Abstract
Current trends in population growth and consumption patterns continue to increase the demand for wheat, a key cereal for global food security. Further, multiple abiotic challenges due to climate change and evolving pathogen and pests pose a major concern for increasing wheat production globally. Triticeae species comprising of primary, secondary, and tertiary gene pools represent a rich source of genetic diversity in wheat. The conventional breeding strategies of direct hybridization, backcrossing and selection have successfully introgressed a number of desirable traits associated with grain yield, adaptation to abiotic stresses, disease resistance, and bio-fortification of wheat varieties. However, it is time consuming to incorporate genes conferring tolerance/resistance to multiple stresses in a single wheat variety by conventional approaches due to limitations in screening methods and the lower probabilities of combining desirable alleles. Efforts on developing innovative breeding strategies, novel tools and utilizing genetic diversity for new genes/alleles are essential to improve productivity, reduce vulnerability to diseases and pests and enhance nutritional quality. New technologies of high-throughput phenotyping, genome sequencing and genomic selection are promising approaches to maximize progeny screening and selection to accelerate the genetic gains in breeding more productive varieties. Use of cisgenic techniques to transfer beneficial alleles and their combinations within related species also offer great promise especially to achieve durable rust resistance.
Keywords: wheat, genetic diversity, introgressions, disease resistance, pest resistance, cisgenesis, genomic selection, nutritional quality
Introduction
Wheat (Triticum aestivum L.), one of the key cereal crops, is grown on 222 million hectares worldwide and is a major source of calories and proteins globally (USDA, 2016). Wheat production has increased from 235 million tons in 1961 to an estimated 733 million tons in 2015 (FAOSTAT, 2014). The Green Revolution of 1960 and 1970s along with changes in policies, fertilizer use and advances in agronomy has stimulated wheat productivity over past decades (Ziska et al., 2012). A highly cited example is the global success of two semi-dwarf wheat varieties “Sonalika and Kalyan Sona” in the 1960s which helped wheat production advance from deficit to surplus in South Asia.
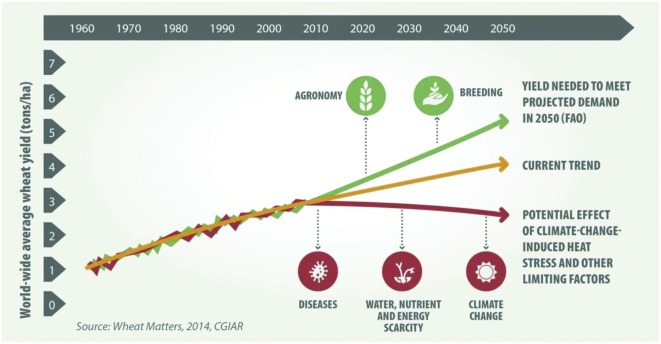
In recent years, changes in population trends, eating habits, and economic and socio-economic conditions, especially in Africa and Asia, have caused an increase in global wheat demand. Under the assumption of favorable growing conditions, the International Grain Council [IGC] (2014) estimated the wheat production and consumption demands till 2020. Based on their predictions, wheat productivity growth was estimated at 1.1% per year for next 5 years, which will make it possible to meet the consumption demands till 2020. However, in recent years, noticeable changes in temperature and rainfall at the global level have had an impact on wheat production. Various crop models have estimated yield reductions of 6–13% in wheat for each °C rise in temperature. Based on the current trends in wheat production, the predicted increase in wheat productivity by 2050 will be short of 1 t/ha which is required to meet the rising global demand (Figure 1). Increased climate variability, frequent extreme weather events, and new variants of pathogens and pests further jeopardize linear productivity growth into the future. Breeding wheat for climatic change tolerance and disease resistance combined with good agronomy can potentially improve wheat productivity to meet the future demands.
FIGURE 1.
Projected demand and yield trends in wheat under several scenarios. Source: CIMMYT (2014).
Wheat is an allopolyploid species that originated from a cross of the tetraploid species Triticum turgidum and the diploid species Aegilop tauschii (Coss) Schmalh. Wild tetraploid emmer wheat evolved from a hybridization of wild Triticum urartu tumanian ex Gandivan and an undiscovered species of the Aegilops speltoides Tausch lineage. During the process of domestication genetic bottlenecks resulted in significant loss of diversity. There has been a keen interest in utilizing the genetic diversity of Triticeae species, which includes the primary, secondary, and tertiary gene pools (Aegilops, Agropyron, Elymus, Hordeum, Leymus, Secale, Thinopyrum, and Triticum). These gene pools are a rich source of genes that can be used to improve diverse traits such as disease resistance, micronutrient availability and abiotic stress adaptation. Novel alleles have been introgressed from nearly 52 species highlighting the genomic plasticity of wheat and the importance of exotic introgressions in wheat improvement (Wulff and Moscou, 2014).
In this review, we highlight the genetic diversity available in wheat for grain yield, adaptation to climate change, disease and insect pest resistance, and nutritional and end-use quality. We also discuss traditional approaches to introgression that are still successful and current technologies that are being used to characterize the genetic diversity and improve the efficiency of the introgression process. We also explore the role of new technologies such as genomic selection (GS) and cisgenesis to integrate diverse genes/alleles and accelerate the breeding process.
Diversity in Wheat For:
Grain Yield Improvement and Climate Resilience
Grain yield per se is a polygenic trait, and yield improvements from alien introgressions are due to their positive impact on phenology, yield components (that is grain size, grain number, floret number, etc.), or through adaptive traits for abiotic stresses (such as heat, drought, and alkaline/acid soils) and resistance to biotic stresses. Landraces, a crucial germplasm pool has been reported to contribute genes for grain yield improvement in irrigated environments or, in heat and drought stress environments (Reynolds et al., 2007a; Lopes et al., 2015). Direct varietal releases from simple crosses with landraces are rare, though a Turkish variety ‘Gerek 79’ is an exception (Smale and McBride, 1996). One of the best examples is the Rht dwarfing gene that was available through the Japanese variety ‘Norin10’ originating from a Japanese landrace Shiro Daruma (Reitz and Salmon, 1968; Dreisigacker et al., 2005). These dwarfing genes were utilized by Dr. Norman E. Borlaug to develop the high-yielding semi-dwarf wheat varieties that triggered the Green Revolution. Several other landraces also have had an impact on improving the germplasm pool: for example, ‘Cheyenne,’ a selection from landrace Crimea, founded the Nebraska wheat gene pool while ‘Turkey Red’ was used for winter wheat breeding in the USA Great Plains (Lopes et al., 2015). Studies on landraces from different regions of the world have identified potential sources for improvement of grain yield and climate resilience, for instance the drought tolerant variety ‘Aragon 03’ was developed from a selection of a landrace population ‘Catalan de Monte’ (Royo and Briceño-Félix, 2011). The potential of Mexican landraces to adapt to temperature and drought stress has been reported (Hede et al., 1999; Vikram et al., 2016). Further, allelic variation for specific plant traits such as improved 1000 kernel weight, biomass, and photosynthesis has also been identified in landraces (Lopes et al., 2015).
The development of synthetic hexaploid wheats has allowed the use of wild relatives such as tetraploid species (e.g., Triticum dicoccum) and the diploid species A. tauschii to transfer adaptive traits in to modern wheat. Genomic regions in A. tauschii can contribute to nearly 10% increase in grain weight (Röder et al., 2008) and improve grain yield (Börner et al., 2015). Synthetic wheats can be used to transfer such useful genetic variations. Studies have reported synthetic wheat lines that can extract more water from deeper soil, which under drought stress is an excellent adaptive trait (Reynolds et al., 2007b). Similarly, other synthetic derivatives with improved tolerance to water logging, high temperatures, and freezing have also been identified (Maes et al., 2001; Villareal et al., 2001; Yang et al., 2002).
Wild relatives of wheat also present a rich source of diversity. Species such as Agropyron elongatum (Host) Beauv. and Agropyron cristatum Gaertn. are reported to contribute to higher grain yields in wheat growing under optimal conditions. In certain wheat backgrounds, chromosome 7 Ag from A. elongatum increases grain yield up to 8% and carries leaf (Lr) and stem rust (Sr) resistance genes Lr19 and Sr25, respectively (Singh et al., 1998). On further study this yield increase from A. elongatum was attributed to a better allocation of assimilates to the reproductive organs (Miralles et al., 2007). Another example is the 6P chromosome from the tetraploid species A. cristatum, which has been reported to increase number of florets, kernels and grain weight in wheat, in addition to improving resistance to the barley yellow-dwarf virus and powdery mildew resistance alleles (Wu et al., 2006; Wang et al., 2011).
One of the most widely used wheat relatives is rye (Secale cereale L.), which is well-documented as a rich source of biotic and abiotic resistance/tolerance. Rye (2n = 2x = 14), is a diploid species, originating from the Near East (Hillman, 1978; Salamini et al., 2002), belongs to the tertiary gene pool of wheat, along with Thinopyrum and Elymus species (Harlan and de Wet, 1971). The first attempts to hybridize wheat and rye were conducted by Stephen Wilson (Wilson, 1873). The first stable amphiploid triticale (Triticosecale Wittmack) is attributed to Rimpau in 1888; thereafter, efforts were dedicated to producing wheat-rye hybrids (Ammar et al., 2004).
Several 100s of cultivars with the (1B)1R substitution or 1BL.1RS and 1AL.1RS translocations from Petkus rye were deployed between 1960 and1990 (Rabinovich, 1998). During the 1990s, the 1BL.1RS translocation was present in 60% of wheat descending from lines developed at the International Maize and Wheat Improvement Center (CIMMYT) and nearly half of the commercial varieties (Rabinovich, 1998). In China, which is one of the major wheat growing countries, about 42% of the wheat cultivars released between 1960 and 2000 were (1B)1R genotypes, and the consistent yield gains over the years were partially attributed to the translocation (Zhou et al., 2007). Most of the desirable characteristics translocated from rye to wheat have been found in chromosome 1R that contributes to yield advantage (Villareal et al., 1998). Translocations from chromosomes 1RL and 1RS improve water use efficiency by promoting root and above ground biomass growth (Ehdaie et al., 2003; Hoffmann, 2008; Karki et al., 2014). Other rye chromosomes such as 3R, 4R, and 6R are also potential donors; introgressions from these regions could improve aluminum and acid soil tolerance in wheat.
Disease Resistance
Diseases, caused by both fungi and fungi-like pathogens pose a major threat to wheat production. Evolution of new virulence through migration, mutation, selection, and recombination of virulence genes occurs in all pathogens, but has been more frequent in those causing rust and powdery mildew. Yield losses due to diseases can be up to 70% in susceptible varieties (Singh et al., 2008). For example, in 1998, stem rust infections were reported in Uganda caused by a new race designated as UG99. A series of reviews by Singh et al. (2006, 2008, 2011, 2015) has documented the significance, emergence, evolution and geographical spread of the Ug99 group as time progressed. Since its first discovery, 13 races within the Ug99 group have been identified across several countries in Africa and Middle East1. Another example in recent years is of the stripe or yellow rust pathogen. Yellow rust (Yr) is found primarily in the Northern latitudes or cooler environments, however, Hovmøller et al. (2015) found ‘Warrior’ and ‘Kranish,’ two aggressive races of yellow rust originating from sexual recombination in the near-Himalayan region of Asia which can infect host under warmer temperatures.
One of the strategies to mitigate the threat from diseases is to identify and utilize diverse sources of durable resistance. Globally important fungal diseases of wheat caused by biotrophs (obligate parasites), include the three rusts; leaf or brown rust, stripe or yellow rust and stem or black rust, caused by Puccinia triticina, Puccinia striiformis f. sp. Tritici, and Puccinia graminis f. sp. Tritici, respectively, powdery mildew caused by Blumeria graminis f. sp. tritici; whereas, those caused by hemibiotrophs (facultative parasites) include Fusarium head blight, Septoria tritici blotch, leaf blotch, spot blotch, and tan spot.
