Abstract
Non-classical human leucocyte antigen (HLA)-G class I molecules have an important role in tumor immune escape mechanisms. We investigated HLA-G expression in lymphonode biopsies taken from 8 controls and 20 patients with advanced-stage classical Hodgkin lymphoma (cHL), in relationship to clinical outcomes and the HLA-G 14-basepair (14-bp) deletion-insertion (del-ins) polymorphism. Lymphnode tissue sections were stained using a specific murine monoclonal HLA-G antibody. HLA-G protein expression was higher in cHL patients than controls. In the group of PET-2 positive (positron emission tomography carried out after 2 cycles of standard chemotherapy) patients with a 2-year progression-free survival rate (PFS) of 40%, we observed high HLA-G protein expression within the tumor microenvironment with low expression on Hodgkin and Reed-Sternberg (HRS) cells. Conversely, PET-2 negative patients with a PFS of 86% had higher HLA-G protein expression levels on HRS cells compared to the microenvironment. Lower expression on HRS cells was significantly associated with the HLA-G 14-bp ins/ins genotype.
These preliminary data suggest that the immunohistochemical pattern of HLA-G protein expression may represent a useful tool for a tailored therapy in patients with cHL, based on the modulation of HLA-G expression in relation to achievement of negative PET-2.
Introduction
Although cure rates in classical Hodgkin lymphoma (cHL) range from 70 to 90%, a significant proportion of patients fail to respond to standard courses of chemotherapy and need to be treated with intensified regimens that carry an increased risk of long-term toxicities and secondary cancer.1 It is of utmost importance to identify immune-biomarkers able to predict poor responders patients to conventional therapy that require intensification regimens. cHL offers an interesting study model for the identification of immunologic and immunogenetic factors that may confer susceptibility to tumor or influence response to treatment.2 The peculiar architecture of malignant lymphogranuloma is characterized by the presence of few neoplastic Hodgkin and Reed-Sternberg (HRS) cells growing within a microenvironment rich in immune system cells incapable of mounting an effective antitumor response.3,4
The aim of our study was to explore the role of non-classical human leucocyte antigen (HLA)-G class I molecules in tumor immune escape mechanisms. These molecules are encoded by a gene located on chromosome 6p21.3 of the major histocompatibility complex (MHC).5 Alternative splicing can generate 7 different isoforms: 4 membrane isoforms (G1-G4) and 3 soluble (s) isoforms (G5-G7). HLA-G molecules are tolerogenic molecules expressed in a restricted number of healthy tissues. They exert their activity through interaction with the human inhibitory receptors immunoglobulin (Ig)-like transcript 2 (ILT2) and ILT4 expressed on natural killer (NK) cells, T and B lymphocytes, dendritic cells and neutrophils.5
The role of HLA-G molecules in tumor-escape have been described in several tumor tissues and has been associated to cancer progression and an unfavorable outcome or prognosis.6-8 In hematologic malignancies, enhanced sHLA-G plasma levels, have been found in B-cell malignancies, such as multiple myeloma (MM), non-Hodgkin’s B-lymphoma (NHL-B) and B-cell chronic lymphatic leukemia (B-CLL).9,10 Different to what has been observed in solid tumors, the derived B and T cells hematologic malignancies express receptors recognized by HLA-G molecules. Hence, the role played by HLA-G in oncohematologic diseases is apparently more complex. Some authors have shown that HLA-G inhibits the proliferation of human B-cell lymphoma, myelomas and B-cell leukemia through binding with ILT2 receptors.11 A correlation between HLA-G expression, tumor onset and clinical outcome has also been investigated in MM, NHL-B and B-CLL.9,10,12,13 HRS cells are ascribed to the B cell lineage and it is possible that these cells express inhibitory receptors capable of interacting with HLA-G molecules as well as cells from the surrounding microenvironment. To the best of our knowledge, only one study has addressed HLA-G expression in cHL, but with no data on clinical outcomes.14 In this study we investigated the expression of HLA-G in lymphonode biopsies from patients diagnosed with advanced-stage cHL, using a specific murine monoclonal antibody. Moreover, we evaluated the impact of HLA-G expression on the tumor microenvironment and HRS cells in patients who achieved negative results for [18F]-fluoro-2-deoxy-d-glucose positron emission tomography carried out after 2 cycles of standard chemotherapy (PET-2). PET-2 is currently the most powerful predictor of treatment outcome in advanced-stage cHL patients.15 Finally, we evaluated HLA-G expression in relation to HLA-G allelic variants characterized by a 14-basepair (14-bp) deletion-insertion polymorphism located in exon 8 of the 3’-untranslated (UT) region of HLA-G, that has been reported associated to different levels of sHLA-G both in normal and pathological conditions.16-18
