Summary
Insulin/IGF‐1‐like signalling (IIS) and dietary restriction (DR) are the two major modulatory pathways controlling longevity across species. Here, we show that both pathways license a common chromatin modifier, ZFP‐1/AF10. The downstream transcription factors of the IIS and select DR pathways, DAF‐16/FOXO or PHA‐4/FOXA, respectively, both transcriptionally regulate the expression of zfp‐1. ZFP‐1, in turn, negatively regulates the expression of DAF‐16/FOXO and PHA‐4/FOXA target genes, apparently forming feed‐forward loops that control the amplitude as well as the duration of gene expression. We show that ZFP‐1 mediates this regulation by negatively influencing the recruitment of DAF‐16/FOXO and PHA‐4/FOXA to their target promoters. Consequently, zfp‐1 is required for the enhanced longevity observed during DR and on knockdown of IIS. Our data reveal how two distinct sensor pathways control an overlapping set of genes, using different downstream transcription factors, integrating potentially diverse and temporally distinct nutritional situations.
Keywords: C. elegans, dietary restriction, daf‐16, insulin signalling, incoherent feed‐forward loop, lifespan, pha‐4, zfp‐1
Introduction
Organisms may possess an intrinsic ability to modulate lifespan in response to the nutritional status of the environmental niche they reside in. This is reflected in the fact that modulations of the conserved insulin/IGF‐1 signalling (IIS) pathway or dietary/caloric restriction (DR) lead to dramatic increases in longevity across species (Kenyon, 2005, 2010). Perfunctorily, it would appear that the modulatory signalling through the IIS pathway that couples nutrient status to aging will generally overlap with the mechanisms by which DR affects lifespan. However, earlier research in Caenorhabditis elegans as well as in Drosophila melanogaster has shown that their relationship is rather complicated. Lifespan extension in the case of the IIS signalling mutants is strictly dependent on the FOXO transcription factor DAF‐16, while different DR regimes have varied requirements for downstream transcription factors (Greer et al., 2007a; Panowski et al., 2007; Greer & Brunet, 2009; Kenyon, 2010). For example, DAF‐16 is required for lifespan extension when DR is initiated by bacterial dilution on solid plates (sDR) or by the dilution of peptone (Greer & Brunet, 2009). On the other hand, in C. elegans, a FOXA transcription factor PHA‐4 is required for two other types of DR‐induced longevity, namely the bacterial dilution in liquid culture (bDR) and in the eat‐2 mutant (Panowski et al., 2007). However, PHA‐4 is not required for IIS pathway mutants to increase lifespan (Panowski et al., 2007). Further, genetically combining an IIS mutant with an eat‐2 mutant increased lifespan over and above that of the individual long‐lived mutants, suggesting independent mechanisms (Lakowski & Hekimi, 1998). Additionally, these pathways seem to have developed extensive mechanisms of crosstalks involving other transcriptional regulators. A FOXO/DAF‐16 coregulator SMK‐1 is required for the DR‐mediated lifespan extension in C. elegans, although the mechanism has not been elucidated (Wolff et al., 2006). Also, the TFEB orthologue HLH‐30 and NRF2‐like transcription factor SKN‐1 function downstream of both the pathways (Bishop & Guarente, 2007; Tullet et al., 2008; Lapierre et al., 2013) to regulate lifespan through distinct mechanisms. Thus, there may be other possible common players integrating IIS and DR signalling that needs to be deciphered in order to understand their complex relationship.
Here, we identify a chromatin‐associated factor ZFP‐1/AF10 as a common mediator of IIS‐ and DR‐mediated lifespan regulation. ZFP‐1 is the homolog of AF10, a zinc finger protein that has roles in RNA interference (RNAi) and development (Grishok et al., 2008; Avgousti et al., 2013). In mammals, AF10 is best known as a fusion partner of the mixed lineage leukaemia (MLL) or clathrin‐associated lymphoid myeloids (CALM) genes that lead to acute leukaemia, mostly in infants. Fused MLL‐AF10 or CALM‐AF10 results in dysregulation of the HOX gene cluster in conjunction with MEIS1 and may be the putative mechanism of leukaemia (Caudell & Aplan, 2008). AF10/ZFP‐1 is known to interact with H3K79 methyltransferase Dot‐1‐like (DOT1L; DOT‐1 in C. elegans) using its octapeptide motif‐leucine zipper domain (OM‐LZ) as well as with GLIOMA‐AMPLIFIED SEQUENCE‐41 (GAS41; GFL‐1 in C. elegans) (Debernardi et al., 2002; Cecere et al., 2013). The ZFP‐1(AF10)/DOT‐1 complex is involved in RNA polymerase II pausing during development and stress by modifying H3K79 and consequently H2B monoubiquitination patterns (Cecere et al., 2013). ZFP‐1 has also been found to utilize its PHD1–PHD2 domains to interact with lysine 4‐methylated histones and is required during embryogenesis (Avgousti et al., 2013). GAS41 is similar to AF9, another fusion partner of MLL and interacts with the SWI/SNF complex (Debernardi et al., 2002). Interestingly, DAF‐16 has been shown recently to promote stress resistance and longevity by employing the SWI/SNF complex (Riedel et al., 2013).
In an earlier study, ZFP‐1 emerged as a strong direct target of DAF‐16/FOXO (Oh et al., 2006). In this study, we demonstrate that both DAF‐16/FOXO and PHA‐4/FOXA directly bind and regulate the expression of the different isoforms of zfp‐1 as well as gfl‐1. Interestingly, ZFP‐1/GFL‐1 in turn negatively regulates the expression of DAF‐16/FOXO and PHA‐4/FOXA target genes, forming apparent incoherent feed‐forward loops (FFLs). We provide evidence that ZFP‐1/GFL‐1 determines the amplitude and duration of target gene expression during low IIS or DR. We show that ZFP‐1 negatively influences the recruitment of DAF‐16 and PHA‐4 to the promoters of their direct target genes, thereby repressing their transcription. Knocking down zfp‐1 or gfl‐1 may therefore lead to large‐scale deregulation of DAF‐16 or PHA‐4 target gene expression under low IIS or DR, respectively. Consequently, ZFP‐1 and GFL‐1 are required for IIS‐ and DR‐mediated longevity assurance. Our study elucidates how two sensor pathways, processing possibly diverse nutrient information cues, converge on a single chromatin‐associated factor to fine‐tune the expression of an overlapping set of genes. Because DAF‐16, PHA‐4 and ZFP‐1 are highly conserved proteins, it is possible that such ZFP‐1/GFL‐1‐mediated fine‐tuning of downstream target gene expression is commonly used by IIS and DR in higher mammals.
Results
DAF‐16/FOXO regulates different isoforms of zfp‐1 as well as gfl‐1
The zfp‐1 gene encodes three distinct isoforms (Figure 1A). The ZFP‐1(2a) protein has two conserved domains, the PHD1–PHD2 zinc finger and the OM‐LZ motif, while ZFP‐1(2c) lacks the PHD1–PHD2 domain (Mansisidor et al., 2011; Avgousti et al., 2013). A third isoform, ZFP‐1(2b) also exists but lacks both the domains and was not considered in this study. Each isoform has distinct SL1 sites (Figure S1A) and their individual promoters can drive the expression of GFP in a tissue‐specific manner (Fig. 1B). While the zfp‐1(2c) promoter drove GFP expression uniformly in the worm, the zfp‐1(2a) promoter‐driven expression was noticeably absent from the pharynx, germline and the tail regions. A genome‐wide endogenous DAF‐16/FOXO ChIP sequencing study in our laboratory (Kumar et al., 2015) showed that the transcription factor binds to the promoters of both the zfp‐1 isoforms in the temperature‐sensitive daf‐2(e1370) allele of daf‐2 (Fig. 1C; see supplementary Materials and methods for analysis details and data access links). Three peaks each were observed on the promoters of both zfp‐1(2a) and zfp‐1(2c) (Fig. 1C). The daf‐2 gene codes for the IIS receptor in worms that negatively regulates DAF‐16/FOXO through a conserved signalling cascade (Kenyon, 2010). The daf‐2(e1370) allele, where DAF‐16 is in an activated state, has extended lifespan, enhanced stress tolerance and at 25 °C arrests as dauers, an alternative developmental stage controlled by the IIS, while at 15 or 20 °C, it enters reproductive development. We validated the binding of DAF‐16 to the individual promoter regions by ChIP‐PCR (Fig. 1D). We further investigated whether each of these isoforms are transcriptionally dependent on DAF‐16. It is not possible to separately detect the zfp‐1(2c) isoform as it has 100% overlap with zfp‐1(2a); the 2a isoform has additional 5′ exons [Fig. 1A,D (lower panel)]. We therefore designed primers specific to the zfp‐1(2a) isoform and a pair that detected the zfp‐1(2a) together with the zfp‐1(2c) isoform (referred to as 2ac). The expression of zfp‐1(2c) may then be deduced by subtracting the zfp‐1(2a) expression values from that of zfp‐1(2ac). We compared the expression of these isoforms between wild‐type, daf‐2(e1370) [represented from now on as daf‐2(‐) for convenience although it is not a null mutant at 20 °C], daf‐16(mgDf50) [daf‐16(‐)] and daf‐16(mgDf50);daf‐2(e1370) [daf‐16(‐); daf‐2(‐)], grown at 20 °C using quantitative RT–PCR (QRT–PCR). We found that both the isoforms are upregulated in daf‐2(‐) compared to wild‐type, in a daf‐16‐dependent manner (Fig. 1E,F). The basal expression of zfp‐1(2a) and zfp‐1(2ac) genes is also dependent on daf‐16. Together, the isoforms of zfp‐1 are direct transcriptional targets of DAF‐16.