Resistance genes can be characterized as race specific and race non-specific, this classification dates back to 1962 when Van der Plank proposed the first theoretical concepts of disease resistance. Race specific genes confer resistance to one or a few races of a pathogen and are known to be based on ‘gene for gene’ interaction. Also known as ‘major genes,’ they usually have large phenotypic effects, but may not confer complete resistance. Although incorporation of race-specific resistance genes may be promising, it increases the risk of faster breakdown. Some examples of major genes for rust resistance include Lr19, Lr26, and Lr42 effective against leaf rust, Yr5, Yr10, and Yr15 against yellow rust and Sr22, Sr26, and Sr35 against stem rust. Race non- specific resistance, is usually effective in the post-seedling growth stage, thus commonly referred to as adult plant resistance (APR). Race-non specific resistance is generally quantitatively inherited and ranges from moderate resistance/moderate susceptibility to nearly complete resistance and interact additively with other non-specific resistance genes. Varieties with high levels of durable resistance to multiple pathogens can be developed by combining multiple race non-specific resistance loci, especially to those which are known to confer resistance to multiple diseases (Singh et al., 2008). Examples of these pleiotropic resistance genes are Lr34, Lr46, and Lr67 which provide resistance to leaf, yellow and stem rust and powdery mildew. Because race non-specific resistance can provide broader and robust resistance to fight pathogen evolution it has been recommended for the high risk areas, for instance in East African highlands where wheat cultivation and pathogen evolution is continuous (Singh et al., 2008).
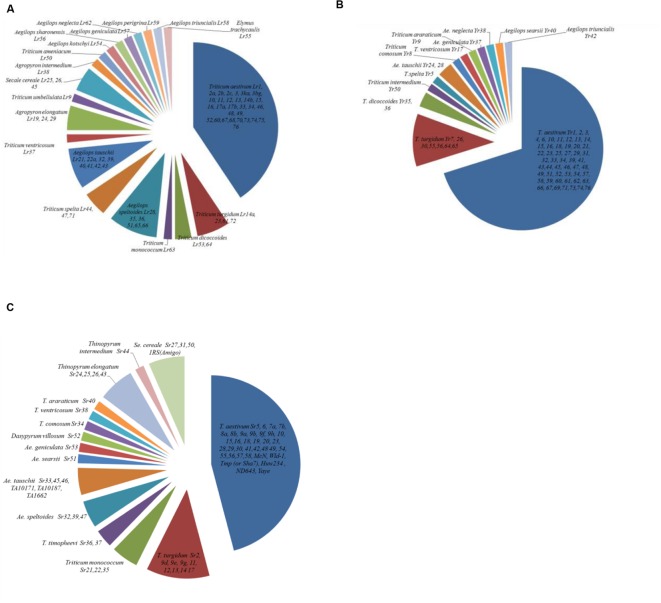
Though most rust resistance genes originated from hexaploid wheat, there are also many genes that originated from the wild relatives and other genera such as Aegilops, Dasypyrum, Thinopyrum, and Secale (Figure 2). As early as 1920 and 1930s, introgression of stem rust resistance from T. turgidum subsp. durum and T. dicoccum subsp. Dicoccum Schrank ex Schubler into bread wheat was reported (Hayes et al., 1920; McFadden, 1930). Both race-specific and non-specific genes have been identified from diverse genetic sources. For instance, Lr9 from Aegilops umbellulata Zhuk, Yr5 from Triticum spelta L., Yr28 from A. tauschii, Sr9e from tetraploids and Sr35 from Triticum monococcum L. are race-specific genes. Examples of race non-specific genes/APR include Lr22a from A. tauschii, Yr36 from Triticum diccocoides (Korn. Ex Asch. and Graebn) Schweinf, Yr48 from synthetic hexaploid wheat PI610750 and Yr52, 56, 57, and 62 from landraces. Introgressions are also associated with multiple disease resistance as well, such as Pm8/Sr31/Lr26/Yr9 from rye, Sr36/Pm6 from Triticum timopheevi (Zhuk.) Zhuk., Pch1 and Sr38/Lr37/Lr17 from Aegilops ventricosa Tausch, and Lr19/Sr25, Sr24/Lr24, and Sr26 from A. elongatum (Host) P. Beauy (Sears, 1956; Friebe et al., 1996; Mago et al., 2005; Wulff and Moscou, 2014). Some genes introgressed from wild relatives have been associated with negative linkage drag and therefore have not been widely deployed in breeding: examples include Sr32 and Sr37 identified in A. speltoides (McIntosh et al., 1995) and T. timopheevi (McIntosh and Gyarfas, 1971) respectively. Other temporarily designated genes that are common in high yielding wheat germplasm offer additional possibilities for combining resistance genes combinations.
FIGURE 2.
Origin of designated (A) leaf rust, (B) stripe rust, and (C) stem rust genes conferring seedling and/or adult plant resistance.
Novel alleles from genetically diverse sources have also been identified for other important wheat diseases. For example, Fusarium head blight resistance genes are from genera Roegneria, Hystrix, Elymus, Kengyilia, and Agropyron (Wan et al., 1997) and other related species, e.g., T. timopheevi, T. monococcum, Triticum karamyschevii Neyski, and T. militinae Zhuk and Migush (Cai et al., 2005). Genes conferring powdery mildew resistance have been reported from T. dicoccoides (Moseman et al., 1984), Triticum carthlicum Nevski, T. monococcum and T. timopheevi (Tomerlin et al., 1984). Some of the designated genes for resistance to powdery mildew, fusarium head blight and Septoria tritici blotch are given in Table 1. Wheat blast caused by Magnaporthe oryzae (anamorph. Pyricularia oryzae) is an emerging disease in the tropical parts of the Southern Cone of South Americas and was reported in Bangladesh as well. Though wheat blast is a recent disease, resistance has been identified in A. tauschii (Bockus et al., 2012) and in synthetic wheats (Cruz et al., 2010). The 2NS/2AS translocation from A. ventricosa was recently found to confer wheat blast resistance (Cruz et al., 2016), though unpublished reports from Paraguay have documented the emergence of new isolates virulent to this resistance. Both qualitative and quantitative resistance have been observed and the former has been validated at the seedling stage (Maciel et al., 2014). So far, eight resistance genes have been identified (i.e., Rmg1 to Rmg8), of which only Rmg2, Rmg3, Rmg7, and Rmg8 are host resistance genes against Triticum isolates of Pyricularia oryzae; the rest are non-host resistance genes (Anh et al., 2015). It is noteworthy that only Rmg7 was identified in T. dicoccum (Tagle et al., 2015) whereas all are from bread wheat (Anh et al., 2015). Thus diverse resistant sources are available for both the existing and the emerging diseases in wheat.
Table 1.
Known genes for resistance to Powdery mildew, Fusarium head blight and Septoria tritici blotch from landraces, wild relatives and synthetic wheat.
| Diseases | Source of Resistance | Genes |
|---|---|---|
| Powdery mildew (Blumeria graminis f. sp. tritici) | Triticum monococcum | Pm4d, Pm1b, and Pm1c |
| Triticum urartu | PmU | |
| Triticum boeoticum | Pm25, PmTb7A.1, and PmTb7A.2 | |
| Triticum dicoccoides | Pm16, Pm26, Pm30, Pm31, Pm36, Pm41, Pm42, and MeIW72 | |
| Triticum dicoccum | Pm4a, Pm5a, Pm49, and Pm50 | |
| Triticum carthlicum | Pm4b, Pm33, and Pm46 | |
| Triticum spelta | Pm1d, Pm10, and Pm11 | |
| Triticum sphaerococum | Pm3b and Pm36 | |
| Triticum timopheevi | Pm6, Pm27, and Pm37 | |
| Aegilops tauschii | Pm2, Pm19, Pm34, and Pm35 | |
| Aegilops speltoides | Pm12 and Pm32 | |
| Aegilops longissimi | Pm13 | |
| Aegilops ovata | Pm29 | |
| Secale cereale | Pm7, Pm8, Pm17, and Pm20 | |
| Thinopyrum intermedium | Pm40 and Pm43 | |
| Haynaldia villosum | Pm21 | |
| Fusarium head blight (Fusarium graminearum) | Triticum spp. | Fhb1, Fhb2, Fhb4, and Fhb5 |
| Leymus rasomsus | Fhb3 | |
| Elymus tsukushiensis | Fhb6 | |
| Thinopyrum ponticum | Fhb7 | |
| Septoria tritici blotch (Mycosphaerellla graminicola) | Synthetic Wheat (Synthetic 6x, W7984, M3) | Stb5, Stb8, Stb16q |
| Triticum monococcum (W7984) | TmStb1 |
Insect Pest Resistance
It is estimated that global yield losses due to insect pests in the pre-green revolution era were about 5.1%, however, the losses increased to 9.3% in the post-green revolution in 1990s (Dhaliwal et al., 2010). Insect pests are dynamic and highly adaptable. Changes in environmental temperature can modify their physiology, behavior, voltinism, and distribution. For instance, with warmer winters, the number of aphid generations per wheat growing cycle may increase (Hullé et al., 2010) and extend their distribution further (Macfadyen and Kriticos, 2012). It has also been proven that aphids can modify their behavior in response to either high or low temperature stress (Ma and Ma, 2012; Alford et al., 2014), enabling them to adapt in the presence of natural selection if genetic variation exists for such traits. While the work on disease resistance has tremendously contributed to protect wheat yields, control of arthropod pests has largely depended on the use of chemicals. A dramatic positive impact could be achieved through the introduction of new resistance genes (either singly or in combination with of multiple genes) to provide a broad spectrum of protection against multiple pathogens and insect biotypes.
There are several examples where genes from alien sources have been found to confer resistance to some of the most important wheat pests such as aphids Schizaphis graminum (Rondani), Diuraphis noxia (Mordvilko), Rhopalosiphum padi L. and Sitobion avenae (F.), the cecidomyid Mayetiola destructor (Say), the nematode Heterodera avenae (Wollenweber) and the mite Aceria tosichell Keifer. Several wheat-related species have been found to be resistant to aphids; however, efforts to incorporate such resistance sources into wheat breeding pipelines are limited and there are only few a specific cases in which aphid resistant cultivars are purposely bred (i.e., D. noxia in the USA and South Africa, and S. graminum in the USA). To determine the utility of such genetic resources for aphid resistance, Smith et al. (2004) evaluated 21 accessions from six species of Aegilops and one accession of Triticum araraticum Jakubz that were previously identified to be resistant to R. padi and found antibiotic effects on S. avenae and D. noxia in an Aegilops neglecta accession. Migui and Lamb (2003) evaluated resistance to R. padi, S. avenae, and S. graminum in 19 species related to wheat, and found that species such as Triticum boeoticum Boiss., A. tauschii and T. araraticum had the higher levels of resistance to R. padi, whereas A. tauschii and T. turgidum had higher levels of overall resistance to S. graminum, and T. araraticum and T. dicoccoides had higher levels of overall resistance to S. avenae. However, for other destructive pests such as, Eurygaster integriceps Puton, more work is required to find adequate resistance levels that can be incorporated in wheat cultivars (El Bouhssini et al., 2009). Friebe et al. (1996) made a comprehensive review of wheat-alien translocations that confer resistance to wheat biotic stresses. Here, some examples of resistance to diseases and pests translocated from rye are reviewed (see Table 2, where we summarize resistance sources by rye chromosome and diseases/pests).
Table 2.
