ranging between 5 µM to 20 µM as determined by amino acid analysis. A scan containing only buffer/buffer with cross-linker was subtracted from sample scans. Helicity was calculated using mean residue ellipticity at 221 nm and maximum ellipticity as reported elsewhere27.
Supplementary Material
Acknowledgments
We thank the University of Michigan Biointerfaces Institute for assistance with CD measurements and Dr. Tim Scott’s lab for assistance with gathering NMR data. Funding was provided by NIH grant 1K01DK093766 (GMT).
Footnotes
Supporting Information Available: The thermodynamic model of helicity and binding is provided in the supporting section. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature. 2008;452:580–589. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao BS, Schwartz LH, Larson SM. Imaging Surrogates of Tumor Response to Therapy: Anatomic and Functional Biomarkers. Journal of Nuclear Medicine. 2009;50:239–249. doi: 10.2967/jnumed.108.056655. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt MM, Wittrup KD. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Molecular Cancer Therapeutics. 2009;8:2861. doi: 10.1158/1535-7163.MCT-09-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akers WJ, Zhang ZR, Berezin M, Ye YP, Agee A, Guo K, Fuhrhop RW, Wickline SA, Lanza GM, Achilefu S. Targeting of alpha(v)beta(3)-integrins expressed on tumor tissue and neovasculature using fluorescent small molecules and nanoparticles. Nanomedicine. 2010;5:715–726. doi: 10.2217/nnm.10.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura RH, Cheng Z, Gambhir SS, Cochran JR. Engineered Knottin Peptides: A New Class of Agents for Imaging Integrin Expression in Living Subjects. Cancer Research. 2009;69:2435–2442. doi: 10.1158/0008-5472.CAN-08-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbonstapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bird GH, Madani N, Perry AF, Princiotto AM, Supko JG, He XY, Gavathiotis E, Sodroski JG, Walensky LD. Hydrocarbon double-stapling remedies the proteolytic instability of a lengthy peptide therapeutic. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14093–14098. doi: 10.1073/pnas.1002713107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sia SK, Carr PA, Cochran AG, Malashkevich VN, Kim PS. Short constrained peptides that inhibit HIV-1 entry. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14664–14669. doi: 10.1073/pnas.232566599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schafmeister CE, Po J, Verdine GL. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. Journal of the American Chemical Society. 2000;122:5891–5892. [Google Scholar]
- 10.Neumann H, Wang KH, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 11.Torres O, Yuksel D, Bernardina M, Kumar K, Bong D. Peptide tertiary structure nucleation by side-chain crosslinking with metal complexation and double "click" cycloaddition. Chembiochem. 2008;9:1701–1705. doi: 10.1002/cbic.200800040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu CX, Taylor JW. Synthesis and study of peptides with semirigid i and i+7 side-chain bridges designed for alpha-helix stabilization. Bioorganic & Medicinal Chemistry. 1999;7:161–175. doi: 10.1016/s0968-0896(98)00232-6. [DOI] [PubMed] [Google Scholar]
- 13.Galande AK, Bramlett KS, Trent JO, Burris TP, Wittliff JL, Spatola AF. Potent inhibitors of LXXLL-based protein-protein interactions. Chembiochem. 2005;6:1991–1998. doi: 10.1002/cbic.200500083. [DOI] [PubMed] [Google Scholar]
- 14.Jackson DY, King DS, Chmielewski J, Singh S, Schultz PG. General- Approach to the Synthesis of Short Alpha-Helical Peptides. Journal of the American Chemical Society. 1991;113:9391–9392. [Google Scholar]
- 15.Bird GH, Gavathiotis E, LaBelle JL, Katz SG, Walensky LD. Distinct BimBH3 (BimSAHB) stapled peptides for structural and cellular studies. ACS Chem Biol. 2014;9:831–837. doi: 10.1021/cb4003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamoto T, Segal D, Zobel K, Fedorova A, Yang H, Fairbrother WJ, Huang DC, Smith BJ, Deshayes K, Czabotar PE. Further insights into the effects of preorganizing the BimBH3 helix. ACS Chem Biol. 2014;9:838–839. doi: 10.1021/cb400638p. [DOI] [PubMed] [Google Scholar]
