Abstract
Aim:
Bufalin is one of the active components in the traditional Chinese medicine ChanSu that is used to treat arrhythmia, inflammation and cancer. BF211 is a bufalin derivative with stronger cytotoxic activity in cancer cells. The aim of this study was to identify the putative target proteins of BF211 and the signaling pathways in cancer cells.
Methods:
A549 human lung cancer cells were treated with BF211. A SILAC-based proteomic analysis was used to detect the protein expression profiles of BF211-treated A549 cells. Cellular proteasome activities were examined using fluorogenic peptide substrates, and the binding affinities of BF211 to recombinant proteasome subunit proteins were evaluated using the Biacore assay. The expression levels of proteasome subunits were determined using RT-PCR and Western blotting, and the levels of the integral 26S proteasome were evaluated using native PAGE analysis.
Results:
The proteomic analysis revealed that 1282 proteins were differentially expressed in BF211-treated A549 cells, and the putative target proteins of BF211 were associated with various cellular functions, including transcription, translation, mRNA splicing, ribosomal protein synthesis and proteasome function. In A549 cells, BF211 (5, 10, and 20 nmol/L) dose-dependently inhibited the enzymatic activities of proteasome. But BF211 displayed a moderate affinity in binding to proteasome β1 subunit and no binding affinity to the β2 and β5 subunits. Moreover, BF211 (0.1, 1, and 10 nmol/L) did not inhibit the proteasome activities in the cell lysates. BF211 (5, 10, and 20 nmol/L) significantly decreased the expression level of proteasome β1 subunit and the levels of integral 26S proteasome in A549 cells. Similarly, knockdown of the β1 subunit with siRNA in A549 cells significantly decreased integral 26S proteasome and proteasome activity.
Conclusion:
BF211 inhibits proteasome activity in A549 cells by decreasing β1 subunit expression and disrupting proteasome assembly.
Keywords: bufalin, ChanSu, cardiac steroids, proteasome β1 subunit, proteasome assembly, A549 human lung cancer cells, proteomics
Introduction
Bufalin (BF) is one of the key active components of the traditional Chinese medicine ChanSu, which is derived from secretions from the skin and parotid venom glands of Bufo bufo gargarizans Cantor or Bufo melanostictus Schneider1,2. ChanSu has been used for centuries to treat arrhythmia, inflammation and cancer in China and other Asian countries based on its cardiotonic, anti-inflammatory and anti-cancer effects3. Medicines containing ChanSu, such as Huachansu injection, are still widely used in the clinical setting to treat various cancers, including lung cancer4,5. BF211 is a derivative of bufalin, and the synthesis and therapeutic use of BF211 as an anti-cancer agent was granted patent protection rights in China (Authorized Announcement No CN 102532235B). In a previous paper, we reported that BF211 exhibited stronger cytotoxic activity in cancer cells than bufalin6.
Multiple papers have described the activities of BF and other bufadienolides in cancer cells7,8,9,10,11,12; however, the effects of bufadienolides have not been fully clarified. In the present study, to identify the possible signaling network activated by BF211 in cancer cells, we conducted a SILAC-based proteomic analysis and compared the protein expression profiles of A549 human lung cancer cells treated with either BF211 or a solvent control. Our findings suggested that BF211 influences proteasome function, and we further evaluated the effects and potential mechanisms mediating this phenomenon.
Materials and methods
Cell culture
The A549 human lung cancer cell line, PC-3 prostate cancer cell line and HeLa cervical cancer cell line were purchased from the Cell Resource Center of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). A549 cells and PC-3 cells were cultured in RPMI-1640 medium supplemented with 10% (v/v) fetal bovine serum, 100 units/mL penicillin and 100 μg/mL streptomycin. HeLa cells were cultured in minimum essential medium supplemented with 10% fetal bovine serum, 100 units/mL penicillin and 100 mg/L streptomycin. All of the materials used for cell culture were purchased from GE Healthcare Life Sciences Hyclone Laboratories (South Logan, Utah, USA).
Cytotoxicity assay
The cytotoxic activity of BF211 and BF in A549 cells was quantified using the MTT assay as previously described13. Briefly, cells were seeded into 96-well cell culture plates at an initial density of 2×104 cells/mL. After the cells were allowed to adhere overnight, they were treated with 0.1% DMSO (solvent control), various concentrations of BF or various concentrations of BF211 for 72 h. Then, cell viability was determined using MTT assays.
SILAC-based proteomic analysis
To label the A549 cells with the “heavy” or “light” label, the cells were maintained in complete RPMI-1640 culture medium supplemented with either the heavy (H) form of 13C6-L-arginine (R6) and 13C6-L-lysine (K6) or the light (L) form of 12C6-L-arginine (R0) and 12C6-L-lysine (K0) for 6 generations. The labeling efficiency was >99% as determined using mass spectrometry analysis. To compare the protein expression profiles of BF211- and control-treated A549 cells, cells with the heavy label were treated with BF211 (10 nmol/L) for 3 h, and cells with the light label were treated with the solvent control (0.1% DMSO). Three independent experiments were conducted. After the treatment, the cells were harvested and washed twice in PBS by centrifugation, and the pellets were re-suspended in RIPA buffer [50 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS and 1 mmol/L EDTA] with protease inhibitor cocktail (Roche Diagnostics Corporation, Indianapolis, USA) and incubated at 4 °C for 30 min. The supernatant containing predominantly total soluble protein was collected after a 5-min centrifugation at 13 000×g at 4 °C. The protein concentration of the supernatant was determined using the A280 method using a UV-Vis Spectrophotometer Q5000 (Quawell Technology, San Jose, CA, USA). Then, the lysates of the heavy-labeled cells and light-labeled cells were combined 1:1 (protein content) for LC-MS/MS analysis. Samples from three independent experiments were used for the LC-MS/MS analysis. A total of 30 μg of the combined protein sample was reduced with 100 mmol/L dithiothreitol dissolved in 100 mmol/L ammonium bicarbonate and heated at 56 °C for 1 h. After the samples cooled to room temperature, the proteins were alkylated using 200 mmol/L iodoacetamide (250 μL) to achieve a final concentration of 100 mmol/L and incubated for 30 min at room temperature. After alkylation, 1 μg/μL trypsin (10 μL) was added to each vial, and the digestion was allowed to proceed overnight at 37 °C. To reduce the volume, the sample was dried at room temperature in a vacuum concentrator/centrifugal evaporator, reconstituted to 30 μL with 0.1% formic acid in water. The samples were stored at 4 °C until the LC-MS/MS analysis. The LC-MS/MS analysis was conducted as described in previous reports14,15. For the first dimension LC analysis (strong cation exchange prefractionation), the trypsin digests were reconstituted using strong cation exchange (SCX) buffer A (10 mmol/L monobasic potassium phosphate (pH 2.75) and 25% ACN) and the pH of the samples was adjusted to 2.5–3 using phosphoric acid. The peptide mixtures were fractionated using a SCX column (100×4.6 mm, 5 mm, 300 Å) on a LabAlliance HPLC system (China). After a 20 min wash at a rate of 0.5 mL/min in 100% buffer A, an 80 min linear gradient