Materials and Methods
Patients, controls and treatment protocols
Twenty patients with advanced-stage cHL were recruited for the study. All patients were enrolled in the HD607 multicenter clinical trial investigating early treatment intensification in patients with high-risk cHL, identified by positive PET-2 scan after two conventional chemotherapy courses. The study was performed in accordance with the 1975 guidelines of the Declaration of Helsinki and written informed consent was obtained from all participating patients. A complete data set containing all the following parameters was collected at baseline and are reported in Table 1. The patients received 2 courses of standard ABVD (Doxorubicin, Bleomycin, Vinblastine, Dacarbazine) treatment before undergoing PET-2 to assess tumor chemosensitivity. Patients were stratified into 2 groups according to the results of PET-2. Patients with positive PET-2 were assigned to receive 4 courses of escalated chemotherapy, followed by auto or allotransplantation of hematopoietic stem cells (HSC). Patients with negative PET-2 scan continued with 4 more courses of standard treatment.
Table 1.
Characteristics of 20 patients with advanced-stage classical Hodgkin lymphoma.
| Patients (n=20) | ||
|---|---|---|
| Sex (n, %) | ||
| Male | 6 (30.0) | |
| Female | 14 (70.0) | |
| Age at diagnosis (mean, range) | 33 (18-61) | |
| Disease subtype (n, %) | ||
| Nodular sclerosing | 14 (70.0) | |
| Mixed-cellularity | 4 (12.5) | |
| Lymphocyte-rich | 2 (10.0) | |
| Lymphocyte-depleted | 0 (0.0) | |
| Albumin, g/dL, (mean, range) | 3.9 (2.51-4.68) | |
| Hemoglobin, g/dL, (mean, range) | 12.9 (8.7-14.8) | |
| Lymphocytes, x103/uL, (mean, range) | 1.70 (0.5-3.3) | |
| Neutrophils, x103/uL, (mean, range) | 8.6 (2.7-18.8) | |
| Bulky mass (n, %) | 12 (60.0) | |
| B symptoms (n, %) | 19 (95.0) | |
| Extranodal extension (n, %) | 9 (45.0) | |
| IPS score ≥4 | 2 (10.0) | |
| Ann Arbor | ||
| Stage 2b (n, %) | 9 (45.0) | |
| Stage 3 (n, %) | 7 (35.0) | |
| Stage 4 (n, %) | 4 (12.5) | |
Controls
Tissue sections from lymphonodes diagnosed as normal or reactive obtained from 8 deceased subjects undergoing standard immunologic characterization procedures for organ donation were also used for control.
HLA-G 14-basepair polymorphism
The presence of the 14-bp deletion/insertion polymorphism in exon 8 of the 3’UT region of HLA-G was determined as previously described.16 The patients were divided into three groups according to HLA-G 14-bp genotype: homozygotes for the 14-bp insertion (in/in), heterozygotes for the 14-bp deletion/insertion (del/in) and homozygotes for the 14-bp deletion (del/del). The 14-bp polymorphism of the HLA-G gene was analyzed using polymerase chain reaction (PCR).
Staining procedures
Tissue samples were fixed in 10% buffered formalin, routinely processed and embedded in paraffin. Serial sections 3 µm thick were obtained from each paraffin block. Tissue sections were then dewaxed, rehydrated and pretreated for immunohistochemical analysis with a 10-min heat-induced epitope retrieval in buffer pH 9.00 (EnVision™ FLEX Target Retrieval Solution High pH - Dako Denmark A/S, Glostrup, Denmark, Code K8004). Slides were then incubated for 20 minutes at room temperature with anti-human MHC Class I HLA-G (LifeSpan BioSciences, Inc., Seattle, WA, USA, Code LS-B3734) mouse monoclonal antibody clone 4H84 at 1:100 dilution. Staining procedures were performed with the Envision™ FLEX+ (Dako, Code K8002) Detection System and the AutostainerLink 48 instrument according to the manufacturer’s instructions. The immunohistochemical evaluation of HLA-G protein expression in stained lymph node biopsy specimens was performed by 3 independent pathologists according to the following scores: 0 points for negative staining (-), 1 for staining <25% of examined cells (- +), 2 between 26 and 50% (+), 3 between 51 and 75% (++), 4 between 76 and 100% (+++). The average agreement for the 3 assessed variables among the three pathologists (DF, GS, GF) was 80%, ranging from 70% for histiocytes, through 80% for lymphocytes, to 90% for HRS cells.
