Abstract
Hepatocellular carcinoma (HCC) represents a major global health problem, since more than 90% of primary liver cancers worldwide are HCC. Most cases of HCC are secondary to viral hepatitis infection (hepatitis B or C), alcoholism and cirrhosis. Sorafenib, an oral tyrosine kinase inhibitor that suppresses tumor proliferation and angiogenesis, emerged as the first effective systemic treatment for HCC after 30 years of research, and is currently the standard-of-care for patients with advanced HCC. Sorafenib is metabolized by cytochrome P450 (CYP450), particularly from the 3A4 isoform, producing two main metabolites: the N-oxide and the N-hydroxymethyl metabolite. We studied 11 HCC sample showing the presence of CYP3A4 and CYP3A7 in most of the samples analysed. Specifically, the immunoreactivity of CYP3A4 was stronger and more widespread than that of CYP3A7. The CYP3A4 immunoreactivity was observed in surrounding hepatocytes in 8 out of 11 cases; while the CYP3A7 immunostaining was found in normal liver cells, in 7 out of 11 cases. These results suggest the existence of a marked inter-individual variability regarding the presence of the isoforms of CYP3A. In addition, since sorafenib is metabolized by CYP3A4, but not by CYP3A7, an overexpression of CYP3A4 may lead to an increase in the degradation of the drug and then to clinical ineffectiveness. These results might implicate the necessity of an individualized approach in the treatment of HCC as positivity to CYP3A4 in HCC liver samples might predict a scarce response to sorafenib.
Key words: HCC, CYP3A4, CYP3A7, immunohistochemistry, sorafenib
Introduction
Liver cancer is a highly common form of tumour accounting for 7% of all cancers. In addition, it is associated with substantial mortality being the third cause of cancer-related death.1 Among liver cancers, hepatocellular carcinoma (HCC) represents a major global health problem given that more than 90% of primary liver cancers worldwide are HCC. Most cases of HCC are secondary to viral hepatitis infection (hepatitis B or C), alcoholism and cirrhosis. However, the mechanisms of liver carcinogenesis remain largely unknown.1,2 Treatment and prognosis of HCC is based on the Barcelona-Clinic Liver Cancer (BCLC) classification, established on validated prognostic variables, such as those related to tumour status (size, number, vascular invasion, N1, M1), liver function (Child-Pugh’s) and health status.1 HCC is included among the most chemo-resistant tumour types, and until 2007 no systemic drug was recommended for patients with advanced tumours, an unparalleled situation in oncology. Sorafenib, an oral tyrosine kinase inhibitor that suppresses tumour proliferation and angiogenesis, emerged as the first effective systemic treatment for HCC after 30 years of research, and is currently the standard of care for patients with advanced disease.3,4 Sorafenib has been the first drug, and remains the only one, to increase survival in patients with advanced HCC, receiving marketing authorization from European Medicines Agency (EMEA) and Food and Drug Administration (FDA) in 2007.1 Sorafenib is metabolited by cytochrome P450 (CYP450), particularly from the CYP3A4 isoform, producing two main metabolites: the N-oxide and the N-hydroxymethyl metabolite.5 In addition, sorafenib undergoes glucuronidation. This pharmacokinetic profile has been shown to determine significant drug-drug interactions with other concomitant therapies, including those with medicinal herbs,6 or with chemotherapeutic agent such as rifampicin.7 Importantly, recent evidence showed that sorafenib metabolism might be impaired in HCC tumour cells.8 This latter finding might depend on the cancer-related alteration of the physiological pattern of CYP450 expression in the liver.
The expression of CYP450 isoenzymes in the human body is substrate and tissue specific.9 Of note, the largest proportion of CYP450 isoenzymes for drug metabolism are expressed in the liver.10-16 Of the several CYP450 isoform, the CYP3A4 metabolizes about 30% of clinically used drugs,9 while the CYP3A7 is mostly expressed in the foetal liver although it converts to express CYP3A7.17 In a previous study, we showed the overlapping expression between CYP3A4 and CYP3A7 in the foetal human liver during development.18
There is paucity of data available on CYP450 expression in the adult liver both in physiological as well as in pathological conditions, including HCC. Here, we aimed to present novel findings on the expression patterns of CYP3A4 and CYP3A7 in HCC. We hypothesize that the presence of these CYP450 isoforms might have implications for personalized treatment of HCC.