- 17.Okamoto T, Zobel K, Fedorova A, Quan C, Yang H, Fairbrother WJ, Huang DC, Smith BJ, Deshayes K, Czabotar PE. Stabilizing the pro-apoptotic BimBH3 helix (BimSAHB) does not necessarily enhance affinity or biological activity. ACS Chem Biol. 2013;8:297–302. doi: 10.1021/cb3005403. [DOI] [PubMed] [Google Scholar]
- 18.Giordanetto F, Revell JD, Knerr L, Hostettler M, Paunovic A, Priest C, Janefeldt A, Gill A. Stapled Vasoactive Intestinal Peptide (VIP) Derivatives Improve VPAC2 Agonism and Glucose-Dependent Insulin Secretion. ACS medicinal chemistry letters. 2013;4:1163–1168. doi: 10.1021/ml400257h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiner T, Thurber G, Gaglia J, Vinegoni C, Liew CW, Upadhyay R, Kohler RH, Li L, Kulkarni RN, Benoist C, et al. Accurate measurement of pancreatic islet beta-cell mass using a second-generation fluorescent exendin-4 analog. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12815–12820. doi: 10.1073/pnas.1109859108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brand C, Abdel-Atti D, Zhang Y, Carlin S, Clardy SM, Keliher EJ, Weber WA, Lewis JS, Reiner T. In vivo imaging of GLP-1R with a targeted bimodal PET/fluorescence imaging agent. Bioconjug Chem. 2014;25:1323–1330. doi: 10.1021/bc500178d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keliher EJ, Reiner T, Thurber GM, Upadhyay R, Weissleder R. Efficient 18F-Labeling of Synthetic Exendin-4 Analogues for Imaging Beta Cells. ChemistryOpen. 2012 doi: 10.1002/open.201200014. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao H, Niu G, Yang M, Quan Q, Ma Y, Murage EN, Ahn J-M, Kiesewetter DO, Chen X. PET of Insulinoma Using 18F-FBEM-EM3106B, a New GLP-1 Analogue. Molecular Pharmaceutics. 2011 doi: 10.1021/mp200141x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikkola K, Yim CB, Fagerholm V, Ishizu T, Elomaa VV, Rajander J, Jurttila J, Saanijoki T, Tolvanen T, Tirri M, et al. 64Cu- and 68Ga-labelled [Nle(14),Lys(40)(Ahx-NODAGA)NH2]-exendin-4 for pancreatic beta cell imaging in rats. Molecular imaging and biology : MIB : the official publication of the Academy of Molecular Imaging. 2014;16:255–263. doi: 10.1007/s11307-013-0691-2. [DOI] [PubMed] [Google Scholar]
- 24.Wicki A, Wild D, Storch D, Seemayer C, Gotthardt M, Behe M, Kneifel S, Mihatsch MJ, Reubi JC, Macke HR, et al. [Lys(40)(Ahx-DTPA-In-111)NH2]-Exendin-4 is a highly efficient radiotherapeutic for glucagon-like peptide-1 receptor-targeted therapy for insulinoma. Clinical Cancer Research. 2007;13:3696–3705. doi: 10.1158/1078-0432.CCR-06-2965. [DOI] [PubMed] [Google Scholar]
- 25.Wild D, Wicki A, Mansi R, Behe M, Keil B, Bernhardt P, Christofori G, Ell PJ, Macke HR. Exendin-4-Based Radiopharmaceuticals for Glucagonlike Peptide-1 Receptor PET/CT and SPECT/CT. Journal of Nuclear Medicine. 2010;51:1059–1067. doi: 10.2967/jnumed.110.074914. [DOI] [PubMed] [Google Scholar]
- 26.Underwood CR, Garibay P, Knudsen LB, Hastrup S, Peters GH, Rudolph R, Reedtz-Runge S. Crystal Structure of Glucagon-like Peptide-1 in Complex with the Extracellular Domain of the Glucagon-like Peptide-1 Receptor. Journal of Biological Chemistry. 2010;285:723–730. doi: 10.1074/jbc.M109.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen NH, Brodsky Y, Neidigh JW, Prickett KS. Medium-dependence of the secondary structure of exendin-4 and glucagon-like-peptide-1. Bioorganic & Medicinal Chemistry. 2002;10:79–85. doi: 10.1016/s0968-0896(01)00263-2. [DOI] [PubMed] [Google Scholar]
- 28.Runge S, Thogersen H, Madsen K, Lau J, Rudolph R. Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. Journal of Biological Chemistry. 2008;283:11340–11347. doi: 10.1074/jbc.M708740200. [DOI] [PubMed] [Google Scholar]
- 29.Adelhorst K, Hedegaard BB, Knudsen LB, Kirk O. Structure-Activity Studies of Glucagon-Like Peptide-1. Journal of Biological Chemistry. 1994;269:6275–6278. [PubMed] [Google Scholar]
- 30.Clardy SM, Keliher EJ, Mohan JF, Sebas M, Benoist C, Mathis D, Weissleder R. Fluorescent exendin-4 derivatives for pancreatic beta-cell analysis. Bioconjug Chem. 2014;25:171–177. doi: 10.1021/bc4005014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker JC, Andrews KM, Rescek DM, Massefski W, Jr, Andrews GC, Contillo LG, Stevenson RW, Singleton DH, Suleske RT. Structure-function analysis of a series of glucagon-like peptide-1 analogs. The journal of peptide research : official journal of the American Peptide Society. 1998;52:398–409. doi: 10.1111/j.1399-3011.1998.tb00664.x. [DOI] [PubMed] [Google Scholar]