Statistical analysis
The probability of achieving progression free survival (PFS) was calculated using the Kaplan-Meier method. The log-rank test was used to compare 2 groups of patients (negative or positive PET). The magnitude of HLA-G staining in lymphocytes, histiocytes and HRS cells was compared between patients and controls. Pearson’s chi-squared test and a two-sided likelihood ratio test were applied. With P-values below 5%, the results were confirmed using the Fisher two-sided exact test at the same level. Variables with a P-value lower than 0.2 in univariate analysis were included in a multivariate analysis using a forward stepwise binary logistic regression model, where negative PET-2 scan was considered a dependent variable. Only P-values ≤0.05 were considered to be statistically significant.
Results
Characteristics of patients
The demographic and clinical characteristics of 20 cHL patients (6 males and 14 females, mean follow up 35 months, range 8-60) are shown in Table 1. The mean age at diagnosis was 33 years (range 18-61). The large majority of patients (70%) had a nodular sclerosing histological subtype. All patients belonged to advanced Ann Arbor stages (IIb-IV).
Characteristics of controls
Eight deceased subjects (4 females and 4 males) who underwent standard immunologic characterization procedures for organ donation, were used as controls. The median age was 32 years, ranging from 17 to 75. The lymphonodes obtained from each subject varied in number, from one to three, and in size, from 1 mm to 15 mm.
Progression-free survival following treatment and according to PET-2 outcome
In the cohort of 20 patients, 4-year PFS was 70.5%. PFS was significantly higher in patients with negative PET-2 compared to positive PET-2 (85.6% vs 40%, P=0.003) Of the five intensified patients: 4 underwent to autologous and 1 allogeneic HST. In univariate analysis, an HLA-G HRS cell staining score of >1 was independently associated with the probability of achieving a negative PET-2 result (P<0.01).
HLA-G expression in lymphonode sections from cHL patients and controls
Table 2 shows the expression of HLA-G in lymphonodes from 20 cHL patients and 8 controls. The median score of HLA-G expression in lymphocytes was 2 (26-50% of lymphocytes showed HLA-G+) in cHL patients compared to 1 (1-25% of lymphocytes showed HLA-G+) in controls. An HLA-G score of ≥2 was present in 13/20 patients in comparison to 1/8 donors (65% vs 12,5%, p= 0.033) (Figure 1). The median score of HLA-G expression in histiocytes was 1 (1-25% of histiocytes showed HLA-G+) both in patients and controls; an HLA-G score of ≥2 was found in 9/20 patients compared to 3/8 donors (45% vs 37.5%, P=NS). 11/20 patients (55%) showed HLA-G+ HRS cells. The median score for HRS cells HLA-G expression was 1 (1-25% of HRS cells were HLA-G+).
Table 2.
HLA-G staining in patients with advanced-stage classical Hodgkin lymphoma and controls. According to HLA-G diffusion, the following scores were assigned: 0 for negative staining (-), 1 for staining <25% (-+), 2 between 26 and 50% (+), 3 between 51 and 75% (++), 4 between 76 and 100% (+++).