Materials and Methods
The positivity to immunostaining for CYP3A4 and CYP3A7 was evaluated in liver samples from 11 HCC carriers, 6 males and 5 females, aged 44-80 years. All the foetuses included in this study had no congenital malformation. All procedures performed were approved by the Ethics Human Studies Committee of the University Medical Centre of Cagliari (according to the instructions of the Declaration of Helsinki). Samples were fixed in 10% formalin, dehydrated and embedded in paraffin. Tissue section were then dewaxed, rehydrated through graded alcohols and pre-treated for 10 min with the heat-induced epitope retrieval in buffer pH 6.00 (EnVision™ FLEX Target Retrieval Solution Low pH- Dako Denmark A/S, Glostrup, Denmark, Code: K8004) for immunohistochemical analysis of CYP3A7. Slides were then incubated for 20 min at room temperature with anti-CYP450 3A4 rabbit polyclonal antibody at 1.1000 dilution and with anti-CYP450 3A7 mouse monoclonal antibody clone F19 P2 H2 at 1:50 dilution (all produced by Abcam, Cambrige, UK, catalogue numbers: ab3572, ab108624 and ab55840, respectively). Staining procedures were performed by EnVision™ FLEX+ (Dako, code: K8002) Detection System and Autostainerlink 48 instrument following dealer’s instructions. Negative control samples were incubated without mouse anti-CYP3A4 and anti-CYP3A7 polyclonal antibody.
The histological examination was performed by three observers (DF, FL and RA), who independently assigned two different scores. The first score regarded the distribution of CYP3A4 and CYP3A7 in HCC and in the surrounding parenchyma: score 1 for focal distribution, score 2 for zonal distribution and score 3 for diffuse distribution. The second score concerned the intensity of immunostaining, which was defined as weak (score 1), medium (score 2) and strong (score 3). In both score systems, score 0 was assigned for negativity. The sum between the distribution score and the intensity score gave rise to the following final grading system: grade 0 for negative stain, grade 1 for a final score from 1 to 3, grade 2 from 4 to 5 and grade 3 for a final score >5. All cases were then discussed at multisession microscope until a consensus between all three observers was obtained.
Bionformatic and statistical analysis
In light of the evidence that sorafenib is metabolized through CYP3A4 and UDP glucuronosyltransferase 1 Family, Polypeptide A9 (UGT1A9), we used publicly available bioinformatics resources, such as SuperCYP and Transformer databases,19-20 to investigate its pharmacokinetic profile. Furthermore, we tested whether other tyrosine kinases analogues of sorafenib underwent the same pharmacokinetic pathway. We also analysed whether the degree of immunostaining of CYP3A4 and CYP3A7 correlated significantly in the HCC samples using polychoric correlation coefficient. This analysis was performed using the polychoric21 package implemented in STATA 10.0 SE.
Results
Immunoreactivity for CYP3A4 and CYP3A7 was detected in all cases of HCC immunostained, except one (case number 11) for CYP3A7. Most cases showed a diffuse and strong positivity for CYP3A4 (resulting in a final grading score equal to 3). Immunostaining for CYP3A7 was observed in a much lower number of cells than for CYP3A4, with the majority of cases reporting the final grading score equal to 1. CYP3A4 and CYP3A7 positivity in the surrounding liver was detected in all but 3 cases for CYP3A4 and all but 4 cases for CYP3A7.
CYP3A4
Cases number 1, 3 and 4 showed a diffuse distribution of CYP3A4 (score 3) in tumour cells (Figure 1). Cases number 2, 5, 6, 10 and 11 presented a score of 2 for CYP3A4 in HCC. A score of 1 for CYP3A4 was found in cases number 7, 8 and 9 (Table 1). Positivity for CYP3A4 was present also in the surrounding liver of 11 samples (Table 1): samples number 1 and 10 showed a diffuse distribution of CYP3A4 (score 3); number 2, 3, 6, 7 and 9 showed a medium distribution of CYP3A4 in cases number 4, 5 and 8 not classified (NC).
Figure 1.