Examples of resistance genes for diseases and pests from rye (Secale cereale).
| Diseases | Gene | Description | Germplasm |
|---|---|---|---|
| Leaf rust (Puccinia triticina) | Lr26 | 1BL.1RS | Petkus rye; Kavkaz and Veery wheat derived |
| Lr25 | 4BS.4BL-2RL | Transec | |
| Lr45 | 2AS-2RS.2RL | RL6144 | |
| Stripe Rust (Puccinia striiformis var. striiformis) | Yr9 | 1BL.1RS | Petkus rye; Kavkaz and Veery wheat derives |
| YrCN17† | 1BL.1RS | R14, Chuan-nong 17 | |
| YrR212† | 1BL.1RS | R212 | |
| Stem rust (Puccinia graminis f. sp. tritici) | Sr31 | 1BL.1RS | Petkus rye; Kavkaz and Veery wheat derives |
| Sr50/SrR | 1BL.1RS | Imperial rye derives | |
| Sr1RSAmigo | 1AL.1RS | Amigo wheat | |
| Sr27 | 3AL.3RS | WRT238 | |
| Powdery mildew (B. graminis f. sp. tritici) | Pm8 | 1BL.1RS | Petkus rye; Kavkaz and Veery wheat derives |
| Pm17; allelic to Pm8 | 1AL.1RS | Insave rye derives; Amigo wheat derives | |
| Pm7 | 4BS.4BL-2RL | Transec | |
| Pm20 | 6BS.6RL | WGRC28 | |
| Greenbug (Schizaphis graminum) | Gb2 | 1AL.1RS | Insave rye, Amigo wheat derives |
| Gb6 | 1AL.1RS | Insave rye, GRS1201 | |
| Diuraphis noxia | Dn7 | 1BL.1RS | 94M370 wheat |
| Hessian fly (Mayetiola destructor) | H21 | 2BS.2RL | KS85HF 011-5 |
| H25 | 4BS.4BL-6RL | Balbo rye; 88HF16 wheat | |
| Aceria tosichell | CmC3 | 1AL.1RS | Amigo wheat |
| Cereal cyst nematode (Heterodera avenae) | CreR | 6DS.6RL | T-701 triticale derives |
†Temporary designation.
End-Use Quality and Nutritional Quality
In addition to combating abiotic and biotic stresses while improving grain yield, wheat breeding must improve or at least maintain the nutritional and end-use quality. The wide variety of food products made from wheat flour has resulted in ongoing demand from the wheat processing industry for wheat with specific quality attributes. Additionally, dietary deficiencies of essential micronutrients such as zinc (Zn) and iron (Fe) are a major health concern in developing countries especially for pregnant women and children under age 5. An estimated 17.3% of the world’s population is at risk for inadequate zinc intake, a factor highly correlated with stunted growth in children (Wessells and Brown, 2012). Genetic biofortification with natural genetic variation present in wild relatives, synthetics and landraces for micronutrient uptake from the soil and translocation in to wheat grain is a sustainable solution that can supplement micronutrient-deficient rural inhabitants with limited access to formal markets or health care systems (Velu et al., 2014).
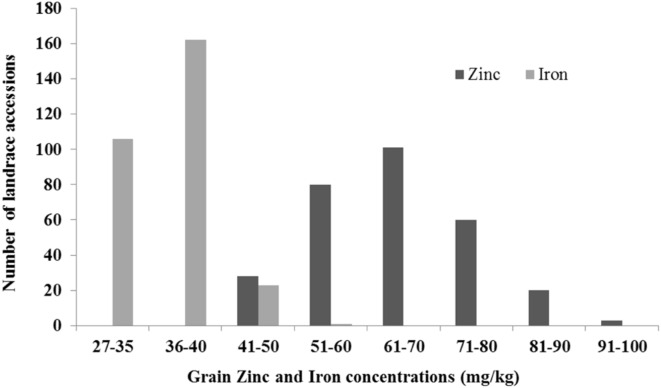
In recent years, the focus has been on “biofortification” of wheat with micronutrients, specifically Zn and Fe. Evaluation of landraces and secondary gene pools (i.e., tetraploid and diploid progenitors of hexaploid wheat) for micronutrient concentration identified T. dicoccoides, A. tauschii, T. monococcum, and T. boeticum Boiss. as the most promising sources for improving Fe and Zn grain concentration (Cakmak et al., 2000; Monasterio and Graham, 2000). Large scale screening of available wheat genetic resources at CIMMYT identified einkorn wheat, wild emmer wheat, and landraces with high amounts of Zn and Fe in grain (Cakmak et al., 2000; Ortiz-Monasterio et al., 2007). The available genetic variation in wild emmer (T. dicoccoides), T. spelta, T. dicoccum species is being used to develop nutrient-enriched wheat germplasm. The stocks (T. turgidum ssp. dicoccum/A. tauschii) are also being used for genetic biofortification of Zn and Fe by CIMMYT’s wheat breeding program (Ortiz-Monasterio et al., 2007; Morgounov et al., 2007). Recently, evaluation of a representative subset of Mexican and Iranian landraces under Zn-enriched soil conditions in Cd. Obregon, Mexico, showed more than a twofold variation for Zn (40–96 mg/kg) and Fe (27–56 mg/kg; Figure 3). A major locus affecting Zn and Fe concentration, Gpc-B1 (250 kb-locu), was mapped, and found to encode a NAC transcription factor (NAM-B1) that accelerates senescence and increases nutrient remobilization from leaves to grain (Uauy et al., 2006; Distelfeld et al., 2007). Interestingly, the favorable allele of Gpc-B1 is from T. dicoccoides and all modern tetraploid and hexaploid wheats possess a non-functional allele of NAM-B1, indicating that the NAM-B1 function was lost during domestication.
FIGURE 3.
Genetic diversity for grain Zinc and Iron in Mexican and Iranian landrace collections.
Apart from micronutrients, wheat grain is also a good source of other beneficial nutrients which could be targeted by breeding programs to improve the nutritional quality of wheat based products. Grain bran is particularly rich in dietary fiber, vitamins (folic acid), and phytochemicals, which have been associated with a protective role for many chronic diseases including cardiovascular diseases and type 2 diabetes (Jacobs et al., 1999; Liu et al., 1999; de Munter et al., 2007). The HEALTHGRAIN cereal diversity screening project reported diversity for dietary fiber and phytochemicals in the wheat primary gene pool. The levels of dietary fiber ranged from 11.5 to 18.3% of dry matter, and more specifically the content of water extractable arabinoxylans (an important source of soluble dietary fiber, which is more readily fermentable in the colon than insoluble one) ranged from 0.3 to 0.85% in bran and from 0.3 to 1.4% in flour (Gebruers et al., 2008; Ward et al., 2008; Kariluto et al., 2010). Various research projects are currently ongoing to screen for genetic variability of the bioactive compounds (Di Silvestro et al., 2012; Giambanelli et al., 2013; Laddomada et al., 2016). High heritability for some of these compounds such as tocols, sterols and arabionoxylan fiber (Shewry et al., 2010) and available genetic diversity increases the chances of utilizing the variation for improving nutritional quality in wheat.
The vast catalog of products prepared from wheat requires genetic variation in traits related to grain composition as well. Exploring novel genetic variation could improve processing and end-use quality. Grain proteins are one of the important components that influence end-use quality. Studies have reported higher grain protein content in landraces than in modern wheat (Rodriguez-Quijano et al., 1994; Dotlacil et al., 2010) which means landraces and wild relatives could be a potential source to improve protein content. In fact, as mentioned above GPC-B1 (also called NAM-B1), the first gene identified for grain protein content variation was transferred from a wild emmer accession (T. dicoccoides) to modern durum wheat background (Avivi, 1978; Joppa and Cantrell, 1990; Joppa et al., 1997). While grain protein content is important, gluten quality is equally important. Gluten, an essential component of dough, is a complex protein network formed mainly by two kinds of proteins, monomeric gliadins and polymeric glutenins, which in turn are divided into high molecular weight glutenins (HMWGs) and low molecular weight glutenins (LMWGs). Although there is allelic variation in modern wheat for the gene Glu-1 encoding HMWGs, use of diversity in the Triticeae pool could potentially contribute to improve processing quality (Xu et al., 2010; Rasheed et al., 2014). The Wheat Gene Catalog currently describes 26 alleles for Glu-A1, 56 for Glu-B1, 24 for Glu-D1, 55 for Glu-A3, 32 for Glu-B3, and 16 for Glu-D3 (McIntosh et al., 2013) Several of those alleles have been detected in modern wheat ancestors and wild relatives, such as Glu-B1q in emmer (Vallega and Waines, 1987), Glu-B1be in wild emmer (Xu et al., 2004), Glu-D1n in spelt (Caballero et al., 2001), or Glu-D1bf in A. Tuaschii (Gianibelli et al., 2001). Genetic resources (for example T. urartu or T. monococcum) can be utilized to introgress Glu-A1 x+y or y active subunits (always silenced in modern durum and bread wheat, respectively), which will lead to new variations (Alvarez et al., 2009). Recently, a novel allele HMW glutenin allele was identified from A. longissimma Schweir and Muschl through the use of a Chinese Spring substitution line CS-1S(1B) that could potentially improve dough and breadmaking quality (Wang et al., 2013).
Other important quality traits such as grain hardness or starch properties are also influenced by diverse proteins and therefore genes. Puroindolines a and b (PINA, PINB), encoded by the genes Pina-D1 and Pinb-D1, are responsible for grain hardness (Morris, 2002). Wild alleles of Pina-D1a and Pinb-D1a are linked to soft grain texture, though several alleles for both Pin-D1 genes have been associated with harder grain in modern wheat (Giroux and Morris, 1997, 1998; Lillemo and Morris, 2000; Ikeda et al., 2010), landraces (Chen et al., 2005, 2007; Ayala et al., 2013) and wild relatives (Massa et al., 2004; Guzmán et al., 2012; Cuesta et al., 2013). It is interesting to note that some of these alleles have been associated with differences in quality traits other than grain hardness (Brites et al., 2008; Tanaka et al., 2008; Chen et al., 2013). While knowledge on the diverse sources for genes to improve end-use quality is available, utilization of the diversity within the breeding programs is not prevalent.
Harnessing Diversity in Wheat
Traditional Breeding Approaches
The success of breeding to introgress beneficial genomic regions into wheat is conditioned by the relatedness between the species (Friebe et al., 1996). Mujeeb-Kazi and Wang (1995) identified certain key requirements for introgression, (1) the genome constitution of the donor species; (2) the genomic relationship between the donor and recipient species; (3) chromosomal location of the loci of interest; (4) whether the gene(s) of interest can be expressed in the recipient species; and (5) whether gene transfer has any negative effect on the recipient species. For instance, introgression can be achieved by direct hybridization, homologous recombination, backcrossing and selection if the donor species belongs to the primary gene pool, e.g., hexaploid landraces, cultivated tetraploids (T. turgidum), wild emmer wheats (T. dicoccoides) or diploids T. monococcum and A. tauschii. If the donor species belongs to the secondary gene pool (e.g., polyploid Aegilops and Triticum species, and the S-genome species of the genus Aegilops) homologous recombination is possible if the loci of interest are transferred in homologous chromosomes. For species belonging to the tertiary gene pool (e.g., Elymus species), gene transfer can be achieved by exploiting the centric breakage-fusion of univalents, induced homoeology and radiation treatment to induce chromosome breaks (Friebe et al., 1996; Feuillet et al., 2008). Synthetic hexaploid wheats carry novel variation for tolerance/resistance to abiotic and biotic stresses but are usually poor in agronomic performance. While they are used for transferring useful genetic variation into common wheat, typically one or two backcrosses to elite germplasm followed by selection are required to identify lines with superior performance.