from 0 to 100% (v/v) buffer B (consisting of buffer A with the addition of 0.5 mol/L potassium chloride) was run, and fractions were collected each minute. Based on the UV absorbance at 220 nm, a total of 10 fractions were collected. The pH values of the fractions were adjusted to 2–3, and the samples were purified using a C18 Sep-Pak® Cartridge (Waters, USA) with 50 μL of 100% ACN wetting solution. Fifty microliters of 0.1% formic acid was used to equilibrate the tips and wash the samples after loading, and the peptides were eluted in 40 μL of 0.1% formic acid and 60% ACN. Then, the samples were completely dried in a vacuum centrifuge and reconstituted in 0.1% formic acid. For the second dimension LC-MS/MS, SCX fractioned peptide mixtures were analyzed using nanoflow liquid chromatography on a Tempo™ nanoMDLC System (AB SCIEX) coupled to an LTQ Orbitrap Velos instrument (Thermo Fisher Scientific, Bremen, Germany) with a nanoelectrospray ion source (Proxeon). The samples were loaded into a trap column (PepMap™100, 300 μm×5 mm), washed for 20 min, and eluted into a C18 reverse phase column (PepMap100 C18, 75 μm×5 μm×15 cm). The chromatography method used a 10 min online trapping and desalting step followed by a 1 h gradient of 5%–40% mobile phase B (mobile B: 98% acetonitrile, 0.1% formic acid). The peptides were eluted at a flow rate of 300 nL/min, and solvent B was used to increase the LC linear gradients from 0% (10 min) to 50% (100 min) and from 50% to 100% (125 min) (mobile phase A: 2% acetonitrile, 0.1% formic acid; mobile phase B: 98% acetonitrile, 0.1% formic acid). The LC eluent was directly nanosprayed into an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) at an ionization voltage of 2.2 kV. During the chromatographic separation, the LTQ Orbitrap Velos was operated in a data-dependent mode switching automatically between full-scan MS and MS/MS acquisition under the direct control of the Xcalibur 2.1.0. software (Thermo Scientific). The MS data were acquired using the following parameters: 10 data-dependent collisional-induced-dissociation (CID) MS/MS scans per full scan in label-free; 6 data-dependent CID MS/MS scans per full scan in metabolic labeling; CID scans acquired in LTQ with two-microscan averaging a 35% normalized collision energy (NCE) in CID, a ±1.5 Da isolation window, dynamic exclusion enabled with a ±1.5 Da exclusion window, 50% to 60% S-lens radio frequency level, a lock mass abundance set to 0%, no sheath and auxiliary gas, and a heated capillary temperature of 200 °C.
MS data interpretation and prediction of a putative BF211 signaling network
The database search of the MS data was conducted using MaxQuant version 1.3.0.5 against the UniProtKB/Swiss-Prot H sapiens proteome database with contaminants and reverse sequences added. The search parameters included enzyme specificity for trypsin and precursor ion mass tolerances of 6 ppm for peptide mass and 20 ppm for fragment ion mass tolerance. Fixed modifications of cysteine carbamidomethylation (Cys 57.021464 Da) and variable modifications for the N-acetylation of proteins (N-term 42.010565 Da) and the oxidation of methionine (Met 15.994915 Da) were specified and allowed for up to two missed cleavage sites. Double or triple labeling was defined accordingly. The parameters allowed for up to two missed cleavages and two labeled amino acids (arginine and lysine). Additional search parameters included the following: full tryptic specificity was required; carbamidomethylation was set as a fixed modification; and oxidation (M) was considered a variable modification. The precursor ion mass tolerances were 10 ppm, and the fragment ion mass tolerance was 0.5 Da for all of the MS2 spectra acquired in the LTQ. The heavy labels were Arg6 and Lys6, and the minimum required peptide length was defined as six amino acids. The normalized H/L ratios, significance and variability (%) were automatically calculated using the MaxQuant program. The peptide and protein false discovery rate (FDR) was set to 1%. A minimum of two ratio counts, including automatic filtering on peptide length, mass error precision estimates, and peptide scores of the forward and reverse peptides were used to determine the normalized protein intensity. The re-quantification feature was enabled. A threshold of 1.50-fold change was used in the experiment comparing heavy (H)- against light (L)-labeled samples. The proteins that were differentially expressed in BF211-treated cells compared with control cells were summarized, and the results of three independent experiments were merged. The list of putative BF211-associated proteins was submitted to version 9.1 of STRING (Search Tool for the Retrieval of Interacting Genes) database (http://string.embl.de/) to predict potential BF211 signaling networks. “Experiments” was selected as the prediction method, and the gene-gene interactions ensured a confidence score greater than 0.7.
RT-PCR analysis of HSP90AA1, HSP90AB1 and PSMA2 expression levels in BF211-treated A549 cells
The proteomic analysis identified HSP90AA1, HSP90AB1 and PSMA2 as differentially expressed proteins. Therefore, we evaluated the expression levels of the genes encoding HSP90AA1, HSP90AB1 and PSMA2 using RT-PCR analysis. Briefly, total RNA was extracted from cells treated for 3 h with 10 nmol/L BF211 or solvent control (0.1% DMSO) using TRIzol reagent (Takara, Dalian, China), and the RNA was reverse-transcribed using the PrimeScriptTM RT reagent Kit (Takara, Dalian, China). Real-time PCR analysis of the cDNA was conducted using DNA primers and the Stratagene Mx3000P Multiplex Quantitative PCR System with SYBR Premix Ex TaqTM (Takara, Dalian, China). Target gene expression was normalized to GAPDH levels, and the comparative cycle threshold (Ct) method was used to calculate the relative mRNA expression levels. The oligonucleotide primers used for RT-PCR are listed in Supplemental Table S1.
Analysis of cellular proteasome activities in BF211-treated cells
The proteasome enzymatic activities, including caspase-like (C-L), trypsin-like (T-L) and chymotrypsin-like (CT-L) activity, were evaluated in cell lysates as previously described13. Briefly, the cells were treated with 5, 10 or 20 nmol/L of BF211 or 1 μmol/L bortezomib (positive control) for 24 h. Then, the cells were harvested, washed with PBS, and lysed in proteasome activity assay buffer (10 mmol/L Tris-HCl (pH 7.8), 0.5 mmol/L DTT, 140 μmol/L ATP and 5 mmol/L MgCl2) for 30 min at 4 °C. After the cells were frozen in liquid nitrogen and thawed twice, the homogenate was centrifuged at 12 000×g for 30 min at 4 °C, and the collected supernatant was used for the activity assays. To evaluate proteasome peptidase activities, 20 μg of supernatant was added to proteasome activity assay buffer with 50 mmol/L EDTA and fluorogenic peptide substrates (25 μmol/L Z-Leu-Leu-Glu-AMC for detecting C-L activity, 100 μmol/L Z-Ala-Arg-Arg-AMC for detecting T-L activity or 100 μmol/L Suc-Leu-Leu-Val-Tyr-AMC for detecting CT-L activity) to achieve a final reaction volume of 200 μL. The reactions were incubated for 120 min, and fluorescence data (excitation wavelength at 360 nm, emission wavelength at 465 nm, gain=60) was collected every 1 min16,17. The data from each sample were plotted as arbitrary fluorescence units (AFU) versus time, and the slope of a line fit to the data was obtained using an appropriate linear regression program. The activities are presented as the percent (%) activity of the control, which was calculated using the following formula: (slope of treated sample/slope of control)×100.
