Abstract
Aim:
MicroRNAs play pivotal roles in regulation of both innate and adaptive immune responses. In the present study, we investigated the effects of microRNA-124 (miR-124) on production of the pro-inflammatory cytokine TNF-α in lipopolysaccharide (LPS)-treated mouse macrophages.
Methods:
Mouse macrophage cell line RAW264.7 was stimulated with LPS (100 ng/mL). The levels of miR-124 and TNF-α mRNA were evaluated using q-PCR. ELISA and Western blotting were used to detect TNF-α protein level in cell supernatants and cells, respectively. 3′-UTR luciferase reporter assays were used to analyze the targets of miR-124. For in vivo experiments, mice were injected with LPS (30 mg/kg, ip).
Results:
LPS stimulation significantly increased the mRNA level of miR-124 in RAW264.7 macrophages in vitro and mice in vivo. In RAW264.7 macrophages, knockdown of miR-124 with miR-124 inhibitor dose-dependently increased LPS-stimulated production of TNF-α protein and prolonged the half-life of TNF-α protein, but did not change TNF-α mRNA levels, whereas overexpression of miR-124 with miR-124 mimic produced the opposite effects. Furthermore, miR-124 was found to directly target two components of deubiquitinating enzymes: ubiquitin-specific proteases (USP) 2 and 14. Knockdown of USP2 or USP14 accelerated protein degradation of TNF-α, and abolished the effect of miR-124 on TNF-α protein stability.
Conclusion:
miR-124, targeting USP2 and USP14, negatively regulates LPS-induced TNF-α production in mouse macrophages, suggesting miR-124 as a new therapeutic target in inflammation-related diseases.
Keywords: microRNA-124, macrophages, LPS, TLR4, TNF-α, USP2, USP14
Introduction
Optimal toll-like receptor (TLR)-mediated production of pro-inflammatory cytokines is necessary for eliminating invading pathogens. However, uncontrolled TLR activation might result in immunological and inflammatory diseases, such as lipopolysaccharide (LPS)/TLR4-induced endotoxin shock. Therefore, TLR signaling must be tightly regulated1,2. Some of the negative regulators of LPS/TLR signaling include heterodimer partner (SHP), mixed-lineage kinase (MLK), disintegrin and metalloproteinase (ADAM) 15, and Dok33,4,5,6.
MicroRNAs (miRNAs) are noncoding transcripts of 18–25 nucleotides (nts) derived from initially long primary transcripts, which are processed in the nucleus to ∼70 nt precursor miRNAs by the RNase III Drosha. These precursors have a hairpin structure that undergoes cleavage by the enzyme Dicer to release the mature miRNA. Mature miRNAs specifically bind to 3′-untranslated regions (3′-UTRs) of target mRNAs, leading to either mRNA degradation or inhibition of translation7. miRNAs have been reported to regulate potentially every aspect of cellular activity, including differentiation and development, metabolism, proliferation, apoptotic cell death, viral infection, and tumorigenesis8,9.
Recently, miRNAs have emerged as key regulators of the TLR pathway, and it is highly likely that they fine-tune signaling during the inflammatory process10,11. miR-146a is rapidly upregulated by LPS in human monocytic cells and regulates TLR4 signaling by targeting tumor necrosis factor (TNF) receptor–associated factor 6 (TRAF6) and interleukin-1 receptor–associated kinase (IRAK)12. miR-92a negatively regulates TLR-triggered inflammatory response in macrophages by targeting mitogen-activated protein kinase kinase 4 (MKK4)13. miR-21 is induced via the MyD88 pathway in macrophages triggered by LPS and controls inflammation by targeting pro-inflammatory tumor suppressor programmed cell death 4 (PDCD4)14. miR-124 is one of the most abundantly expressed miRNAs in the nervous system. It is widely expressed in neurons in the brain, retina, and spinal cord and regulates neuronal differentiation during central nervous system (CNS) development and adult neurogenesis15,16,17. Recently, Ponomarev et al found that miRNA-124 promotes microglia (tissue-resident macrophages in CNS) quiescence and suppresses experimental autoimmune encephalomyelitis (EAE) by deactivating macrophages via the CCAAT/enhanced-binding protein (C/EBP)-α–PU.1 pathway18. These findings prompted the current study in which we investigated the involvement of miR-124 in the regulation of TLR activation in macrophages.
Macrophages serve as the first line of defense against invading pathogens. Pro-inflammatory cytokines, such as TNF-α released from macrophages, further augment systemic inflammation19. In the present study, we found that LPS treatment highly induced miR-124 in RAW264.7 macrophages and septic mice. In RAW264.7 macrophages, knocking down LPS-induced miR-124 increased TNF-α protein production but did not affect its mRNA level. Further experiments demonstrated that miR-124 accelerated the degradation of TNF-α protein by targeting USP2 and USP14. This study has identified miR-124 as a new therapeutic target in inflammation-related diseases.
Materials and methods
Reagents
LPS and cycloheximide (CHX) were purchased from Sigma (St Louis, MO, USA). Antibodies specific to TNF-α, USP2, and USP14 were from Cell Signaling Technology (Danvers, MA, USA). Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (60004-1-lg) was from ProteinTech Group, Inc (Chicago, IL, USA). miR-124 mimic (a chemically synthesized, double-stranded RNA molecule that is intended to mimic the transient duplexed product of Dicer complex processing, which results in a gain of function of miR-124) and inhibitor (chemically synthesized, non-hydrolyzable, single-stranded reverse complement to the mature miRNA, which results in a loss-of-function of miR-124) were from Dharmacon company (Lafayette, CO, USA). Enzyme-linked immunosorbent assay (ELISA) kits for TNF-α were obtained from R&D Systems (Minneapolis, MN, USA).