Figure 1.
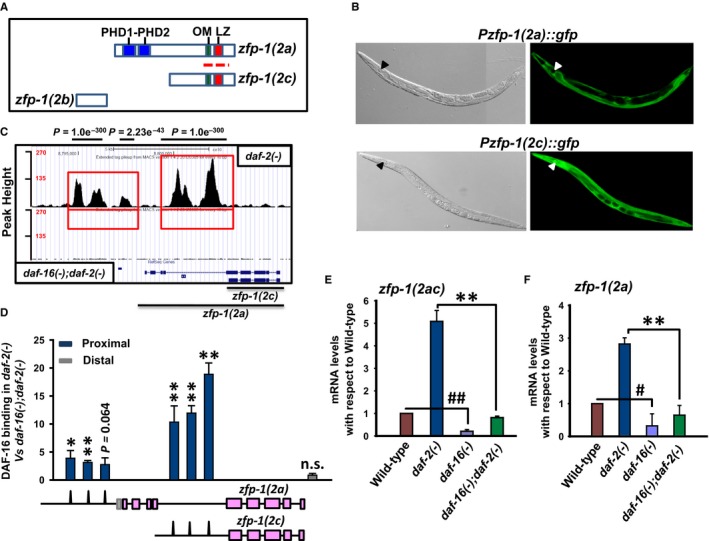
The isoforms of worm AF10 ortholog are direct transcriptional target of DAF‐16/FOXO. (A) Organization of ZFP‐1 isoforms. The polypeptide encoded by zfp‐1(2a) isoform possesses the PHD domain, OM (octapeptide motif) and LZ (leucine zipper) motifs, whereas the shorter isoform encoded by zfp‐1(2c) has the OM‐LZ motif only. The third isoform zfp‐1(2b) lacks all these domains and motifs. Red dotted line indicates region from where RNAi was designed. (B) Expression pattern of GFP driven by zfp‐1(2a) or zfp‐1(2c) promoters (right panels). Corresponding DIC images are shown in the left panels. Pharynx is marked by arrow head. (C) UCSC browser view of DAF‐16/FOXO peaks as determined by ChIP‐seq using anti‐DAF‐16/FOXO antibody (Kumar et al., 2015). Red boxes indicate the promoter regions of zfp‐1(2a) and zfp‐1(2c) where DAF‐16/FOXO peaks are observed. Lower panel shows daf‐16(mgDf50);daf‐2(e1370) [represented as daf‐16(‐);daf‐2(‐)] that lacks specific DAF‐16/FOXO peaks, while upper panel shows peaks obtained in daf‐2(e1370) [represented as daf‐2(‐)] (D) ChIP‐PCR validation of DAF‐16/FOXO binding to zfp‐1 promoters obtained by ChIP‐seq. Binding in daf‐2(‐) is normalized to that of daf‐16(‐);daf‐2(‐). The corresponding peaks on the promoters of zfp‐1, as obtained by ChIP‐seq, are shown pictographically below the graph. (E‐F) quantitative RT (QRT)–PCR detection of transcript levels for zfp‐1(2ac) or zfp‐1(2a) in WT and different mutants as mentioned. The zfp‐1(2c) transcript cannot be detected separately from zfp‐1(2a) as explained in the text. Error bars are standard deviation. **P ≤ 0.01; *P ≤ 0.05; n.s., P not significant by Student's t‐test. ## P ≤ 0.01; # P ≤ 0.05 compared to wild‐type. The graphs were plotted from three experiments.
In mammals, AF10 physically associates with GAS41, a protein that interacts with the human SWI/SNF complex (Debernardi et al., 2002). DAF‐16 ChIP‐seq data indicated that gfl‐1 (the C. elegans gene that codes for the GAS41 homolog) promoter is bound by DAF‐16 (Figure S1B). We verified the binding by ChIP‐PCR using primers designed against the single DAF‐16 binding site (Figure S1C). Additionally, the expression of gfl‐1 in daf‐2(‐) is dependent on daf‐16, although in wild‐type it is not (Figure S1D). Together, zfp‐1 and its interactor, gfl‐1 (that has overlapping expression pattern with zfp‐1 isoforms; Figure S1E), are direct targets of DAF‐16, downstream of the IIS pathway.
Differential regulation of zfp‐1 isoforms by different DAF‐16/FOXO isoforms
ZFP‐1 is a strong target of DAF‐16/FOXO (Oh et al., 2006), indicating that it may carry out important functions downstream of the transcription factor. The daf‐16 gene codes for several distinct and well‐characterized isoforms (Lee et al., 2001; Lin et al., 2001; Kwon et al., 2010). The daf‐16(a) isoform is coded by R13H8.1b, the daf‐16(b) by R13H8.1a and the daf‐16(f) by R13H8.1f. We next asked whether all the DAF‐16 isoforms regulate the ZFP‐1 isoforms, attesting to the importance of this downstream target. We used integrated strains where daf‐16(a) (HT1881), daf‐16(b) (HT1882) or daf‐16(f) (HT1883) is rescued in daf‐16(‐);daf‐2(‐) (Kwon et al., 2010) and determined binding of these isoforms to zfp‐1 or gfl‐1 promoters (Figures S2A and S3A). We found that all the isoforms of DAF‐16 bind to the promoters of zfp‐1 or gfl‐1 to varying extent (Figures S2A, S3A). QRT–PCR analysis revealed that all the zfp‐1 isoforms as well as gfl‐1 are transcriptionally regulated by all the three DAF‐16 isoforms (Figures S2B and S3B). These observations supported our assumption that ZFP‐1 and GFL‐1 may be important DAF‐16/FOXO targets.
The FOXA transcription factor PHA‐4 also regulates zfp‐1 and gfl‐1 gene expression
Forkhead transcription factors play important roles in regulating longevity in C. elegans. While DAF‐16/FOXO acts mostly downstream of the IIS (Kenyon, 2010) and in some forms of DR (Greer & Brunet, 2009), PHA‐4/FOXA is required for two other paradigms of DR‐mediated longevity, but not for IIS (Panowski et al., 2007). As zfp‐1 and gfl‐1 were found to be robust targets of DAF‐16/FOXO, we asked whether they are also regulated by the FOXA transcription factor. In worms, DR regimes that require PHA‐4 can be initiated either by using the eat‐2 mutants or by diluting the bacterial feed (Panowski et al., 2007). The eat‐2 mutants have defective pharyngeal pumping that lead to lower food intake and are considered to mimic DR (Lakowski & Hekimi, 1998). We reanalysed the PHA‐4 ChIP‐seq data available at MODENCODE (Zhong et al., 2010) using our analysis pipeline and found that zfp‐1 and gfl‐1 promoters are directly bound by PHA‐4 (Figs 2A and S4A). While two distinct PHA‐4 binding peaks were observed on the zfp‐1(2a) and zfp‐1(2c) promoters, gfl‐1 promoter possessed a single peak (Figs 2A and S4A). We validated the recruitment of PHA‐4 to the individual promoter regions by ChIP QRT–PCR in unc‐119(ed3) III; wgIs37 (OP37) strain using anti‐GFP antibody (Fig. 2B). Consequently, the expression of the zfp‐1 isoforms and gfl‐1 is elevated in eat‐2(ad1116) as well as in eat‐2(ad1113) and eat‐2(ad465), compared to wild‐type (Figs 2C,D and S4B). This increased expression was dependent on PHA‐4 (Figs 2E, S4C and S4D). Importantly, in another newly identified DR‐like paradigm (Chamoli et al., 2014), knocking down a mekk‐3‐like gene drl‐1 led to an increased expression of the zfp‐1 isoforms, but not gfl‐1 (Figure S4E). Together, these data show that similar to DAF‐16/FOXO, PHA‐4/FOXA also transcriptionally regulates the expression of zfp‐1 and gfl‐1.
Figure 2.