Analysis of proteasome activities in cell lysates treated with BF211 in vitro
To determine whether BF211 can directly inhibit proteasome activities, we evaluated proteasome activity in A549 cell lysates incubated in vitro with various concentrations of BF211, the solvent control (0.1% DMSO) or 10 μmol/L bortezomib (positive control). The cells were lysed and proteasome activities were assayed as described above.
Western blot analysis of ubiquitinated proteins
The Western blot assays used to examine the levels of ubiquitinated proteins in BF211-treated and control cells were conducted as previously described13. The following primary antibodies were used: mouse polyclonal antibody against ubiquitin (#3936) (1:1000) and rabbit polyclonal antibody against GAPDH (#2118) (1:1500). HRP-linked horse anti-rabbit IgG (#7076) (1:1000) and HRP-linked goat anti-rabbit IgG (#7074) (1:1000) were used as secondary antibodies. All of the antibodies were obtained from Cell Signaling Technology (Danvers, MA, USA).
Biacore assay of the binding affinity between BF211 and recombinant proteasome subunit proteins
Similar to the methods described in our previous report13, the recombinant human proteasome β1, β2 and β5 subunits were expressed as His-tagged proteins by Escherichia coli and purified using affinity chromatography. Full-length cDNA constructs encoding the proteasome β1, β2 and β5 subunit was synthesized by Life Technology Ltd (Shanghai, China). The cDNA constructs encoding the proteasome β1 and β2 subunits were subcloned into the pET-28a expression vector (Novagen, Madison, WI, USA), and the cDNA construct encoding the proteasome β5 subunit was subcloned into the pET-32b expression vector (Novagen, Madison, WI, USA). The recombinant plasmids were transformed into E coli BL21 (DE3) cells (Novagen, Madison, WI, USA). The BL21 transformants were cultured in LB medium with ampicillin until the absorbance at 600 nm at 37 °C was measured at 0.8. Then, the cells were induced by adding isopropyl beta-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mmol/L and incubated overnight at 20 °C. The His-tagged fusion protein was isolated from bacterial lysates using Ni2+-chelation affinity chromatography with the Profinia system (BioRad, USA). Finally, the dialyzed protein solution was concentrated to >1 mg/mL for SPR analysis. The Biacore assay used to assess the binding affinity between BF or BF211 and the recombinant proteins was conducted by the Drug Discovery and Design Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences using a Biacore 3000 instrument (Biacore AB, Rapsgatan 7, S-754 50 Uppsala, Sweden) as previously described18. Briefly, the recombinant β1, β2 and β5 proteins were immobilized on CM5 sensor chips as ligands with binding affinities of 11624.5 RU, 8754.0 RU, 5232.0 RU, respectively, using N-ethyl-N'-(3-dimethylaminopropyl) carbodiimide and N-hydroxysuccinimide according to the standard primary amine-coupling procedures. HBS-EP (10 mmol/L HEPES, 150 mmol/L NaCl, 3 mmol/L EDTA, 0.005% (v/v) surfactant P20, pH 7.4) was used as the running buffer. The chips were equilibrated using a continuous flow of HBS-EP through the chip surface for 1 to 2 h. The Biacore data were collected at 25 °C, and the HBS-EP running buffer was run at a constant flow of 20 μL/min. Various concentrations of the BF211 solution were injected into the channels at a flow rate of 20 μL/min, and the channels were subsequently washed with the running buffer. The binding responses were continuously recorded in response units (RU) at a frequency of 1 Hz as sensorgrams, and the data are presented as a function of time. In a preliminary experiment, we evaluated the binding affinity of BF211 at concentrations of 1 and 10 μmol/L. To analyze the binding characteristics between BF211 and the recombinant proteins that demonstrated positive results in the preliminary experiment, the proteins were incubated with 2.4, 3.43, 4.90, 7.0, 10 or 20 μmol/L. The association (ka) and dissociation (kd) rate constants, and the equilibrium dissociation constant (KD=kd/ka) were determined by analyzing the sensorgram curves associated with the various concentrations of BF211 using the BIA evaluation software version 3.1 (Biacore Life Sciences, USA) and the 1:1 Langmuir binding fitting model.
RT-PCR analysis of proteasome subunit mRNA expression
The mRNA levels of PSMB6 (encoding the β1 subunit), PSMB7 (encoding the β2 subunit) and PSMB5 (encoding the β5 subunit) were evaluated using RT-PCR analysis as described above. The oligonucleotide primers used to amplify PSMB6, PSMB7, and PSMB5 are listed in Supplemental Table S1.
Western blot analysis of proteasome subunit protein levels
The protein levels of proteasome subunits were evaluated using Western blot assays as described above. Rabbit polyclonal primary antibodies against the proteasome β1 subunit (#13267) (1:600), the β2 subunit (#13207) (1:1000), the β5 subunit (#12919) 1:1000) and GAPDH (#2118) (1:1500) were used. HRP-linked goat anti-rabbit IgG (#7074) (1:1000) was used as the secondary antibody. All of the antibodies were obtained from Cell Signaling Technology.
Native PAGE analysis of assembly of the 26S proteasome
Native PAGE was conducted as described in a previous report19. Briefly, the cells were harvested and lysed in ice-cold lysate buffer [0.2% (v/v) NP-40, 25 mmol/L Tris-HCl (pH 7.5), 5 mmol/L MgCl2, 1 mmol/L DTT and 2 mmol/L ATP] for 30 min. The lysate was combined with one-fifth the volume of 6×Native Sample Buffer (Shanghai Harmonious One Biotechnology), and the mixture was separated using 4%–12% gradient Tris-Gly gels (Shanghai Hegao Biotechnology). The separated proteins were subsequently transferred to a polyvinylidene difluoride membrane and blotted with the rabbit polyclonal antibody against the proteasome β5 subunit (1:1000) (#12919, Cell Signaling Technology).
siRNA knockdown of the proteasome β1 subunit
The knockdown of proteasome β1 subunit was achieved by transfecting the cells with siRNA targeting PSMB6 (Sigma-Aldrich, St Louis, MO, USA) using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA). Scrambled negative control siRNAs (Sigma-Aldrich, St Louis, MO, USA) were used as the negative controls. To confirm the efficiency of the siRNA transfection, the levels of PSMB6 mRNA and the proteasome β1 subunit protein in siRNA-treated cells were evaluated using RT-PCR and Western blot, respectively. Proteasome assembly and activity in siRNA-treated cells were evaluated as described above.
Statistical analysis
Student's t-test was used to evaluate the differences between the treated and the control groups. The data are expressed as the mean±SEM, and results from a minimum of three independent experiments were used for the statistical analysis. Asterisks indicate a significant difference (P<0.05) compared with the untreated control.