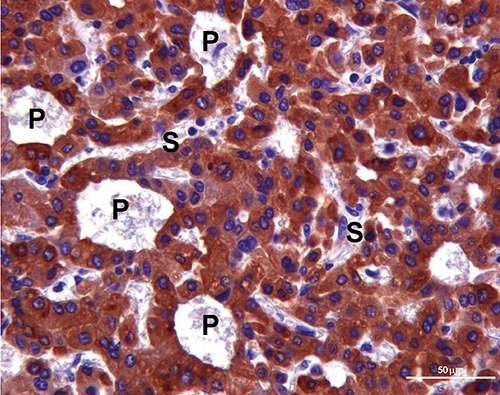
CYP3A4 positivity brown labelled in the cytoplasm of neoplastic hepatocytes characyerized by pseudoglandular architecture (P). S, sinusoids. Scale bar: 50 µm.
Table 1.
Expression of CYP3A4 in hepatocellular carcinoma and in the surrounding parenchyma.
| Cases | Pathology | Hepatocellular carcinoma | Parenchyma | ||||
|---|---|---|---|---|---|---|---|
| D | I | G | D | I | G | ||
| 1 | Cirrhosis | 3 | 3 | 3 | 3 | 3 | 3 |
| 2 | Cirrhosis | 2 | 3 | 2 | 2 | 3 | 2 |
| 3 | Hepatitis | 3 | 3 | 3 | 2 | 3 | 2 |
| 4 | Not classified | 3 | 3 | 3 | NC | NC | NC |
| 5 | Not classified | 2 | 3 | 2 | NC | NC | NC |
| 6 | Hepatitis | 2 | 2 | 2 | 2 | 3 | 2 |
| 7 | Hepatitis | 1 | 2 | 1 | 2 | 2 | 2 |
| 8 | Not classified | 1 | 1 | 1 | NC | NC | NC |
| 9 | Cirrhosis | 1 | 1 | 1 | 2 | 2 | 2 |
| 10 | Not classified | 2 | 1 | 2 | 3 | 3 | 3 |
| 11 | Cirrhosis | 2 | 2 | 2 | 2 | 3 | 2 |
D, distribution: 1, focal; 2, zonal; 3, diffuse; I, intensity: 1, weak; 2, medium; 3, strong; G, grade (calculated by addition of distribution and intensity: grade 0; grade 1, ≤3; grade 2, ≤5; grade 3, >5); NC, not classified.
The surrounding parenchyma was positive for CYP3A4 in 8 out of 11 cases in which it could be evaluated (Table 1). In cases number 1 (Figure 2), 2, 9 and 11 the surrounding area was affected by cirrhosis (C); cases number 3 (Figure 3), 6 and 7 showed chronic hepatitis (H); the remaining 3 livers were NC.
Figure 2.
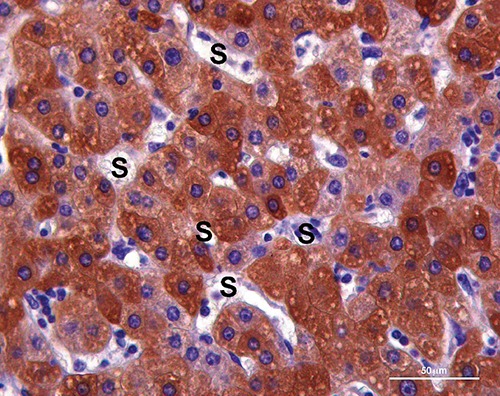
CYP3A4 immunoreactivity (brown label) in the cytoplasm of hyperplastic hepatocytes with plates of two cells thick between sinusoids (S). Scale bar: 50 µm.
Figure 3.
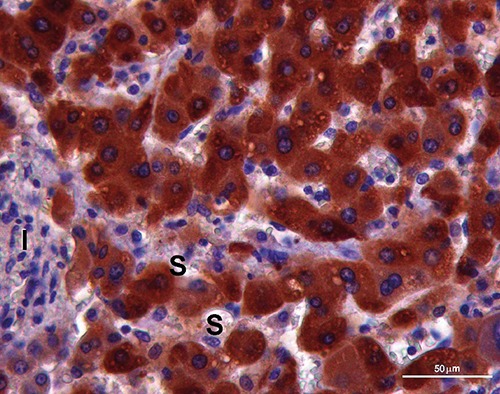
Positivity for CYP3A4 (brown label) in the cytoplasm of hepatocytes with plates of one cell thick between sinusoids (S) near inflammatory focus (I) of hepatitis. Scale bar: 50 µm.