Animals
C57BL/6 mice (18–22 g) were purchased from Sino-British SIPPR/BK Laboratory Animal Ltd (Shanghai, China). Mice were 5 to 6 weeks of age at the start of the experiments and were maintained in animal rooms at 22 °C in a 12-h light/dark cycle with free access to water and a standard rodent diet. All animal experiments were undertaken in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of Second Military Medical University (Shanghai, China).
Cell culture and transfection
Macrophage cell lines RAW264.7 (mouse) and HEK293 (human) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured according to the manufacturer's instructions. The cells were transfected with Lipo2000 (Invitrogen Corp, Carlsbad, CA, USA) according to the manufacturer's instructions.
RNA interference
The USP2-specific siRNAs were 5′-CCA UCG CAA AGA GAG GUU Att-3′ (sense) and 5′-UAA CCU CUC UUU GCG AUG Ggc-3′ (antisense). The USP14-specific siRNAs were 5′-GAA GGU GUA GAA UUG AAU Att-3′ (sense) and 5′-UAU UCA AUU CUA CAC CUU Caa-3′ (antisense). The scrambled control RNA sequences were 5′-UUC UCC GAA CGU GUC ACG UTT-3′ (sense) and 5′-ACG UGA CAC GUU CGG AGA ATT-3′ (antisense). siRNA duplexes were transfected into RAW264.7 cells according to the standard protocol.
3′-UTR luciferase reporter assays
The USP2 and USP14 3′-UTR luciferase reporter constructs were made by polymerase chain reaction (PCR) amplifying the corresponding mouse mRNA 3′-UTR sequences and cloning into the XbaI site of pGL3-promoter construct (Promega, Madison, WI, USA). HEK293 cells were co-transfected with 80 ng of luciferase reporter plasmid, 40 ng of pRL-TK-Renilla-luciferase plasmid, and the indicated miR-124 mimic (final concentration, 20 nmol/L). After 24 h, luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Data were normalized for transfection efficiency by dividing firefly luciferase activity with that of Renilla luciferase.
RNA quantification
Total RNA, containing miRNA, was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Quantitative RT-PCR (qPCR) analysis was performed using Taqman MicroRNA Assays (ABI, Carlsbad, CA, USA) and SYBR RT-PCR kits (Takara Shuzo, Otsu, Japan). The relative expression level of miR-124 was normalized to that of internal control sno234. The primers for TNF-α were: 5′-AAG CCT GTA GCC CAC GTC GTA-3′ (sense) and 5′-GGC ACC ACT AGT TGG TTG TCT TTG-3′ (antisense); for β-actin were: 5′-AGT GTG ACG TTG ACA TCC GT-3′ (sense) and 5′-GCA GCT CAG TAA CAG TCC GC-3′ (antisense). Data were normalized by the level of β-actin expression in each sample.
ELISA
TNF-α level in the culture supernatants was measured with ELISA kit according to the manufacturer's protocol.
Immunoblotting
