Summary
The G allele of the FOXO3 single nucleotide polymorphism (SNP) rs2802292 exhibits a consistently replicated genetic association with longevity in multiple populations worldwide. The aims of this study were to quantify the mortality risk for the longevity‐associated genotype and to discover the particular cause(s) of death associated with this allele in older Americans of diverse ancestry. It involved a 17‐year prospective cohort study of 3584 older American men of Japanese ancestry from the Honolulu Heart Program cohort, followed by a 17‐year prospective replication study of 1595 white and 1056 black elderly individuals from the Health Aging and Body Composition cohort. The relation between FOXO3 genotype and cause‐specific mortality was ascertained for major causes of death including coronary heart disease (CHD), cancer, and stroke. Age‐adjusted and multivariable Cox proportional hazards models were used to compute hazard ratios (HRs) for all‐cause and cause‐specific mortality. We found G allele carriers had a combined (Japanese, white, and black populations) risk reduction of 10% for total (all‐cause) mortality (HR = 0.90; 95% CI, 0.84–0.95; P = 0.001). This effect size was consistent across populations and mostly contributed by 26% lower risk for CHD death (HR = 0.74; 95% CI, 0.64–0.86; P = 0.00004). No other causes of death made a significant contribution to the survival advantage for G allele carriers. In conclusion, at older age, there is a large risk reduction in mortality for G allele carriers, mostly due to lower CHD mortality. The findings support further research on FOXO3 and FoxO3 protein as potential targets for therapeutic intervention in aging‐related diseases, particularly cardiovascular disease.
Keywords: FOXO3, heart disease, longevity, mortality
Introduction
The aging of the world's population has enormous implications for society, social insurance programs, and health care (National Institute on Aging, 2007; US Department of Health and Human Services, 2012). Thus, understanding how humans can age more healthfully is of vital importance. Genetic factors that affect the rate of aging and/or risk for age‐related diseases are important, as they account for approximately one‐third of the variability in human lifespan (Brooks‐Wilson, 2013; Shadyab & LaCroix, 2015). However, despite almost three decades since the first finding of longevity‐associated genotypes in long‐lived persons (Takata et al., 1987), the effects on longevity of only the apolipoprotein E gene (APOE) and the forkhead box O‐3 (FoxO3) gene (FOXO3) have been consistently replicated across multiple, diverse human populations (Flachsbart et al., 2009; Pawlikowska et al., 2009; Murabito et al., 2012; Brooks‐Wilson, 2013; Morris et al., 2015; Shadyab & LaCroix, 2015).
FoxO3 is an evolutionarily conserved transcription factor in the insulin signaling pathway (Kenyon, 2010). It regulates expression of genes controlling a multitude of processes that could enhance health and lifespan (Willcox et al., 2008; Kenyon, 2010; Eijkelenboom & Burgering, 2013). A previous study of American men of Japanese ancestry was the first to find an association of three single nucleotide polymorphisms (SNPs) of FOXO3 with human longevity, the strongest SNP being rs2802292 (Willcox et al., 2008). The association was replicated in 11 other independent studies of populations of diverse ancestry (Bao et al., 2014). A meta‐analysis found these and two other FOXO3 SNPs retained statistical significance, the strongest association with longevity involving the original G allele of rs2802292 in men (odds ratio (OR), 1.54; 95% confidence intervals (CIs), 1.33–1.67) (Bao et al., 2014). A meta‐analysis of genomewide association studies of longevity yielded an OR of 1.17 (P = 1.9 × 10−10) for this allele (Broer et al., 2015). The effect size has not been well quantified in terms of mortality risk over time. The mechanisms by which the protective allele(s) reduce mortality to promote human longevity are also not known. Identifying the cause of death in longevity‐allele carriers vs. noncarriers may provide clues as to why FOXO3 SNPs strongly protect against mortality.
We hypothesized that the longevity‐associated FOXO3 genotype would be associated with a sizable risk reduction for mortality and with one or more major age‐associated clinical causes of death, such as coronary heart disease (CHD), cancer, and stroke. To test this hypothesis we utilized an extensive, prospectively collected dataset from our long‐lived cohort of American men of Japanese ancestry, well characterized for aging phenotypes, drawn from the Honolulu Heart Program (HHP) prospective cohort study (Kagan, 1996). We genotyped this study population to prospectively assess the following: (i) the effect size of the protective (longevity‐associated) FOXO3 genotype on total (all‐cause) mortality in 17 years of follow‐up; and (ii) the effect of the protective FOXO3 genotype on cause‐specific mortality. We then attempted a replication of major findings in a suitable cohort of elderly white and black Americans of both sexes in the Health Aging and Body Composition (ABC) cohort study, which had 17 years of follow‐up.
Results
Study participants
The baseline characteristics of each cohort of subjects (Japanese, white, and black) are shown in Table 1. HHP study participants tended to be older and had higher prevalence of hypertension, diabetes, and CHD than Health ABC participants. There was a higher percentage of G allele carriers in the Health ABC population, particularly in blacks, in whom the G allele was the common allele.
Table 1.
Characteristics of the three study cohorts of older Americans
| Parameter | Japanese (n = 3584) | Whites (n = 1794) | Blacks (n = 1281) |
|---|---|---|---|
| Age (years) | 77.7 ± 4.6 | 73.8 ± 2.9 | 73.4 ± 2.9 |
| Body mass index (kg m−2) | 23.5 ± 3.2 | 26.5 ± 4.1 | 28.6 ± 5.4 |
| History of ever smoking (%) | 62.4 | 56.8 | 55.2 |
| Alcohol (3+ drinks per week) (%) | 40.3 | 23.3 | 9.6 |
| Exercise regularly (%) | 68.3 | 49.7 | 31.2 |
| Glucose (g dL−1) | 113 ± 29.5 | 100.8 ± 27.3 | 109.3 ± 41.8 |
| Total cholesterol (mg dL−1) | 189.8 ± 33.1 | 201.3 ± 37.4 | 204.9 ± 40.1 |
| CHD (%)a | 20.6 | 13.1 | 13.7 |
| Stroke (%)a | 4.5 | 1.4 | 3.7 |
| Cancer (%)a | 15.6 | 24.5 | 11.0 |
| Diabetes (%)a | 28.6 | 10.7 | 21.0 |
| Hypertension (%)a,b | 73.6 | 48.7 | 62.9 |
| FOXO3 G allele frequency (%) | 27.7 | 36.5 | 73.2 |
| FOXO3 G allele carrier (%)c | 47.0 | 58.4 | 92.7 |
Americans of Japanese ancestry tended to be older, had more prevalent CHD and diabetes, and lower frequency of the longevity‐associated FOXO3 G allele at the baseline examination.