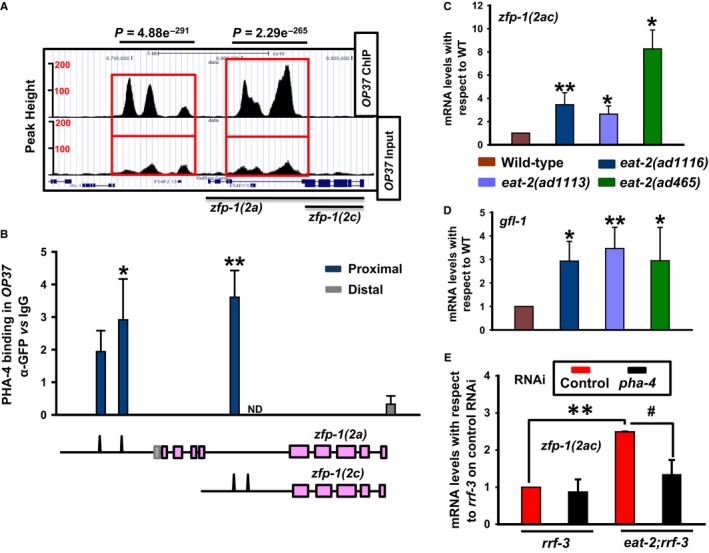
Dietary restriction (DR)‐specific transcription factor PHA‐4/FOXA also directly regulates zfp‐1 and gfl‐1. (A) UCSC browser view of PHA‐4/FOXA peaks on zfp‐1(2a) and zfp‐1(2c) promoters as determined by ChIP‐seq analysis of unc‐119(ed3) III; wgIs37 (OP37) strain; data mined from MODENCODE and reanalysed using our bioinformatic pipeline. Red boxes indicate the promoter regions of zfp‐1(2a) and zfp‐1(2c) where peaks are observed. Lower panel shows peaks in input samples. (B) ChIP‐PCR validation of PHA‐4/FOXA binding to zfp‐1 promoters obtained by ChIP‐seq analysis of OP37 strain. Enrichment using GFP antibody when normalized to that of IgG is plotted in the Y‐axis. The corresponding peaks on the promoters of zfp‐1, as obtained by ChIP‐seq (MODENCODE), are shown pictographically below the graph. ND – not determined. (C,D) Quantitative RT (QRT)–PCR analysis of the transcript levels of zfp‐1(2ac) (C) or gfl‐1 (D) in different eat‐2 mutant strains. The graph is plotted from three or more experiments. (E) QRT–PCR detection of transcript levels for zfp‐1(2ac) in rrf‐3(pk1426) or eat‐2(ad1116);rrf‐3(pk1426) grown on control or pha‐4 RNAi. Error bars are standard deviation. **P ≤ 0.01;*P ≤ 0.05; # P ≤ 0.05 by Student's t‐test.
ZFP‐1 regulates DAF‐16 target gene expression
Why would DAF‐16/FOXO regulate the transcription of chromatin modifiers like ZFP‐1/GFL‐1? We hypothesized that because DAF‐16 and ZFP‐1 may be part of a larger complex (Riedel et al., 2013), they would possibly influence the expression of DAF‐16 target genes. To study this aspect of regulation, we focused on the well‐known DAF‐16 direct target, sod‐3 (Oh et al., 2006; Mukhopadhyay et al., 2008; Kumar et al., 2015). Under conditions of low insulin signalling, as seen in daf‐2(‐), sod‐3 is chronically upregulated in a daf‐16‐dependent manner (Libina et al., 2003). The dynamics of this regulation can be studied by using an integrated Psod‐3::gfp transgenic strain (Libina et al., 2003). We grew Psod‐3::gfp, daf‐16(‐);Psod‐3::gfp, daf‐2(‐);Psod‐3::gfp or daf‐16(‐);daf‐2(‐);Psod‐3::gfp on control, zfp‐1(2ac) or gfl‐1 RNAi and quantified the GFP fluorescence of the worms. The zfp‐1(2ac) RNAi effectively knocks down the expression of both the 2a and the 2c transcripts, while the gfl‐1 RNAi significantly reduced gfl‐1 mRNA levels (Figure S4F, G). We found that knocking down the zfp‐1(2ac) as well as gfl‐1 increased the expression of Psod‐3::gfp in WT as well as in daf‐2(‐) background and this was dependent on daf‐16 as we observed no increase in daf‐16(‐);Psod‐3::gfp or daf‐16(‐);daf‐2(‐);Psod‐3::gfp (Fig. 3A,B,D). QRT–PCR analysis of sod‐3 mRNA expression in WT and daf‐2(‐) also showed a similar upregulation when zfp‐1 was knocked down (Figure S5A). Thus, DAF‐16 positively controls the expression of sod‐3 as well as zfp‐1. However, ZFP‐1/GFL‐1 negatively regulates the expression of sod‐3. Therefore, these genes form a typical type I incoherent feed‐forward loop (FFL) (Mangan & Alon, 2003; Alon, 2007) downstream of the IIS (Fig. 3C). This type I incoherent FFL has two transcriptional regulators, one (DAF‐16) positively regulating the other (ZFP‐1). These factors then regulate a common transcriptional target sod‐3; DAF‐16 controls it positively, while ZFP‐1 negatively modulates it.
Figure 3.
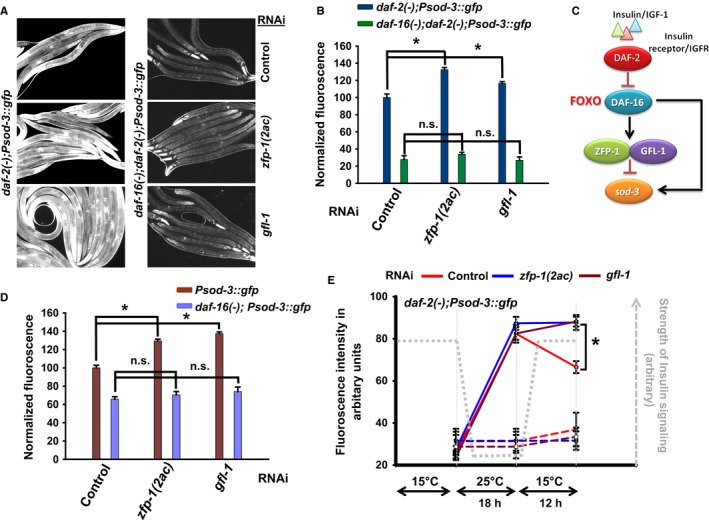
ZFP‐1 and GFL‐1 negatively regulate sod‐3, a positively regulated DAF‐16/FOXO direct target, forming a type 1 incoherent feed‐forward loop. (A) daf‐2(‐);Psod‐3::gfp or daf‐16(‐);daf‐2(‐);Psod‐3::gfp was grown on control, zfp‐1(2ac) or gfl‐1 RNAi and images captured under fluorescence microscope at 100× magnification. (B) Quantification of GFP fluorescence as observed in (A) using ImageJ. Fluorescence was normalized to daf‐2(‐);Psod‐3::gfp on control RNAi. The error bars indicate SEM between two or more experiments. (C) A circuit diagram showing a type I incoherent feed‐forward loop involving ZFP‐1, DAF‐16/FOXO and sod‐3. (D) Quantification of GFP fluorescence in Psod‐3::gfp or daf‐16(‐);Psod‐3::gfp on control, zfp‐1(2ac) or gfl‐1 RNAi. Fluorescence was normalized to Psod‐3::gfp on control RNAi. The error bars indicate SEM between three experiments. (E) daf‐2(‐);Psod‐3::gfp worms were grown at 15 °C on control, zfp‐1(2ac) or gfl‐1 RNAi since hatching. L3 worms were shifted to 25 °C for 18 h and then shifted back to 15 °C. At the mentioned time points, images were acquired using a fluorescence microscope and quantified using ImageJ. Data shown are from one of two experiments. Dotted lines represent the worms maintained at 15 °C throughout on respective RNAi. The error bars indicate standard error mean for n > 25. *P ≤ 0.05 by Student's t‐test, n.s., non‐significant.
To determine whether such a mode of regulation exists only in the case of sod‐3, we checked several other genes regulated by DAF‐16 (Kwon et al., 2010; Zhang et al., 2013) and observed a similar upregulation of expression in the case of mtl‐1, lys‐7, ZK742.4, hsp‐12.6, sod‐5, scl‐1, sip‐1, dod‐11, gpd‐2 as well as hsp‐16.2 when zfp‐1(2ac) was knocked down (Figure S5A–C). This showed that DAF‐16 and ZFP‐1 may regulate many other genes using FFLs. However, we found that gfl‐1 knockdown did not significantly change expression in the case of mtl‐1, hsp‐16.2 and gpd‐2 (Figure S5B), but had a more robust response in the case of dod‐11 (Figure S5C), suggesting that GFL‐1 and ZFP‐1 may collaborate in some contexts, but not in others.