RAW264.7 cells were lysed with M-PER protein extraction reagent (Pierce, Rockford, IL, USA) supplemented with the protease inhibitor mixture (CalBiochem, San Diego, CA, USA). After centrifugation, proteins in the supernatant were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes. Immunoblotting was performed as described previously.
Statistical analysis
Statistical significance was determined by Student's t-test, with P<0.05 considered to be statistically significant.
Results
miR-124 level was elevated in LPS-triggered RAW264.7 macrophages and mice
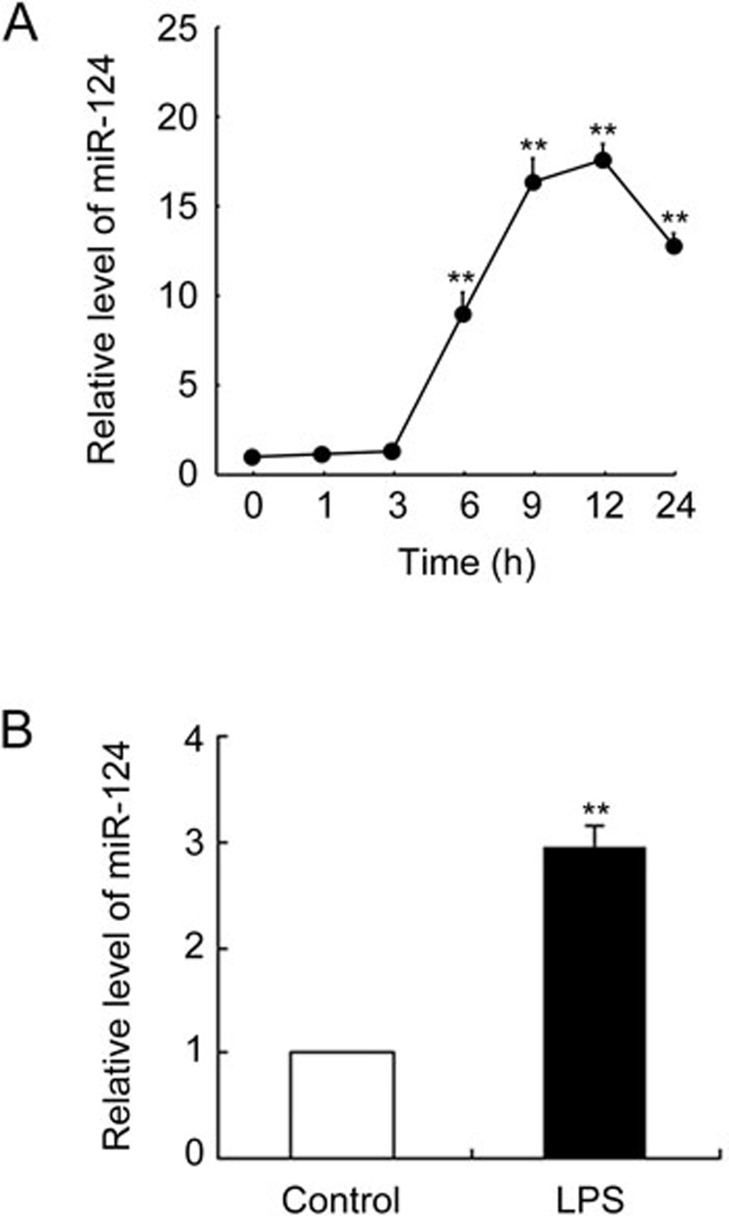
To determine whether LPS induces miR-124 expression in murine macrophages, we stimulated RAW264.7 macrophages with LPS for 24 h and detected miR-124 expression by qPCR. As shown in Figure 1A, levels of miR-124 began to increase 6 h after LPS addition, peaked at approximately 12 h, and then gradually declined. miR-124 expression in serum was also significantly elevated 6 h after a 30 mg/kg LPS intraperitoneal injection in C57BL/6 mice (Figure 1B).
Figure 1.
LPS increased miR-124 levels in vitro and in vivo. (A) RAW264.7 cells were stimulated with 100 ng/mL of LPS for the time indicated. We measured miR-124 by qPCR (n=3 per time point). **P<0.01 vs 0 h. (B) C57BL/6 mice were injected intraperitoneally with vehicle or 30 mg/kg of LPS. We measured serum miR-124 by qPCR after 6 h. Data represent mean±SD. n=6. **P<0.01 vs control.
miR-124 knockdown or overexpression, respectively, exaggerated or attenuated, the production of LPS-induced TNF-α protein in RAW264.7 macrophages
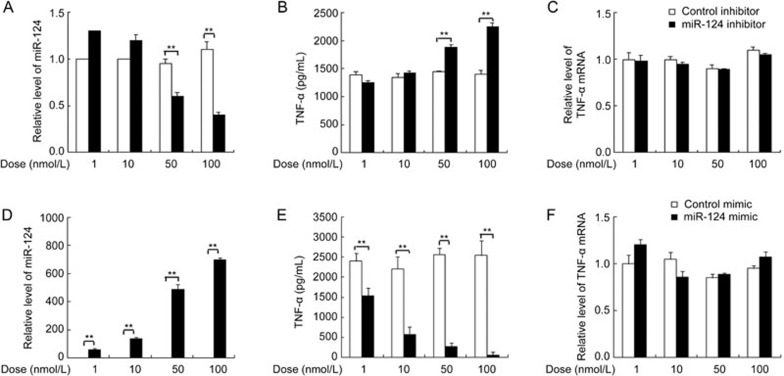
LPS treatment induces the expression of miR-124, indicating that miR-124 may have a regulatory role in LPS/TLR4 signaling in macrophages. To investigate the physiological functions of miR-124 in LPS/TLR4 signaling, RAW264.7 macrophages were transfected with miR-124 inhibitor to knockdown miR-124 (Figure 2A). In LPS-triggered RAW264.7 macrophages, miR-124 inhibitor dose-dependently increased TNF-α protein production compared with control (Figure 2B). However, miR-124 inhibitor did not affect TNF-α mRNA levels under the same condition (Figure 2C). Conversely, miR-124 mimic transfection (Figure 2D) dose-dependently decreased TNF-α protein production (Figure 2E) but did not affect its mRNA level (Figure 2F).
Figure 2.
MiR-124 knockdown or overexpression, respectively, exaggerated or attenuated the production of TNF-α protein in LPS-treated RAW264.7 macrophages. RAW264.7 cells were transfected with indicated doses of miR-124 inhibitor or mimic and their corresponding negative control for 36 h and then stimulated with 100 ng/mL of LPS for 9 h. We measured cellular miR-124 by qPCR (A, D), supernatant TNF-α by ELISA (B, E) and cellular TNF-α mRNA by qPCR (C, F). Data represent mean±SD. n = 3. **P<0.01.