Prevalent disease (not age‐adjusted) was defined and detected based on respective study surveillance programs: CHD: any myocardial infarction, bypass surgery, angioplasty, other heart surgery, or ECG evidence; Diabetes: fasting glucose ≥ 126 mg mL−1 or 2‐h glucose ≥ 200 mg mL−1, or on diabetes medication.
Participants were considered to have hypertension if they had a systolic blood pressure of ≥ 140 mmHg and/or diastolic blood pressure of ≥ 90 mmHg or were on antihypertensive medication.
Carriers were persons who had at least one G allele of FOXO3 SNP rs2802292. Shown are homozygotes and heterozygotes combined.
Meta‐analysis of mortality (all‐cause)
Cox proportional hazards models were used to generate hazard ratios (HRs) over a 17‐year prospective follow‐up for black (Health ABC), white (Health ABC), and Japanese (HHP) ancestry. Tests for heterogeneity for all populations combined yielded P = 0.96, demonstrating that the populations could be used for a combined analysis, utilizing a dominant model. During follow‐up, mortality risk for G allele carriers ranged from 0.87 (blacks) to 0.91 (whites) with a combined (Japanese, white, and black populations) risk reduction of 10% for total mortality (HR = 0.90; 95% CI, 0.84–0.95; P = 0.001) (Fig. 1). Tests of model proportionality were met.
Figure 1.
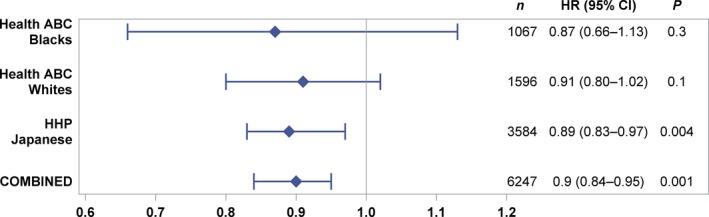
Meta‐analysis of all‐cause mortality for G allele carriers. Hazard ratios (HRs) for mortality over prospective follow‐up periods of 17 years for the cohorts of Americans of black (Health ABC) and white (Health ABC) individuals and 17 years for Americans of Japanese (Honolulu Heart Program (HHP)) ancestry, utilizing a dominant model. Tests for heterogeneity for all populations combined were P = 0.96, demonstrating that the populations are appropriate for a combined meta‐analysis. During the 17‐year prospective follow‐up, G allele carriers (FOXO3 SNP rs2802292) had HR that ranged from 0.87 (blacks) to 0.91 (whites) and a combined risk reduction of 10% for total mortality (HR = 0.90; 95% CI, 0.84–0.95; P = 0.001).
Total (all‐cause) mortality risk for carriers of the FOXO3 G allele
In the HHP cohort, risk of death over a 17‐year prospective follow‐up period for carriers (vs. noncarriers as baseline risk group) of the longevity‐associated G allele of FOXO3 (rs2802292) in Japanese American men, utilizing a multivariable Cox regression model, demonstrated an 11% risk reduction (HR 0.89; 95% CI: 0.83–0.97; P = 0.004) for all‐cause mortality (Table 2A). Five important risk factors for mortality in older persons did not substantially alter mortality risk, suggesting that they are not in the causative pathway between FOXO3 variation and mortality in older Japanese males.
Table 2.
All‐cause mortality risk for carriers of the FOXO3 G allele in multivariable risk factor model. (A) Honolulu Heart Program cohort. (B) Health ABC cohort
| (A) | ||
|---|---|---|
| Risk factor | HR (95% CI) | P |
| Age (years) | 1.11 (1.10–1.12) | < 0.001 |
| Fasting glucose (per 10 mg dL−1) | 1.06 (1.05–1.08) | < 0.001 |
| Smoking historya (per 10 pack‐years) | 1.04 (1.03–1.05) | < 0.001 |
| Systolic blood pressure (per 10 mm Hg) | 1.00 (0.98–1.02) | 0.84 |
| Fasting total cholesterol (per 10 mg dL−1) | 0.98 (0.97–0.99) | 0.007 |
| Body mass index (per kg m−2) | 0.95 (0.94–0.97) | < 0.001 |
| FOXO3 G allele carrier (age‐adjusted) | 0.89 (0.83–0.97) | 0.004 |
| FOXO3 G allele carrierb (age‐ and risk factor‐adjusted) | 0.86 (0.79–0.93) | < 0.001 |
| (B) | ||||
|---|---|---|---|---|
| Blacks | Whites | |||
| Risk factor | HR (95% CI) | P | HR (95% CI) | P |
| Age (years) | 1.06 (1.03–1.09) | < 0.0001 | 1.12 (1.09–1.14) | < 0.0001 |
| Fasting glucose (per 10 mg dL−1) | 1.03 (1.02–1.05) | < 0.0001 | 1.06 (1.04–1.08) | < 0.0001 |
| Smoking history (ever smoking) | 1.26 (1.08–1.46) | 0.003 | 1.37 (1.2–1.55) | < 0.0001 |
| Systolic blood pressure (per 10 mmHg) | 1.06 (1.02–1.1) | 0.002 | 1.01 (0.98–1.05) | 0.39 |
| Fasting total cholesterol (per 10 mg dL−1) | 0.97 (0.95–0.98) | 0.0002 | 0.97 (0.96–0.99) | 0.001 |
| Body mass index (per kg m−2) | 0.97 (0.96–0.98) | < 0.0001 | 0.98 (0.97–1.00) | 0.047 |
| FOXO3 G allele carrier (age‐adjusted) | 0.87 (0.66–1.13) | 0.29 | 0.91 (0.91–1.02) | 0.063 |
| FOXO3 G allele carrierc (age‐ and risk factor‐adjusted) | 0.91 (0.69–1.2) | 0.51 | 0.91 (0.81–1.03) | 0.15 |
(A) Risk of death over a 17‐year follow‐up period for carriers vs. noncarriers of longevity‐associated G allele of FOXO3 (rs2802292) in HHP population of Japanese Americans. (B) Risk of death over a 17‐year follow‐up period for carriers vs. noncarriers of longevity‐associated G allele of FOXO3 (rs2802292) in Health ABC cohorts.