Although such introgressions can be of benefit to wheat, the donor sources often negatively impact previously selected adaptation traits in the recipient germplasm because alien chromatin is usually incorporated as large blocks that may carry alleles associated with undesirable agronomic characteristics. Depending on the wheat genetic background, the rye source and the type of abiotic stress factors, studies have shown that rye transferred into wheat may have both positive and negative effects on wheat performance. Monneveux et al. (2003) reported that depending on the wheat background, 1BL.1RS translocations can negatively impact yield under rainfed conditions and heat stress. However, in general, under non-stressed conditions, 1RS confers higher yield regardless of which wheat chromosome (1A, 1B, or 1D) it is translocated into (Kim et al., 2004). On the other hand, the position of 1RS in the wheat genome can negatively affect baking quality, thus genotypes with 1AL.1RS and 1DL.1RS are preferred over genotypes with 1BL.1RS (Graybosch et al., 1993; Kim et al., 2005). Traditional breeding methods such as repeated backcrossing and selection of desirable genotypes often require extensive efforts and are time consuming. However, with the new advances in phenotyping, QTL mapping, and genetic modification, along with sequencing technologies are expected to improve the precision and speed of alien introgression (Jacobsen and Schouten, 2007; Tiwari et al., 2014).
Modern Breeding Approaches
High Throughput Phenotyping
Phenotypic characterization is important prior to the efficient utilization of genetic diversity. Most phenotypic traits, heading time, photoperiodic responses and vernalization responses are explained by the germplasm’s geographical origin (Kato and Yokoyama, 1991; Cavanagh et al., 2013). Phenotyping for agronomic traits, response to disease and pests and other adaptive traits is crucial for the introduction of new allelic variation in breeding programs. Targeted characterization of germplasm panels such as the Focused Identification of Germplasm Strategy (FIGS), developed based on agro-ecological data enables identification of specific adaptive traits within the genetic resources. For instance, Reynolds et al. (2015) applied FIGS set to evaluate landraces, and found that those from heat and drought stressed regions had 40% greater biomass under heat and drought compared to modern varieties.
Greenhouse based automated phenotyping platforms using robotics and sensor imaging are being used for data acquisition in different crops by a number of institutes globally (e.g., IPK Gatersleben, Germany and The Plant Accelerator, Adelaide, Australia). Though, the high operational cost of such high throughput phenotyping platforms limits their large-scale use in breeding programs. Recent developments in remote sensing and high throughput phenotyping technologies allow characterization of a large number of germplasm in a short amount of time. Spectral imagery can be utilized to measure normalized difference vegetation index (NDVI), canopy temperature, hydration status, and pigment composition (Honsdorf et al., 2014; Rahaman et al., 2015; Reynolds et al., 2015). These spectral indices have already been linked to ground based measurements of yield, biomass, and adaptation (Reynolds et al., 1994). Availability of high-resolution cameras has made it possible to focus on phenotypic characterization at the plot level. For instance, spectral indices estimated by using low level airborne remote sensing showed significant association with those collected at ground level (Tattaris et al., 2013). Along with advances in statistical modeling methods, it is possible to predict plant performances in the field, based on the information obtained from high-throughput phenotyping. Such technologies could be used for characterization of the diverse germplasm pools to identify potential sources for tolerance/resistance to abiotic and biotic stresses.
Genome Wide Association Mapping and Marker Assisted Backcrossing
The use of molecular markers for identifying functional genes and genome wide association studies (GWAS) can greatly facilitate the introgression process. GWAS studies on landraces and wild relatives of wheat have identified quantitative trait loci (QTL) associated with morphological traits in normal irrigated, heat and drought environments and with disease resistance (Kertho et al., 2015; Liu et al., 2015; Sukumaran et al., 2015). If large effect QTL exist for traits of interest and the favorable alleles originate from exotic sources, then marker assisted backcrossing (MABC) can be used to more rapidly introgress such alleles into elite backgrounds compared to conventional backcrossing (Hillel et al., 1990; Tanksley and Nelson, 1996).
Marker assisted backcrossing involves selecting of favorable alleles using QTL linked markers during each backcrossing generation. To reduce the number of backcrossing generations required to recover the recurrent parent genome, markers distributed across the genome can be used to select individuals with the favorable donor QTL and the highest proportion of recurrent parent genome (Young and Tanksley, 1989; Hillel et al., 1990, Hospital et al., 1992). This approach, referred to as MABC with foreground and background selection, can be highly effective with availability of gene based markers and markers tightly linked to QTL determine (Ellis et al., 2014). This approach has been suggested for improving a wide range of traits conferred by large effect genes, including rust resistance genes in wheat. If QTL positions are uncertain (such is the case of positions inferred by QTL mapping studies), then flanking markers located several centimorgans on either side of the QTL are needed to ensure the QTL is not lost during backcrossing (Visscher et al., 1996). This may be problematic if there is linkage drag associated with the QTL, and large flanking segment may inevitably be introgressed. Fine-mapping or cloning the QTL to develop closely linked or functional markers would be ideal for backcross introgression from exotic germplasm. Unfortunately, in wheat, fine mapping and cloning can take several years.
In addition to certainty of QTL positions and availability of tightly linked markers, the number of targeted QTL is another factor that should be considered before attempting MABC. The proportion of single MABC progeny containing donor alleles at all QTL is 0.5n, where n is the number of QTL and assuming QTL are unlinked, and the position of the QTL is known with certainty. For example, to introgress of 5 QTL, approximately 3% of the progeny can be expected to contain all favorable alleles; thus 145 progeny would be required obtain one individual with all three alleles with a 1% risk of failure. Reducing linkage drag when introgressing multiple QTL can be hastened dramatically when using background selection to identify the desired recombinants. However, the probability of observing the desired recombinants remains low, and several generations of backcrossing may ultimately be needed. To introgress multiple QTL, a QTL pyramiding scheme where QTL are first introgressed in the desired background singly and then combined would be more efficient (Hosptial and Charcosset, 1997). An algorithm for designing optimal gene or QTL pyramiding schemes was presented by Servin et al. (2004).
Marker assisted backcrossing is being applied at CIMMYT to improve grain Zn and Fe concentrations. Various studies have reported QTL for high grain Fe and Zn concentrations on chromosomes 1A, 2A, 2B, 3D, 4B, 6A, 6B, and 7A in different species of diploid, tetraploid, and hexaploid wheat (Peleg et al., 2009; Tiwari et al., 2009; Xu et al., 2012; Hao et al., 2014; Srinivasa et al., 2014). A recombinant inbred line (RIL) population developed from the cross between ‘PBW343’ and ‘Kenya Swara’ was used to identify QTL and markers associated with Zn. Two novel large effect QTL on chromosomes 2B and 3A were successfully converted into usable form for marker assisted introgression of this QTL in to an elite background. During the 2014–2015 crop season, selected RILs that showed significantly enhanced Zn compared to either of the parental lines PBW 343 or Kenya Swara was used to transfer the QTL of interest using foreground selection. This strategy will serve to move desirable alleles rapidly and precisely into the adapted background.
Genomic Selection
When the number of QTL is large, MABC and pyramiding schemes may not be feasible. Phenotypic selection is currently the most reliable and widely used method for introgressing of favorable alleles from an exotic, non-adapted parent. GS techniques can also be applied to increase the rate of genetic gain in populations derived from exotic and elite parents. As reviewed by Lorenz et al. (2011), GS is a marker assisted breeding method in which genome wide markers and phenotypes from a reference population are used to train a prediction model. That prediction model is then used to predict breeding values based only on their genome-wide marker data. GS is more effective than MAS or marker assisted recurrent selection for polygenic traits (Bernardo and Yu, 2007). To achieve good prediction accuracy, it is important that the model training population be representative of the selection candidates that are to be predicted (Hayes et al., 2009; Pszczola et al., 2012).
If exotic parents are used in the breeding program, then an existing model training population will not be effective for predicting the progeny of these crosses. If one is to use GS to select among progeny from an exotic by elite cross, then a subset of the progeny will need to be phenotyped for model training. That prediction model could then be used for a few generations of recurrent selection within the bi-parental population. If the objective is to backcross favorable alleles from the exotic parent into an elite background, then GS can be used to identify the backcross progeny to cross to the recurrent parent. A simulation study by Bernardo (2016) found that the most effective GS backcrossing approach to introgress QTL from an exotic into an elite background was to train the GS model using F2 progeny and then apply that model during multiple generations of backcrossing. This approach led to a greater selection compared to phenotypic selection or selection based on QTL linked markers alone.
If crossing with exotic parents without backcrossing or within family recurrent selection, then it would be best to refrain from GS among families where one of the parents is exotic or exotic-derived until a sufficient number of progeny and other relatives descending from the exotic have been phenotyped. GS for allele introgressions has not yet been attempted in wheat; however, the use of GS in breeding with elite germplasm has shown significant potential. There are at least 29 studies on GS in wheat that have been published. Two studies (Heffner et al., 2011a,b) showed the potential of this approach to predict end-use quality traits of soft bread wheat germplasm, and obtained promising results, although forward prediction of quality traits was not carried out. A 5-years study conducted at CIMMYT with elite breeding lines for flour quality reported forward prediction accuracies of 0.68 and 0.49 for aleveograph W and loaf volume respectively (Battenfield et al., 2016). In another GS study a cross-validation of genomic predictions revealed moderately high predictability for grain Zn(0.5) and Fe(0.6) (Velu et al., 2016). Several cross validation studies have assessed the potential to use GS for improving disease resistance (Ornella et al., 2012; Rutkoski et al., 2012, 2015; Daetwyler et al., 2014; Arruda et al., 2015; Mirdita et al., 2015) and for grain yield (Crossa et al., 2010; Poland et al., 2012; Dawson et al., 2013) in wheat.
Next Generation Approaches
The concept of cisgenesis was defined by Schouten et al. (2006) as the transfer of genes within the gene pool of sexually compatible species of same genus. Though similar to classical breeding, this approach has the potential to overcome its two major limitations. Cisgenesis can be used to hasten the transfer of targeted genes between related species and can avoid linkage drag associated with classical breeding. The strategy can also be used to improve traits with limited natural allelic variation in the gene pool. Higher expression of the traits can be obtained by re-introducing the gene with its own promoter and terminator or expression levels can be lowered through silencing constructs (Holme et al., 2013). In the case of wheat, cisgenic transfers are limited within the Triticum genus, though the availability of triticale, a hybrid between rye and wheat, and hybrids between barley and wheat, opens up new opportunities for cisgenesis between two divergent sexually compatible gene pools, Triticum–Secale and Triticum–Hordeum (Holme et al., 2013). There are a few of examples of the use of cisgensis in cereals. The HMW glutenin subunit 1Dy10 associated with superior bread-making quality is present in hexaploid bread wheat but absent in durum wheat. Cisgenesis was used to transfer the 1Dy10 HMW glutenin gene from bread wheat to durum wheat (Gadaleta et al., 2008). Further work is ongoing to improve phytase activity in barley (Kerr et al., 2010) and drought tolerance in ryegrass (Bajaj et al., 2008).
With evolution of technologies, introgression of multiple genes as cassettes through cisgenesis is also being explored. Extensive time and effort is required to transfer multiple genes from genetically diverse sources in to cultivated varieties, requiring multiple backcrosses and selection against undesirable traits. The development of gene cassettes could potentially solve the issues related to sexual incompatibility, linkage drag and introgression from other genera. Wulff and Moscou (2014) described it to be equivalent to the wheat-rye translocation (1BL:1RS) which harbors different genes for disease resistance. Ellis et al. (2014) suggested constructing gene cassettes with multiple resistance genes combined resistance against the three rust diseases will result in durable resistance in wheat. The ability to produce cassettes will allow combining genes that cannot be selected in a normal breeding processes or introgress genes linked in repulsion; this will lead to rapid introgression into cultivars. Though the new technologies show great potential, they have several limitations. Both the cisgenesis and gene cassettes approaches will require genome-editing technologies that are still under development. Furthermore, there may be issues with gene suppression or loss of gene expression due to host gene interaction. For example, Hurni et al. (2014) observed that the powdery mildew gene Pm3 in wheat suppresses its ortholog Pm8 transferred to wheat from diploid rye due to interactions of encoded proteins, thus limiting transfer of multiple genes for resistance. Finally, government regulations and acceptance within the scientific and social community will drive the application of these technologies in wheat breeding.