ZFP‐1 determines the duration of expression of DAF‐16 target genes
Incoherent FFLs typically function to control the amplitude and duration of gene expression (Alon, 2007). We asked whether the DAF‐16:ZFP‐1:SOD‐3 FFL performs similar function downstream of the IIS pathway. By shifting the temperature‐sensitive daf‐2(‐) between 15 and 25 °C, we could modulate the IIS and as a consequence, the sod‐3::gfp expression in the mutant. The daf‐2(‐);Psod‐3::gfp worms were grown at 15 °C on control RNAi, zfp‐1(2ac) or gfl‐1 RNAi from hatching and then shifted to 25 °C for 18 h at L2 stage. This step led to an enhanced sod‐3::gfp expression, possibly due to further lowering of the IIS and/or activation of DAF‐16 (Fig. 3E). Worms that were continuously maintained at 15 °C did not exhibit the surge in GFP expression (Fig. 3E). These worms were then returned to 15 °C, and the expression of Psod‐3::gfp was monitored after 12 h. We found that while the expression of Psod‐3::gfp declined when the worms were restored to 15 °C on control RNAi, expression in the ones grown on zfp‐1(2ac) or gfl‐1 RNAi failed to decrease to similar extent (Fig. 3E). We performed similar experiments using QRT–PCR to detect mRNA levels of sod‐3 and mtl‐1; the fall in the mRNA expression levels was slower in the case of zfp‐1 as well as gfl‐1 knockdown (Figure S5D). Thus, ZFP‐1 and GFL‐1 regulate the duration of sod‐3 gene expression, apart from amplitude, following the activation of DAF‐16/FOXO.
ZFP‐1 regulates the extent of DAF‐16 binding to target promoters
We found that knocking down the zfp‐1(2ac) isoform leads to an increased expression of DAF‐16 direct targets like sod‐3 and mtl‐1. In order to understand the nature of this regulation, we asked whether the recruitment of DAF‐16 is influenced by ZFP‐1. For this, we performed ChIP using anti‐DAF‐16 antibody in wild‐type, zfp‐1(ok554) as well as daf‐16(‐), and evaluated DAF‐16 binding at the sod‐3 promoter by quantitative PCR. We found that binding of DAF‐16 increased significantly in the absence of ZFP‐1, on the sod‐3 promoter (Fig. 4A). However, no increase was seen in the distal region of sod‐3. This correlates well with the increase in sod‐3 expression on zfp‐1(2ac) knockdown and showed that ZFP‐1 negatively regulates the direct binding of DAF‐16 to the sod‐3 promoter. Similar observations were made in the case of other DAF‐16 targets (Fig. 4A). Interestingly, ZFP‐1 also negatively regulates the binding of DAF‐16 to its own promoter, forming a feed‐back loop as shown earlier (Cecere et al., 2013). We were unable to generate a daf‐2(e1370);zfp‐1(ok554) or daf‐2(e1368);zfp‐1(ok554) double mutant due to larval lethality, as reported previously (Mansisidor et al., 2011), but anticipate similar mechanism in a daf‐2(‐) scenario. Thus, ZFP‐1 may fine‐tune DAF‐16‐dependent expression of genes by regulating the binding of the transcription factor to its target promoters.
Figure 4.
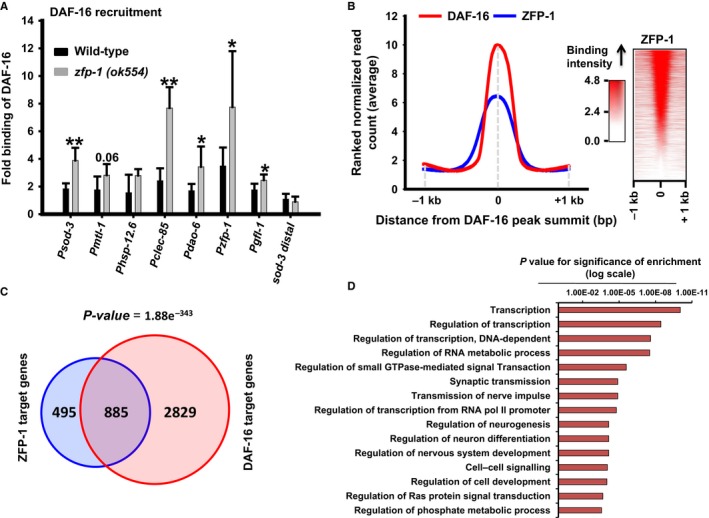
ZFP‐1 influences DAF‐16/FOXO recruitment to its target promoters. (A) ChIP‐PCR to determine DAF‐16 recruitment to different target promoters in WT and zfp‐1(ok554). Binding in WT /zfp‐1(ok554) is normalized to that of daf‐16(‐). Recruitment at a distal region of sod‐3 is taken as control. The graph is plotted from three experiments. Error bars represent standard deviation. **P ≤ 0.01; *P ≤ 0.05 by Student's t‐test. (B) Distribution of ZFP‐1 peaks with respect to DAF‐16/FOXO binding summits as determined by ChIP‐seq experiments (left panel). Distribution of ZFP‐1 reads with respect to DAF‐16 binding summit across all chromosomes (right panel). ZFP‐1 ChIP‐seq data from MODENCODE were reanalysed using our bioinformatic pipeline. (C) Overlap of ZFP‐1 and DAF‐16 direct target genes as determined by ChIP‐seq experiments. (D) Gene Ontology (GO) term analysis of common target genes of DAF‐16/FOXO and ZFP‐1. Only top 15 GO terms that were enriched are shown.
Next, we asked whether this phenomenon is restricted to sod‐3 and a few other DAF‐16 direct targets or is a more global feature. For this, we reanalysed the ChIP‐seq data for ZFP‐1 available at MODENCODE (seq‐JL00006_ZFP1_N2_L3) and compared it with that of our DAF‐16 ChIP‐seq data (Kumar et al., 2015). We found that the ZFP‐1 peaks on the chromatin colocalized with the DAF‐16 peak summits over the entire genome (Fig. 4B, left panel). This indicated that DAF‐16 and ZFP‐1 binds to similar regions on the chromatin. However, not all DAF‐16 peaks overlap with those of ZFP‐1 (Fig. 4B, right panel), suggesting that other DAF‐16 direct targets may be regulated differently. Alternatively, it is possible that because the DAF‐16 and ZFP‐1 ChIP‐seq experiments were performed in different genetic backgrounds and developmental stages, not all peaks show an overlap. Additionally, it appears that the ZFP‐1 footprint on DAF‐16 target promoters may vary depending on the target genes as we observed narrow as well as broad spread of ZFP‐1 binding with respect to DAF‐16 summits (Fig. 4B, right panel). Together, ZFP‐1 may bind to a large number of regions where DAF‐16/FOXO binds in the genome and influences the recruitment of the latter to the chromatin.
Genes targeted by both DAF‐16 and ZFP‐1
Because ZFP‐1, a direct target of DAF‐16/FOXO, also influences the binding of DAF‐16 to its target promoters and forms feed‐forward loops, we asked what genes are cotargeted by the two proteins. For this, we only considered the genes where DAF‐16 and ZFP‐1 peaks are situated within the 2.0 kb of the promoter proximal region. We found that ZFP‐1 binds to the promoters of 1380 genes, while DAF‐16 peaks were found on 3714 gene promoters (Fig. 4C). However, a total of 885 genes were targeted by both DAF‐16 and ZFP‐1 (P = 1.88e‐343 by hypergeometric test) that may constitute the feed‐forward loops. We used the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 package (Dennis et al., 2003) to analyse Gene Ontology (GO) term enrichment in these 885 common genes to predict their biological functions. We found that these genes are mostly enriched for molecular components required for transcriptional regulation, signal transduction and neurogenesis/neural signalling (Fig. 4D, Table S4). In fact, transcriptional regulators like nuclear hormone receptor (P = 9.42E‐11), zinc finger proteins (P = 4.55E‐11) and signalling proteins like serine/threonine protein kinases (P = 5.32E‐07), Pleckstrin homology (PH) domain possessing proteins (P = 6.81E‐05), Src homology‐3 (SH3) domain containing proteins (P = 2.96E‐04) as well as EF‐hand proteins (P = 3.57E‐04) are significantly enriched. Pathway analysis using Kyoto Encyclopaedia of Genes and Genomes (KEGG) database revealed an enrichment of components of MAPK signalling (P = 1.60E‐04), mTOR signalling (P = 1.10E‐03) as well as Jak‐STAT signalling (P = 3.60E‐03) pathways. These data indicate that DAF‐16 and ZFP‐1 coregulate genes that may be important for longevity effects seen in daf‐2(‐). In fact, 62 genes among the common DAF‐16 and ZFP‐1 targets have known function in aging (P = 4.34 e‐7) according to the GenAge database (http://genomics.senescence.info/genes/), while genes involved in aging and lifespan regulation were enriched in the GO term analysis (P = 3.90E‐02) (Table S4).