Number of packs smoked per day × number of years smoked (pack‐years).
Multivariable Cox regression model for subjects with complete risk factor data (n = 3200; total deaths = 2345). HR = hazard ratio for all‐cause mortality.
Multivariable Cox regression model for subjects with complete risk factor data. HR = hazard ratio for all‐cause mortality.
In the Health ABC cohort, a similar analysis of risk of death over 17 years of follow‐up demonstrated a similarly large (9%) risk reduction for all‐cause mortality in whites (HR 0.91; 95% CI: 0.80–1.02; P = 0.06) and 13% in blacks (HR 0.87; 95% CI: 0.66–1.13; P = 0.29), respectively(Table 2B). When entering five important mortality risk factors for mortality at older ages into the model, there was a reduction in statistical significance and effect size (HR) in whites and blacks, suggesting that one or more risk factor variables are in the causative pathway between FOXO3 variation and mortality in these populations.
Specific causes of death and HR for mortality (age‐adjusted)
In the HHP cohort, the G allele conferred a highly statistically significant protective effect against CHD mortality in Japanese American men (Table 3A). Hazard ratio in G (longevity allele) carriers (HR: 0.75; 95% CI 0.63–0.90; P = 0.001) was 25% lower than noncarriers.
Table 3.
Specific causes of death and hazard ratio, age‐adjusted, for carriers of the FOXO3 G allele. (A) Honolulu Heart Program (HHP) cohort. (B) Health ABC cohort
| (A) | |||
|---|---|---|---|
| Cause of death | No. of deaths | HRa (95% CI) | P‐value |
| Cancer | 546 | 1.01 (0.85–1.19) | 0.93 |
| CHD (coronary heart disease) | 524 | 0.75 (0.63–0.90) | 0.001 |
| Stroke | 315 | 0.97 (0.77–1.21) | 0.76 |
| Dementia | 221 | 1.01 (0.78–1.32) | 0.93 |
| Other cardiovascular disease (CVD) | 213 | 0.85 (0.65–1.11) | 0.23 |
| Infectious disease | 188 | 0.91 (0.68–1.21) | 0.52 |
| Chronic obstructive pulmonary disease (COPD) | 117 | 0.83 (0.57–1.19) | 0.31 |
| Renal failure | 45 | 0.86 (0.48–1.55) | 0.61 |
| GI (gastrointestinal disease) | 39 | 1.22 (0.65–2.30) | 0.54 |
| Other deaths | 480 | 0.86 (0.72–1.03) | 0.09 |
| (B) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Whites | Blacks | Combined | |||||||
| Cause of deatha | n | HRa (95% CI) | P | n | HRa (95% CI) | P | n | HR (95% CI) | P |
| 927 | 687 | 1614 | |||||||
| Cancer | 220 | 0.87 (0.67–1.14) | 0.31 | 179 | 1.67 (0.82–3.41) | 0.16 | 399 | 0.97 (0.76–1.24) | 0.81 |
| CHD | 224 | 0.76 (0.58–0.98) | 0.04 | 140 | 0.61 (0.35–1.04) | 0.07 | 364 | 0.73 (0.57–0.92) | 0.0088 |
| Stroke | 80 | 0.87 (0.56–1.35) | 0.53 | 67 | 0.55 (0.26–1.15) | 0.11 | 147 | 0.78 (0.53–1.15) | 0.21 |
| Other CVD | 41 | 0.96 (0.52–1.8) | 0.91 | 47 | 1.04 (0.32–3.38) | 0.95 | 88 | 0.98 (0.57–1.7) | 0.96 |
| COPD | 42 | 0.82 (0.44–1.5) | 0.51 | 29 | 2.07 (0.28–15.3) | 0.47 | 71 | 0.91 (0.52–1.6) | 0.75 |
| Infection | 36 | 1.21 (0.62–2.4) | 0.58 | 24 | 0.54 (0.16–1.84) | 0.33 | 60 | 1.03 (0.56–1.91) | 0.92 |
| Renal failure | 17 | 0.61 (0.24–1.59) | 0.32 | 25 | aN/A | N/A | 42 | 1.01 (0.45–2.26) | 0.98 |
| GI | 7 | 0.88 (0.2–3.94) | 0.87 | 9 | 0.46 (0.05–3.85) | 0.47 | 16 | 0.72 (0.2–2.59) | 0.61 |
| Other | 260 | 1.11 (0.87–1.43) | 0.40 | 167 | 0.69 (0.4–1.18) | 0.18 | 427 | 1.02 (0.81–1.29) | 0.84 |
HR shown is for carriers of the G allele. (A) Number of deaths are all deaths in each disease category from n = 2688 total deaths in HHP subjects (mean age 77.7 ± 4.6 years at baseline) followed up for 17 years. Cause of death was coded according the International Classification of Diseases (ICD) system (version 8) by an expert morbidity and mortality committee after comprehensive review of multiple data sources including death certificates, hospital discharge records and interviews with physicians, among other data. All the study participants were ascertained for vital status at the end of study; in only 17 (1.3%) deaths could a cause not be ascertained. There was a large (25%) risk reduction in CHD mortality. Bonferroni correction of CHD death yielded P = 0.01. (B) Number of deaths are all deaths in each disease category from n = 1614 total deaths in Health ABC subjects followed up for 17 years. Surveillance for cause of death was similar to HHP methodology. There was a large (24%) risk reduction in CHD mortality for whites (HR: 0.76; 0.58–0.98; P = 0.036) and a 39% risk reduction for blacks (HR: 0.61; 95% CI 0.35–1.04; P = 0.068 for trend). White males and females were aged 73.8 ± 2.9 years at baseline. Black males and females were aged 73.4 ± 2.9 years at baseline. Bonferroni correction of CHD death yielded P = 0.32 for blacks and P = 0.08 for whites. CVD, cardiovascular disease; GI, gastrointestinal.