Results
BF211 and BF inhibited proliferation on A549 cells
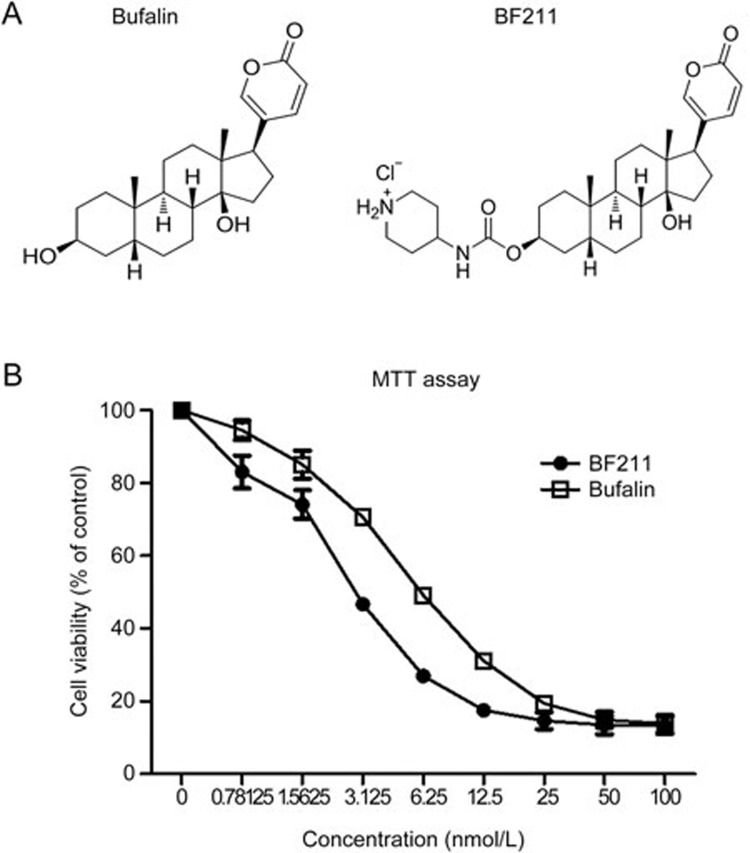
The chemical structures of BF and BF211 are presented in Figure 1A, and the effects of BF and BF211 on the proliferation in A549 cells are presented in Figure 1B. As shown in Figure 1B, both BF and BF211 inhibited cell proliferation in a dose-dependent manner. The IC50 values of BF and BF211 were 8.15±0.69 nmol/L and 3.72±0.62 nmol/L, respectively.
Figure 1.
The structure and anti-proliferative effects of BF211. (A) Structures of BF and BF211. (B) Viability of A549 cells after 72 h of exposure to various concentrations of BF or BF211. Data are represented as the mean±SEM. n=3 independent experiments.
Putative BF211-associated proteins and signaling networks
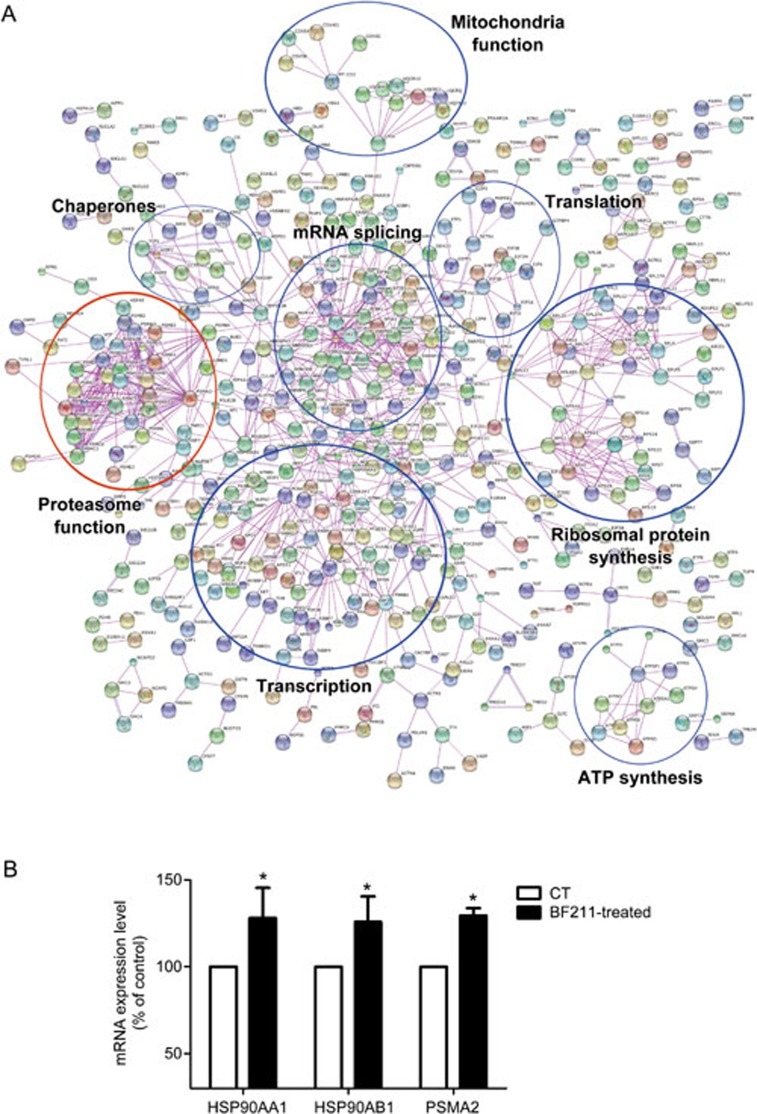
A total of 2453 proteins were identified in the initial proteomic analysis. Among these, 1282 proteins were differently expressed in BF211-treated cells compared with control cells. The differentially expressed proteins are summarized in Supplemental Table S2. As shown in Figure 2A, a potential BF211 signaling network was predicted based on the differentially expressed proteins identified in the proteomic analysis. In addition, we analyzed the effects of BF211 on the expression of HSP90AA1, HSP90AB1 and PSMA2, which were identified as up-regulated proteins. As shown in Figure 2B, the mRNA expression levels of HSP90AA1, HSP90AB1 and PSMA2 increased in BF211-treated cells compared with control cells. These results were in accordance with the results of the proteomic analysis. As shown in Figure 2A, the findings from the proteomic analysis suggested that BF211 is associated with critical cellular functions, including transcription, translation, mRNA splicing, ribosomal protein synthesis, and proteasome function. As the influence of cardiac steroids on proteasome function has not been systematically studied, we analyzed the effects of BF211 on proteasome activity and investigated the mechanisms underlying these effects.
Figure 2.
(A) The BF211 signaling network predicted according to the proteins identified in the proteomic analysis. (B) The expression levels of HSP90AA1, HSP90AB1 and PSMA2 in control cells and cells treated with 10 nmol/L BF211 for 3 h. Data are presented as the mean±SEM. n=3 independent experiments. *P<0.05 vs the solvent control.