Conclusion
The rich genetic diversity available in wheat is a source of numerous novel alleles for grain yield, disease resistance and tolerance to abiotic stress. While scientists realized the importance of genetic diversity decades ago, there is still a huge gap in characterization of the available genetic resources and their utilization in breeding programs. Over the years traditional breeding strategies have successfully incorporated novel alleles into elite germplasm, which has had significant impacts on production globally. A recent example is the development and release of biofortified wheat ‘Zinc Shakti (Chitra),’ developed by introgressing synthetic hexaploid (A. tauschii background) with elite germplasm, which has 40% higher grain Zn (Velu et al., 2015). Technologies such as GWAS and MABC are currently being used to explore the diversity and incorporate novel alleles into elite lines, though the lack of well-characterized genes and their closely linked markers impedes the process. GS and cisgenesis are promising technologies that could help harness large numbers of favorable exotic alleles and subsequently transfer them to elite backgrounds. Initiatives for genotyping and phenotyping of genetic resources through the gene banks are required to harness diversity efficiently and utilize in the breeding for improved wheat varieties.
Author Contributions
RS: invited author; contributed to genetic diversity in grain yield and disease resistance, breeding approaches for harnessing diversity; SM: corresponding author; Tasks: compiling the review manuscript and contribution to following topics, genetic diversity for grain yield, climate resilience, traditional breeding approaches, high throughput phenotyping and cisgenesis; JR: harnessing genetic diversity through Genome wide association studies, marker assisted backcrossing, and genomic selection; GV: genetic diversity for Zinc and Iron and breeding approaches for Biofortified wheat; PS: genetic diversity for disease resistance (other than Wheat rust); XH: genetic diversity for disease resistance (other than Wheat rust); LC-H: genetic diversity for pest resistance, grain yield and breeding approaches; CG: genetic diversity for grain composition and end-use quality; SB: genetic diversity for rust resistance (Stem rust, yellow rust); CL: genetic diversity for rust resistance (Yellow rust, leaf rust).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Editing assistance from CIMMYT Communications is highly appreciated.
Footnotes
References
- Alford L., Andrade T. O., Georges R., Burel F., van Baaren J. (2014). Could behavior and not physiological thermal tolerance determine winter survival of aphids in cereal fields? PLoS ONE 9:e114982 10.1371/journal.pone.0114982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J. B., Caballero L., Nadal S., Ramírez M. C., Martín A. (2009). Development and gluten strength evaluation of introgression lines of Triticum urartu in durum wheat. Cereal Res. Commun. 37 243–248. 10.1556/CRC.37.2009.2.11 [DOI] [Google Scholar]
- Ammar K., Mergoum M., Rajaram S. (2004). “History of triticale,” in Triticale: Improvement and Production eds Mergoum M., Gomez-Macpherson H. (Rome: FAO; ) 154. [Google Scholar]
- Anh V. L., Anh N. T., Tagle A. G., Vy T. T., Inoue Y., Takumi S., et al. (2015). Rmg8, a new gene for resistance to Triticum isolates of Pyricularia oryzae in hexaploid wheat. Phytopathology 105 1568–1572. 10.1094/PHYTO-02-15-0034-R [DOI] [PubMed] [Google Scholar]
- Arruda M. P., Brown P. J., Lipka A. E., Krill A. M., Thurber C., Kolb F. L. (2015). Genomic selection for predicting Fusarium head blight resistance in a wheat breeding program. Plant Genome 8 10.3835/plantgenome2015.01.0003 [DOI] [PubMed] [Google Scholar]
- Avivi L. (1978). “High protein content in wild tetraploid Triticum dicoccoides Korn,” in Proceedings of the 5th International Wheat Genetics Symposium ed. Ramanujam S. (New Dehli: The Indian Society of Genetics and Plant Breeding; ) 372–380. [Google Scholar]
- Ayala M., Guzmán C., Alvarez J., Peña R. (2013). Characterization of genetic diversity of puroindoline genes in Mexican wheat landraces. Euphytica 190 53–63. 10.1007/s10681-012-0773-2 [DOI] [Google Scholar]
- Bajaj S., Puthigae S., Templeton K., Bryant C., Gill G., Lomba P., et al. (2008). “Towards engineering drought tolerance in perennial ryegrass using its own genome,” in Proceedings of the 6th Canadian Plant Genomics Workshop Toronto, ON: 62. [Google Scholar]
- Battenfield S. D., Guzmán C., Gaynor R. C., Singh R. P., Peña R. J., Dreisigacker S., et al. (2016). Genomic selection for processing and end-use quality traits in the CIMMYT spring bread wheat breeding program. Plant Genome 9 10.3835/plantgenome2016.01.0005 [DOI] [PubMed] [Google Scholar]
- Bernardo R. (2016). Genomewide predictions for backcrossing a quantitative trait from an exotic to an adapted line. Crop Sci. 56 1067–1075. 10.2135/cropsci2015.09.0586 [DOI] [Google Scholar]
- Bernardo R., Yu J. (2007). Prospects for genome-wide selection for quantitative traits in maize. Crop Sci. 47 1082–1090. 10.2135/cropsci2006.11.0690 [DOI] [Google Scholar]
- Bockus W., Cruz C., Kalia B., Gill B., Stack J., Pedley K., et al. (2012). Reaction of selected accessions of Aegilops tauschii to wheat blast, 2011. Plant Dis. Manage. Rep. 6:CF005. [Google Scholar]
- Börner A., Ogbonnaya F. C., Röder M. S., Rasheed A., Periyannan S., Lagudah E. S. (2015). “Aegilops tauschii introgressions in wheat,” in Alien Introgression in Wheat eds Molar-Lang M., Ceoloni C., Dolezel J. (Cham: Springer International Publishing; ), 245–271. 10.1007/978-3-319-23494-6_10 [DOI] [Google Scholar]
- Brites C. M., dos Santos C. A. L., Bagulho A. S., Beirao-da-Costa M. L. (2008). Effect of wheat puroindoline alleles on functional properties of starch. Eur. Food Res. Technol. 226 1205–1212. 10.1007/s00217-007-0711-z [DOI] [Google Scholar]
- Caballero L., Martin L. M., Alvarez J. B. (2001). Allelic variation of the HMW glutenin subunits in Spanish accessions of spelt wheat (Triticum aestivum ssp. spelta L. em. Thell.). Theor. Appl. Genet. 103 124–128. 10.1007/s001220100565 [DOI] [PubMed] [Google Scholar]
- Cai X., Chen P. D., Xu S. S., Oliver R. E., Chen X. (2005). Utilization of alien genes to enhance Fusarium head blight resistance in wheat: a review. Euphytica 142 309–318. 10.1007/s10681-005-2437-y [DOI] [Google Scholar]
- Cakmak I., Ozkan H., Braun H.-J., Welch R. M., Romheld V. (2000). Zinc and iron concentrations in seeds of wild, primitive and modern wheats. Food Nutr. Bull. 21 401–403. 10.1177/156482650002100411 [DOI] [Google Scholar]
- Cavanagh C. R., Chao S., Wang S., Huang B. E., Stephen S., Kiani S., et al. (2013). Genome wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. U.S.A. 110 8057–8062. 10.1073/pnas.1217133110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., He Z., Xia X., Lillemo M., Morris C. (2005). A new puroindoline b mutation present in Chinese winter wheat cultivar Jingdong 11. J. Cereal Sci. 42 267–269. 10.1016/j.jcs.2005.03.004 [DOI] [Google Scholar]
- Chen F., Li H., Li X., Dong Z., Zuo A., Shang X., et al. (2013). Alveograph and mixolab parameters associated with puroindoline-D1 genes in Chinese winter wheats. J. Sci. Food Agric. 93 2541–2548. 10.1002/jsfa.6073 [DOI] [PubMed] [Google Scholar]
- Chen F., Yu Y., Xia X., He Z. (2007). Prevalence of a novel puroindoline b allele in Yunnan endemic wheats (Triticum aestivum ssp. yunnanense King). Euphytica 156 39–46. 10.1007/s10681-006-9347-5 [DOI] [Google Scholar]
- Crossa J., Campos G. D. L., Pérez P., Gianola D., Burgueño J., Araus J. L., et al. (2010). Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186 713–724. 10.1534/genetics.110.118521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz C. D., Peterson G. L., Bockus W. W., Kankanala P., Dubcovsky J., Jordan K. W., et al. (2016). The 2NS translocation from Aegilops ventricosa confers resistance to the Triticum pathotype of Magnaporthe oryzae. Crop Sci. 56 990–1000. 10.2135/cropsci2015.2107.0410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz M. F. A., Prestes A. M., Maciel J. L., Scheeren P. L. (2010). Partial resistance to blast on common and synthetic wheat genotypes in seedling and in adult plant growth stages. Trop. Plant Pathol. 35 24–31. [Google Scholar]
- Cuesta S., Guzmán C., Alvarez J. B. (2013). Allelic diversity and molecular characterization of puroindoline genes in five diploid species of the Aegilops genus. J. Exp. Bot. 64 5133–5143. 10.1093/jxb/ert299 [DOI] [PubMed] [Google Scholar]
- Daetwyler H. D., Bansal U. K., Bariana H. S., Hayden M. J., Hayes B. J. (2014). Genomic prediction for rust resistance in diverse wheat landraces. Theor. Appl. Genet. 127 1795–1803. 10.1007/s00122-014-2341-8 [DOI] [PubMed] [Google Scholar]
- Dawson J. C., Endelman J. B., Heslot N., Crossa J., Poland J., Dreisigacker S., et al. (2013). The use of unbalanced historical data for genomic selection in an international wheat breeding program. Field Crops Res. 154 12–22. 10.1016/j.fcr.2013.07.020 [DOI] [Google Scholar]
- de Munter J. S. L., Hu F. B., Spiegelman D., Franz M., van Dam R. M. (2007). Whole grain, bran, and germ intake and risk of type 2 diabetes: a prospective cohort study and systematic review. PLoS Med. 4:e261 10.1371/journal.pmed.0040261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaliwal G. S., Jindal V., Dhawan A. K. (2010). Insect pest problems and crop losses: changing trends. Indian J. Ecol. 37 1–7. [Google Scholar]
- Di Silvestro R., Marotti I., Bosi S., Bregola V., Segura-Carretero A., Sedej I., et al. (2012). Health-promoting phytochemicals of Italian common wheat varieties grown under low-input agricultural management. J. Sci. Food Agric. 92 2800–2810. 10.1002/jsfa.5590 [DOI] [PubMed] [Google Scholar]
- Distelfeld A., Cakmak I., Peleg Z., Ozturk L., Yazici A. M., Budak H. (2007). Multiple QTL-effects of wheat Gpc-B1 locus on grain protein and micronutrient concentrations. Physiol. Plant. 129 635–643. 10.1111/j.1399-3054.2006.00841.x [DOI] [Google Scholar]
- Dotlacil L., Hermuth J., Stehno Z., Dvoracek V., Bradova J., Leisova L. (2010). How can wheat landraces contribute to present breeding? Czech J. Genet. Plant Breed. 46 70–74. [Google Scholar]
- Dreisigacker S., Zhang P., Warburton M. L., Skovmand B., Hoisington D., Melchinger A. W. (2005). Genetic diversity among and within CIMMYT wheat landrace accessions investigated with SSRs and implications for plant genetic resources management. Crop Sci. 45 653–661. 10.2135/cropsci2005.0653 [DOI] [Google Scholar]