In the Health ABC cohort, there was a similarly large (24%) risk reduction in CHD mortality for whites (HR: 0.76; 95% CI 0.58–0.98; P = 0.036) and even larger for blacks (39%; HR 0.61; 95% CI 0.35–1.04; P = 0.068 for trend; Table 3B). While certain other diseases (such as cancer in whites and stroke in blacks) showed lower HRs for mortality, none reached statistical significance.
Meta‐analysis of CHD mortality
Cox proportional hazards models were used to generate hazard ratios over the 17‐year prospective follow‐up period for individuals of black (Health ABC), white (Health ABC), and Japanese (HHP) ancestry. Tests for heterogeneity for all populations combined yielded P = 0.75, demonstrating that the populations could be used for a combined analysis, utilizing a dominant model. During follow‐up, CHD mortality risk for G allele carriers ranged from 0.61 (blacks) to 0.76 (whites) with a combined (Japanese, white, and black populations) risk reduction of 26% for total CHD mortality (HR = 0.74; 95% CI, 0.64–0.86; P = 0.00004) (Fig. 2). Tests of model proportionality were met.
Figure 2.
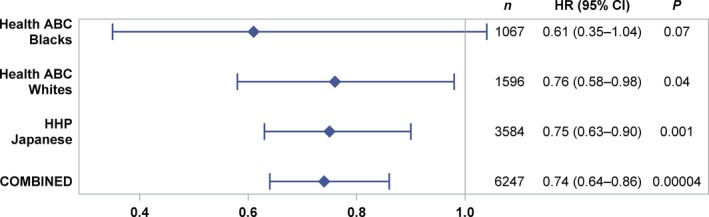
Meta‐analysis of CHD mortality. Hazard ratios (HRs) for CHD mortality over prospective follow‐up periods of 17 years for the cohorts of Americans of black (Health ABC; n = 140 CHD deaths), white (Health ABC; n = 224 CHD deaths) and 17 years for Americans of Japanese (Honolulu Heart Program (HHP; n = 524 CHD deaths)) ancestry, utilizing a dominant model. Tests for heterogeneity for all populations combined yielded P = 0.75, demonstrating that the populations could be used for a combined analysis, utilizing a dominant model. CHD mortality risk for G allele carriers ranged from 0.61 (blacks) to 0.76 (whites) with a combined (Japanese, white and black populations) risk reduction of 26% for total mortality (HR = 0.74; 95% CI, 0.64–0.86; P = 0.00004). Tests of model proportionality were met.
FOXO3 genotype and age at death
Carrier status for the protective G allele increased from age in the 70s to age of ≥ 90 years, consistent with protection against mortality for carriers in the three populations (Fig. 3).
Figure 3.
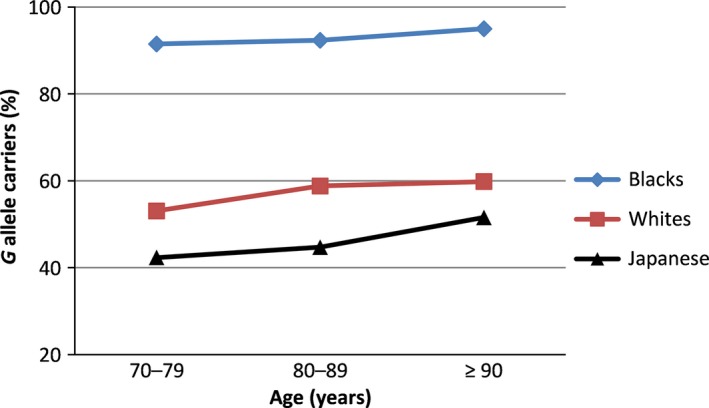
FOXO3 G allele carrier status (as % of cohort) by attained age. Percentage of carriers of the longevity‐associated FOXO3 G allele of SNP rs2802292 increased with age in the pooled population (P < 0.0001; logistic regression; adjusted for race).
Serum TNF‐α level by FOXO3 G allele carrier status
In a pilot analysis of HHP Japanese men, carriers of the longevity‐associated G allele of FOXO3 SNP rs2802292 had lower blood levels of the inflammatory cytokine TNF‐α (Table S1).
Discussion
In this, the first large, comprehensive, multiethnic prospective cohort study to quantify the risk reduction associated with the minor allele (G) of the FOXO3 longevity‐associated genotype, we demonstrated a large (10%) protective effect against all‐cause mortality and 26% for CHD mortality over 17 years of follow‐up. The protective effect was, moreover, observed in three genetically different populations. Our study contrasts with the vast majority of prior investigations of FOXO3 variants and longevity, which have been case:control studies that did not quantify risk over time, but rather simply tested for association with an outcome.
The magnitude of the impact of an absence of the FOXO3 G allele was comparable to the increase in risk of death from smoking a pack of cigarettes a day for 25 years in Japanese men. In black males and females, it was equivalent to having a 20 mmHg higher systolic blood pressure, and in white men and women to a 20 mg dL−1 elevation in fasting blood glucose.
Of particular interest, in multivariable models of mortality risk factors, five of the top risk factors for mortality at older ages (i.e. smoking, hypertension, fasting glucose, low BMI, low cholesterol) had little influence on the effect on mortality of FOXO3 genotype in Japanese men. In the Health ABC cohort of white and black men and women, there did appear to be mediator variables, in that the effect was partially attenuated when risk variables were entered into a multivariable model. The data suggest a possible mediator role for hypertension, which we found to be less prevalent in middle‐aged women carrying the FOXO3 G allele (Morris et al., 2015).