BF211 inhibited cellular proteasome activity and induced the accumulation of ubiquitinated proteins
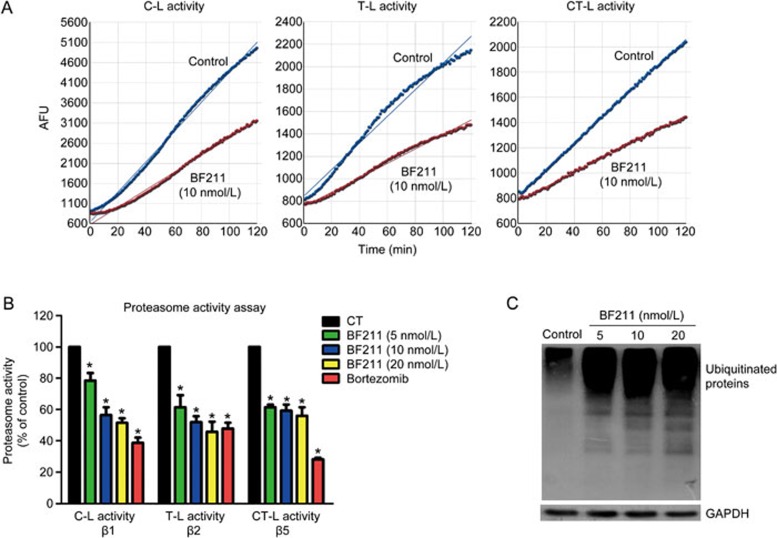
Figure 3A presents representative results of the experiments evaluating proteasome enzymatic activities. The fluorescence was measured at 1 min intervals. The data were plotted as arbitrary fluorescence units (AFU) versus time, and the slope of the curve (the relative velocity of the reaction), which represents enzymatic activity, was calculated. Enzymatic activity in the BF211-treated group is presented as the percentage (%) of enzymatic activity in the control group (Figure 3B). As shown in Figure 3B, BF211 significantly inhibited the three cellular proteasome activities, C-L, T-L and CT-L activity, mediated by the proteasome β1, β2 and β5 subunits, respectively. In addition, Western blot assays demonstrated that ubiquitinated proteins, resulting from the inhibition of proteasome function, accumulated in A549 cells treated with BF211 (Figure 3C). Furthermore, similar effects on the level of ubiquitinated proteins in other cell lines were observed. As shown in Supplemental Figure 1, BF211 induced the accumulation of ubiquitinated proteins in PC-3 and HeLa cells. These results suggested that BF211 inhibits cellular proteasome activities.
Figure 3.
BF211 inhibited proteasome activity in A549 cells. (A) Representative results of C-L, T-L and CT-L activity in cell lysates. (B) Cellular proteasome activities in A549 cells treated with 0.1% DMSO (negative control), bortezomib (positive control) or various concentrations of BF211 for 24 h. Data are presented as the mean±SEM. n=3 independent experiments. *P<0.05 vs the solvent control. (C) Western blot analysis of ubiquitinated proteins in BF211-treated cells and control cells. The levels of ubiquitinated proteins increased in BF211-treated cells in a dose-dependent manner.
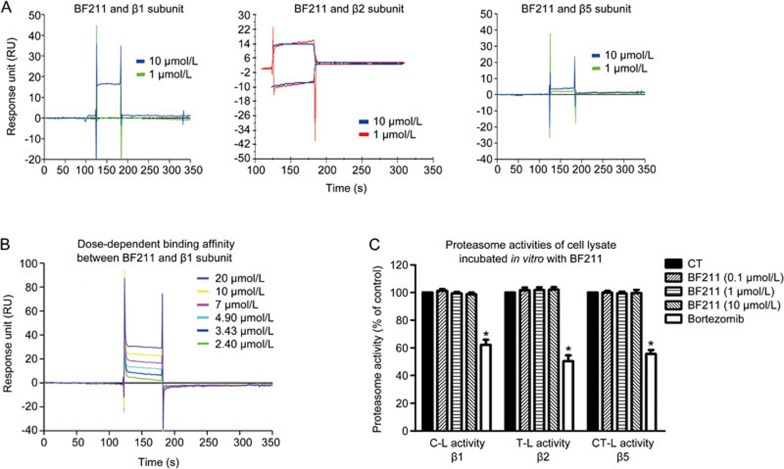
BF211 exhibited weak binding affinity to the recombinant proteasome β1 subunit
The results of the preliminary experiment examining the binding affinity of BF211 to the recombinant proteins are shown in Figure 4A. The ability of BF211 to bind the recombinant proteins was represented by the response unit (RU) values directly recorded by a Biacore 3000 instrument. As shown in Figure 4A, BF211 exhibited no binding affinity to the β2 or β5 subunit and exhibited weak binding affinity to the β1 subunit. The result of the dose-dependent analysis of the binding affinity between BF211 and the β1 subunit is shown in Figure 4B. The association (ka), dissociation (kd), and equilibrium dissociation (KD) constants of BF211 binding to the immobilized β1 subunit protein were 1.32×104 [(mol/L)−1·s−1], 0.23 [s−1] and 1.74×10−5 [mol/L], respectively. The Chi2 value was 11.7. These results indicated that the binding affinity between BF211 and the recombinant β1 subunit was weak.
Figure 4.
Binding affinity between BF211 and the proteasome β subunits. (A) Results of a preliminary experiment examining the binding affinity of BF211 (1 and 10 μmol/L) to the recombinant proteasome β1, β2 and β5 subunit proteins. (B) Real-time binding affinity measurements of 2.40, 3.43, 4.9, 7, 10 and 20 μmol/L BF211 with the recombinant proteasome β1 subunit protein. (C) Proteasome activities in cell lysate incubated in vitro with various concentrations of BF211 or bortezomib (positive control). Data are presented as the mean±SEM. n=3 independent experiments. *P<0.05 vs the solvent control.
BF211 did not inhibit proteasome activities in cell lysates in vitro
To determine whether BF211 can directly inhibit proteasome activity, we analyzed enzymatic activities in cell lysates from untreated cells incubated with BF211 in vitro. As shown in Figure 4C, bortezomib, a known proteasome inhibitor that we used as a positive control, directly inhibited proteasome activities in cell lysates from untreated cells. By contrast, BF211 did not inhibit proteasome activities in cell lysates. These results suggested that BF211 cannot directly inhibit proteasome activities.