miR-124 deficiency attenuated the degradation of TNF-α protein
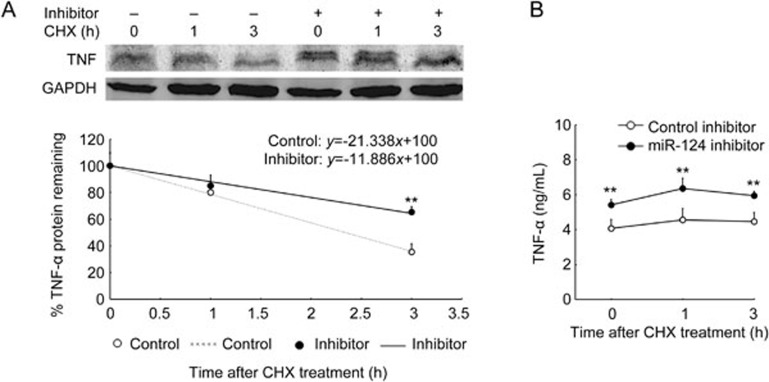
Because miR-124 post-transcriptionally regulated TNF-α-production, we determined whether miR-124 could affect TNF-α protein stability. RAW264.7 cells were transfected with miR-124 inhibitor for 36 h. Following treatment with 100 ng/mL of LPS for 9 h, cells were incubated with the protein synthesis inhibitor CHX (30 μg/mL) for the time indicated. We measured TNF-α protein levels in cells by Western blotting and in the supernatant by ELISA. As shown in Figure 3A, the half-life of TNF-α protein in the cells was approximately 2.3 h in the control group. When LPS-induced miR-124 was knocked down, the half-life of TNF-α protein in these cells was extended to 4.2 h, and the level of TNF-α protein in the supernatant was significantly increased compared with control (Figure 3B).
Figure 3.
MiR-124 deficiency attenuated TNF-α protein degradation. RAW264.7 cells were transfected with miR-124 inhibitor or negative control for 36 h. Following treatment with 100 ng/mL of LPS for 9 h, cells were incubated with protein synthesis inhibitor CHX (30 μg/mL) for the time indicated. We quantified cellular TNF-α protein level by Western blot and generated an x–y graph and associated graph equation. TNF-α/GAPDH ratio at time 0 h was considered 100% expression and used to calculate the protein half-life at all other time points (A, representative of three independent experiments). We measured supernatant TNF-α protein by ELISA (B). Data represent mean±SD. n=3. **P<0.01 vs control.
miR-124 overexpression accelerated the degradation of TNF-α protein
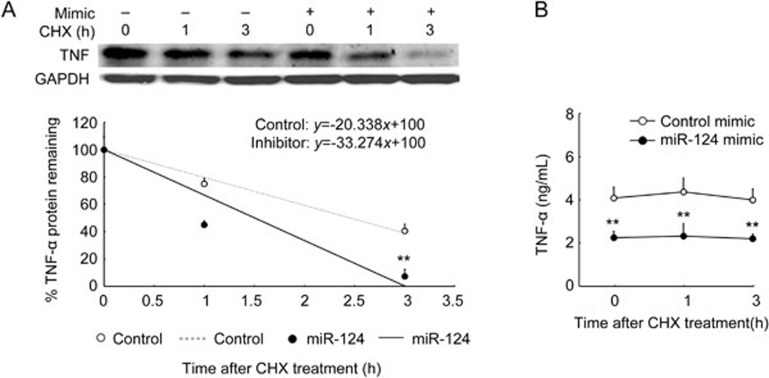
To further study the role of miR-124 overexpression in TNF-α protein stability, we transfected RAW264.7 cells with the miR-124 mimic for 36 h, stimulated with LPS for 9 h, and then incubated with CHX for the time indicated. Contrary to miR-124 deficiency, miR-124 overexpression significantly shortened TNF-α protein half-life from 2.5 h to 1.5 h (Figure 4A) and decreased the level of TNF-α protein in the supernatant compared with control (Figure 4B).
Figure 4.
MiR-124 overexpression accelerated the degradation of TNF-α protein. RAW264.7 cells were transfected with miR-124 mimic or negative control for 36 h, and then treated and analyzed as described in Figure 3. We measured cellular TNF-α protein with Western blot and generated an x–y graph and associated graph equation (A, representative of three independent experiments). We measured TNF-α protein in the supernatant with ELISA (B). Data represent mean±SD. n=3. **P<0.01 vs control.
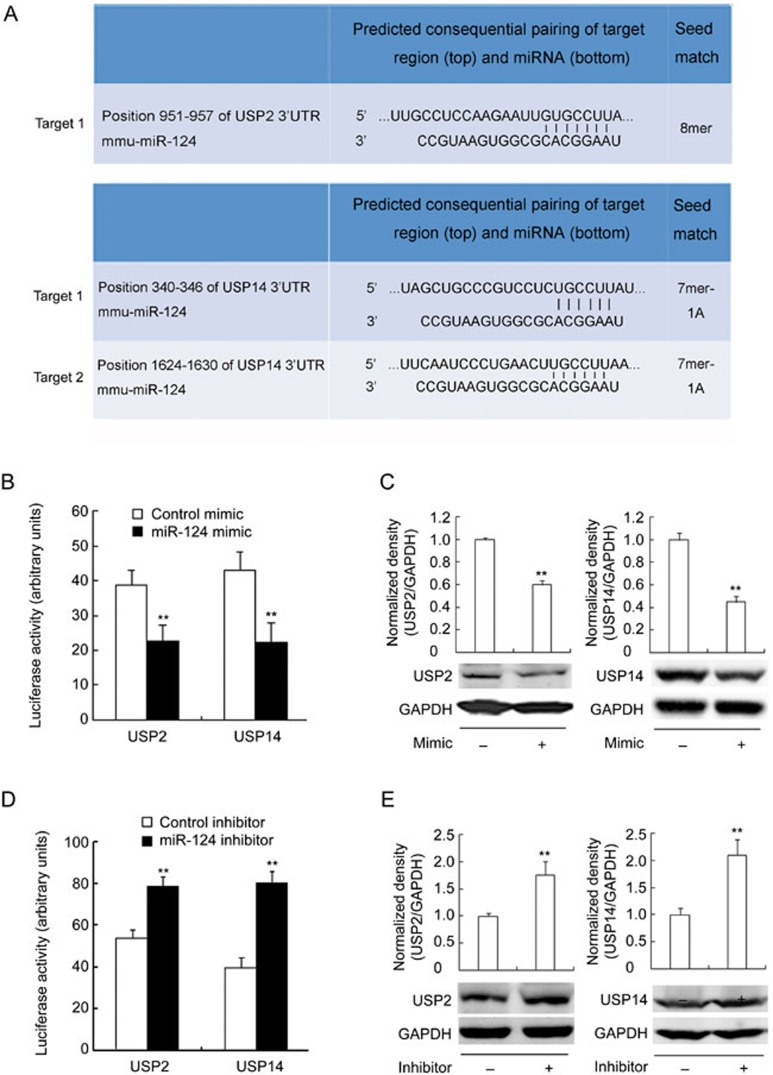
miR-124 targets USP2 and USP14