CYP3A7
Ten out of 11 HCC samples showed a medium or weak distribution for CYP3A7 (score 2 and 1), while 1 was negative (Table 2). Cases number 4, 5 and 10 were characterized by a zonal distribution of CYP3A7 (score 2) (Figure 4), while the remaining cases showed a weak distribution for CYP3A7 (score 1). Case 11 did not show any expression of CYP3A7.
Table 2.
Expression of CYP3A7 in hepatocellular carcinoma and in the surrounding parenchyma.
| Cases | Pathology | Hepatocellular carcinoma | Parenchyma | ||||
|---|---|---|---|---|---|---|---|
| D | I | G | D | I | G | ||
| 1 | Cirrhosis | 1 | 1 | 1 | 1 | 1 | 1 |
| 2 | Cirrhosis | 1 | 1 | 1 | 1 | 1 | 1 |
| 3 | Hepatitis | 1 | 1 | 1 | 1 | 1 | 1 |
| 4 | Not classified | 2 | 2 | 2 | NC | NC | NC |
| 5 | Not classified | 2 | 2 | 2 | NC | NC | NC |
| 6 | Hepatitis | 1 | 2 | 1 | 2 | 2 | 2 |
| 7 | Hepatitis | 1 | 1 | 1 | 2 | 2 | 2 |
| 8 | Not classified | 1 | 1 | 1 | NC | NC | NC |
| 9 | Cirrhosis | 1 | 1 | 1 | 2 | 2 | 2 |
| 10 | Not classified | 1 | 2 | 2 | NC | NC | NC |
| 11 | Cirrhosis | 0 | 0 | 0 | 1 | 1 | 1 |
D, distribution: 1, focal; 2, zonal; 3, diffuse; I, intensity: 1, weak; 2, medium; 3, strong; G, grade (calculated by addition of distribution and intensity: grade 0; grade 1, ≤3; grade 2, ≤5; grade 3, >5); NC, not classified.
Figure 4.

Weaker brown labelled immunoreactivity for CYP3A7 in the cytoplasm of neoplastic hepatocytes with thickening of hepatocellular plate (T) between sinusoids (S) and pseudoglandular architecture (P); in the left down corner of the picture a mitosis (M) is also present. Scale bar: 50 µm.
The surrounding parenchyma was positive for CYP3A7 in 8 out of 11 cases where it could be evaluated (Table 2). Cases number 6, 7 and 9 showed moderate positivity of CYP3A7 (score 2), cases number 1, 2, 3 and 11 showed a weak distribution of the CYP3A7 (score 1), while the remaining were NC.
Bioinformatics and statistical analyses
The search in the SuperCYP and Transformer databases did not show any interaction between sorafenib and CYP isoform CYP3A4 and CYP3A7. However, Transformer databases confirmed that sorafenib undergoes glucuronidation through UGT1A9. Interestingly, the bioinformatics analysis showed that other three tyrosine kinase inhibitors -imatinib, gefitinib and pazopanibare substrates of CYP3A4 (with imatinib and pazopanib acting as inhibitors), while imatinib acts also as a substrate of CYP3A7. No other metabolic pathways were found through the bioinformatic search.
The polychoric correlation coefficient showed a moderate concordance in the expression patterns of CYP3A4 and CYP3A7 in HCC samples (Rho= 0.26, standard error= 0.2; goodness of fit test: Pearson G2 = 4.64).