| Patients | Disease subtype | Lymphocytes HLA-G + (%) | Lymphocytes HLA-G + (Score) | Histiocytes HLA-G + (%) | Histiocytes HLA-G + (Score) | HRS cell HLA-G + (%) | HRS cell HLA-G + (Score) |
|---|---|---|---|---|---|---|---|
| 1 | LR | +++ | 4 | ++ | 3 | +++ | 4 |
| 2 | NS | - | 0 | - | 0 | - | 0 |
| 3 | NS | - | 0 | - | 0 | + | 2 |
| 4 | NS | + | 2 | ++ | 3 | -+ | 1 |
| 5 | NS | - | 0 | -+ | 1 | -+ | 1 |
| 6 | NS | + | 2 | -+ | 1 | - | 0 |
| 7 | NS | -+ | 1 | -+ | 1 | -+ | 1 |
| 8 | NS | -+ | 1 | -+ | 1 | + | 2 |
| 9 | NS | ++ | 3 | -+ | 1 | + | 2 |
| 10 | NS | +++ | 4 | + | 2 | -+ | 1 |
| 11 | NS | -+ | 1 | -+ | 1 | - | 0 |
| 12 | NS | + | 2 | -+ | 1 | - | 0 |
| 13 | NS | + | 2 | + | 2 | - | 0 |
| 14 | MC | + | 2 | +++ | 4 | - | 0 |
| 15 | NS | + | 2 | + | 2 | - | 0 |
| 16 | MC | +++ | 4 | - | 0 | -+ | 1 |
| 17 | LR | ++ | 3 | -+ | 1 | -+ | 1 |
| 18 | NS | -+ | 1 | + | 2 | + | 2 |
| 19 | MC | ++ | 3 | + | 2 | - | 0 |
| 20 | NS | +++ | 4 | + | 2 | - | 0 |
| Median | 2 | 1 | 1 | ||||
| Controls | |||||||
| 1 | -+ | 1 | -+ | 1 | |||
| 2 | -+ | 1 | -+ | 1 | |||
| 3 | -+ | 1 | - | 0 | |||
| 4 | - | 0 | - | 0 | |||
| 5 | -+ | 1 | + | 2 | |||
| 6 | + | 2 | + | 2 | |||
| 7 | -+ | 1 | + | 2 | |||
| 8 | -+ | 1 | -+ | 1 | |||
Figure 1.
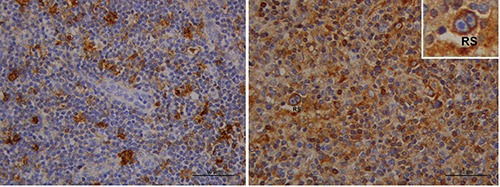
Immunohistochemical staining of HLA-G expression (brown color) in a control (left) and in an advanced-stage cHL patient (right) with a larger number of immunopositive reactive cells and a Reed-Sternberg cell (RS). Scale bars: 50 μm.
Relationship between HLA-G expression in lymphonode sections from cHL patients and PET-2 results
We found that 80% of patients with PET-2 positivity had an HLA-G staining score for lymphocytes of ≥2 compared to 60% of PET-2 negative patients; this difference did not reach statistical significance (P=0.08); conversely, none of the PET-2 positive patients showed HLA-G+ HRS cells with a staining score of ≥2, in comparison to 40% of PET-2 negative patients (P=0.09, not significant). Figure 2 shows HLA-G expression in 2 patients with different PET-2 outcomes.
Figure 2.
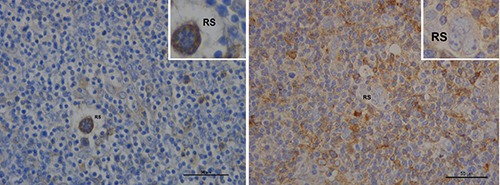
HLA-G expression (brown color) according to PET-2 results. Left: a 45 year-old female with negative PET-2. The patient shows low HLA-G protein expression in lymphocytes; conversely, Reed-Srernberg cells (RS) are stained. Right: HLA-G is more expressed in reactive cells then in Reed-Srernberg cells (RS) in a 37 year-old female with positive PET 2. Scale bars: 50 μm.
HLA-G expression in lymphonode sections from cHL patients according to the HLA-G 14-bp polymorphism
Patients homozygotes for the 14-bp deletion polymorphism (del/del) showed a trend, although not statistically significant, for higher HLA-G expression with a staining score of ≥2, compared to in/in patients (100% vs 50% of cases in lymphocytes; 75% vs 0% in histiocytes; 50% vs 25% in HRS cells). Fisher exact test showed a significant association between homozygosity in/in and lower expression of HLA-G on HRS cells (P=0.01). Figure 3 shows HLA-G expression in 2 patients homozygous for the insertion (in/in) and deletion (del/del) polymorphism, respectively.