PHA‐4/FOXA and ZFP‐1 also constitute FFLs downstream of DR signalling
Next, we determined whether ZFP‐1 is also a part of a regulatory loop involving PHA‐4/FOXA, similar to the case of DAF‐16/FOXO. For this, we chose a few published targets of PHA‐4 like sod‐1, sod‐2, sod‐4 (Panowski et al., 2007), cyp‐32B1, cyp‐33C8, cyp‐34A4, cyp‐37B1 and ugt‐16 (Chamoli et al., 2014). Knocking down zfp‐1(2ac) significantly increased the expression of several of these genes in the eat‐2 mutant, thereby showing that these genes may also form FFLs downstream of DR (Fig. 5A). We found that knocking down zfp‐1 also leads to a greater enrichment of PHA‐4 at its target promoters, as in the case of DAF‐16 (Fig. 5B). Although we show that ZFP‐1/PHA‐4 FFLs controlled the amplitude of target gene expression, duration control experiments could not be performed in the eat‐2 mutant. However, similar to DAF‐16, we found that ZFP‐1 peaks overlapped with PHA‐4 peaks in the ChIP‐seq experiments (Fig. 5C, left panel) although not all the PHA‐4 peaks are occupied by ZFP‐1 (Fig. 5C, right panel). Finally, a significant number of genes having PHA‐4 binding sites in the 2.0 kb promoter region overlapped with targets of ZFP‐1 (P = 5.54e‐133 by hypergeometric test), showing that ZFP‐1 may also fine‐tune the expression of a large number of PHA‐4 direct targets (Fig. 5D).
Figure 5.
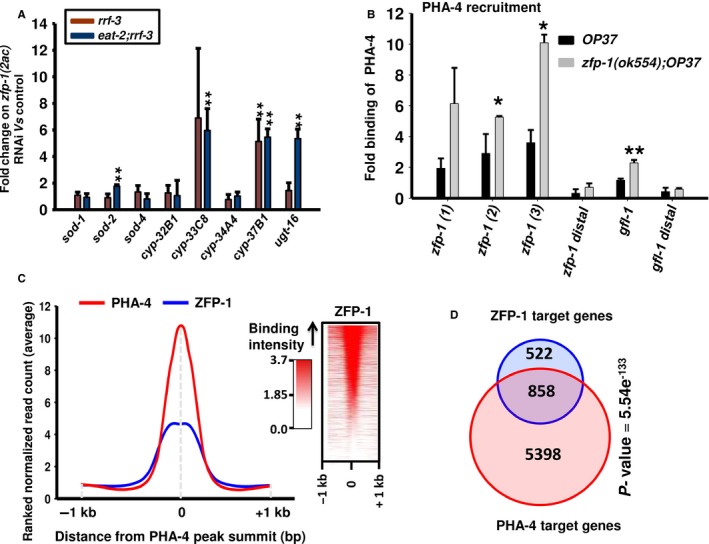
ZFP‐1 regulates expression of PHA‐4/FOXA target genes, forming incoherent feed‐forward loops (FFLs). (A) Relative expression levels of PHA‐4 target genes in rrf‐3(pk1426) or eat‐2(ad1116);rrf‐3(pk1426) grown on control vs zfp‐1(2ac) RNAi. (B) ChIP‐PCR to determine PHA‐4 recruitment to different target promoters in unc‐119(ed3) III; wgIs37 (OP37) and zfp‐1(ok554);OP37. Enrichment using anti‐GFP antibody when normalized to that of IgG control is plotted on the Y‐axis. Recruitment at zfp‐1 (three peak regions) or gfl‐1 promoters was quantified, while distal regions of zfp‐1 and gfl‐1 were taken as control. The graph is plotted from three experiments. Error bars are standard deviation. **P ≤ 0.01; *P ≤ 0.05 by Student's t‐test. (C) Distribution of ZFP‐1 peaks with respect to PHA‐4/FOXA binding summits as determined by reanalysis of previously published ChIP‐seq data available at MODENCODE (left panel). Distribution of ZFP‐1 reads with respect to PHA‐4/FOXA binding summit across all chromosomes (right panel). (D) Overlap of ZFP‐1 and PHA‐4/FOXA direct target genes as determined by the ChIP‐seq experiments.
ZFP‐1 and GFL‐1 are required for IIS‐ as well as DR‐mediated lifespan extension
Considering the role of DAF‐16/FOXO and PHA‐4/FOXA in regulating the amplitude/duration of their target genes using ZFP‐1/GFL‐1, knocking down the latter may deregulate a large part of the transcriptome during low IIS or DR. So we asked whether knocking down zfp‐1/gfl‐1 will influence the longevity associated with these pathways.
The daf‐2(‐) worms live more than double that of wild‐type (Kenyon et al., 1993). We grew the wild‐type or mutant worms on control, zfp‐1 (2ac), gfl‐1 or daf‐16 RNAi since hatching and performed lifespan analysis. We found that zfp‐1(2ac) RNAi suppressed the daf‐2(‐) lifespan by ~32% (Fig. 6B, Tables S1, S3). This shortening of daf‐2(‐) lifespan by zfp‐1 RNAi was not simply because of sickness as the RNAi did not shorten lifespan of wild‐type, daf‐16(‐) or daf‐16(‐);daf‐2(‐) to such extent (Figs 6A, S6A and B, Tables S1, S3). The gfl‐1 RNAi had a small but statistically significant effect on the daf‐2(‐) as well as WT lifespans.
Figure 6.
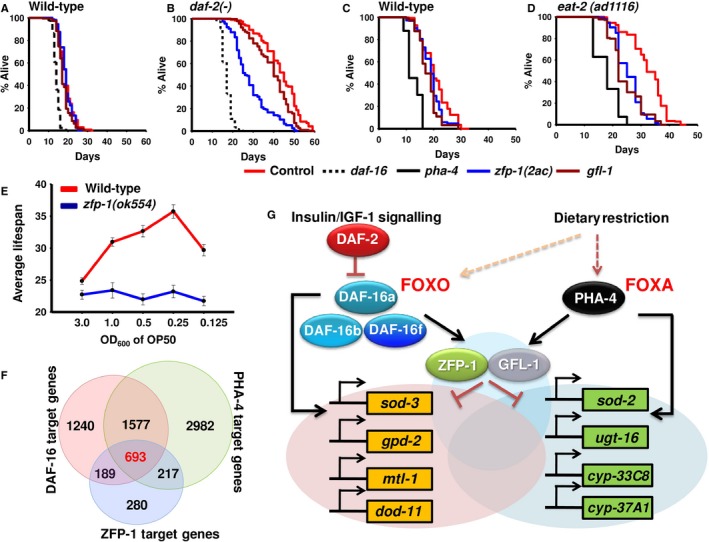
ZFP‐1 acts as a converging point for the IIS and dietary restriction (DR) pathways to regulate longevity and development. Effects of zfp‐1(2ac), gfl‐1 or daf‐16 knockdown on lifespan of WT (A) and daf‐2(‐)(B). The daf‐16 RNAi was taken as control. Effects of zfp‐1(2ac), gfl‐1 or pha‐4 knockdown on lifespan of wild‐type (C) and eat‐2(ad1116) (D). The pha‐4 RNAi was taken as control. (E) In zfp‐1(ok554), bacterial dilution‐induced DR (bDR) failed to produce a typical bell‐shaped curve as seen in WT. Error bars are SEM between two experiments. Lifespans were determined at 20 °C. (F) Overlap of direct target genes of DAF‐16, PHA‐4 and ZFP‐1 as determined by ChIP‐seq analysis. ChIP‐seq data for PHA‐4 and ZFP‐1 were obtained from MODENCODE, while DAF‐16 data were generated in the laboratory (Kumar et al., 2015). (G) Different isoforms of DAF‐16 and PHA‐4 transcriptionally regulate zfp‐1 and gfl‐1. ZFP‐1, along with GFL‐1, in turn, may fine‐tune the expression of a large number of genes downstream of the two pathways and controlled by DAF‐16 and/or PHA‐4, thereby modulating lifespan and development.
The daf‐16(a) and daf‐16(f) isoforms regulate lifespan differentially with the daf‐16(f) as the major contributor (Kwon et al., 2010). We knocked down the zfp‐1 and gfl‐1 in daf‐16(‐);daf‐2(‐);daf‐16(a) or daf‐16(‐);daf‐2(‐);daf‐16(f) and found that zfp‐1(2ac) RNAi was mainly able to suppress the daf‐16(‐);daf‐2(‐);daf‐16(f) lifespan (Figure S6D, Tables S1, S3) with no effect on daf‐16(‐);daf‐2(‐);daf‐16(a) (Figure S6C). To determine whether this effect was indeed specific, we evaluated another phenotype that is controlled by IIS, that is dauer formation. Interestingly for this phenotype, although both DAF‐16 isoforms (a and f) contribute to the phenotype, zfp‐1(2ac) RNAi was found to enhance only the DAF‐16(a)‐mediated dauer formation (Figure S6E–G). The gfl‐1 RNAi decreased lifespan in a small but statistically significant manner in both daf‐16(‐);daf‐2(‐);daf‐16(a)and daf‐16(‐);daf‐2(‐);daf‐16(f) (Figure S6C,D), similar to wild‐type. This suggests that in daf‐2(‐), zfp‐1(2ac) RNAi may be suppressing the effect of daf‐16(f) isoform. Together, zfp‐1 differentially affects lifespan and dauer phenotypes associated with the IIS pathway that are regulated by DAF‐16. In line with previous reports that DAF‐16 regulates longevity of germline‐ablated worms and TOR pathway mutants (Berman & Kenyon, 2006; Robida‐Stubbs et al., 2012), we observed that the increased lifespans of glp‐1(e2141) and let‐363(ok3018) were also partially dependent on zfp‐1 and gfl‐1 (Figure S7A–D).