Although we found a lower risk for stroke in all three populations, this did not reach statistical significance. The only prior studies by others of FOXO3 genotype and cardiovascular disease led to conflicting results. One, a 4‐year prospective cohort study of Dutch men and women aged ≥ 85 years found that a particular FOXO3 haplotype conferred an increased risk for incident stroke and also an increased risk of mortality from CHD, stroke, and all‐cause (total) mortality (Kuningas et al., 2007). Another investigation, a case:control study of Danish men and women aged ≥ 90 years, failed to find an association of FOXO3 genotype with CHD or stroke (Soerensen et al., 2015). A case:control study of Han Chinese found no relation with CHD (Zhao et al., 2014), while two other case:control studies found FOXO3 longevity‐allele carriage was associated with lower CHD prevalence (Japanese American males) (Willcox et al., 2008) and with lower CHD and stroke mortality (white males and females) (Pawlikowska et al., 2009).
Although FOXO3 is a tumor suppressor (Lam et al., 2013; Fruman & Rommel, 2014), we could find only one study that reported an association of FOXO3 genotype with all‐cancer mortality (Pawlikowska et al., 2009) and another study in which a borderline association with cancer prevalence was found (Willcox et al., 2008). Both were case:control studies of longevity‐associated alleles. The current study found no detectable protection against cancer mortality, although larger studies of particular cancer subtypes may be useful.
It should, however, be noted that the prior studies were limited by small sample sizes, short follow‐up time, a case:control study design, monoethnic populations, and/or very old age of the study subjects. The current study did not suffer from these challenges. Therefore, while a contributory role for other diseases and disease processes cannot be ruled out, protection against CHD mortality seems paramount.
Based on studies of FoxO3 in model organisms we suspect immune dysregulation may be particularly important. In aging humans, adaptive immunity decreases, leading to immunosenescence, but innate immunity increases, leading to a pro‐inflammatory phenotype termed ‘inflamm‐aging’ (Franceschi et al., 2007; Peng, 2007; Salminen et al., 2008). FoxO3 inhibits the production of several inflammatory cytokines linked to human aging. These include interleukin‐2 (Oh et al., 2011), interleukin‐6 (Dejean et al., 2009), and TNF‐α (Lee et al., 2013), each of which has been linked to both aging and cardiovascular disease.
In support of this premise, the minor allele of FOXO3 SNP rs12212067 (which is in linkage disequilibrium with rs2802292 and could act as a proxy) was associated with a milder clinical course of seemingly unrelated inflammatory conditions (Crohn's disease and rheumatoid arthritis) (Lee et al., 2013). Minor allele carriage limited inflammatory responses in monocytes through TGF‐β1‐induced reduction of pro‐inflammatory cytokines, including TNF‐α. It also increased production of anti‐inflammatory cytokines. Our finding of lower TNF‐α in FOXO3 G allele carriers is consistent with protection against inflammation and thence CHD. More work is needed to confirm a major influence of reduced inflammation conferred by FOXO3 genotype in protection against CHD, as an explanation for its ability to confer increased lifespan.
Interestingly, the G allele is the ancestral allele as it is the most prevalent allele in African black populations and it persists as the common allele in the American black population (dbSNP, 2016). A question arises as to why the nonprotective (T) allele has become the common allele in whites and Asians. This is likely the result of population bottle necks that have pruned the G allele frequencies in “out of Africa” populations. This process could have fixed the rs2802292 G allele in a haplotype that contains a functional variant, such as one that results in better old age health. For most of human history infectious disease, of which inflammation is a major component, was likely the major cause of death (Finch, 2010). Perhaps TT homozygotes may not be able to defend against a high inflammatory load, which can lead to sepsis (among other potentially fatal maladies), and thus, fewer of these survived to pass on T alleles. It is plausible that there might be an unknown antagonistic pleiotropic effect where the allele protects early in life and becomes a risk later in life (Blagosklonny, 2010). There are examples of antagonistic pleiotropy involving the FoxO3 gene in model organism studies (Shen & Tower, 2010). As yet, we do not know if FOXO3 genotype protects against mortality at young or middle age, only that it protects against mortality in later life, as no studies have assessed the effects of FOXO3 genotype over the adult life course in humans. There are also several other plausible explanations of why the G allele has persisted as the common allele in blacks but has decreased in frequency in white and Asian populations. FoxO is critical in maintaining stem cell populations (Miyamoto et al., 2007). Perhaps human populations have only recently been able to survive long enough for the beneficial effects of the FOXO3 G allele on stem cell populations to be realized. Another potential explanation may be the fact that FoxO3 regulates stress resistance and protects against ultraviolet damage (Yang et al., 2006). Ultraviolet radiation would have been a major biological stressor in African populations. Future work may be able to answer such questions.
Our study has several strengths, including rigorous ascertainment of cause of death, large sample size that was adequate for detection of dominant genetic effects of FOXO3 on cause‐specific mortality, and replication of the main findings across cohorts. Another major strength is the prospective, population‐based cohort design, with virtually complete mortality ascertainment in near‐extinct cohorts. This eliminated recall bias concerning prevalent diseases and risk factors and also allowed us to quantify cause‐specific mortality risk over time. The relative homogeneity of the HHP population was also an advantage, and prior work has found no evidence of a population stratification artifact in this cohort (Willcox et al., 2008). Population substructure was also controlled for in the Health ABC population. The replication we saw in a methodologically similar cohort of elderly white and black Americans demonstrates that the findings are robust and generalizable to other genetic populations.
Summary and conclusions
The present study is the first to quantify the protection against mortality in a large, well‐powered, prospective cohort study of populations of contrasting ethnicities, and to demonstrate that CHD is the main clinical cause of death driving the strong survival advantage seen in carriers of the major longevity‐associated FOXO3 allele. As CHD is the primary cause of death in most industrialized countries, and death at older ages is increasingly dominated by inflammation‐related diseases, the fact that allelic variation in FOXO3 is now known to significantly modify this risk has important clinical implications, especially in high risk patient populations, such as those with type 2 diabetes (Gregg et al., 2014) and obesity (Olshansky et al., 2005). It has already become apparent that FoxO3 plays in important role in disease progression in age‐related diseases, such as cancer, and may be useful for monitoring responses to treatment (Monsalve & Olmos, 2011; Morris et al., 2015). Moreover, therapeutic interventions that modulate biological pathways associated with aging are now becoming a plausible approach to help people age more healthfully (Kennedy & Pennypacker, 2015). Our novel findings prompt consideration of FOXO3 and its protein (Foxo3) as therapeutic targets in healthy aging with a focus on CHD and possibly other inflammation‐related diseases. More study is needed to determine molecular/cellular mechanisms.