Figure 3.
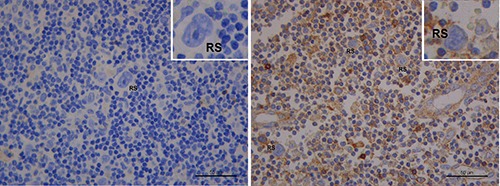
HLA-G expression (brown color) according to the HLA-G 14-bp polymorphism in cHL. Left: a 32 year-old female, homozygous for the 14-bp insertion (in/in). Right: a 46 year-old male, homozygous for the 14-bp deletion (del/del). HLA-G expression is higher in the del/del patient sample. RS, Reed-Sternberg cells. Scale bars: 50 μm.
Discussion
Our study, albeit limited by the relatively small number of patients, allows us to make some considerations. Firstly, HLA-G protein expression was higher in lymphonodes from cHL patients than controls. Moreover, HRS cells were immunoreactive for HLA-G in 55% of the patients, which is in agreement with previously published data.14 Conversely, our findings do not confirm the very low percentage of HLA-G staining previously observed for histiocytes and lymphocytes in the tumor microenvironment.14 Indeed, we found significantly higher staining scores (≥2) for HLA-G protein expression in lymphocytes of patients compared to the controls (65% vs 12,5%, P=0.033) (Table 2, Figure 1). This finding is coherent with the hypothesis that HLA-G molecule determines a tolerogenic status through down-regulation of immune response by cells of the tumor microenvironment, leading to survival of the neoplastic cell.
A second important finding emerging from our study was that different patterns of HLA-G protein expression in both the tumor microenvironment and HRS cells were associated with different clinical outcomes. In line with other studies,15 Patients with negative PET-2 had significantly higher PFS rates than patients with a positive PET-2 scan (85.6% vs 40%, P=0.003). In the group of PET-2 positive patients, we observed a higher HLA-G protein expression on lymphocytes and a lower expression on HRS cells (Figure 2). Conversely, PET-2 negative patients had higher HLA-G protein expression levels on HRS cells compared to the tumor microenvironment (Figure 2). Classical HL represents an interesting study model for the assessment of immunologic and immunogenetic factors that may confer susceptibility to tumors or facilitate tumor immune escape mechanisms.4,19-22 HLA-G molecules might have a key role in impaired NK cell function through interaction with ILT2 and KIR2DL4 receptors expressed on NK cell surface that inhibit their cytotoxicity, IFN- secretion and chemotaxis.20,23,24 However, since HRS cells pertain to the B-cell lineage, it is also possible that they express HLA-G inhibitory receptors, that would imply a direct anti-proliferative effect on the tumor. In-vitro data have shown that negative signaling mediated by HLA-G down-regulates proliferation of the tumor in human B-cell lymphomas, myelomas and B-cell leukemia.11 The different immunohistochemical expression of HLA-G protein observed in our study for HRS cells compared to the surrounding microenvironment may suggest a model similar to that hypothesized by Carosella et al.5 (Figure 4a). Higher HLA-G expression in the tumor microenvironment surrounding HRS cells could favor an immune-escape mechanism (Figure 4b), whereas higher HLA-G expression in HRS cells could offer better control of tumor proliferative activity
Figure 4.
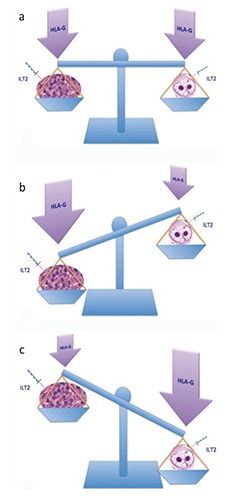
a) Possible dual role of HLA-G expression in HRS cells and the tumor microenvironment in classical Hodgkin lymphoma; adapted from: Rouas-Freiss et al., J Immunol Res 2014;2014:359748. b) HLA-G balancing model, promoting tumor immune-escape. c) HLA-G balancing model, promoting anti-tumor activity.