To identify potential miR-124 targets in TNF-α protein stability regulation, we used the miRNA prediction program TargetScan (http://www.targetscan.org). We found that the 3′-UTR of mouse USP2 mRNA contains one miR-124 target site and USP14 mRNA contains two putative miR-124 target sites (Figure 5A). To determine whether miR-124 regulates the stability of USP2 and USP14 mRNA, we constructed a reporter plasmid with cDNA corresponding to the 3′-UTR of mouse USP2 and USP14 mRNA inserted into the 3′-region of the firefly luciferase gene. In HEK293 cells transfected with the reporter plasmid containing the 3′-UTR of USP2 and USP14 mRNA, the miR-124 mimic significantly decreased luciferase activity (Figure 5B). We then examined the effects of miR-124 on USP2 and USP14 expression in RAW264.7 cells. As shown in Figure 5C, the level of USP2 and USP14 protein was significantly decreased when the cells were treated with the miR-124 mimic for 36 h. Conversely, miR-124 inhibitor showed enhanced effects in USP2/USP14 reporter activity (Figure 5D) and protein expression (Figure 5E). This result suggests that miR-124 may be a modulator of USP2 and USP14. Interestingly, USP14 was very recently reported to be a target of miR-124 in neurons20.
Figure 5.
MiR-124 targets USP2 and USP14. (A) The alignment of miR-124 and its target sites in 3'-UTR of UPS2 and USP14 based on TargetScan (http://www.targetscan.org) data. (B, D) Luciferase activity in HEK293 cells transfected with the reporter containing 3'UTR of USP2 or USP14 mRNA and miR-124 mimic (B) or inhibitor (D). Data represent mean±SD. n=6. **P<0.01 vs control. (C, E) Representative Western blots and quantification of the levels of USP2 or USP14 in RAW264.7 cells transfected with miR-124 mimic (C) or inhibitor (E) for 36 h. Data represent mean±SD. n=3. **P<0.01 vs control.
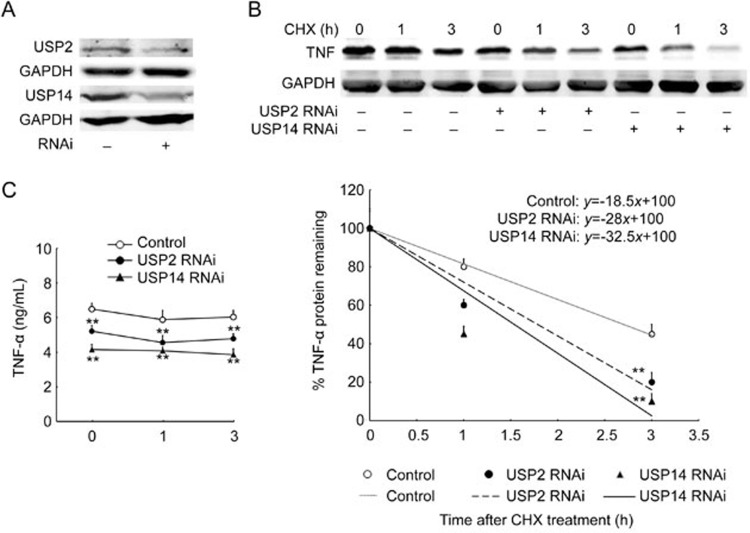
Knocking down USP2 and USP14 accelerated the degradation of TNF-α protein and abolished the effect of miR-124 on TNF-α protein stability
To study further the involvement of USP2 and USP14 in TNF-α protein stability, we knocked down USP2 and USP14 with specific siRNA (Figure 6A). As shown in Figure 6B, when USP2 and USP14 were knocked down, the half-life of TNF-α protein shortened from 2.7 h to 1.7 h and 1.5 h, respectively. As expected, TNF-α level in the supernatant was significantly decreased when we knocked down USP2 and USP14 (Figure 6C). These results suggest that USP2 and USP14 promote TNF-α protein degradation.
Figure 6.
Knocking down USP2 and USP14 accelerated the degradation of TNF-a protein. (A) RAW264.7 macrophages were transfected with USP2, USP14, and control RNAi for 36 h, followed by Western blot detection of USP2 and USP14 protein. Results are representative of three independent experiments. (B–C) RAW264.7 cells were transfected with USP2 or USP14 siRNA for 36 h, followed by incubation with 100 ng/mL LPS treatment for 9 h, and then 30 μg/mL of CHX for the time indicated. TNF-α protein in the cells (B) and in the supernatant (C) was detected and analyzed as described in Figure 3 (n=3). **P<0.01 vs control.