Experimental procedures
Study design
We utilized data from the Kuakini Hawaii Lifespan Study, an ancillary study of the genetic and environmental antecedents of healthy aging and longevity, drawn from the Kuakini HHP cohort of aging men (Willcox et al., 2006, 2008; Donlon et al., 2012). The HHP began in 1965 as a population‐based prospective study of cardiovascular diseases and included 8006 American men of Japanese ancestry (Kagan, 1996). All study participants were recruited from World War II service records of 9877 men who had valid contact information, were born in 1900–1919, and who were living on the island of Oahu. For the current study, we utilized DNA samples and data collected in 1991–93, the first examination that focused on aging‐related measures, which were added at that time to the ongoing HHP study (White et al., 1996). We genotyped 3584 (~ 80% of available HHP survivors) stored blood samples collected at the 1991–1993 examination. Follow‐up monitoring total mortality and cause‐specific mortality for 17 years was conducted. All examinations, and the collection of biological samples, were approved by the Institutional Review Board of Kuakini Medical Center. Written informed consent was obtained at each examination from all study participants.
The Health ABC cohort was utilized for a replication study. Health ABC is a prospective observational study of 3075 well‐functioning, community‐dwelling, male and female, white and black Americans, aged 70–79 years at baseline, recruited from field centers at the University of Pittsburgh, Pennsylvania, and the University of Tennessee, Memphis, and followed up for 17 years.
Genotyping
For HHP subjects, total cellular DNA was isolated from buffy coat of blood using the PureGene system (Gentra Systems, Inc., Big Lake, MN, USA) and quantified using PicoGreen staining (Molecular Probes, Eugene, OR, USA). We selected 32 SNPs that reflected the major genetic variability within intron 2 of FOXO3, based on degree of linkage disequilibrium and allele frequencies of ≥ 5%. We chose variant rs2802292 (alleles T/G) as representative of the strongest longevity‐associated SNPs and genotyped an extended sample set of 3584 subjects. Genotyping for rs2802292 involved an allelic discrimination assay.
TaqMan® (Applied Biosystems, Inc. [ABI], Carlsbad, CA, USA) reagents (purchased from Life Technologies, Carlsbad, CA, USA) were used for PCR amplification under standard conditions with AmpliTaq Gold® DNA polymerase (Perkin‐Elmer Corp., Waltham, MA, USA). PCR products were detected by TaqMan® assay, using a 6‐FAM‐labeled FRET probe for one allele and a VIC‐labeled probe for the other allele, with minor groove binding (MGB) quenchers to enhance assay signal. PCR products were measured using an ABI Prism 7000 Sequence Detection System. Genotype data were managed through a sample processing and database system with previously proven reliability. All positive controls on each genotyping plate were evaluated for consistency. We have found that these methods are exceptionally robust and have an accuracy of > 99.9% on retesting and a success rate of 99.6% (data not shown). We regenotyped 1% of samples to monitor quality.
For Health ABC samples, genomewide SNP genotyping was performed by the Center for Inherited Disease Research using the Illumina Human1M‐Duo BeadChip system. Samples were excluded from the dataset for reasons of sample failure, genotypic sex mismatch, and first‐degree relative of an included individual based on genotype data. Health ABC genotyped 1794 self‐described white participants at baseline with available DNA and consent to genetics testing. Of these, 1661 passed quality control benchmarks (call rate > 97%, no sex mismatch, and cryptic relatedness). In addition, 1139 self‐described African Americans had genotype information available after quality control. Principal component analysis (PCA) was performed using Eigenstrat (Price et al., 2006). PCA was first performed on genotype data from all Health ABC participants and HapMap samples as anchor points to identify population outliers. PCA was then performed on genotype data separately for black and white participants who were not genetic outliers, and the resulting eigenvectors were used as covariates in regression models to adjust for population structure. Imputation was performed for the autosomes using MACH software version 1.0.16. SNPs with minor allele frequency ≥ 1%, call rate ≥ 97% and Hardy–Weinberg equilibrium P ≥ 10−6 were used for imputation. HapMap II phased haplotypes were used as reference panels.
For whites, 914 263 SNPs passed quality control (QC) and were available for imputation using the HapMap CEPH reference panel (phase 2, release 22, build 36). For blacks, 1 007 948 SNPs passed QC and were available for imputation based on a 1:1 mixture of the CEPH:Yoruban (YRI) HapMap phase‐2 release‐22 build‐36 reference panel. A total of 2 543 887 SNPs in whites and 1 958 375 SNPs in blacks were imputed. SNP rs2802292 was imputed. All fractional values within 0.1 of an integer were rounded to the nearest integer to create the number of rs2802292 G alleles. The imputation quality measured by MACH r 2 was 0.99 in whites and blacks.
Outcome measures
Total (all‐cause) mortality and cause‐specific mortality were the principal outcomes for the study. Death occurrence was ascertained by review of obituaries in local newspapers, through the medical examiner's office, by phone calls to family members or primary care physicians, and search of the National Death Index. Cause of death was determined by an expert morbidity and mortality committee, consisting of physicians who adjudicated the cause of death using standardized criteria, through continuous comprehensive surveillance of data collected from multiple sources (Kagan, 1996). These sources included death certificates, medical history from prior examinations, interview of healthcare proxies, hospital discharge records, cancer registry data, and autopsy reports, when available. Similar procedures were utilized in Health ABC.
Statistics
Genotypes were evaluated and found to be in Hardy–Weinberg equilibrium. The relation of FOXO3 genotype with total mortality and cause‐specific mortality was determined by Cox regression, adjusting for age, in carriers vs. noncarriers of the G allele of rs2802292 in Japanese, whites, and blacks. For meta‐analysis of total mortality, age‐adjusted Cox regression models for Japanese, whites, and blacks were utilized, using both dominant and additive models for the effects of carrier status on total mortality. A dominant model was found to be most appropriate. Tests for heterogeneity were performed to assess suitability for a combined analysis. All statistical analyses were performed using the Statistical Analysis System (SAS) version 9.3 (SAS Institute, Cary, NC, USA).