Although gfl‐1 is a strong target of DAF‐16, we failed to observe any dramatic effect on lifespan of the low IIS mutant. It is possible that GFL‐1 is required downstream of the IIS to regulate phenotypes other than lifespan. This was indeed true as knocking down gfl‐1 in daf‐2(‐) suppressed dauer formation significantly (Figure S6E,F). Similar to zfp‐1(2ac), this effect on dauer formation was due to the contribution of daf‐16(a) and not the daf‐16(f) isoform (Figure S6G). Together, ZFP‐1 and GFL‐1 are required differentially for phenotypes controlled by the IIS pathway that are dependent on different isoforms of DAF‐16.
Next, we studied the role of ZFP‐1 and GFL‐1 in DR‐mediated longevity enhancement. As mentioned earlier, DR‐like condition can be initiated either by using the eat‐2 mutants or by knocking down drl‐1 or else by diluting the bacterial feed, all these interventions requiring PHA‐4. Lifespan analysis showed that knocking down zfp‐1(2ac) or gfl‐1 significantly decreased lifespan of eat‐2(ad1116), eat‐2(ad1113) and eat‐2(ad1116);rrf‐3(pk1426) (Figs 6D, S8B, C, Tables S1, S3). We did not observe a significant suppression in the case of eat‐2(ad465) (Figure S8A, Tables S1, S3). This shortening was not a nonspecific effect of sickness as we did not observe similar effects in the WT or RNAi‐hypersensitive mutant rrf‐3(pk1426) (Figs 6A, C, and S8D). Importantly, lifespan increase observed under drl‐1 (Chamoli et al., 2014) knockdown was completely dependent on zfp‐1 (Figure S8E), but independent of gfl‐1 (Figure S8F). For the latter case, it may also be noted that the expression of gfl‐1 does not change significantly on drl‐1 knockdown (Figure S4E), supporting the fact that different paradigms of DR may function through distinct mechanisms. This dependence was not due to the fact that zfp‐1 deletion prevents RNAi from working as we found that zfp‐1(ok554) failed to grow on pos‐1 RNAi, similar to WT (Figure S8G) (Dudley et al., 2002; Grishok et al., 2005). The pos‐1 gene codes for a CCCH‐type zinc finger protein that is required for specification of germ cells, intestine, pharynx and hypodermis (Tabara et al., 1999); knocking it down leads to larval lethality. Finally, in zfp‐1(ok554), the nongenetic mode of dietary restriction was unable to generate the typical bell‐shaped curve when average lifespan was plotted against the bacterial concentration (OD600), as seen in WT (Fig. 6E, Tables S1, S3). Together, these data suggest that zfp‐1 and gfl‐1 are also regulated by PHA‐4/FOXA and are differentially required for longevity in multiple paradigms of DR or in conditions that mimic DR.
Discussion
IIS and DR pathways commonly employ transcriptional feed‐forward loops involving ZFP‐1/AF10
DAF‐16/FOXO is a central regulator of longevity, stress tolerance, metabolism and development downstream of the IIS cascade (Kenyon, 2010). It is also required for longevity assurance through germline and neuronal signalling as well as for some DR regimes (Greer & Brunet, 2009; Kenyon, 2010). On the other hand, PHA‐4/FOXA that is an important factor in pharynx development (Gaudet & Mango, 2002) is also required for select DR‐induced lifespan extension (Panowski et al., 2007). How these transcription factors can regulate such a diverse array of phenotypes is an interesting and emerging area of research. Extensive research over the last decade has shown that DAF‐16 regulates a large number of genes, directly or indirectly (McElwee et al., 2003; Murphy et al., 2003; Oh et al., 2006; Schuster et al., 2010; Riedel et al., 2013; Tepper et al., 2013; Kumar et al., 2015). The activity of DAF‐16 is also regulated by multiple kinases including MST‐1, JNK‐1 and AMPK apart from the IIS pathway kinases (Oh et al., 2005; Lehtinen et al., 2006; Greer et al., 2007a,b; Kwon et al., 2010). Several modifying enzymes also interact with DAF‐16 to regulate its function under different conditions, including SIR‐2, RLE‐1, PRMT‐1 and p300/CBP‐1 (Wolff et al., 2006; Takahashi et al., 2011; Chiang et al., 2012; Riedel et al., 2013). Recently, DAF‐16 was found to employ chromatin‐modifying enzymes SWI/SNF to regulate gene expression (Riedel et al., 2013). On the other hand, very little is known about how PHA‐4 functions (Pandit et al., 2014). However, none of the regulators of these transcription factors have been reported to be a transcriptional direct target. Our study provides a glimpse of an additional level of sophistication in DAF‐16/FOXO biology and a new insight into PHA‐4 regulation. We show that DAF‐16 as well as PHA‐4 directly regulates the transcription of ZFP‐1, a protein involved in chromatin remodelling. ZFP‐1, on the other hand, modulates the transcription of DAF‐16‐ as well as PHA‐4‐regulated genes, possibly by preventing access of the transcription factors to chromatin, forming incoherent feed‐forward loops (Fig. 6G). ZFP‐1 is known to recruit DOT1.1 on actively transcribed genes to deposit H3K79 methylation, opposing H2B ubiquitination, to initiate a negative feedback regulation that modulates gene expression (Cecere et al., 2013). Thus, in the presence of ZFP‐1, chromosome compaction may ensue at the promoters that possibly prevent the access of DAF‐16 or PHA‐4 to the chromatin; in the absence of ZFP‐1, these transcription factors thus show a higher recruitment to the promoter of target genes.
Isoform‐specific interaction between DAF‐16 and ZFP‐1
Our data reveal an intricate relationship between the different isoforms of DAF‐16 and ZFP‐1. Previous studies have pointed at overlapping as well as distinct functions of the DAF‐16 isoforms that differentially affect lifespan (Kwon et al., 2010), with the DAF‐16(f) as the major contributor. Although all the isoforms of DAF‐16 regulate the expression of the zfp‐1 isoforms during low IIS condition, we found that zfp‐1 knockdown only affected the enhanced lifespan of the daf‐16(‐);daf‐2(‐);daf‐16(f) strain and not the one rescued with DAF‐16(a) isoform. On the other hand, ZFP‐1 interacted with DAF‐16(a) for dauer regulation. Thus, ZFP‐1 may influence the recruitment of different DAF‐16 isoforms, thereby differentially affecting their downstream genes and the resulting phenotypes.
ZFP‐1 fine‐tunes gene expression downstream of IIS and DR
The IIS and DR pathways are critical signal transducers that promptly respond to changes in nutritional status of an organism (Kenyon, 2010). Tight control of these pathways is essential for survival as evidenced by the lethality and/or arrest seen in insulin receptor‐knockout animals (Gems et al., 1998; Kitamura et al., 2003) and lifespan shortening when DR proceeds towards starvation (Mair & Dillin, 2008). Interestingly however, lowering the signals through IIS pathway or optimum nutrition can increase lifespan across species (Kenyon, 2010). Because DAF‐16/FOXO and PHA‐4/FOXA are the major outputs of these pathways, they need to be under strict regulation. While these proteins act as transcriptional activators, the system must build in safeguards to reduce the amplitude and duration of gene expression following the activation of the transcription factors by upstream kinases. One possible way of achieving that is through degradation by a ubiquitin‐proteasome pathway. But in the case of IIS, the RLE‐1‐mediated removal of DAF‐16 may have been designed to eliminate inactive DAF‐16 accumulating in the cytoplasm following AKT phosphorylation (Li et al., 2007). Alternatively, microRNA may be utilized effectively for this purpose. Although not known for DAF‐16/FOXO, we have recently shown that PHA‐4/FOXA may utilize microRNA FFLs to fine‐tune gene expression during DR (Pandit et al., 2014). By using such FFLs, the system builds in robustness that may reduce phenotypic variations. Here, we identify a mechanism that is common to both the pathways. We propose that the ZFP‐1‐mediated FFLs may have been devised to control the amplitude and duration of DAF‐16‐ or PHA‐4‐mediated gene expression, within the nucleus, following the changes in the nutrition available to the organism. By effectively coupling the activation of downstream genes with simultaneous upregulation of the negative regulator (ZFP‐1), IIS and DR pathways ensure that target genes are not overactivated. Thus, knocking down zfp‐1 in daf‐2(‐) or during DR may lead to the deregulation of a large number of DAF‐16 or PHA‐4 target genes, ultimately affecting longevity and other phenotypes. This type of interaction may not be limited to IIS or DR and may be commonly employed by other signalling pathways that culminate on DAF‐16 or PHA‐4.