- 32.Martin SF. Preorganization in biological systems: Are conformational constraints worth the energy? Pure and Applied Chemistry. 2007;79:193–200. [Google Scholar]
- 33.Reiner T, Kohler RH, Liew CW, Hill JA, Gaglia J, Kulkarni RN, Weissleder R. Near-Infrared Fluorescent Probe for Imaging of Pancreatic beta Cells. Bioconjugate Chemistry. 2010;21:1362–1368. doi: 10.1021/bc100184w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James ML, Gambhir SS. A molecular imaging primer: modalities, imaging agents, and applications. Physiol Rev. 2012;92:897–965. doi: 10.1152/physrev.00049.2010. [DOI] [PubMed] [Google Scholar]
- 35.Lee S, Xie J, Chen X. Peptide-based probes for targeted molecular imaging. Biochemistry. 2010;49:1364–1376. doi: 10.1021/bi901135x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobsen O, Maekawa H, Ge NH, Gorbitz CH, Rongved P, Ottersen OP, Amiry-Moghaddam M, Klaveness J. Stapling of a 3(10)-Helix with Click Chemistry. J. Org. Chem. 2011;76:1228–1238. doi: 10.1021/jo101670a. [DOI] [PubMed] [Google Scholar]
- 37.Scrima M, Le Chevalier-Isaad A, Rovero P, Papini AM, Chorev M, D'Ursi AM. Cu-I-Catalyzed Azide-Alkyne Intramolecular i-to-(i+4) Side-Chain-to-Side-Chain Cyclization Promotes the Formation of Helix-Like Secondary Structures. European Journal of Organic Chemistry. 2010:446–457. [Google Scholar]
- 38.Johnson JA, Lu YY, Van Deventer JA, Tirrell DA. Residue-specific incorporation of non-canonical amino acids into proteins: recent developments and applications. Curr. Opin. Chem. Biol. 2010;14:774–780. doi: 10.1016/j.cbpa.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muppidi A, Wang ZY, Li XL, Chen JD, Lin Q. Achieving cell penetration with distance-matching cysteine cross-linkers: a facile route to cell-permeable peptide dual inhibitors of Mdm2/Mdmx. Chemical Communications. 2011;47:9396–9398. doi: 10.1039/c1cc13320a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahren B, Schmitz O. GLP-1 receptor agonists and DPP-4 inhibitors in the treatment of type 2 diabetes. Hormone and Metabolic Research. 2004;36:867–876. doi: 10.1055/s-2004-826178. [DOI] [PubMed] [Google Scholar]
- 41.Tyndall JD, Fairlie DP. Conformational homogeneity in molecular recognition by proteolytic enzymes. Journal of molecular recognition : JMR. 1999;12:363–370. doi: 10.1002/(SICI)1099-1352(199911/12)12:6<363::AID-JMR478>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 42.Moore SJ, Gephart MGH, Bergen JM, Su YRS, Rayburn H, Scott MP, Cochran JR. Engineered knottin peptide enables noninvasive optical imaging of intracranial medulloblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14598–14603. doi: 10.1073/pnas.1311333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern LA, Case BA, Hackel BJ. Alternative Non-Antibody Protein Scaffolds for Molecular Imaging of Cancer. Current opinion in chemical engineering. 2013;2 doi: 10.1016/j.coche.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldstein R, Sosabowski J, Livanos M, Leyton J, Vigor K, Bhavsar G, Nagy-Davidescu G, Rashid M, Miranda E, Yeung J, et al. Development of the designed ankyrin repeat protein (DARPin) G3 for HER2 molecular imaging. Eur J Nucl Med Mol Imaging. 2014 doi: 10.1007/s00259-014-2940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau YH, de Andrade P, Quah ST, Rossmann M, Laraia L, Skold N, Sum TJ, Rowling PJE, Joseph TL, Verma C, et al. Functionalised staple linkages for modulating the cellular activity of stapled peptides. Chemical Science. 2014;5:1804–1809. [Google Scholar]
- 46.Lau YH, de Andrade P, McKenzie GJ, Venkitaraman AR, Spring DR. Linear Aliphatic Dialkynes as Alternative Linkers for Double-Click Stapling of p53-Derived Peptides. Chembiochem. 2014 doi: 10.1002/cbic.201402374. [DOI] [PubMed] [Google Scholar]
- 47.Li H, Aneja R, Chaiken I. Click chemistry in peptide-based drug design. Molecules. 2013;18:9797–9817. doi: 10.3390/molecules18089797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White CJ, Yudin AK. Contemporary strategies for peptide macrocyclization. Nat. Chem. 2011;3:509–524. doi: 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.