Discussion
The CYP450 superfamily in humans is formed by genes and pseudogenes encoding for functional mono-oxygenase. These are enzymes critical for the metabolism of several exogenous and endogenous substances.10-20 The most important function of CYP450 super-family are related to: i) the detoxification of exogenous xenobiotics, such as carcinogens and foreign chemicals; ii) the decomposition of most of the currently used drugs; iii) the metabolism of endogenous growth factors, such as hormones, fatty acids, prostaglandins, cholesterol, vitamin D and bile acids.22,23 Genetic polymorphisms of CYPs have been studied as risk markers for HCC development in cell lines, in mouse and human patients.24 In previous studies the expression of CYP3A7 in foetal liver has been reported to start at the ninth week of gestation25 and to be abolished within the first postnatal week. CYP3A7 has been reported to be the dominant enzyme in the foetal liver, given that a subsequent age-related shift between CYP3A7 and CYP3A4 occurs after birth.16 However, CYP3A4 as well as CYP3A7 immunohistochemical positivity has been recently detected in foetal livers.26 It is conceivable that the reported changes in the expression of CYP3A7 (up-regulation) and CYP3A4 (down-regulation) occurring after birth, as well as the restriction of CYP3A7 expression just in the prenatal life,16 might not be as solid as previously established. In fact, the immunohistochemical patterns of CYP3A4 and CYP3A7 during development showed that the relationships between CYP3A4 and CYP3A7 are more complex then previously thought.26 The analysis of CYP genes in end-stage livers compared with normal donor liver showed that the expression of CYP3A4, CYP3A5 and CYP3A7 was decreased in HCC.27 Immunohistochemical CYP3A4 positivity was previously reported in HCC, hepatocellular adenomas (HCA) and focal nodular hyperplasia (FNH), suggesting that the expression of CYP3A4 may change in HCA and HCC and indicating specific metabolic characterization in the subgroup of HCA and HCC.28 No evidence points to similar patterns of expression for CYP3A7 in HCC. Here, we reported the immunohistochemical positivity of both CYP3A4 and CYP3A7 in HCC. Our findings showed that both CYP3A4 and CYP3A7 were present in the vast majority of the HCC samples analysed, being detected in 10 out of 11 HCCs. CYP3A7 was negative in just one HCC sample. Regarding the comparison of the patterns of positivity of the two isoforms, we found that immunoreactivity for CYP3A4 was more diffuse and stronger than CYP3A7 in HCC cells. Significant differences were found regarding CYP expression between tumour cells and the surrounding parenchyma. Immunoreactivity for CYP3A4 was observed in the surrounding hepatocytes in 8 out of 11 cases, whereas immunostaining for CYP3A7 was found in the normal liver cells in 7 out of 11 cases.
Taken together, these findings suggest the existence of a marked inter-individual variability regarding CYP3A isoforms both in physiological tissue, as shown by differences among the surrounding livers, and in pathological one, as evidenced by differences observed among tumour cells. This finding confirms previous data on the expression of CYP3A4 and CYP3A7 in foetal human livers26 and suggests the need for a tailored individualized medicine in the therapeutic approach to HCC. Of interest, the polychoric correlation confirmed to some extent this qualitative analysis by showing a low to moderate degree of concordance in the patterns of CYP3A4 and CYP3A7 immunostaining in HCC. The potential implications for HCC treatment might be relevant. As previously described, sorafenib undergoes selective CYP3A4-dependent biotransformation to its major oxidized metabolites.5,29 Conversely, our bioinformatic search did not highlight any interaction of sorafenib with CYP3A4 as well with CYP3A7. The latter finding, however, appears to be consistent with the literature, with no reports of sorafenib CYP3A7-mediated metabolism. Of interest is the evidence that other tyrosine kinase inhibitors analogues of sorafenib do undergo metabolism through CYP3A7. The pharmacokinetic profile of sorafenib allows prioritizing the expression patterns identified in HCC in our sample. Indeed, the absence of interaction between sorafenib and CYP3A7, at least according to the current knowledge, renders less probable that specific alterations of CYP3A7 expression might be pharmacokinetically, and thus clinically, relevant. On the contrary, CYP3A4 increased expression is particularly significant for sorafenib metabolism. An overexpression of CYP3A4 could lead to increased degradation of the drug and thus to clinical ineffectiveness. This could be clinically relevant given that sorafenib response rate remains still quite low at around 2%.30
The finding of a moderate concordance in the expression patterns of the two CYP isoform deserves a final comment. This finding is based on a small sample size and as such should be interpreted with caution. However, if the pattern of correlated positivity to immunostaining will be confirmed in larger cohort of patients, it will be possible to use immunostaining of either CYP isoforms in liver biopsies to study the pharmacokinetic profile on an individual basis.