- Ehdaie B., Whitkus R. W., Waines J. G. (2003). Root biomass, water-use efficiency, and performance of wheat-rye translocations of chromosomes 1 and 2 in spring bread wheat “Pavon”. Crop Sci. 43 710–717. 10.2135/cropsci2003.0710 [DOI] [Google Scholar]
- El Bouhssini M., Street K., Joubi A., Ibrahim Z., Rihawi F. (2009). Sources of wheat resistance to Sunn pest, Eurygaster integriceps Puton, in Syria. Genet. Resour. Crop Evol. 56 1065–1069. 10.1007/s10722-009-9427-1 [DOI] [Google Scholar]
- Ellis J. G., Lagudah E. S., Spielmeyer W., Dodds P. N. (2014). The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 5:641 10.3389/fpls.2014.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT (2014). Food and Agriculture Organization of the United Nations Statistics Division. Rome: FAO. [Google Scholar]
- Feuillet C., Langridge P., Waugh R. (2008). Cereal breeding takes a walk on the wild side. Trends Genet. 24 24–32. 10.1016/j.tig.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Friebe B., Jiang J., Raupp W. J., McIntosh R. A., Gill B. S. (1996). Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91 59–87. 10.1007/BF00035277 [DOI] [Google Scholar]
- Gadaleta A., Giancaspro A., Blechl A. E., Blanco A. (2008). A transgenic durum wheat line that is free of marker genes and expresses 1DY10. J. Cereal Sci. 48 439–445. 10.1016/j.jcs.2007.11.005 [DOI] [Google Scholar]
- Gebruers K., Dornez E., Boros D., Fras A., Dynkowska W., Bedo Z., et al. (2008). Variation in the content of dietary fiber and components thereof in wheats in the HEALTHGRAIN diversity screen. J. Agric. Food. Chem. 56 9740–9749. 10.1021/jf800975w [DOI] [PubMed] [Google Scholar]
- Giambanelli E., Ferioli F., Koçaoglu B., Jorjadze M., Alexieva I., Darbinyan N., et al. (2013). A comparative study of bioactive compounds in primitive wheat populations from Italy, Turkey, Georgia, Bulgaria and Armenia. J. Sci. Food Agric. 93 3490–3501. 10.1002/jsfa.6326 [DOI] [PubMed] [Google Scholar]
- Gianibelli M. C., Gupta R. B., Lafiandra D., Margiotta B., MacRitchie F. (2001). Polymorphism of high Mrglutenin subunits in Triticum tauschii: characterization by chromatography and electrophoretic methods. J. Cereal Sci. 33 39–52. 10.1006/jcrs.2000.0328 [DOI] [Google Scholar]
- Giroux M., Morris C. (1997). A glycine to serine change in puroindoline b is associated with wheat grain hardness and low levels of starch-surface friabilin. Theor. Appl. Genet. 95 857–864. 10.1007/s001220050636 [DOI] [Google Scholar]
- Giroux M., Morris C. (1998). Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Proc. Nat. Acad. Sci. U.S.A. 95 6262–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybosch R. A., Peterson C. J., Hansen L. E., Worrall D., Shelton D. R., Lukaszewski A. (1993). Comparative flour quality and protein characteristics of 1BL/1RS and 1AL/1RS wheat-rye translocation lines. J. Cereal Sci. 17 95–106. 10.1006/jcrs.1993.1010 [DOI] [Google Scholar]
- Guzmán C., Caballero L., Martín L. M., Alvarez J. B. (2012). Waxy genes from spelt wheat: new alleles for modern wheat breeding and new phylogenetic inferences about the origin of this species. Ann. Bot. 110 1161–1171. 10.1093/aob/mcs201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Velu G., Peña R. J., Singh S., Singh R. P. (2014). Genetic loci associated with high grain zinc concentration and pleiotropic effect on kernel weight in wheat (Triticum aestivum L.). Mol. Breed. 34 1893–1902. 10.1007/s11032-014-0147-7 [DOI] [Google Scholar]
- Harlan J. R., de Wet J. M. J. (1971). Toward a rational classification of cultivated plants. Taxon 20 509–517. 10.2307/1218252 [DOI] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. C., Verbyla K., Goddard M. E. (2009). Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet. Sel. Evol. 41 51 10.1186/1297-9686-41-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes H. K., Parker J. H., Kurtzweil C. (1920). Genetics of rust resistance in crosses of varieties of Triticum vulgare with varieties of T. durum and T. dicoccum. J. Agric. Res. 19 523–542. [Google Scholar]
- Hede A. R., Skovmand B., Reynolds M. P., Crossa J., Vilhelmsen A. L., Stølen O. (1999). Evaluating genetic diversity for heat tolerance traits in Mexican wheat landraces. Genet. Resour. Crop Evol. 46 37–45. 10.1023/A:1008684615643 [DOI] [Google Scholar]
- Heffner E. L., Jannink J.-L., Iwata H., Souza E., Sorrells M. E. (2011a). Genomic selection accuracy for grain quality traits in biparental wheat populations. Crop Sci. 51 2597–2606. 10.2135/cropsci2011.05.0253 [DOI] [Google Scholar]
- Heffner E. L., Jannink J.-L., Sorrells M. E. (2011b). Genomic selection accuracy using multifamily prediction models in a wheat breeding program. Plant Genome 4 1–11. 10.3835/plantgenome.2010.12.0029 [DOI] [Google Scholar]
- Hillel J., Schaap T., Haberfeld A., Jeffreys A. J., Plotzky Y., Cahaner A., et al. (1990). DNA fingerprints applied to gene introgression in breeding programs. Genetics 124 783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman G. C. (1978). On the origins of domestic rye: secale cereale: the finds from aceramic can Hasan III in Turkey. Anatol. Stud. 28 157–174. 10.2307/3642748 [DOI] [Google Scholar]
- Hoffmann B. (2008). Alteration of drought tolerance of winter wheat caused by translocation of rye chromosome segment 1RS. Cereal Res. Commun. 36 269–278. 10.1556/crc.36.2008.2.7 [DOI] [Google Scholar]
- Holme I. B., Wendt T., Holm P. B. (2013). Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biot. J. 11 395–407. 10.1111/pbi.12055 [DOI] [PubMed] [Google Scholar]
- Honsdorf N., March J. T., Berger B., Tester M., Pillen K. (2014). High-throughput phenotyping to detect drought tolerance QTL in wild barley introgression lines. PLoS ONE 9:e97047 10.1371/journal.pone.0097047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F., Chevalet C., Mulsant P. (1992). Using markers in gene introgression breeding programs. Genetics 132 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosptial F. F., Charcosset A. (1997). Marker-assisted introgression of quantitative trait loci. Genetics 147 1469–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovmøller M. S., Walter S., Bayles R. A., Hubbard A., Flath K., Sommerfeldt N., et al. (2015). Replacement of the European wheat yellow rust population by new races from the centre of diversity in the near-Himalayan region. Plant Pathol. 65 402–411. 10.1111/ppa.12433 [DOI] [Google Scholar]
- Hullé M., Coeur d’Acier A., Bankhead-Dronnet S., Harrington R. (2010). Aphids in the face of global changes. C. R. Biol. 333 497–503. 10.1016/j.crvi.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Hurni S., Brunner S., Stirnweis D., Herren G., Peditto D., McIntosh R. A., et al. (2014). The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J. 79 904–913. 10.1111/tpj.12593 [DOI] [PubMed] [Google Scholar]
- Ikeda T. M., Cong H., Suzuki T., Takata K. (2010). Identification of new Pina null mutations among Asian common wheat cultivars. J. Cereal Sci. 51 235–237. 10.1016/j.jcs.2010.01.012 [DOI] [Google Scholar]
- International Grain Council [IGC] (2014). Five-Year Global Supply and Demand Projections. London: International Grains Council. [Google Scholar]
- Jacobs D. R., Jr., Meyer K. A., Kushi L. H., Folsom A. R. (1999). Is whole grain intake associated with reduced total and cause specific death rates in older women? The Iowa women’s health study. Am. J. Public Health 89 322–329. 10.2105/AJPH.89.3.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen E., Schouten H. J. (2007). Cisgenesis strongly improves introgression breeding and induced translocation breeding of plants. Trends Biotechnol. 25 219–223. 10.1016/j.tibtech.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Joppa L. R., Cantrell R. G. (1990). Chromosomal location of genes for grain protein content of wild tetraploid wheat. Crop Sci. 30 1059–1064. 10.2135/cropsci1990.0011183X003000050021x [DOI] [Google Scholar]
- Joppa L. R., Du C., Hart G. E., Hareland G. A. (1997). Mapping gene(s) for grain protein in tetraploid wheat (Triticum turgidum L.) using a population of recombinant inbred chromosome lines. Crop Sci. 37 1586–1589. 10.2135/cropsci1997.0011183X003700050030x [DOI] [Google Scholar]
- Kariluto S., Edelmann M., Piironen V. (2010). Effects of environment and genotype on folate contents in wheat in the HEALTHGRAIN Diversity Screen. J. Agric. Food Chem. 58 9324–9331. 10.1021/jf100251j [DOI] [PubMed] [Google Scholar]
- Karki D., Wyant W., III, Berzonsky W. A., Glover K. D. (2014). Investigating physiological and morphological mechanisms of drought tolerance in wheat (Triticum aestivum L.) lines with 1RS translocation. Am. J. Plant Sci. 05 1936–1944. 10.4236/ajps.2014.513207 [DOI] [Google Scholar]
- Kato K., Yokoyama H. (1991). Geographical variation in heading characters among wheat landraces, Triticum aestivum L., and its implication for their adaptability. Theor. Appl. Genet. 84 259–265. 10.1007/BF00229480 [DOI] [PubMed] [Google Scholar]
- Kerr B. J., Weber T. E., Miller P. S., Southern L. L. (2010). Effect of phytase on apparent total tract digestibility of phosphorus in corn-soybean meal diets fed to finishing pigs. J. Anim. Sci. 88 238–247. 10.2527/jas.2009-2146 [DOI] [PubMed] [Google Scholar]
- Kertho A., Mamidi S., Bonman J. M., McClean P. E., Acevedo M. (2015). Genome-wide association mapping for resistance to leaf and stripe rust in winter-habit hexaploid wheat landraces. PLoS ONE 10:e0129580 10.1371/journal.pone.0129580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Johnson J. W., Baenziger P. S., Lukaszewski A. J., Gaines C. S. (2004). Agronomic effect of wheat-rye translocation carrying rye chromatin (1R) from different sources. Crop Sci. 44 1254–1258. 10.2135/cropsci2004.1254 [DOI] [Google Scholar]
- Kim W., Johnson J. W., Baenziger P. S., Lukaszewski A. J., Gaines C. S. (2005). Quality effect of wheat-rye (1R) translocation in “Pavon 76”. Plant Breed. 124 334–337. 10.1111/j.1439 [DOI] [Google Scholar]
- Laddomada B., Durante M., Mangini G., D´Amico L., Lenucci M. S., Simeone R. (2016). Genetic variation for phenolic acids concentration and composition in a tetraploid wheat (Triticum turgidum L.) collection. Genet. Resour. Crop Evol. 10.1007/s10722-016-0386-z [Epub ahead of print]. [DOI] [Google Scholar]