(Figure 4c). Hence, HLA-G could exert a dual role: i) tumor escape mediated by HLA-G-driven inhibitory mechanisms of antitumor response; and ii) tumor antiproliferative function. According with this hypothesis, the final effect on tumor progression might derive from the balance between these two mechanisms.23 Regarding the potential influence of the HLA-G 14-bp polymorphism on tissue expression, although not statistically significant, patients homozygous for the del/del polymorphism showed a trend to a higher expression of HLA-G protein both in the microenvironment and neoplastic cells as compared to patients with the in/in genotype (Figure 3). The del/del genotype was associated with more stable mRNA and higher HLA-G expression, reduced overall survival in CLL and diffuse large B-cell lymphomas (DLBCL).12,13,25,26 Our data revealed a significant association between the HLA-G in/in genotype and lower expression of HLA-G on HRS cells.
Finally, the present study is the first to provide evidence of different patterns of HLA-G protein expression in HRS cells and the surrounding tumor microenvironment of lymphonodes from cHL patients. In our opinion, our data suggest that the immunoreactive pattern for HLA-G in the lymphnodes of patients affected by cHL might represent a useful tool for therapeutic purposes. Eventhough survival of advanced-stage cHL patients has increased to 70-85%, the optimal balance of the risks and benefits of treatment is still a matter of debate. The main dilemma remains whether frontline intensive chemotherapy should be given to all patients or be restricted to patients who are poor responders to standard treatment approaches. For this reason, new predictive factors for prognosis and response to treatment in cHL are important goals. Within this contest, HLA-G expression may represent a potential biomarker able to predict a response to treatment and our results may enforce the rationale for developing innovative and targeted treatments in cHL patients, based on the modulation of HLA-G function.
Acknowledgments
We are deeply grateful to all the patients who participated in this study. We are also grateful to Anna Maria Koopmans for professional writing assistance.
References
- 1.Diefenbach C, Advani R. Customized targeted therapy in Hodgkin lymphoma: hype or hope? Hematol Oncol Clin North Am 2014;28:105-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott DW, Steidl C. The classical Hodgkin lymphoma tumor microenvironment: macrophages and gene expression-based modeling. ASH Education Program Book 2014;2014:144-50. [DOI] [PubMed] [Google Scholar]
- 3.Romano A, Vetro C, Caocci G, Greco M, Parrinello NL, Di Raimondo F, et al. Immunological deregulation in classic hodgkin lymphoma. Mediterr J Hematol Infect Dis 2014;6:e2014039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Berg A. Microenvironment, CrossTalk, and Immune Escape Mechanisms. Hodgkin Lymphoma. 2011. [Google Scholar]
- 5.Carosella ED, Rouas-Freiss N, Roux DT, Moreau P, LeMaoult J. HLA-G: An Immune Checkpoint Molecule. Adv Immunol 2015;127:33-144. [DOI] [PubMed] [Google Scholar]
- 6.Karagoz B, Haholu A, Ozgun A, Bilgi O, Tuncel T, Emirzeoglu L, et al. HLA-G in testicular germ cell tumors. Oncol Res Treat 2014;37:245-8. [DOI] [PubMed] [Google Scholar]
- 7.Lau DT, Norris MD, Marshall GM, Haber M, Ashton LJ. HLA-G polymorphisms, genetic susceptibility, and clinical outcome in childhood neuroblastoma. Tissue Antigens 2011;78:421-7. [DOI] [PubMed] [Google Scholar]
- 8.Yan W-H. HLA-G expression in cancers: potential role in diagnosis, prognosis and therapy. Endocr Metab Immune Disord Drug Targets 2011;11:76-89. [DOI] [PubMed] [Google Scholar]
- 9.Leleu X, Le Friec G, Facon T, Amiot L, Fauchet R, Hennache B, et al. Total soluble HLA class I and soluble HLA-G in multiple myeloma and monoclonal gammopathy of undetermined significance. Clin Cancer Res 2005;11:7297-303. [DOI] [PubMed] [Google Scholar]