Funding
The work in Honolulu was funded by the Kuakini Medical Center, the US National Institutes of Health (contract N01‐AG‐4‐2149, Grants 5 U01 AG019349‐05, 5R01AG027060 [Kuakini Hawaii Lifespan Study], 5R01AG038707 [Kuakini Hawaii Healthspan Study]), contract N01‐HC‐05102 from the National Heart, Lung, and Blood Institute, and Kuakini Medical Center. We acknowledge the expert assistance of the Hawaii State Department of Health, particularly for supplying death certificate data. Research involving the Health ABC study was supported by National Institutes of Aging contracts N01AG62101, N01AG62103, and N01AG62106 and in part by the Intramural Research Program of the NIH, National Institute on Aging (Health ABC study). The Health ABC genomewide association study was funded by NIA grant 1R01AG032098‐01A1 to Wake Forest University Health Sciences, and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract (number HHSN268200782096C) from the National Institutes of Health to The Johns Hopkins University. We thank all study participants and their families for their cooperation and the Hawaii State Department of Health for death certificate data. We thank Dr. Eunjung Lim for assistance with statistical analysis.
Conflict of interest
None declared.
Author contributions
BJW and TAD contributed to the study concept, data interpretation, study design, and writing the manuscript. KHM supervised recruitment and data collection on the HHP subjects and researchers at the Health ABC study sites recruited and characterized the Health ABC subjects. TAD and YL performed the HHP and Health ABC genotyping, respectively; MG, RA and PD performed cytokine analysis; TAD, YL, RC, EL, QH, NP, GJT, and DE managed data and contributed results; and RC, QH, YL, DES, and NP were responsible for data and statistical analyses. BJW, BJM, and TAD drafted the manuscript. All authors contributed to, read, and approved the final version of the manuscript.
Supporting information
Table S1. Serum TNF‐α level by FOXO3 G allele carrier status.
References
- Bao JM, Song XL, Hong YQ, Zhu HL, Li C, Zhang T, Chen W, Zhao SC, Chen Q (2014) Association between FOXO3A gene polymorphisms and human longevity: a meta‐analysis. Asian J. Androl. 16, 446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV (2010) Revisiting the antagonistic pleiotropy theory of aging: TOR‐driven program and quasi‐program. Cell Cycle 9, 3151–3156. [DOI] [PubMed] [Google Scholar]
- Broer L, Buchman AS, Deelen J, Evans DS, Faul JD, Lunetta KL, Sebastiani P, Smith JA, Smith AV, Tanaka T, Yu L, Arnold AM, Aspelund T, Benjamin EJ, De Jager PL, Eirkisdottir G, Evans DA, Garcia ME, Hofman A, Kaplan RC, Kardia SL, Kiel DP, Oostra BA, Orwoll ES, Parimi N, Psaty BM, Rivadeneira F, Rotter JI, Seshadri S, Singleton A, Tiemeier H, Uitterlinden AG, Zhao W, Bandinelli S, Bennett DA, Ferrucci L, Gudnason V, Harris TB, Karasik D, Launer LJ, Perls TT, Slagboom PE, Tranah GJ, Weir DR, Newman AB, van Duijn CM, Murabito JM (2015) GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J. Gerontol. A Biol. Sci. Med. Sci. 70, 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks‐Wilson AR (2013) Genetics of healthy aging and longevity. Hum. Genet. 132, 1323–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Database of Single Nucleotide Polymorphisms (dbSNP). Bethesda (MD): National Center for Biotechnology Information, National Library of Medicine. dbSNP accession on 2/9/2016:{rs2802292}, (dbSNP Build ID: {100/146). Available from: http://www.ncbi.nlm.nih.gov/SNP/.
- Dejean AS, Beisner DR, Ch'en IL, Kerdiles YM, Babour A, Arden KC, Castrillon DH, DePinho RA, Hedrick SM (2009) Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat. Immunol. 10, 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlon TA, Curb JD, He Q, Grove JS, Masaki KH, Rodriguez B, Elliott A, Willcox DC, Willcox BJ (2012) FOXO3 gene variants and human aging: coding variants may not be key players. J. Gerontol. A Biol. Sci. Med. Sci. 67, 1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelenboom A, Burgering BM (2013) FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 14, 83–97. [DOI] [PubMed] [Google Scholar]
- Finch CE (2010) Evolution in health and medicine Sackler colloquium: evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc. Natl. Acad. Sci. USA 107(Suppl 1), 1718–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanché H, von Eller‐Eberstein H, Nikolaus S, Schreiber S, Nebel A (2009) Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc. Natl. Acad. Sci. USA 106, 2700–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Capri M, Monti D, Giunta S, Olivieri F, Sevini F, Panourgia MP, Invidia L, Celani L, Scurti M, Cevenini E, Castellani GC, Salvioli S (2007) Inflammaging and anti‐inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech. Ageing Dev. 128, 92–105. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Rommel C (2014) PI3K and cancer: lessons, challenges and opportunities. Nat. Rev. Drug Discov. 13, 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg EW, Li Y, Wang J, Burrows NR, Ali MK, Rolka D, Williams DE, Geiss L (2014) Changes in diabetes‐related complications in the United States, 1990–2010. N. Engl. J. Med. 370, 1514–1523. [DOI] [PubMed] [Google Scholar]
- Kagan A, ed. (1996) The Honolulu Heart Program: An Epidemiological Study of Coronary Heart Disease and Stroke. Amsterdam, The Netherlands: Harwood Academic Publishers. [Google Scholar]
- Kennedy BK, Pennypacker JK (2015) Aging interventions get human. Oncotarget 6, 590–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ (2010) The genetics of ageing. Nature 464, 504–512. [DOI] [PubMed] [Google Scholar]
