Summary
Many studies have focused on the mechanisms underlying length and width determination in rod-shaped bacteria. Here, we focus instead on cell surface area to volume ratio (SA/V), and demonstrate that SA/V homeostasis underlies size determination. We propose a model whereby the instantaneous rates of surface and volume synthesis both scale with volume. This model predicts that these relative rates dictate SA/V and that cells approach a new steady-state SA/V exponentially, with a decay constant equal to the volume growth rate. To test this, we exposed diverse bacterial species to sublethal concentrations of a cell wall biosynthesis inhibitor and observed dose-dependent decreases in SA/V. Furthermore, this decrease was exponential and had the expected decay constant. The model also quantitatively describes SA/V alterations induced by other chemical, nutritional, and genetic perturbations. We additionally present evidence for a surface material accumulation threshold underlying division, sensitizing cell length to changes in SA/V requirements.
Introduction
Genetically identical rod-shaped bacterial cells adopt a remarkably narrow range of lengths and widths under constant growth conditions (Schaechter et al., 1962). However, rapidly growing cells in nutrient-rich medium are typically much larger, both in width and length, than isogenic cells growing slowly in minimal medium (Schaechter et al., 1958). These classic observations raise questions that remain open and whose answers will be critical for a thorough understanding of bacterial physiology: what principles set and maintain this narrow range of cellular dimensions, and how are these dimensions modulated in response to a change in the environment?
In most bacteria, the cell wall plays a deterministic role in setting the size and shape of cells (for reviews, see Typas et al., 2011; Young, 2010). This covalent network is composed of cross-linked peptidoglycan (PG) that surrounds the cell and counteracts turgor pressure. The synthesis of new PG begins in the cytoplasm, where a series of cytosolic enzymes catalyze successive steps in PG precursor biosynthesis, and eventually precursors are incorporated into the growing cell wall. In rod-shaped bacteria, growth is traditionally divided into two alternating modes: elongation and septation, although these may overlap in time. During elongation, new PG is inserted into the lateral wall and cells become longer while maintaining a relatively constant width; during septation, cells constrict and form two new poles, which eventually resolve to form two daughter cells. Different PG insertion machineries coordinate these two modes of growth and are active at different times during the cell cycle, but both draw from the same pool of PG precursors.
Due to the alternating modes of elongation and division, cell length in rod-shaped cells is primarily determined by how much cells typically elongate before dividing (Typas et al., 2011; Young, 2010). Many models of division timing – and thus length control – have been proposed. Historically, it was thought that cells initiate chromosome replication after reaching a critical mass and divide a fixed amount of time later (Cooper and Helmstetter, 1968). Recently, an “adder” model has been proposed, where cells add a constant amount of volume during each cell cycle before dividing (Amir, 2014; Campos et al., 2014; Deforet et al., 2015; Jun and Taheri-Araghi, 2015; Taheri-Araghi et al., 2015; Tanouchi et al., 2015). How cells are able to “measure” a constant increase in volume, however, remains unknown, and the “adder” model does not address length differences across different growth rates. Several nutrient-sensing proteins have been tied to changes in cell length in response to the availability of certain nutrients (Hill et al., 2013; Weart et al., 2007; Yao et al., 2012), though these are insufficient to explain how restricting different nutrients leads to similar changes in growth rate and cell size (Schaechter et al., 1958), nor do they address the gradual, growth rate-dependent nature of this transition (Volkmer and Heinemann, 2011).
In addition to studies based on measurement of cell length, much work has focused on how rod-shaped bacteria adopt a specific width. Several factors have been implicated in this process, including MreB, which is thought to coordinate the insertion of lateral cell wall material (reviewed in Chastanet and Carballido-Lopez, 2012). MreB depletion leads to the loss of rod-shape, and mutations in MreB can lead to wider or thinner cells (Dye et al., 2011; Kruse et al., 2003; Monds et al., 2014). These results raise the possibility that MreB can determine bacterial cell width. However, as with length, the fluid modulation of cell width in response to changing physiological conditions (Volkmer and Heinemann, 2011) implies that genetic control cannot be the only force at play. Indeed, when we analyzed the growth patterns of an MreB mutant with a variable-width phenotype (Harris et al., 2014), we found that cell surface area to volume ratio (SA/V) was still conserved; cells modified their width in order to achieve and maintain a specific, condition-dependent SA/V, suggesting that attaining a target SA/V could lie upstream of width determination.