Despite recent advances in diagnosis and management of HCC, the prognosis and the median survival of patients is still poor. Therefore, the identification of new putative markers to improve the HCC prognosis became indispensable and opened a new research field. Our group previously evaluated the expression of thymosin beta 4 (Tβ4) and thymosin beta 10 (Tβ10) in HCC tissues.31 Moreover, Guerriero et al. showed an increase of SELM expression32 and GPX4 and GPX7 over-expression33 in human HCC tissues by immunohistochemistry.
Our data suggest that immunohistochemistry for CYP3A4 and CYP3A7 should be introduced in clinical practice to further elucidate its relation with the clinical response to chemotherapy with sorafenib. Immunohistochemistry for CYP isoenzymes might represent a prognostic marker for current management of HCC and may play a role along the way towards a personalized therapy for HCC.
References
- 1.European Association for the study of the liver, European Organisation for Research and treatment of cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2012;56:908-43. [DOI] [PubMed] [Google Scholar]
- 2.Fanni D, Gerosa C, Faa G. The histomorphological and immunohistochemical diagnosis of hepatocellular carcinoma. In: Joseph WY. (ed.) Hepatocellular Carcinoma. Intech, Croatia: 2012. [Google Scholar]
- 3.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378-90. [DOI] [PubMed] [Google Scholar]
- 4.Tabrizian P, Roayaie S, Schwartz ME. Current management of hepatocellular carcinoma. World J Gastroenterol 2014;20:10223-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghassabian S, Rawling T, Zhou F, Doddareddy MR, Tattam BN, Hibbs DE, et al. Role of human CYP3A4 in the biotrasformation of Sorafenib to its major oxidized metabolites. Biochemical Pharmacol 2012;84:215-23. [DOI] [PubMed] [Google Scholar]
- 6.Thomas-Schoemann A, Blanchet B, Bardin C, Noe G, Boudou-Rouquette P, Vidal M, et al. Drug interactions with solid tumour-targeted therapies. Crit Rev Oncol Hematol 2014;89:179-96. [DOI] [PubMed] [Google Scholar]
- 7.Keating G.M., Santoro A. Sorafenib: a review of its use in advanced hepatocellular carcinoma. Drugs 2009;69:223-40. [DOI] [PubMed] [Google Scholar]
- 8.Ye L, Yang X, Guo E, Chen W, Lu L, Wang Y, et al. Sorafenib metabolism is significantly altered in the liver tumor tissue of hepatocellular carcinoma patient. PLoS One 2014;9:96664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 2013;138:103-41. [DOI] [PubMed] [Google Scholar]
- 10.Yang X, Zhang B, Molony C, Chudin E, Hao K, Zhu J, et al. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 2010;20:1020-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson TN, Rostami-Hodjegan A, Tucker GT. Prediction of the clearance of eleven drugs and associated variability in neonates, infants and children. Clin Pharmacokinetics 2006;45:931-56. [DOI] [PubMed] [Google Scholar]
- 12.Bjorkman S. Prediction of cytochrome p450-mediated hepatic drug clearance in neonates, infants and children: how accurate are available scaling methods? Clin Pharmacokinetics 2006;45:1-11. [DOI] [PubMed] [Google Scholar]
- 13.Kearns GL, Robison PK, Wilson JT, Wilson-Costello D, Knight GR, Ward RM, et al. Cisapride disposition in neonates and infants: in vivo reflection of cytochrome P450 3A4 ontogeny. Clin Pharmacol Ther 2003;74:312-25. [DOI] [PubMed] [Google Scholar]
- 14.De Wildt SN, Kearns GL, Hop WC, Murry DJ, Abdel-Rahman SM, van den Anker JN. Pharmacokinetics and metabolism of oral midazolam in preterm infants. Br J Clin Pharmacol 2002;53:390-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smits A, Kulo A, de Hoon JN, Allegaert K. Pharmacokinetics of drugs in neonates: pattern recognition beyond compound specific observations. Curr Pharm Des 2012;18:3119-46. [DOI] [PubMed] [Google Scholar]