- Kuningas M, Mägi R, Westendorp RG, Slagboom PE, Remm M, van Heemst D (2007) Haplotypes in the human Foxo1a and Foxo3a genes; impact on disease and mortality at old age. Eur. J. Hum. Genet. 15, 294–301. [DOI] [PubMed] [Google Scholar]
- Lam EW, Brosens JJ, Gomes AR, Koo CY (2013) Forkhead box proteins: tuning forks for transcriptional harmony. Nat. Rev. Cancer 13, 482–495. [DOI] [PubMed] [Google Scholar]
- Lee JC, Espéli M, Anderson CA, Linterman MA, Pocock JM, Williams NJ, Roberts R, Viatte S, Fu B, Peshu N, Hien TT, Phu NH, Wesley E, Edwards C, Ahmad T, Mansfield JC, Gearry R, Dunstan S, Williams TN, Barton A, Vinuesa CG, Parkes M, Lyons PA, Smith KG (2013) Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3‐regulated pathway. Cell 155, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, Chen C, Hosokawa K, Nakauchi H, Nakayama K, Nakayama KI, Harada M, Motoyama N, Suda T, Hirao A (2007) Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1, 101–112. [DOI] [PubMed] [Google Scholar]
- Monsalve M, Olmos Y (2011) The complex biology of FOXO. Curr. Drug Targets 12, 1322–1350. [DOI] [PubMed] [Google Scholar]
- Morris BJ, Willcox DC, Donlon TA, Willcox BJ (2015) FOXO3: a major gene for human longevity – A Mini‐Review. Gerontology 61, 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris BJ, Chen R, Donlon TA, Evans DS, Tranah GJ, Parimi N, Ehret GB, Newton‐Cheh C, Seto T, Willcox DC, Masaki KH, Kamide K, Ryuno H, Oguro R, Nakama C, Kabayama M, Yamamoto K, Sugimoto K, Ikebe K, Masui Y, Arai Y, Ishizaki T, Gondo Y, Rakugi H (2016) Association analysis of FOXO3 longevity variants with blood pressure and essential hypertension. Am. J. Hypertens. (Epub ahead of print Oct 16, 2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murabito JM, Yuan R, Lunetta KL (2012) The search for longevity and healthy aging genes: insights from epidemiological studies and samples of long‐lived individuals. J. Gerontol. A Biol. Sci. Med. Sci. 67, 470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute on Aging . (2007) Living Long and Well in the 21st Century Strategic Directions for Research on Aging: National Institute on Aging [WWW document]. URL http://www.nia.nih.gov/about/living-long-well-21st-century-strategic-directions-research-aging [accessed on 20 December 2014].
- Oh HM, Yu CR, Golestaneh N, Amadi‐Obi A, Lee YS, Eseonu A, Mahdi RM, Egwuagu CE (2011) STAT3 protein promotes T‐cell survival and inhibits interleukin‐2 production through up‐regulation of class O forkhead transcription factors. J. Biol. Chem. 286, 30888–30897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshansky SJ, Passaro DJ, Hershow RC, Layden J, Carnes BA, Brody J, Hayflick L, Butler RN, Allison DB, Ludwig DS (2005) A potential decline in life expectancy in the United States in the 21st century. N. Engl. J. Med. 352, 1138–1145. [DOI] [PubMed] [Google Scholar]
- Pawlikowska L, Hu D, Huntsman S, Sung A, Chu C, Chen J, Joyner AH, Schork NJ, Hsueh WC, Reiner AP, Psaty BM, Atzmon G, Barzilai N, Cummings SR, Browner WS, Kwok PY, Ziv E (2009) Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell 8, 460–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SL (2007) Immune regulation by Foxo transcription factors. Autoimmunity 40, 462–469. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome‐wide association studies. Nat. Genet. 38, 904–909. [DOI] [PubMed] [Google Scholar]
- Salminen A, Huuskonen J, Ojala J, Kauppinen A, Kaarniranta K, Suuronen T (2008) Activation of innate immunity system during aging: NF‐kB signaling is the molecular culprit of inflamm‐aging. Ageing Res. Rev. 7, 83–105. [DOI] [PubMed] [Google Scholar]
- Shadyab AH, LaCroix AZ (2015) Genetic factors associated with longevity: a review of recent findings. Ageing Res. Rev. 19, 1–7. [DOI] [PubMed] [Google Scholar]
- Shen J, Tower J (2010) Drosophila foxo acts in males to cause sexual‐dimorphism in tissue‐specific p53 life span effects. Exp. Gerontol. 45, 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerensen M, Nygaard M, Dato S, Stevnsner T, Bohr VA, Christensen K, Christiansen L (2015) Association study of FOXO3A SNPs and aging phenotypes in Danish oldest‐old individuals. Aging Cell 14, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata H, Suzuki M, Ishii T, Sekiguchi S, Iri H (1987) Influence of major histocompatibility complex region genes on human longevity among Okinawan‐Japanese centenarians and nonagenarians. Lancet 2, 824–826. [DOI] [PubMed] [Google Scholar]
- US Department of Health and Human Services . (2012) Office of Disease Prevention and Health Promotion. Healthy People 2020 [WWW document]. URL. http://www.healthypeople.gov [accessed on 20 December 2014].
- White L, Petrovitch H, Ross GW, Masaki KH, Abbott RD, Teng EL, Rodriguez BL, Blanchette PL, Havlik RJ, Wergowske G, Chiu D, Foley DJ, Murdaugh C, Curb JD (1996) Prevalence of dementia in older Japanese‐American men in Hawaii: the Honolulu‐Asia Aging Study. JAMA 276, 955–960. [PubMed] [Google Scholar]
- Willcox BJ, He Q, Chen R, Yano K, Masaki KH, Grove JS, Donlon TA, Willcox DC, Curb JD (2006) Midlife risk factors and healthy survival in men. JAMA 296, 2343–2350. [DOI] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD (2008) FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. USA 105, 13987–13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Xia W, Hu MC (2006) Ionizing radiation activates expression of FOXO3a, Fas ligand, and Bim, and induces cell apoptosis. Int. J. Oncol. 29, 643–648. [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu Y, Tian X, Yang X, Li X, Jiang F, Chen Y, Shi M (2014) Association study to evaluate FoxO1 and FoxO3 gene in CHD in Han Chinese. PLoS One 9, e86252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Serum TNF‐α level by FOXO3 G allele carrier status.