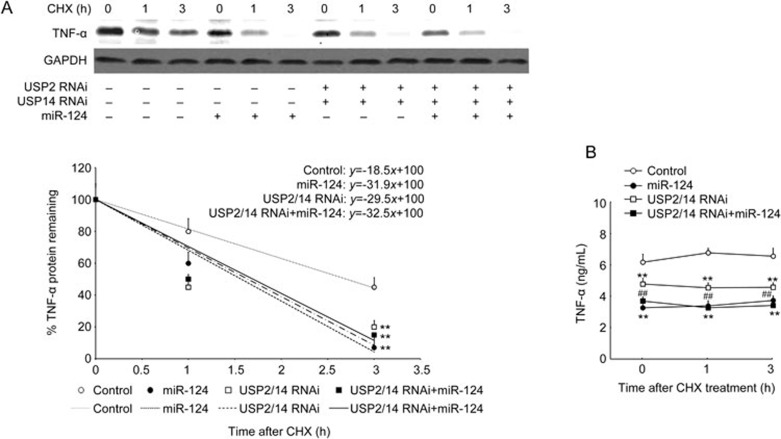
Furthermore, we confirmed that miR-124 affects TNF-α protein stability by targeting USP2 and USP14. As shown in Figure 7, the miR-124 mimic significantly shortened TNF-α protein half-life from 2.7 h to 1.6 h and decreased TNF-α protein levels in the supernatant. siRNA mediated knockdown of USP2 and USP14 had a similar effect as transfection of miR-124 on the half-life of TNF-α (1.7 h and 1.6 h, respectively), but the effect of USP2 and USP14 knockdown on reducing supernatant TNF-α protein was weaker than that of miR-124 transfection. In addition, the miR-124 mimic did not affect TNF-α protein stability when USP2 and USP14 were both knocked down (half-life is 1.6 h), but it did significantly decrease TNF-α protein levels in the supernatant under the same condition (Figure 7B).
Figure 7.
MiR-124 failed to promote degradation TNF-α protein when USP2 and USP14 were both knocked down. RAW264.7 cells were transfected control RNAi or USP2 and USP14 RNAi in the absence or presence of miR-124 for 36 h. For co-transfection, RAW264.7 cells were first transfected with USP2 and USP14 RNAi for 16 h, then transfected with miR-124 mimic for another 24 h. Cells were then stimulated with 100 ng/mL of LPS for 9 h, followed by incubation with 30 μg/mL of CHX for the time indicated. TNF-α protein in the cells (A) and in the supernatant (B) was detected and analyzed as described in Figure 3. Mean±SD. (n=3). **P<0.01 control. ##P<0.01 USP2/14 RNAi+miR-124 vs USP2/USP14 RNAi.
Discussion
The goal of inflammation is to resolve injury and return the body to homeostasis. Recently, it has been shown that multiple miRNAs, such as miR-155, miR-146, miR-147, miR-21, and miR-9, modulate TLR/NF-κB activation10,21,22. In this study, we provide evidence that miR-124 is associated with LPS/TLR4 activation. Importantly, we demonstrate that LPS-induced miR-124 functions as a negative regulator in LPS-induced TNF-α expression by targeting USP2 and USP14 and decreasing TNF-α protein stability in RAW264.7 macrophages. Thus, miR-124 acts as a negative feedback regulator in the LPS/TLR4 signaling pathway.
Many targets of miR-124 have been identified. Notably, miR-124 can regulate the expression of I kappa B (IκB), which has been found to be necessary for the transactivation of a subset of nuclear factor kappa B (NF-κB) target genes23,24. miR-124 can target transcription factor C/EBP- and promote quiescence in microglia that function as macrophages in the CNS18. miR-124 has also been reported to decrease embryonic stem cell signal transducer and activator of transcription (STAT)3 phosphorylation and affect neural lineage differentiation25. However, IκBα, C/EBP-α, and phosphorylated-STAT3, which all act as transcriptional factors, may not be involved in the post-transcriptional regulation of TNF-α induced by miR-124.
Post-translational protein modification by ubiquitination represents a central mechanism for modulating a wide range of cellular functions, such as protein stability26. Ubiquitination is a reversible process counteracted by de-ubiquitinating enzymes (DUBs)27,28. USPs, with more than 50 members, comprise the largest class of DUBs. USP enzymes are likely to have a role in protein stability control because they cleave ubiquitin adducts from substrate proteins, thereby protecting the substrate from proteasome-mediated degradation29. It has been reported that USP10 regulates p53 localization and stability by deubiquitinating p5330, USP22 regulates cell proliferation by de-ubiquitinating the transcriptional regulator FBP131, USP2 enhances tumor progression by impairing ubiquitination and stabilizing an important cell cycle regulator cyclin A132, and USP15 regulates human papillomavirus type 16 E6 protein stability33. In the present study, we found that miR-124 could decrease TNF-α protein stability and knocking down USP2 and USP14, two targets of miR-124, could significantly abolish this effect. These data suggest that miR-124 decreases TNF-α protein stability by targeting USP2 and USP14. Interestingly, the level of supernatant TNF-α protein remained relatively stable after CHX addition during the observation period. One possible explanation is that most of the TNF-α was released into the supernatant before the CHX addition. The changes in the degradation of the remaining TNF-α in the cells after CHX addition did not produce enough effect on the total TNF-α content in the supernatant to be detectable. Other unknown mechanisms may be involved as well. More time-point observations are needed to confirm this phenomenon, and the detailed mechanism underlying this process should also be further clarified.