ZFP‐1 represents another converging point for IIS and DR
The IIS and the DR pathways may respond to distinct nutritional cues to modulate gene expression, animal development and lifespan. While insulin signalling is typically sensitive to glucose availability, DR can be initiated even by restricting the intake of a single amino acid (Zimmerman et al., 2003; MacDonald et al., 2005; Miller et al., 2005; Grandison et al., 2009; Fontana & Partridge, 2015). Perfunctorily, it may appear that DR increases lifespan by modulating the IIS pathway, but these pathways often work independently. The transcription factor output of one of these signal transduction pathways may not always be licensed by the other, especially in the case of the IIS. These pathways, however, converge on core transcription factors like DAF‐16, SKN‐1 and HLH‐30 or coregulators like SMK‐1 to regulate DR‐ or IIS‐specific gene expression. Here, we provide new evidence that these two pathways also converge onto a conserved chromatin modifier to possibly fine‐tune the expression of an overlapping set of genes. In fact, we found that a considerable number of genes are commonly bound by DAF‐16, PHA‐4 as well as ZFP‐1 that may constitute important longevity genes (Fig. 6F). This observation raises an interesting conundrum; if such a large number of genes are shared between IIS and DR, why knocking down daf‐16 does not affect all forms of DR or why PHA‐4 is not required for daf‐2(e1370) lifespan. Although both the pathways involve ZFP‐1‐mediated FFLs to regulate the expression of overlapping sets of genes, they are probably activated under different temporal or spatial windows of nutritional cues. It is also possible that the IIS and DR pathways have different thresholds and are activated independently. Nonetheless, by using a common downstream factor like ZFP‐1/AF10, the signalling cascades may have designed a possible mechanism for synergism if both cascades are activated simultaneously.
Together, our study shows the intricate nature of gene regulatory modules downstream of IIS and DR pathways that may help fine‐tune gene expression for an enhanced longevity (Fig. 6G). Due to the conserved nature of the proteins, we posit that similar mechanisms may exist in mammals.
Materials and methods
Detailed as well as additional materials and methods are provided as a supplementary file. Unless otherwise mentioned, all strains were maintained at 20 °C using standard C. elegans techniques (Stiernagle, 2006). The zfp‐1(2ac) (752 bp) was amplified from wild‐type cDNA and cloned into the pL4440 vector (Addgene, Cambridge, Massachusetts, USA) for RNAi. Lifespan analysis was performed as described previously (Chamoli et al., 2014). Lifespan graph was plotted with percentage alive on Y‐axis and the number of days on the X‐axis. Statistical analyses for survival were conducted using Mantel–Cox log‐rank test through OASIS software available at http://sbi.postech.ac.kr/oasis (Yang et al., 2011). Lifespans are expressed as average lifespan ± SEM consolidated for all the lifespan experiments. Consolidated lifespan data with the number of experiments (N) and the number of animals (n) are reported in Table S1 and individual lifespan experiments are recorded in Table S3. For dauer assay, daf‐2(‐), daf‐16(‐);daf‐2(‐);daf‐16a, or daf‐16(‐);daf‐2(‐);daf‐16f animals were grown at 15 °C, and following hypochlorite treatment, eggs were placed on RNAi plates. The eggs were then incubated at 22 °C for 72 h after which they were scored for dauers. Dauers were confirmed by treating the animals with 1% SDS for an hour. For promoter‐gfp reporter assays, the eggs were grown on different RNAi bacteria at 20 °C till they reached L4‐YA. The worms were photographed using Axioimager M2 (Zeiss, Oberkochen, Germany), and fluorescence intensity was quantified using NIH ImageJ software (http://imagej.nih.gov/ij/index.html). Fluorescence intensity is represented as percentage fluorescence of control RNAi. For duration control experiments, the daf‐2(‐);Psod‐3::gfp worms were maintained and grown at 15 °C till L2 and then shifted to 25 °C for 18 h to lower the flux in the IIS pathway. Subsequently, they were shifted back to 15 °C for 12 h before being photographed under fluorescence microscope. Fluorescence intensity is represented in arbitrary units. RNA isolation and QRT–PCR were performed as previously reported (Chamoli et al., 2014). ChIP was performed as described previously (Oh et al., 2006; Kumar et al., 2015).
Conflict of interest
The authors declare no conflict of interest.
Author contributions
AM conceived the project and wrote the manuscript with the help from AS. NK and AG performed ChIP experiments; LM generated transgenic lines; VJ analysed ChIP‐seq data along with NK; AS performed all other experiments and analysis.
Funding
Department of Biotechnology, Ministry of Science and Technology, (Grant/Award Number: ‘BT/HRD/35/02/12/2008’, ‘BT/PR13720/BAB/10/779/2010’)
Indian Council of Medical Research, (Grant/Award Number: ‘54/3/CFP/GER/2011‐NCD‐II’).
Supporting information
Fig. S1 (A) Positions of SL1 (splice leader) sites in the zfp‐1(2a) and zfp‐1(2c) transcripts.
Fig. S2 (A) ChIP‐PCR analysis of the binding of DAF‐16/FOXO isoforms (a‐upper, b‐middle or f‐lower panel) on the promoter of gfl‐1.
Fig. S3 (A) ChIP‐PCR analysis of binding of DAF‐16/FOXO isoforms (a‐upper, b‐middle or f‐lower panel) to the different regions on the promoters of zfp‐1(2a) and zfp‐1(2c).
Fig. S4 (A) UCSC browser view of PHA‐4/FOXA peak on gfl‐1 promoter as determined by reanalysis of ChIP‐seq data of OP37 strain; data mined from MODENCODE.
Fig. S5 (A) QRT–PCR detection of mRNA levels of DAF‐16 targets in wild‐type or daf‐2(‐).
Fig. S6 (A–D) Lifespan analysis of indicated strains on control, zfp‐1(2ac), gfl‐1 or daf‐16 RNAi.
Fig. S7 (A–D) Lifespan analysis of indicated strains on control, zfp‐1(2ac), gfl‐1 or daf‐16 RNAi.
Fig. S8 (A,B) Lifespan of different eat‐2 alleles on control, zfp‐1(2ac), gfl‐1 or pha‐4 RNAi.
Table S1 Details of life span experiments performed (consolidated data).
Table S2 List of primers used in the study.
Table S3 Details of life span experiments performed (Individual experiments used for consolidation).
Table S4 GO term analysis of the 885 genes that are common targets of DAF‐16 and ZFP‐1 when C. elegans whole genome was used as a background.
Data S1 Materials and methods.