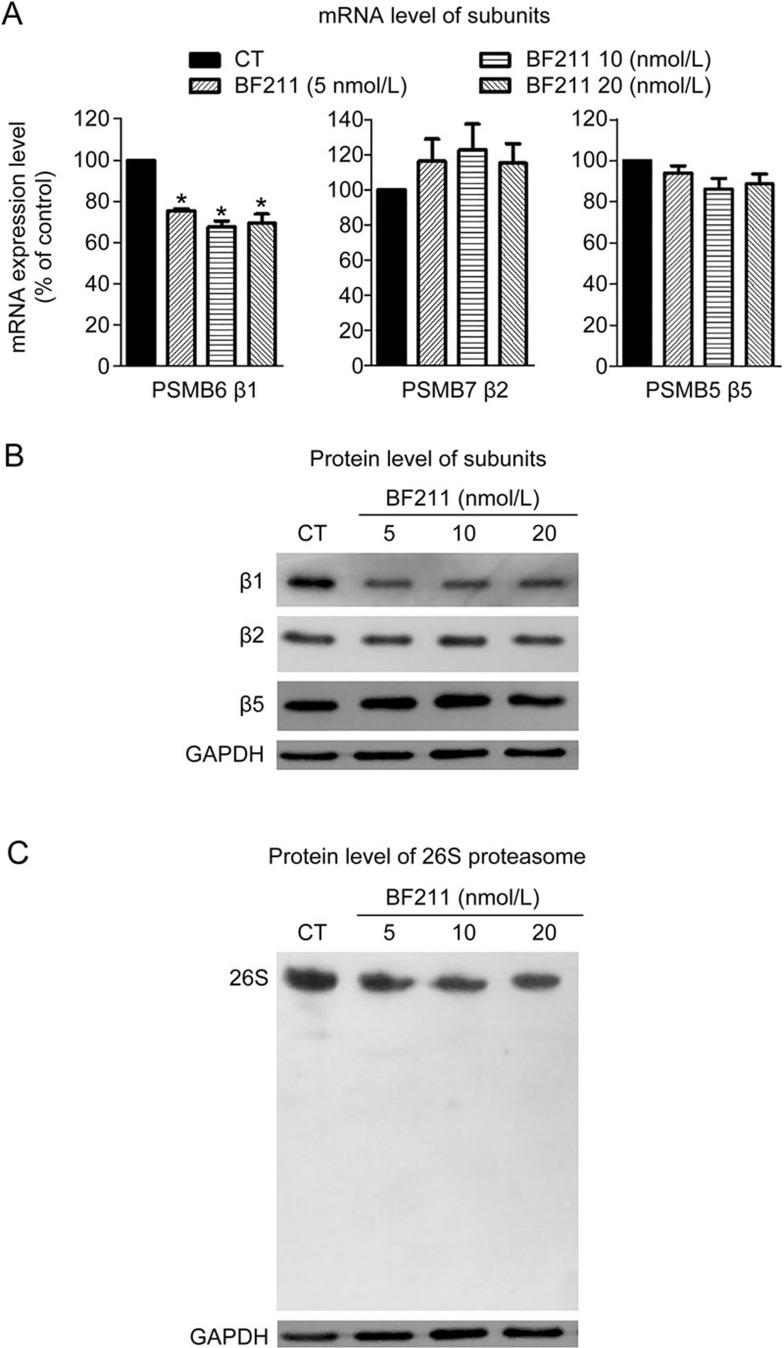
BF211 down-regulated the proteasome β1 subunit
The mRNA level of PSMB6, the gene encoding the proteasome β1 subunit, significantly decreased in cells treated with BF211 compared with the control cells (Figure 5A). By contrast, the mRNA levels of PSMB7 and PSMB5, the genes encoding the β2 and β5 subunit, respectively, were not significantly changed by BF211 (Figure 5A). Consistent with these findings, Western blot assays demonstrated that protein levels of the β1 subunit decreased in BF or BF211-treated cells.
Figure 5.
BF211 down-regulated the expression of the proteasome β1 subunit and inhibited proteasome assembly. (A) Results of the RT-PCR analysis of PSMB6, PSMB7 and PSMB5 mRNA expression levels in BF211-treated cells and control cells. Data are presented as the mean±SEM. n=3 independent experiments. *P<0.05 vs the solvent control. (B) Results of Western blot analysis of β1, β2 and β5 proteasome subunit protein levels in BF211-treated cells and control cells. (C) Results of the native PAGE analysis of integral 26S proteasome levels in BF211-treated cells and control cells.
BF211 inhibited the assembly of the 26S proteasome
As shown in Figure 5C, levels of the integral 26S proteasome decreased in cells treated with BF211. These results indicated that BF211 might inhibit the assembly of the 26S proteasome, which contains the sites of the three distinct hydrolytic activities20.
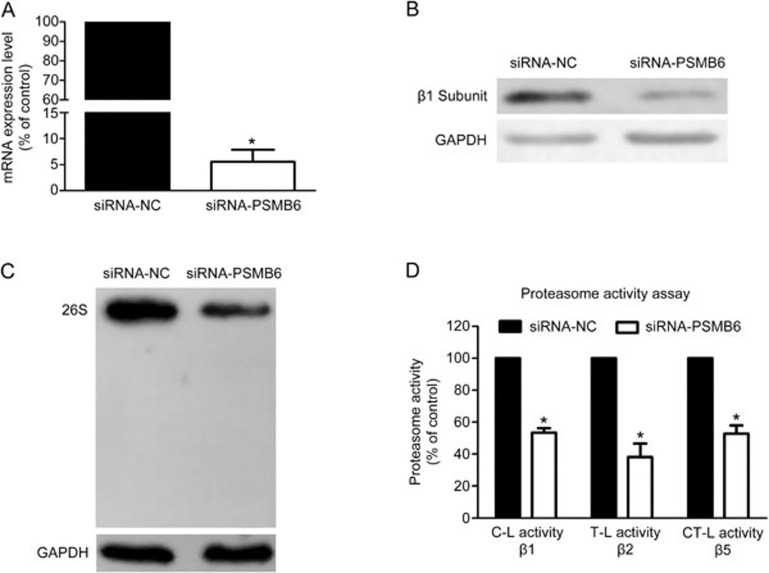
PSMB6 depletion disrupted the assembly of the 26S proteasome and activities of the proteasome
To determine whether the proteasome β1 subunit (PSMB6) is required for the assembly and activities of the proteasome, levels of the 26S proteasome and proteasome activities were evaluated in PSMB6 knockdown cells. As shown in Figure 6A and Figure 6B, transfection of siRNAs against PSMB6 efficiently reduced the mRNA and protein levels of the β1 subunit compared with the negative control. Furthermore, knockdown of the proteasome β1 subunit disrupted the assembly of the 26S proteasome and inhibited the three protease activities evaluated.
Figure 6.
Knockdown of the proteasome β1 subunit inhibited the assembly of the 26S proteasome and cellular proteasome activities. (A) Results of the RT-PCR analysis of PSMB6 mRNA levels in negative control cells and PSMB6-knockdown cells. Data are presented as the mean±SEM. n=3 independent experiments. *P<0.05 vs the negative control. (B) Results of the Western blot analysis of proteasome β1 subunit protein levels in negative control cells and PSMB6-knockdown cells. (C) Results of native PAGE analysis of the integral 26S proteasome in the negative control cells and PSMB6-knockdown cells. (D) Results of the enzymatic activity assays in negative control cells and PSMB6-knockdown cells. Data are presented as the mean±SEM. n=3 independent experiments. *P<0.05 vs the negative control.
Discussion
BF211, a derivative of BF, belongs to the family of bufadienolides. In the past two decades, the anti-cancer effects of cardiac steroids, including bufadienolides and cardenolides, have been a major focus of attention in anti-cancer drug research21,22. However, the complex signaling cascades activated by cardiac steroids in cancer cells have not been fully clarified. In the present study, we searched for target-related proteins of BF211 in A549 cells to predict the mechanism underlying the effects of BF211. Our findings indicated that BF211 might influence multiple cellular functions, including transcription, translation and protein synthesis. Our results were consistent with previous reports demonstrating that bufalin influences transcription, translation and protein synthesis in cancer cells7,8,9,10. Interestingly, we found that BF211 disrupted proteasome function in the present study. There are few reports describing the effect of cardiac steroids on the ubiquitin-proteasome system. In one genomic study, the gene encoding the ubiquitin-conjugating enzyme E2M was differentially expressed in HL-60 cells treated with bufalin23. In addition, a proteomic study demonstrated that the expression of the proteasome subunit 5 was regulated by digoxin in endothelial cells24. The influence of ouabain on AMPA receptor levels suggested that ouabain influenced proteasome-mediated proteolysis of AMPA receptor25. Furthermore, bufalin was shown to reduce SRC-3 levels by promoting proteasome-mediated SRC-3 degradation26. Protein degradation by the ubiquitin-proteasome system includes two distinct steps: protein ubiquitination and proteasome-mediated degradation of ubiquitinated proteins. The effects of cardiac steroids on the ubiquitination of different proteins might vary due to the specificity of ubiquitinating and deubiquitinating enzymes for different proteins27,28. To date, the effects of cardiac steroids on the enzymatic activities have not been systematically evaluated. Therefore, we investigated the influence of BF211 on proteasome activities in the present study.