- Lillemo M., Morris F. (2000). A leucine to proline mutation in puroindoline b is frequently present in hard wheats from Northern Europe. Theor. Appl. Genet. 100 1100–1107. 10.1007/s001220051392 [DOI] [Google Scholar]
- Liu S., Stampfer M. J., Hu F. B., Giovannucci E., Rimm E. B., Manson J. E., et al. (1999). Whole grain consumption and risk of coronary heart disease: results from the nurse’s health study. Am. J. Clin. Nutr. 3 412–419. [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang L., Mao S., Liu K., Lu Y., Wang J., et al. (2015). Genome-wide association study of 29 morphological traits in Aegilops tauschii. Nat. Sci. Rep. 5:15562 10.1038/srep15562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes S. M., El-Basyoni I., Baenziger P. S., Singh S., Royo C., Ozbek K., et al. (2015). Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 66 3625–3638. 10.1093/jxb/erv122 [DOI] [PubMed] [Google Scholar]
- Lorenz A. J., Chao S., Asoro F. G., Heffner E. L., Hayashi T., Iwata H., et al. (2011). Genomic selection in plant breeding: knowledge and prospects. Adv. Agron. 110 77–123. 10.1016/B978-0-12-385531-2.00002-5 [DOI] [Google Scholar]
- Ma G., Ma C. S. (2012). Climate warming may increase aphids’ dropping probabilities in response to high temperatures. J. Insect Physiol. 58 1456–1462. 10.1016/j.jinsphys.2012.08.012 [DOI] [PubMed] [Google Scholar]
- Macfadyen S., Kriticos D. J. (2012). Modelling the geographical range of a species with variable life-history. PLoS ONE 7:e40313 10.1371/journal.pone.0040313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciel J. L. N., Ceresini P. C., Castroagudin V. L., Zala M., Kema G. H. J., McDonald B. A. (2014). Population structure and pathotype diversity of the wheat blast pathogen Magnaporthe oryzae 25 years after its emergence in Brazil. Phytopathology 104 95–107. 10.1094/PHYTO-11-12-0294-R [DOI] [PubMed] [Google Scholar]
- Maes B., Trethowan R. M., Reynolds M. P., van Ginkel M., Skovmand B. (2001). The influence of glume pubescence on spikelet temperature of wheat under freezing conditions. J. Plant Physiol. 28 141–148. [Google Scholar]
- Mago R., Bariana H. S., Dundas I. S., Spielmeyer W., Lawrence G. J., Pryor A. J., et al. (2005). Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor. Appl. Genet. 111 496–504. 10.1007/s00122-005-2039-z [DOI] [PubMed] [Google Scholar]
- Massa A. N., Morris C. F., Gill B. S. (2004). Sequence diversity of puroindoline-a, puroindoline-b and the grain softness protein genes in Aegilops tauschii Coss. Crop Sci. 44 1808–1816. 10.2135/cropsci2004.1808 [DOI] [Google Scholar]
- McFadden E. S. (1930). A successful transfer of emmer characters to vulgare wheat. J. Am. Soc. Agron. 22 1020–1034. 10.2134/agronj1930.00021962002200120005x [DOI] [Google Scholar]
- McIntosh R. A., Gyarfas J. (1971). Triticum timopheevii as a source of resistance to wheat stem rust. Z. Pflanzenzüchtung 66 240–248. [Google Scholar]
- McIntosh R. A., Wellings C. R., Park R. F. (1995). Wheat Rusts: An Atlas of Resistance Genes. Clayton, VIC: CSIRO Publications. [Google Scholar]
- McIntosh R. A., Yamazaki Y., Dubcovsky J., Rogers W. J., Morris C., Appels R., et al. (2013). Catalogue of Gene Symbols for Wheat. Available at: http://www.shigen.nig.ac.jp/wheat/komugi/genes/macgene/2013/GeneSymbol.pdf (accessed June 1 2016). [Google Scholar]
- Migui S. M., Lamb R. J. (2003). Patterns of resistance to three cereal aphids among wheats in the genus Triticum (Poaceae). Bull. Entomol. Res. 93 323–333. 10.1079/ber2003246 [DOI] [PubMed] [Google Scholar]
- Miralles D. J., Resnicoff E., Carretero R. (2007). “Yield improvement associated with Lr19 translocation in wheat: which plant attributes are modified?,” in Scale and Complexity in Plant Systems Research: Gene-Plant-Crop Relations eds Spiertz J. H. J., Struik P. C., van Laar H. H. (Dordrecht: Springer; ) 169–176. [Google Scholar]
- Mirdita V., He S., Zhao Y., Korzun V., Bothe R., Ebmeyer E., et al. (2015). Potential and limits of whole genome prediction of resistance to Fusarium head blight and Septoria tritici blotch in a vast Central European elite winter wheat population. Theor. Appl. Genet. 128 2471–2481. 10.1007/s00122-015-2602-1 [DOI] [PubMed] [Google Scholar]
- Monasterio I., Graham R. D. (2000). Breeding for trace minerals in wheat. Food Nutr. Bull. 21 393–396. 10.1177/156482650002100409 [DOI] [Google Scholar]
- Monneveux P., Reynolds P. M., Zaharieva M., Mujeeb-Kazi A. (2003). Effect of T1BL.1RS chromosome translocation on bread wheat grain yield and physiological related traits in a warm environment. Cereal Res. Commun. 31 3–4. [Google Scholar]
- Morgounov A., Gómez-Becerra H. F., Abugalieva A., Dzhunusova M., Yessimbekova M., Muminjanov H., et al. (2007). Iron and zinc grain density in common wheat grown in Central Asia. Euphytica 155 193–203. 10.1007/s10681-006-9321-2 [DOI] [Google Scholar]
- Morris C. F. (2002). Puroindolines: the molecular genetic basis of wheat grain hardness. Plant Mol. Biol. 48 633–647. 10.1023/A:1014837431178 [DOI] [PubMed] [Google Scholar]
- Moseman J. G., Nevo E., Morshidy M. A. E., Zohary D. (1984). Resistance of Triticum dicoccoides to infection with Erysiphe graminis tritici. Euphytica 33 41–47. 10.1007/BF00022748 [DOI] [Google Scholar]
- Mujeeb-Kazi A., Wang R. R. C. (1995). “Perennial and annual wheat relatives in the triticeae,” in Utilizing Wild Grass Biodiversity in Wheat Improvement: 15Years of Wide Cross Research at CIMMYT eds Mujeeb-Kazi A., Hettel G. P. (Mexico city: CIMMYT; ), 5–14. [Google Scholar]
- Ornella L., Singh S., Perez P., Burgueño J., Singh R., Tapia E., et al. (2012). Genomic prediction of genetic values for resistance to wheat rusts. Plant Genome 5 136–148. 10.3835/plantgenome2012.07.0017 [DOI] [Google Scholar]
- Ortiz-Monasterio I., Palacios-Rojas N., Meng E., Pixley K., Trethowan R., Pena R. J. (2007). Enhancing the mineral and vitamin content of wheat and maize through plant breeding. J. Cereal Sci. 46 293–307. 10.1016/j.jcs.2007.06.005 [DOI] [Google Scholar]
- Peleg Z., Cakmak I., Ozturk L., Yazici A., Jun Y., Budak H., et al. (2009). Quantitative trait loci conferring grain mineral nutrient concentrations in durum wheat × wild emmer wheat RIL population. Theor. Appl. Genet. 119 353–369. 10.1007/s00122-009-1044-z [DOI] [PubMed] [Google Scholar]
- Poland J., Endelman J., Dawson J., Rutkosk J., Wu S., Manes Y., et al. (2012). Genomic selection in wheat breeding using Genotyping-by-Sequencing. Plant Genome 5 103–113. 10.3835/plantgenome2012.05.0005 [DOI] [Google Scholar]
- Pszczola M., Strabel T., Mulder H. A., Calus M. P. L. (2012). Reliability of direct genomic values for animals with different relationships within and to the reference population. J. Dairy Sci. 95 389–400. 10.3168/jds.2011-4338 [DOI] [PubMed] [Google Scholar]
- Rabinovich S. V. (1998). Importance of wheat-rye translocations for breeding modern cultivars of Triticum aestivum L. (Reprinted from Wheat: prospects for global improvement, 1998). Euphytica 100 323–340. 10.1023/A:1018361819215 [DOI] [Google Scholar]
- Rahaman M. M., Chen D., Gillani Z., Klukas C., Chen M. (2015). Advanced phenotyping and phenotype data analysis for the study of plant growth and development. Front. Plant Sci. 6:619 10.3389/fpls.2015.00619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed A., Xia X., Yan Y., Appels R., Mahmood T. (2014). Wheat seed storage proteins: advances in molecular genetics, diversity and breeding applications. J. Cereal Sci. 60 11–24. 10.1016/j.jcs.2014.01.020 [DOI] [Google Scholar]
- Reitz L. P., Salmon S. C. (1968). Origin, history, and use of Norin 10 Wheat1. Crop Sci. 8 686–689. 10.2135/cropsci1968.0011183X000800060014x [DOI] [Google Scholar]
- Reynolds M., Balota M., Delgado M., Fischer R. A. (1994). Physiological and morphological traits associated with spring wheat yield under hot, irrigated conditions. Aust. J. Plant Physiol. 21 717–730. 10.1071/PP9940717 [DOI] [Google Scholar]
- Reynolds M., Dreccer F., Trethowan R. (2007a). Drought adaptive traits derived from wheat wild relatives and landraces. J. Exp. Bot. 58 177–186. 10.1093/jxb/erl250 [DOI] [PubMed] [Google Scholar]
- Reynolds M., Saint Pierre C., Abu S. I. S., Condon A. G. (2007b). Evaluating potential genetic gains in wheat associated with stress-adaptive trait expression in diverse germplasm under drought and heat stress. Crop Sci. 47 172–189. 10.2135/cropsci2007.10.0022IPBS [DOI] [Google Scholar]
- Reynolds M., Tattaris M., Cossani M. C., Ellis M., Yamaguchi-Shinozaki K., Saint Pierre C. (2015). “Exploring genetic resources to increase adaptation of wheat to climate change,” in Advances in Wheat Genetics: From Genome to Field eds Ogihara Y., Takumi S., Handa H. (Tokyo: Springer; ), 335–368. [Google Scholar]
- Röder M. S., Huang X., Börner A. (2008). Fine mapping of the region on wheat chromosome 7D controlling grain weight. Funct. Integr. Genomics 1 79–86. 10.1007/s10142-007-0053-8 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Quijano K., Vazquez J. F., Garillo J. M. (1994). Variation of high molecular weight glutenin subunits in Spanish landraces of Triticum aestivum ssp. vulgare and ssp. spelta. J. Genet. Breed. 44 121–126. [Google Scholar]
- Royo C., Briceño-Félix G. A. (2011). “Spanish wheat pool,” in The World Wheat Book. A History of Wheat Breeding eds Bojean A. P., Angus W. J., van Ginkel M. (Paris: Lavoisier Publishing; ), 121–154. [Google Scholar]
- Rutkoski J., Benson J., Jia Y., Brown-Guedira G., Jannink J.-L., Sorrells M. (2012). Evaluation of genomic prediction methods for Fusarium head blight resistance in wheat. Plant Genome 5 51–61. 10.3835/plantgenome2012.02.0001 [DOI] [Google Scholar]
- Rutkoski J., Singh R. P., Huerta-Espino J., Bhavani S., Poland J., Jannink J. L., et al. (2015). Genetic gain from phenotypic and genomic selection for quantitative resistance to stem rust of wheat. Plant Genome 8 1–10. 10.3835/plantgenome2014.10.0074 [DOI] [PubMed] [Google Scholar]
- Salamini F., Özkan H., Brandolini A., Schäfer-Pregl R., Martin W. (2002). Genetics and geography of wild cereal domestication in the Near East. Nat. Rev. Genet. 3 429–441. [DOI] [PubMed] [Google Scholar]
- Schouten H. J., Krens F. A., Jacobsen E. (2006). Cisgenic plants are similar to traditionally bred plants. EMBO Rep. 7 750–753. 10.1038/sj.embor.7400769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears E. R. (1956). The transfer of leaf-rust resistance from Aegilops umbellulata to wheat. Brookhaven Symp. Biol. 9 1–22. [Google Scholar]