TNF-α is synthesized as a membrane-anchored precursor (pro-TNF-α) that is processed by TNF-α-converting enzyme (TACE) at the cell membrane to generate mature TNF-α34. TNF-α is then secreted into the extracellular space or remains inside the cells in the form of secretary vesicles. Studies of other intracellular vesicles have shown that the destination of these vesicles is regulated by ubiquitination at their cytoplasmic surface35. It is conceivable that the miR-124-mediated decrease of USP2 and USP14 resulted in increased ubiquitination of vesicle protein(s), which perhaps led to fusion of the vesicles with lysosomes and degradation of TNF-α. This hypothesis could be tested in the future through studies of the ubiquitination and intracellular trafficking of TNF-α and TNF-α--containing vesicles.
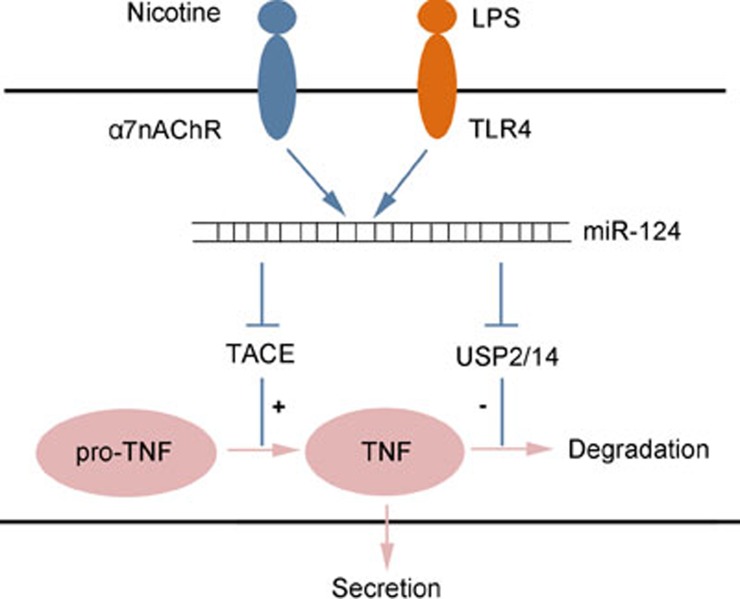
Interestingly, we previously reported that the cholinergic anti-inflammatory pathway could activate miR-124, which can target TACE. Furthermore, TACE knockdown by specific siRNAs could abolish the inhibitory effect of miR-124 on the production of mature TNF-α36. Here, we also found that miR-124 decreased the protein stability of intracellular TNF-α by targeting USP2 and USP14, demonstrating that miR-124 regulated TNF-α production at a minimum of two levels, controlling the maturation at the pro-TNF-α level by targeting TACE and promoting protein degradation at the TNF-α level by targeting USP2 and USP14 (Figure 8). These findings explain the results shown in Figure 7B. When USP2 and USP14 were both knocked down, miR-124 no longer affected cellular TNF-α protein stability, but it still decreased TNF-α protein levels in the supernatant via targeting TACE.
Figure 8.
MiR-124 regulates TNF-α production at a minimum of two levels. Cholinergic agonist nicotine and LPS both can induce miR-124, which can regulate TNF-α production at a minimum of two levels, controlling the maturation at the pro-TNF-α level by targeting TACE (Cell Res 2013; 23: 1270–83) and promoting protein degradation at the TNF-α level by targeting USP2 and USP14.
Taken together, the present study identified miR-124 as a negative feedback player in LPS/TLR4 signaling. Our previous and current findings suggest that multiple miRNAs can modulate one signaling pathway and that a single miRNA can affect multiple molecules in the same pathway. These miRNAs represent a complex network involved in the restriction of LPS-induced excessive pro-inflammatory responses. Our current findings add to a growing body of evidence on how multiple miRNAs can collaborate to properly balance inflammatory responses.
Conclusion
Our data show that miR-124 is an important feedback negative regulator for LPS-induced production of TNF-α by targeting USP2 and USP14. The level of miR-124 was rapidly increased in LPS-treated RAW264.7 macrophages and mice. miR-124 knockdown or overexpression significantly increased or decreased the protein stability of TNF-α, respectively. miR-124 directly targeted USP2 and USP14, whereas knockdown of USP2 and USP14 abolished the effect of miR-124 on protein degradation of TNF-α. These results indicate a new mechanism for fine-tuning the TLR-triggered inflammatory response.
Author contribution
Xia LIU and Jian-guang YU designed and supervised the experiments. Yang SUN, Zhen QIN, Qi LI, Jing-jing WAN, Ming-he CHENG, and Peng-yuan WANG performed the experiments and data analysis. Xia LIU drafted the manuscript. Ding-feng SU participated in the discussion and drafting of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No 81273606, 81473259 and 81230083) and the National Science and Technology Major Project (No 2014ZX09J14103-08C).