Acknowledgments
We thank all members of the Molecular Aging laboratory along with Drs. S. Rath, K. Chakraborty and S. Basak for their inputs, C. Kenyon and H. Tissenbaum for strains. We are grateful to Department of Biotechnology (DBT), Government of India, for a generous infrastructure grant for the establishment of the NII NGS core facility. This project is partly funded by the Ramalingaswami fellowship to AM (BT/HRD/35/02/12/2008), DBT Grant No. BT/PR13720/BAB/10/779/2010, ICMR Grant No. 54/3/CFP/GER/2011‐NCD‐II and core funding from National Institute of Immunology. AS and LM are supported by CSIR fellowships, while AG is a DBT JRF. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
References
- Alon U (2007) Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450. [DOI] [PubMed] [Google Scholar]
- Avgousti DC, Cecere G, Grishok A (2013) The conserved PHD1‐PHD2 domain of ZFP‐1/AF10 is a discrete functional module essential for viability in Caenorhabditis elegans. Mol. Cell. Biol. 33, 999–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman JR, Kenyon C (2006) Germ‐cell loss extends C. elegans life span through regulation of DAF‐16 by kri‐1 and lipophilic‐hormone signaling. Cell 124, 1055–1068. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L (2007) Two neurons mediate diet‐restriction‐induced longevity in C. elegans . Nature 447, 545–549. [DOI] [PubMed] [Google Scholar]
- Caudell D, Aplan PD (2008) The role of CALM‐AF10 gene fusion in acute leukemia. Leukemia 22, 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere G, Hoersch S, Jensen M, Dixit S, Grishok A (2013) The ZFP‐1(AF10)/DOT‐1 Complex Opposes H2B Ubiquitination to Reduce Pol II Transcription. Mol. Cell 50, 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamoli M, Singh A, Malik Y, Mukhopadhyay A (2014) A novel kinase regulates dietary restriction‐mediated longevity in Caenorhabditis elegans . Aging Cell 13, 641–655. doi:10.1111/acel.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Tishkoff DX, Yang B, Wilson‐Grady J, Yu X, Mazer T, Eckersdorff M, Gygi SP, Lombard DB, Hsu AL (2012) C. elegans SIRT6/7 homolog SIR‐2.4 promotes DAF‐16 relocalization and function during stress. PLoS Genet. 8, e1002948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi S, Bassini A, Jones L, Chaplin T, Linder B, de Bruijn D, Meese E, Young B (2002) The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood 99, 275. [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- Dudley NR, Labbe JC, Goldstein B (2002) Using RNA interference to identify genes required for RNA interference. Proc. Natl Acad. Sci. USA 99, 4191–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L (2015) Promoting health and longevity through diet: from model organisms to humans. Cell 161, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE (2002) Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA‐4. Science 295, 821–825. [DOI] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL (1998) Two pleiotropic classes of daf‐2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans . Genetics 150, 129–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandison RC, Piper MD, Partridge L (2009) Amino‐acid imbalance explains extension of lifespan by dietary restriction in Drosophila . Nature 462, 1061–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A (2009) Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans . Aging Cell 8, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A (2007a) An AMPK‐FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans . Curr. Biol. 17, 1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A (2007b) The energy sensor AMP‐activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 282, 30107–30119. [DOI] [PubMed] [Google Scholar]
- Grishok A, Sinskey JL, Sharp PA (2005) Transcriptional silencing of a transgene by RNAi in the soma of C. elegans . Genes Dev. 19, 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Hoersch S, Sharp P (2008) RNA interference and retinoblastoma‐related genes are required for repression of endogenous siRNA targets in Caenorhabditis elegans . Proc. Natl Acad. Sci. USA 105, 20386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C (2005) The plasticity of aging: insights from long‐lived mutants. Cell 120, 449–460. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ (2010) The genetics of ageing. Nature 464, 504–512. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Kahn CR, Accili D (2003) Insulin receptor knockout mice. Annu. Rev. Physiol. 65, 313–332. [DOI] [PubMed] [Google Scholar]
- Kumar N, Jain V, Singh A, Jagtap U, Verma S, Mukhopadhyay A (2015) Genome‐wide endogenous DAF‐16/FOXO recruitment dynamics during lowered insulin signalling in C. elegans . Oncotarget 6, 41418–41433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon ES, Narasimhan SD, Yen K, Tissenbaum HA (2010) A new DAF‐16 isoform regulates longevity. Nature 466, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S (1998) The genetics of caloric restriction in Caenorhabditis elegans . Proc. Natl Acad. Sci. USA 95, 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, Dillin A, Hansen M (2013) The TFEB orthologue HLH‐30 regulates autophagy and modulates longevity in Caenorhabditis elegans . Nat. Commun. 4, 2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RY, Hench J, Ruvkun G (2001) Regulation of C. elegans DAF‐16 and its human ortholog FKHRL1 by the daf‐2 insulin‐like signaling pathway. Curr. Biol. 11, 1950–1957. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A (2006) A conserved MST‐FOXO signaling pathway mediates oxidative‐stress responses and extends life span. Cell 125, 987–1001. [DOI] [PubMed] [Google Scholar]
- Li W, Gao B, Lee S‐M, Bennett K, Fang D (2007) RLE‐1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF‐16 polyubiquitination. Dev. Cell 12, 235. [DOI] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C (2003) Tissue‐specific activities of C. elegans DAF‐16 in the regulation of lifespan. Cell 115, 489–502. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C (2001) Regulation of the Caenorhabditis elegans longevity protein DAF‐16 by insulin/IGF‐1 and germline signaling. Nat. Genet. 28, 139–145. [DOI] [PubMed] [Google Scholar]
- MacDonald PE, Joseph JW, Rorsman P (2005) Glucose‐sensing mechanisms in pancreatic beta‐cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 2211–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Dillin A (2008) Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 77, 727–754. [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U (2003) Structure and function of the feed‐forward loop network motif. Proc. Natl Acad. Sci. USA 100, 11980–11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansisidor AR, Cecere G, Hoersch S, Jensen MB, Kawli T, Kennedy LM, Chavez V, Tan MW, Lieb JD, Grishok A (2011) A conserved PHD finger protein and endogenous RNAi modulate insulin signaling in Caenorhabditis elegans . PLoS Genet. 7, e1002299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH (2003) Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF‐16. Aging Cell 2, 111–121. [DOI] [PubMed] [Google Scholar]
- Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, Smith‐Wheelock M (2005) Methionine‐deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF‐I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 4, 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA (2008) Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real‐time PCR to study transcription factor binding to DNA in Caenorhabditis elegans . Nat. Protoc. 3, 698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C (2003) Genes that act downstream of DAF‐16 to influence the lifespan of Caenorhabditis elegans . Nature 424, 277–283. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA (2005) JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF‐16. Proc. Natl Acad. Sci. USA 102, 4494–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA (2006) Identification of direct DAF‐16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat. Genet. 38, 251–257. [DOI] [PubMed] [Google Scholar]
- Pandit A, Jain V, Kumar N, Mukhopadhyay A (2014) PHA‐4/FOXA‐regulated microRNA feed forward loops during Caenorhabditis elegans dietary restriction. Aging 6, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A (2007) PHA‐4/Foxa mediates diet‐restriction‐induced longevity of C. elegans . Nature 447, 550–555. [DOI] [PubMed] [Google Scholar]
- Riedel C, Dowen R, Lourenco G, Kirienko N, Heimbucher T, West J, Bowman S, Kingston R, Dillin A, Asara J, Ruvkun G (2013) DAF‐16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 15, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida‐Stubbs S, Glover‐Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann‐Haefelin E, Sabatini DM, Blackwell TK (2012) TOR signaling and rapamycin influence longevity by regulating SKN‐1/Nrf and DAF‐16/FoxO. Cell Metab. 15, 713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster E, McElwee JJ, Tullet JM, Doonan R, Matthijssens F, Reece‐Hoyes JS, Hope IA, Vanfleteren JR, Thornton JM, Gems D (2010) DamID in C. elegans reveals longevity‐associated targets of DAF‐16/FoxO. Mol. Syst. Biol. 6, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T (2006) Maintenance of C. elegans . WormBook, 11, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y (1999) pos‐1 encodes a cytoplasmic zinc‐finger protein essential for germline specification in C. elegans . Development 126, 1–11. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Daitoku H, Hirota K, Tamiya H, Yokoyama A, Kako K, Nagashima Y, Nakamura A, Shimada T, Watanabe S, Yamagata K, Yasuda K, Ishii N, Fukamizu A (2011) Asymmetric arginine dimethylation determines life span in C. elegans by regulating forkhead transcription factor DAF‐16. Cell Metab. 13, 505–516. [DOI] [PubMed] [Google Scholar]
- Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ (2013) PQM‐1 complements DAF‐16 as a key transcriptional regulator of DAF‐2‐mediated development and longevity. Cell 154, 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK (2008) Direct inhibition of the longevity‐promoting factor SKN‐1 by insulin‐like signaling in C. elegans . Cell 132, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A (2006) SMK‐1, an essential regulator of DAF‐16‐mediated longevity. Cell 124, 1039–1053. [DOI] [PubMed] [Google Scholar]
- Yang JS, Nam HJ, Seo M, Han SK, Choi Y, Nam HG, Lee SJ, Kim S (2011) OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE 6, e23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Judy M, Lee SJ, Kenyon C (2013) Direct and indirect gene regulation by a life‐extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab. 17, 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HY, Preston E, Slightham C, Hillier LW, Brock T, Agarwal A, Auerbach R, Hyman AA, Gerstein M, Mango SE, Kim SK, Waterston RH, Reinke V, Snyder M (2010) Genome‐wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA‐4/FOXA in development and environmental response. PLoS Genet. 6, e1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JA, Malloy V, Krajcik R, Orentreich N (2003) Nutritional control of aging. Exp. Gerontol. 38, 47–52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 (A) Positions of SL1 (splice leader) sites in the zfp‐1(2a) and zfp‐1(2c) transcripts.
Fig. S2 (A) ChIP‐PCR analysis of the binding of DAF‐16/FOXO isoforms (a‐upper, b‐middle or f‐lower panel) on the promoter of gfl‐1.
Fig. S3 (A) ChIP‐PCR analysis of binding of DAF‐16/FOXO isoforms (a‐upper, b‐middle or f‐lower panel) to the different regions on the promoters of zfp‐1(2a) and zfp‐1(2c).
Fig. S4 (A) UCSC browser view of PHA‐4/FOXA peak on gfl‐1 promoter as determined by reanalysis of ChIP‐seq data of OP37 strain; data mined from MODENCODE.
Fig. S5 (A) QRT–PCR detection of mRNA levels of DAF‐16 targets in wild‐type or daf‐2(‐).
Fig. S6 (A–D) Lifespan analysis of indicated strains on control, zfp‐1(2ac), gfl‐1 or daf‐16 RNAi.
Fig. S7 (A–D) Lifespan analysis of indicated strains on control, zfp‐1(2ac), gfl‐1 or daf‐16 RNAi.
Fig. S8 (A,B) Lifespan of different eat‐2 alleles on control, zfp‐1(2ac), gfl‐1 or pha‐4 RNAi.
Table S1 Details of life span experiments performed (consolidated data).
Table S2 List of primers used in the study.
Table S3 Details of life span experiments performed (Individual experiments used for consolidation).
Table S4 GO term analysis of the 885 genes that are common targets of DAF‐16 and ZFP‐1 when C. elegans whole genome was used as a background.
Data S1 Materials and methods.