- 10.Sebti Y, Le Maux A, Gros F, De Guibert S, Pangault C, Rouas-Freiss N, et al. Expression of functional soluble human leucocyte antigen-G molecules in lymphoproliferative disorders. Br J Haematol 2007;138:202-12. [DOI] [PubMed] [Google Scholar]
- 11.Naji A, Menier C, Maki G, Carosella ED, Rouas-Freiss N. Neoplastic B-cell growth is impaired by HLA-G/ILT2 interaction. Leukemia 2012;26:1889-92. [DOI] [PubMed] [Google Scholar]
- 12.Bielska M, Bojo M, Klimkiewicz-Wojciechowska G, Jesionek-Kupnicka D, Borowiec M, Kalinka-Warzocha E, et al. Human leukocyte antigen-G polymorphisms influence the clinical outcome in diffuse large B-cell lymphoma. Genes Chromosomes Cancer 2015;54:185-93. [DOI] [PubMed] [Google Scholar]
- 13.Rizzo R, Audrito V, Vacca P, Rossi D, Brusa D, Stignani M, et al. HLA-G is a component of the chronic lymphocytic leukemia escape repertoire to generate immune suppression: impact of the HLA-G 14 base pair (rs66554220) polymorphism. Haematologica 2014;99:888-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diepstra A, Poppema S, Boot M, Visser L, Nolte IM, Niens M, et al. HLA-G protein expression as a potential immune escape mechanism in classical Hodgkin’s lymphoma. Tissue Antigens 2008;71:219-26. [DOI] [PubMed] [Google Scholar]
- 15.Gallamini A, Barrington SF, Biggi A, Chauvie S, Kostakoglu L, Gregianin M, et al. The predictive role of interim positron emission tomography for Hodgkin lymphoma treatment outcome is confirmed using the interpretation criteria of the Deauville five-point scale. Haematologica 2014;99:1107-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen MH, Hviid TV. Human leukocyte antigen-G polymorphism in relation to expression, function, and disease. Hum Immunol 2009;70:1026-34. [DOI] [PubMed] [Google Scholar]
- 17.Hviid TV, Hylenius S, Rorbye C, Nielsen LG. HLA-G allelic variants are associated with differences in the HLA-G mRNA isoform profile and HLA-G mRNA levels. Immunogenetics 2003;55:63-79. [DOI] [PubMed] [Google Scholar]
- 18.Littera R, Piredda G, Pani A, Frongia M, Onano B, Michittu MB, et al. Role of human leukocyte antigen-G 14-base pair polymorphism in kidney transplantation outcomes. J Nephrol. 2013;26(6):1170-8. [DOI] [PubMed] [Google Scholar]
- 19.Locafaro G, Amodio G, Tomasoni D, Tresoldi C, Ciceri F, Gregori S. HLA-G expression on blasts and tolerogenic cells in patients affected by acute myeloid leukemia. J Immunol Res 2014;2014:636292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purdy AK, Campbell KS. Natural killer cells and cancer: regulation by the killer cell Ig-like receptors (KIR). Cancer Biol Ther 2009;8:2211-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cencini E, Fabbri A, Rigacci L, Lazzi S, Gini G, Cox MC, et al. Evaluation of the prognostic role of tumour-associated macrophages in newly diagnosed classical Hodgkin lymphoma and correlation with early FDG-PET assessment. Hematol Oncol 2015. doi: 10.1002/hon.2249 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 22.Molin D. Bystander cells and prognosis in Hodgkin lymphoma. Review based on a doctoral thesis. Ups J Med Sci 2004;109:179-228. [DOI] [PubMed] [Google Scholar]
- 23.Rouas-Freiss N, Moreau P, LeMaoult J, Carosella ED. The dual role of HLA-G in cancer. J Immunol Res 2014;2014:359748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiners KS, Kessler J, Sauer M, Rothe A, Hansen HP, Reusch U, et al. Rescue of impaired NK cell activity in hodgkin lymphoma with bispecific antibodies in vitro and in patients. Mol Ther 2013;21:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rousseau P, Le Discorde M, Mouillot G, Marcou C, Carosella ED, Moreau P. The 14 bp deletion-insertion polymorphism in the 3’ UT region of the HLA-G gene influences HLA-G mRNA stability. Hum Immunol 2003;64:1005-10. [DOI] [PubMed] [Google Scholar]
- 26.La Nasa G, Littera R, Locatelli F, Lai S, Alba F, Caocci G, et al. The human leucocyte antigen-G 14-basepair polymorphism correlates with graft-versus-host disease in unrelated bone marrow transplantation for thalassaemia. Br J Haematol 2007;139:284-8. [DOI] [PubMed] [Google Scholar]