We demonstrated that BF211 inhibited the 3 types of protease activities of the cellular proteasome. The 3 protease activities, C-L, T-L and CT-L activity, are mediated by the three proteolytically active subunits, the β1, β2 and β5 subunits, respectively29. In general, proteasome activities are inhibited by direct binding interactions between putative proteasome inhibitors and the active proteasome β subunits. For example, bortezomib, the positive control compound used in the present study, directly binds the proteasome β5 subunit and inhibits CT-L activity and other activities of the proteasome30. As cardiac steroids have been shown to bind to intracellular targets31, the possibility that BF211 directly binds proteasome subunits cannot be excluded. However, BF211 exhibited no binding affinity to the β2 or β5 subunits and only weakly bound to the β1 subunit. Furthermore, in vitro incubation of BF211 with cell lysates did not inhibit activities of the proteasome. In our preliminary experiments, BF211 also did not inhibit activities of purified cellular proteasomes (data not shown). These results suggest that the inhibitory effects of BF211 on cellular proteasome activities in cultured cells might be indirect rather than due to the direct binding of BF211 to proteasome subunits.
Interestingly, BF211 treatment significantly decreased the mRNA and protein levels of the proteasome β1 subunit. The 26S proteasome is a 2.6 MDa protein complex that consists of two subcomplexes: the 19S regulatory particle and the 20S catalytic core particle. The 20S catalytic core particle comprises two outer α rings and two inner β rings (αββα). The four heptameric stacked rings form a conserved, cylindrical structure. Each of the α and β rings consists of seven subunits, referred to as α1–α7 and β1–β7, respectively. The α subunits maintain the structure, whereas the core β rings contains the three proteolytically active β subunits32. Notably, previous studies reported that the active proteasome β subunits exist in an immature precursor form before being assembled into the integral 26S proteasome33,34,35,36,37. The β subunits mature once the proteasome assembles38,39. The propeptides of β1, β2 and β5 subunits contribute to the efficient assembly and maturation of the proteasome. Deletion of the β1 subunit disrupts β5 processing, and the simultaneous deletion of the β1 and β2 propeptides reduced proteasome levels and caused a pronounced defect in yeast growth40. Therefore, decreasing the expression levels of the β1 subunit might disrupt proteasome assembly. In this study, the results of native PAGE analysis confirmed that BF211 decreased the levels of the integral 26S proteasome in cells, and this decrease might contribute to the inhibitory effects of BF211 on the 3 protease activities of the cellular proteasome. The observation that the knockdown of the β1 subunit in A549 cells reduced 26S proteasome levels and inhibited activities of the proteasome confirmed that the proteasome β1 subunit is critical to the assembly of the integral 26S proteasome.
In summary, we demonstrated that the bufalin derivative BF211 inhibited cellular proteasome activities in A549 cells and that this effect might be mediated by a decrease in β1 subunit expression levels and a disruption of the assembly of the 26S proteasome. Notably, the inhibitory effects of BF211 on proteasome activity were only moderate. The BF211 signaling network predicted by the proteomic analysis indicated that BF211 participates in multiple functions, including transcription, translation and protein synthesis, processes that play important roles in the survival and proliferation of cancer cells. However, there is still no direct evidence that the anti-proliferative effects of BF211 are mediated by proteasome inhibition, and it is possible that the contribution of proteasome inhibition to the cytotoxicity of BF211 is minimal. Together, our finding regarding the effects of BF211 on the proteasome sheds new light onto the functional role of BF211 and cardiac steroids.
Author contribution
Peng SUN conducted the experiments evaluating BF211 cytotoxicity and proteasome function, analyzed the data and wrote a draft of the manuscript. Li-xing FENG generated the recombinant proteins and conducted the Biacore assay. Dong-mei ZHANG conducted the proteomic analysis, and Miao LIU conducted the bioinformatics analysis. Wang LIU and Tian MI conducted the RT-PCR analysis and Western blot assays. Wan-ying WU, Bao-hong JIANG, and Min YANG supervised the experiments and analyzed the data. Li-hong HU synthesized BF211 and revised the manuscript. De-an GUO initiated the research project and revised the manuscript. Xuan LIU designed the experiments and revised the manuscript.
Acknowledgments
This work was supported in part by Shanghai Science & Technology Support Program (No 13431900401), Shanghai Science & Technology Innovation Action program (No 15140904800), the National Natural Science Foundation of China (No 81373964), and the National Science & Technology Major Project of China (No 2014ZX09301-306-03).
Footnotes
Supplementary information is available at the website of Acta Pharmacologica Sinica.
Supplementary Information
BF211 induce accumulation of ubiquitinated proteins in PC-3 cells and HeLa cells.
Primers used in RT-PCR assay of mRNA levels of HSP90AA1, HSP90AB1, PSMA2, PSMB6, PSMB7, PSMB5 and GAPDH.
List of proteins differentially expressed in BF211-treated cells compared with control cells.