- 16.Lacroix D, Sonnier M, Moncion A, Cheron G, Cresteil T. Expression of CYP3A in the human liver-evidence that shift between CYP3A7 and CYP3A4 occurs immediately after birth. Eur J Biochem 1997;247:625-34. [DOI] [PubMed] [Google Scholar]
- 17.Burk O, Tegude H, Koch I, Hustert E, Wolbold R, Glaeser H, et al. Molecular mechanisms of polymorphic CYP3A7 expression in adult human liver and intestine. J Biol Chem 2002;277:24280-8. [DOI] [PubMed] [Google Scholar]
- 18.Fanni D, Fanos V, Ambu R, Lai F, Gerosa C, Pampaloni P, et al. Overlapping between CYP3A4 and CYP3A7 expression in the fetal human liver during development. Matern Fetal Neonatal Med 2014;10:1-5. [DOI] [PubMed] [Google Scholar]
- 19.Preissner SC, Hoffmann MF, Preissner R, Dunkel M, Gewiess A, Preissner S. Polymorphic cytochrome P450 enzymes (CYPs) and their role in personalized therapy. PLoS One 2013;8:e82562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preissner S, Kroll K, Dunkel M, Senger C, Goldsobel G, Kuzman D, et al. SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucleic Acids Res 2010;38:D237-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolenikov S, Angeles G. The use of discrete data in principal component analysis with applications to socio-economic indices. CPC. MEASURE Working paper No. WP-04-85, 2004. [Google Scholar]
- 22.Wang D, Jiang Z, Shen Z, Wang H, Wang B, Shou W, et al. Functional evaluation of genetic and environmental regulators of p450 mRNA levels PLoS One. 2011;6:e24900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson DR. Cytochrome P450 nomenclature, 2004. Methods Mol Biol 2006;320:1-10. [DOI] [PubMed] [Google Scholar]
- 24.Blum HE. Hepatocellular carcinoma: susceptibility markers. IARC Sci Publ 2001;154:241-4. [PubMed] [Google Scholar]
- 25.Hakkola J, Raunio H, Purkunen R, Saarikoski S, Vahakangas K, Pelkonen O, et al. Cytochrome P450 3A expression in the human fetal liver: evidence that CYP3A5 is expressed in only a limited number of fetal livers. Biol Neonate 2001;80:193-201. [DOI] [PubMed] [Google Scholar]
- 26.Fanni D, Fanos V, Ambu R, Lai F, Gerosa C, Pampaloni P, et al. Overlapping between CYP3A4 and CYP3A7 expression in the fetal human liver during development. Matern Fetal Neonatal Med 2014;10:1-5. [DOI] [PubMed] [Google Scholar]
- 27.Chen H, Shen ZY, Xu W, Fan TY, Li J, Lu YF, et al. Expression of P450 and nuclear receptors in normal and end-stage Chinese livers. World J Gastroenterol 2014;20:8681-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas S, Merkelbach-Bruse S, Justenhoven C, Brauch H, Fischer HP. Expression of xenobiotic and steroid hormone metabolizing enzymes in hepatocellular tumors of the non-cirrhotic liver. Pathol Res Pract 2009;205:716-25. [DOI] [PubMed] [Google Scholar]
- 29.Zimmerman EI, Roberts JL, Li L, Finkelstein D, Gibson A, Chaudhry AS, et al. Ontogeny and sorafenib metabolism. Clin Cancer Res 2012;18:5788-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin 2012;62:394-9. [DOI] [PubMed] [Google Scholar]
- 31.Theunissen W, Fanni D, Nemolato S, Di Felice E, Cabras T, Gerosa C, et al. Thymosin beta 4 and thymosin beta 10 expression in hepatocellular carcinoma. Eur J Histochem 2014;58:2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerriero E, Accardo M, Capone F, Colonna G, Castello G, Costantini S. Assessment of the Selenoprotein M (SELM) over-expression on human hepatocellular carcinoma tissues by immunohistochemistry. Eur J Histochem 2014;58:2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guerriero E, Capone F, Accardo M, Sorice A, Costantini M, Colonna G, et al. GPX4 and GPX7 over-expression in human hepatocellular carcinoma tissues. Eur J Histochem 2015;59:2540. [DOI] [PMC free article] [PubMed] [Google Scholar]