References
- Medzhitov R. Origin and physiological roles of inflammation. Nature 2008; 454: 428–35. [DOI] [PubMed] [Google Scholar]
- Sabroe I, Parker LC, Dower SK, Whyte MK. The role of TLR activation in inflammation. J Pathol 2008; 214: 126–35. [DOI] [PubMed] [Google Scholar]
- Yuk JM, Shin DM, Lee HM, Kim JJ, Kim SW, Jin HS, et al. The orphan nuclear receptor SHP acts as a negative regulator in inflammatory signaling triggered by Toll-like receptors. Nat Immunol 2011; 12: 742–51. [DOI] [PubMed] [Google Scholar]
- Seit-Nebi A, Cheng W, Xu H, Han J. MLK4 has negative effect on TLR4 signaling. Cell Mol Immunol 2012; 9: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Maratha A, Butt AQ, Shevlin E, Miggin SM. TRIF-mediated TLR3 and TLR4 signaling is negatively regulated by ADAM15. J Immunol 2013; 190: 2217–28. [DOI] [PubMed] [Google Scholar]
- Peng Q, O'Loughlin JL, Humphrey MB. DOK3 negatively regulates LPS responses and endotoxin tolerance. PLoS One 2012; 7: e39967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 2012; 19: 586–93. [DOI] [PubMed] [Google Scholar]
- Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem 2011; 67: 129–39. [DOI] [PubMed] [Google Scholar]
- Gommans WM, Berezikov E. Controlling miRNA regulation in disease. Methods Mol Biol 2012; 822: 1–18. [DOI] [PubMed] [Google Scholar]
- Nahid MA, Satoh M, Chan EK. MicroRNA in TLR signaling and endotoxin tolerance. Cell Mol Immunol 2011; 8: 388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol 2012; 30: 295–312. [DOI] [PubMed] [Google Scholar]
- Hou J, Wang P, Lin L, Liu X, Ma F, An H, et al. MicroRNA-146a feedback inhibits RIG-I-dependent type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol 2009; 183: 2150–8. [DOI] [PubMed] [Google Scholar]
- Lai L, Song Y, Liu Y, Chen Q, Han Q, Chen W, et al. MicroRNA-92a negatively regulates Toll-like receptor (TLR)-triggered inflammatory response in macrophages by targeting MKK4 kinase. J Biol Chem 2013; 288: 7956–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol 2010; 11: 141–7. [DOI] [PubMed] [Google Scholar]
- Mishima T, Mizuguchi Y, Kawahigashi Y, Takizawa T. RT-PCR-based analysis of microRNA (miR-1 and -124) expression in mouse CNS. Brain Res 2007; 1131: 37–43. [DOI] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 2007; 21: 744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K, Otto W, Johannes S, Baumgart J, Nitsch R, Schumacher S. miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J 2012; 31: 2908–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med 2011; 17: 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoy N. Macrophage biology and pathobiology in the evolution of immune responses: a functional analysis. Pathobiology 2001; 69: 179–211. [DOI] [PubMed] [Google Scholar]
- Doeppner TR, Doehring M, Bretschneider E, Zechariah A, Kaltwasser B, Muller B, et al. MicroRNA-124 protects against focal cerebral ischemia via mechanisms involving Usp14-dependent REST degradation. Acta Neuropathol 2013; 126: 251–65. [DOI] [PubMed] [Google Scholar]
- Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB signaling. J Mol Cell Biol 2011; 3: 159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam MM, O'Neill LA. MicroRNAs and the resolution phase of inflammation in macrophages. Eur J Immunol 2011; 41: 2482–5. [DOI] [PubMed] [Google Scholar]
- Lindenblatt C, Schulze-Osthoff K, Totzke G. IkappaBzeta expression is regulated by miR-124a. Cell Cycle 2009; 8: 2019–23. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Matsuo S, Muta T, Yamamoto M, Akira S, Takeshige K. Gene-specific requirement of a nuclear protein, IkappaB-zeta, for promoter association of inflammatory transcription regulators. J Biol Chem 2008; 283: 32404–11. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 2006; 24: 857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM. Regulating protein degradation by ubiquitination. Immunol Today 1997; 18: 189–98. [DOI] [PubMed] [Google Scholar]
- Hutchins AP, Liu S, Diez D, Miranda-Saavedra D. The repertoires of ubiquitinating and deubiquitinating enzymes in eukaryotic genomes. Mol Biol Evol. 2013; 30: 1172–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005; 123: 773–86. [DOI] [PubMed] [Google Scholar]
- Faesen AC, Luna-Vargas MP, Sixma TK. The role of UBL domains in ubiquitin-specific proteases. Biochem Soc Trans 2012; 40: 539–45. [DOI] [PubMed] [Google Scholar]
- Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 2010; 140: 384–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov BS, Dent SY. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep 2011; 12: 924–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim WJ, Liu Z, Loda M, Freeman MR. The ubiquitin-specific protease USP2a enhances tumor progression by targeting cyclin A1 in bladder cancer. Cell Cycle 2012; 11: 1123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos RM, Altreuter J, White EA, Howley PM. The ubiquitin-specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. J Virol 2009; 83: 8885–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature 1997; 385: 733–6. [DOI] [PubMed] [Google Scholar]
- Lu A, Pfeffer SR. A CULLINary ride across the secretory pathway: more than just secretion. Trends Cell Biol 2014; 24: 389–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li Q, Gui H, Xu DP, Yang YL, Su DF, et al. MicroRNA-124 mediates the cholinergic anti-inflammatory action through inhibiting the production of pro-inflammatory cytokines. Cell Res 2013; 23: 1270–83. [DOI] [PMC free article] [PubMed] [Google Scholar]