- Servin B., Martin O. C., Mézard M., Hospital F. (2004). Toward a theory of marker-assisted gene pyramiding. Genetics 168 513–523. 10.1534/genetics.103.023358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry P. R., Piironen V., Lampi A., Eldelmann M., Kariluoto S., Nurmi T., et al. (2010). The HEALTHGRAIN wheat diversity screen: effects of genotype and environment on Phytochemicals and dietary fiber component. J. Agric. Food Chem. 58 9291–9298. 10.1021/jf100039b [DOI] [PubMed] [Google Scholar]
- Singh R. P., Hodson D. P., Huerta-Espino J., Jin Y., Bhavani S., Njau P., et al. (2011). The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 49 465–481. 10.1146/annurev-phyto-072910-095423 [DOI] [PubMed] [Google Scholar]
- Singh R. P., Hodson D. P., Huerta-Espino J., Jin Y., Njau P., Wanyera R., et al. (2008). Will stem rust destroy the world’s wheat crop? Adv. Agron. 98 271–309. 10.1016/S0065-2113(08)00205-8 [DOI] [Google Scholar]
- Singh R. P., Hodson D. P., Jin Y., Huerta-Espino J., Kinyua M., Wanyera R., et al. (2006). Current status, likely migration and strategies to mitigate the threat to wheat production from race Ug99 (TTKS) of stem rust pathogen. CAB Rev. 1 1–13. 10.1079/PAVSNNR20061054 [DOI] [Google Scholar]
- Singh R. P., Hodson D. P., Jin Y., Lagudah E. S., Ayliffe M. A., Bhavani S., et al. (2015). Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology 105 872–884. 10.1094/PHYTO-01-15-0030-FI [DOI] [PubMed] [Google Scholar]
- Singh R. P., Huerta-Espino J., Rajaram S., Crossa J. (1998). Agronomic effects from chromosome translocations 7DL.7AG and 1BL.1RS in spring wheat. Crop Sci. 38 27–33. 10.2135/cropsci1998.0011183X003800010005x [DOI] [Google Scholar]
- Smale M., McBride T. (1996). “Understanding global trends in the use of international flows of wheat genetic resources,” in CIMMYT 1995/6 Trends: Understanding Global Trends in the Use of Wheat Diversity and Wheat Genetic Resources (Mexico City: Cimmyt; ). [Google Scholar]
- Smith C. M., Havlickova H., Starkey S., Gill B. S., Holubec V. (2004). Identification of Aegilops germplasm with multiple aphid resistance. Euphytica 135 265–273. 10.1023/B:EUPH.0000013306.15133.33 [DOI] [Google Scholar]
- Srinivasa J., Arun B., Mishra V. K., Singh G. P., Velu G., Babu R., et al. (2014). Zinc and Iron concentration QTL mapped in a Triticum spelta x T. aestivum cross. Theor. Appl. Genet. 127 1643–1651. 10.1007/s00122-014-2327-6 [DOI] [PubMed] [Google Scholar]
- Sukumaran S., Dreisigacker S., Lopes M., Chavez P., Reynolds M. P. (2015). Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor. Appl. Genet. 128 353–363. 10.1007/s00122-014-2435-3 [DOI] [PubMed] [Google Scholar]
- Tagle A. G., Chuma I., Tosa Y. (2015). Rmg7, a new gene for resistance to Triticum isolates of Pyricularia oryzae identified in tetraploid wheat. Phytopathology 105 495–499. 10.1094/PHYTO-06-14-0182-R [DOI] [PubMed] [Google Scholar]
- Tanaka H., Morris C. F., Haruna M., Tsujimoto H. (2008). Prevalence of puroindoline alleles in wheat varieties from eastern Asia including the discovery of a new SNP in puroindoline b. Plant Genet. Resour. 6 142–152. [Google Scholar]
- Tanksley S. D., Nelson J. C. (1996). Advanced backcross QTL analysis: a method for the 12 simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into13 elite breeding lines. Theor. Appl. Genet. 92 191–203. 10.1007/BF00223376 [DOI] [PubMed] [Google Scholar]
- Tattaris M., Reynolds M., Pietragalla J., et al. (2013). “Airborne remote sensing for high throughput phenotyping of wheat,” in Proceedings of the workshop UAV-based remote sensing methods for monitoring vegetation Cologne. [Google Scholar]
- Tiwari V. K., Rawat N., Chhuneja P., Neelam K., Aggarwal R., Randhawa G. S., et al. (2009). Mapping of quantitative trait loci for grain iron and zinc concentration in diploid A genome wheat. J. Heredity 100 771–776. 10.1093/jhered/esp030 [DOI] [PubMed] [Google Scholar]
- Tiwari V. K., Wang S., Sehgal S., Vrána J., Friebe B., Kubaláková M., et al. (2014). SNP discovery for mapping alien introgressions in wheat. BMC Genomics 15:273 10.1186/1471-2164-15-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomerlin J., El-Morshidy M., Moseman J., Baenziger P., Kimber G. (1984). Resistance to Erysiphe graminis f. sp. tritici, Puccinia recondita f. sp. tritici, and Septoria nodorum in wild Triticum species. Plant Dis. 68 10–13. 10.1094/PD-68-10 [DOI] [Google Scholar]
- Uauy C., Distelfeld A., Fahima T., Blechl A., Dubcovsky J. (2006). A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314 1298–1301. 10.1126/science.1133649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA (2016). World Agricultural Production. USDA Foreign Agricultural Service, Washington DC 20250. Available at: http://apps.fas.usda.gov/psdonline/circulars/grain-wheat.pdf (accessed April 1 2016) [Google Scholar]
- Vallega V., Waines J. G. (1987). High molecular weight glutenin subunit variation in Triticum turgidum var. dicoccum. Theor. Appl. Genet. 74 706–710. 10.1007/BF00247545 [DOI] [PubMed] [Google Scholar]
- Velu G., Crossa J., Singh R. P., Hao Y., Rodriguez-Perez P., Joshi A. K., et al. (2016). Genomic prediction for grain zinc and iron in wheat. Theor. Appl. Genet. 10.1007/s00122-016-2726-y [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Velu G., Ortiz-Monasterio I., Cakmak I., Hao Y., Singh R. P. (2014). Biofortification strategies to increase grain zinc and iron concentrations in wheat. J. Cereal Sci. 59 365–372. 10.1016/j.jcs.2013.09.001 [DOI] [Google Scholar]
- Velu G., Singh R., Arun B., Mishra V. K., Tiwari C., Joshi A., et al. (2015). Reaching out to farmers with high Zinc wheat varieties through public-private partnerships – An Experience from Eastern-Gangetic Plains of India. Adv. Food Technol. Nutr. Sci. 1 73–75. 10.17140/AFTNSOJ-1-112 [DOI] [Google Scholar]
- Vikram P., Franco J., Burgueno-Ferreira J., Li H., Sehgal D., Saint Pierre C., et al. (2016). Unlocking the genetic diversity of creole wheats. Nat. Sci. Rep. 6 23092 10.1038/srep23092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal R., Bañuelos O., Mujeeb-Kazi A., Rajaram S. (1998). Agronomic performance of chromosomes 1B and T1BL.1RS near-isolines in the spring bread wheat Seri M82. Euphytica 103 195–202. 10.1023/a:1018392002909 [DOI] [Google Scholar]
- Villareal R. L., Sayre K., Bañuelos O., Mujeeb-Kazi A. (2001). Registration of four synthetic hexaploid wheat (Triticum turgidum/Aegilops tauschii) germplasm lines tolerant to waterlogging. Crop Sci. 41 274 10.2135/cropsci2001.411274x [DOI] [Google Scholar]
- Visscher P. M., Haley C. S., Thompson R. (1996). Marker-assisted introgression in backcross breeding programs. Genetics 144 1923–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y. F., Yen C., Yang J. L. (1997). Sources of resistance to head scab in Triticum. Euphytica 94 31–36. 10.1023/A:1002982005541 [DOI] [Google Scholar]
- Wang J., Liu W., Wang H., Li L., Wu J., Yang X., et al. (2011). QTL mapping of yield-related traits in the wheat germplasm 3228. Euphytica 177 277–292. 10.1007/s10681-010-0267-z [DOI] [Google Scholar]
- Wang S., Yu Z., Cao M., Shen X., Li N., Li X., et al. (2013). Molecular mechanisms of HMW glutenin subunit from 1S genome of Aegilops longissima positively affecting wheat breadmaking quality. PLoS ONE 8:58947 10.1371/journal.pone.0058947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. L., Poutanen K., Gebruers K., Piironen V., Lampi A. M., Nystrom L., et al. (2008). The HEALTHGRAIN cereal diversity screen: concept, results, and prospects. J. Agric. Food Chem. 56 9699–9709. 10.1021/jf8009574 [DOI] [PubMed] [Google Scholar]
- Wessells K. R., Brown K. H. (2012). Estimating the global prevalence of zinc deficiency: results based on zinc availability in national food supplies and the prevalence of stunting. PLoS ONE 7:e50568 10.1371/journal.pone.0050568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S. (1873). Wheat and rye hybrids. Trans. Bot. Soc. Edinb. 12 286–288. 10.1080/03746607309469536 [DOI] [Google Scholar]
- Wu J., Yang X., Wang H., Li H., Li L., Li X., et al. (2006). The introgression of chromosome 6P specifying for increased numbers of florets and kernels from Agropyron cristatum into wheat. Theor. Appl. Genet. 114 13–20. 10.1007/s00122-006-0405-0 [DOI] [PubMed] [Google Scholar]
- Wulff B. B. H., Moscou M. J. (2014). Strategies for transferring resistance to wheat: from wide crosses to GM cassettes. Front. Plant Sci. 5:692 10.3389/fpls.2014.00692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. S., Khan K., Klindworth D. L., Faris J. D., Nygard G. (2004). Chromosome location of genes from novel glutenin subunits and gliadins in wild emmer (Triticum turgidum var. dicoccoides). Theor. Appl. Genet. 108 1221–1228. 10.1007/s00122-003-1555-y [DOI] [PubMed] [Google Scholar]
- Xu S. S., Khan K., Klindworth D. L., Nygard G. (2010). Evaluation and characterization of high-molecular weight 1D glutenin subunits from Aegilops tauschii in synthetic hexaploid wheats. J. Cereal Sci. 52 333–336. 10.1016/j.jcs.2010.05.004 [DOI] [Google Scholar]
- Xu Y. F., An D. G., Liu D. C., Zhang A. M., Xu H. X., Li B. (2012). Molecular mapping of QTLs for grain zinc, iron and protein concentration of wheat across two environments. Field Crops Res. 138 57–62. 10.1016/j.fcr.2012.09.017 [DOI] [Google Scholar]
- Yang J., Sears R. G., Gill B. S., Paulsen G. M. (2002). Growth and senescence characteristics associated with tolerance of wheat-alien amphiploids to high temperature under controlled conditions. Euphytica 126 185–193. 10.1023/A:1016350509689 [DOI] [Google Scholar]
- Young N. D., Tanksley S. D. (1989). RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theor. Appl. Genet. 77 353–359. 10.1007/BF00305828 [DOI] [PubMed] [Google Scholar]
- Zhou Y., He Z. H., Sui X. X., Xia X. C., Zhang X. K., Zhang G. S. (2007). Genetic improvement of grain yield and associated traits in the northern China winter wheat region from 1960 to 2000. Crop Sci. 47 245–253. 10.2135/cropsci2006.03.0175 [DOI] [Google Scholar]
- Ziska L. H., Bunce J. A., Shimono H., Gealy D. R., Baker J. T., Newton P. C. D., et al. (2012). Food security and climate change: on the potential to adapt global crop production by active selection to rising atmospheric carbon dioxide. Proc. Biol. Sci. 279 4097–4105. 10.1098/rspb.2012.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]