References
- Zhao L, Liu S, Che X, Hou K, Ma Y, Li C, et al. Bufalin inhibits TGF-beta-induced epithelial-to-mesenchymal transition and migration in human lung cancer A549 cells by downregulating TGF-beta receptors. Int J Mol Med 2015; 36: 645–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Zhao MN, Liu TY, Wu XS, Weng H, Ding Q, et al. Bufalin induces cell cycle arrest and apoptosis in gallbladder carcinoma cells. Tumour Biol 2014; 35: 10931–41. [DOI] [PubMed] [Google Scholar]
- Li C, Hashimi SM, Cao S, Qi J, Good D, Duan W, et al. Chansu inhibits the expression of cortactin in colon cancer cell lines in vitro and in vivo. BMC Complementary Alter Med 2015; 15: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Li A, Inagaki Y, Kokudo N, Tamura S, Nakata M, et al. Antitumor activity of extracts and compounds from the skin of the toad Bufo bufo gargarizans Cantor. Int Immunopharmacol 2011; 11: 342–9. [DOI] [PubMed] [Google Scholar]
- Meng Z, Yang P, Shen Y, Bei W, Zhang Y, Ge Y, et al. Pilot study of huachansu in patients with hepatocellular carcinoma, nonsmall-cell lung cancer, or pancreatic cancer. Cancer 2009; 115: 5309–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Xiao Z, Ma B, Chen Y, Liu M, Liu J, et al. Synthesis and biological evaluation of bufalin-3-yl nitrogen-containing-carbamate derivatives as anticancer agents. Steroids 2016; 108: 56–60. [DOI] [PubMed] [Google Scholar]
- Wu SH, Wu TY, Hsiao YT, Lin JH, Hsu SC, Hsia TC, et al. Bufalin induces cell death in human lung cancer cells through disruption of DNA damage response pathways. Am J China Med 2014; 42: 729–42. [DOI] [PubMed] [Google Scholar]
- Wu SH, Hsiao YT, Chen JC, Lin JH, Hsu SC, Hsia TC, et al. Bufalin alters gene expressions associated DNA damage, cell cycle, and apoptosis in human lung cancer NCI-H460 cells in vitro. Molecules 2014; 19: 6047–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin PH, Liu X, Qiu YY, Cai JF, Qin JM, Zhu HR, et al. Anti-tumor activity and apoptosis-regulation mechanisms of bufalin in various cancers: new hope for cancer patients. Asian Pac J Cancer Prev 2012; 13: 5339–43. [DOI] [PubMed] [Google Scholar]
- Sun L, Chen T, Wang X, Chen Y, Wei X. Bufalin induces reactive oxygen species dependent bax translocation and apoptosis in astc-a-1 cells. Evid Based Complement Alternat Med 2011; 2011: 249090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Mu L, Jin H, Zhang P, Wang Y, Ma X, et al. The effects of bufadienolides on HER2 overexpressing breast cancer cells. Tumor Biol 2016; 37: 7155–63. [DOI] [PubMed] [Google Scholar]
- Kai S, Lu JH, Hui PP, Zhao H. Pre-clinical evaluation of cinobufotalin as a potential anti-lung cancer agent. Biochem Biophys Res Commun 2014; 452: 768–74. [DOI] [PubMed] [Google Scholar]
- Feng L, Zhang D, Fan C, Ma C, Yang W, Meng Y, et al. ER stress-mediated apoptosis induced by celastrol in cancer cells and important role of glycogen synthase kinase-3beta in the signal network. Cell Death Disease 2013; 4: e715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Angelopoulos N, Xu Y, Grothey A, Nunes J, Stebbing J, et al. Proteomic profile of KSR1-regulated signalling in response to genotoxic agents in breast cancer. Breast Cancer Res Treatment 2015; 151: 555–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounap K, Leetsi L, Matsoo M, Kurg R. The stability of ribosome biogenesis factor WBSCR22 is regulated by interaction with TRMT112 via ubiquitin-proteasome pathway. PLoS One 2015; 10: e0133841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack TA, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, Plamondon L, et al. Kinetic studies of the branched chain amino acid preferring peptidase activity of the 20S proteasome: development of a continuous assay and inhibition by tripeptide aldehydes and clasto-lactacystin beta-lactone. Biochemistry 1998; 37: 7792–800. [DOI] [PubMed] [Google Scholar]
- Stein RL, Melandri F, Dick L. Kinetic characterization of the chymotryptic activity of the 20S proteasome. Biochemistry 1996; 35: 3899–908. [DOI] [PubMed] [Google Scholar]
- Yue QX, Cao ZW, Guan SH, Liu XH, Tao L, Wu WY, et al. Proteomics characterization of the cytotoxicity mechanism of ganoderic acid D and computer-automated estimation of the possible drug target network. Mol Cell Proteomics 2008; 7: 949–61. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Murata S. Using siRNA techniques to dissect proteasome assembly pathways in mammalian cells. Methods Mol Biol 2012; 832: 433–42. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fruh K, Ahn K, Peterson PA. In vivo assembly of the proteasomal complexes, implications for antigen processing. J Biol Chem 1995; 270: 27687–94. [DOI] [PubMed] [Google Scholar]
- Alevizopoulos K, Calogeropoulou T, Lang F, Stournaras C. Na+/K+ ATPase inhibitors in cancer. Curr Drug Targets 2014; 15: 988–1000. [PubMed] [Google Scholar]
- Calderon-Montano JM, Burgos-Moron E, Orta ML, Maldonado-Navas D, Garcia-Dominguez I, Lopez-Lazaro M. Evaluating the cancer therapeutic potential of cardiac glycosides. BioMed Res Int 2014; 2014: 794930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Yu J, Zhang L, Sun Y, Zhang Y, Guo H, et al. Microarray and biochemical analysis of bufalin-induced apoptosis of HL-60 cells. Biotechnol Lett 2009; 31: 487–94. [DOI] [PubMed] [Google Scholar]
- Qiu J, Gao HQ, Liang Y, Yu H, Zhou RH. Comparative proteomics analysis reveals role of heat shock protein 60 in digoxin-induced toxicity in human endothelial cells. Biochim Biophys Acta 2008; 1784: 1857–64. [DOI] [PubMed] [Google Scholar]
- Zhang D, Hou Q, Wang M, Lin A, Jarzylo L, Navis A, et al. Na,K-ATPase activity regulates AMPA receptor turnover through proteasome-mediated proteolysis. J Neurosci 2009; 29: 4498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lonard DM, Yu Y, Chow DC, Palzkill TG, Wang J, et al. Bufalin is a potent small-molecule inhibitor of the steroid receptor coactivators SRC-3 and SRC-1. Cancer Res 2014; 74: 1506–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletr ZM, Wilkinson KD. Regulation of proteolysis by human deubiquitinating enzymes. Biochim Biophys Acta 2014; 1843: 114–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans 2009; 37: 937–53. [DOI] [PubMed] [Google Scholar]
- Vangala JR, Dudem S, Jain N, Kalivendi SV. Regulation of PSMB5 protein and beta subunits of mammalian proteasome by constitutively activated signal transducer and activator of transcription 3 (STAT3): potential role in bortezomib-mediated anticancer therapy. J Biol Chem 2014; 289: 12612–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, et al. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res 1999; 59: 2615–22. [PubMed] [Google Scholar]
- Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgamma activity. Nature 2011; 472: 486–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S proteasome complex during oxidative stress. Sci Signaling 2010; 3: ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HM, Yu Y, Cheng Y. Structure characterization of the 26S proteasome. Biochim Biophys Acta 2011; 1809: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L, Paine S, Sheppard PW, Mayer RJ, Roelofs J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol 2010; 20: 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Ann Rev Biochem 1996; 65: 801–47. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Kaneko T, Okamoto K, Bai M, Yashiroda H, Furuyama K, et al. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J 2008; 27: 2204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y. Structure, assembly and homeostatic regulation of the 26S proteasome. J Mol Cell Biol 2010; 2: 308–17. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol 2014; 15: 122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz P, Unverdorben P, Beck F, Pfeifer G, Schweitzer A, Forster F, et al. Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci U S A 2013; 110: 7264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjappu MJ, Hochstrasser M. Assembly of the 20S proteasome. Biochim Biophys Acta 2014; 1843: 2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BF211 induce accumulation of ubiquitinated proteins in PC-3 cells and HeLa cells.
Primers used in RT-PCR assay of mRNA levels of HSP90AA1, HSP90AB1, PSMA2, PSMB6, PSMB7, PSMB5 and GAPDH.
List of proteins differentially expressed in BF211-treated cells compared with control cells.