As noted above, previous studies of cell size have generally focused on width and length as independently controlled variables. In this work, we demonstrate that SA/V is the key variable that cells control, and in doing so they coordinately modulate length and width. We furthermore propose a predictive, quantitative model for SA/V maintenance in bacteria that not only addresses the tight distribution of cell sizes observed in a given physiological condition, but also explains why slight changes in growth rate give rise to changes in cell size. Finally, we present a model for how cells modulate their length in response to SA/V requirements, potentially revealing a general mechanism for division triggering in bacteria.
Results
A “Relative Rates” Model Predicts SA/V Homeostasis and Bacterial Cell Size Determination
We have previously shown that individual Caulobacter crescentus cells move toward and maintain a target SA/V over time, even changing their radius when necessary (Harris et al., 2014). Here, we develop a model for SA/V determination that could explain this ability of cells to adopt a steady-state SA/V over time without the need to invoke a specific SA/V sensing system. As has been demonstrated (Godin et al., 2010; Monod, 1949; Wang et al., 2010), we begin with the assumption that individual bacterial cells increase both mass and volume exponentially. Thus, dV/dt can be written as a linear function of the current volume, V(t), related by the growth rate α (with units of time−1) (Figure 1A). By analogy, we could also assume that SA grows exponentially – that dA/dt is a function of A(t) (Gánti, 1975). This, however, produces an unstable SA/V. Rather, we postulate that the instantaneous rate of SA growth is a function of the current volume. This is the central hypothesis we explore below.
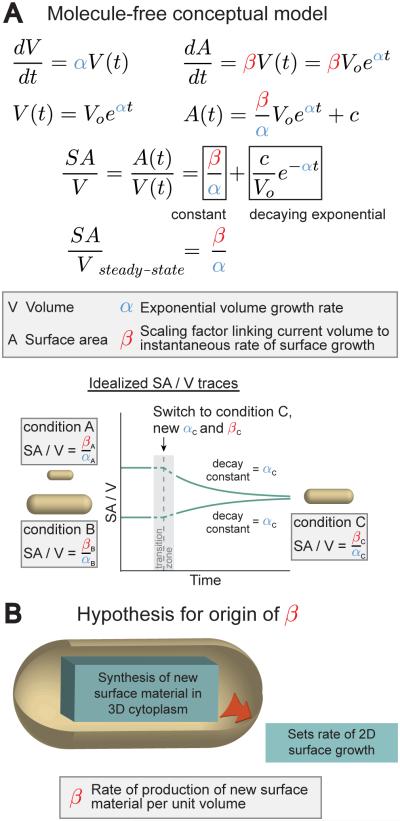
Figure 1. A “Relative Rates” Model Can Underlie SA/V Determination and Size Control in Bacteria.
A. A molecule-free conceptual model for SA/V maintenance during exponential growth of cells. Making only the assumption that the instantaneous rates of surface and volume synthesis both scale with volume, we predict that SA/V will move toward a steady-state value over time, and that the approach to steady state will be a decaying exponential with decay constant equal to the volume growth rate.
B. A hypothesis for why SA growth rate scales with volume. We propose that this could arise from the synthesis of new SA material in the cytoplasm setting the rate of SA growth.
Under this hypothesis, dA/dt is a linear function of V(t), related by the scaling factor β (with units of length−1 time−1). Both α and β are expected to vary with growth condition. Integrating both dA/dt and dV/dt to express SA and volume as functions of time and taking the ratio of these expressions gives SA/V as a function of time (Figure 1A). This makes the specific prediction that over time SA/V approaches a steady-state value equal to the ratio β/α (note that both ratios have units of length−1). Another non-trivial prediction is that, upon a change in growth condition, individual cells alter their size to the new steady-state SA/V exponentially, with a decay constant equal to the volume growth rate α. Satisfied that this conceptual model could give rise to SA/V homeostasis, we wondered: what could be the molecular origin of such scaling between volume and SA growth rate? Because the synthesis of new surface material begins in the cytoplasm, we hypothesized that the rate of production of new SA material might scale with volume and also limit the rate of SA expansion, producing the scaling between volume and SA growth rate (Figure 1B).
Inhibition of Peptidoglycan Biosynthesis Increases Cell Size and Decreases SA/V
If the scaling between volume and SA growth rate does in fact arise from the synthesis of new SA material in the cytoplasm setting the rate of SA expansion, decreasing this synthesis rate by only a few percent should decrease β, while having a smaller effect on the mass and volume growth rate (α), and therefore lower the steady-state SA/V, which is equal to β/α in the model. To test this, we chose to inhibit cell wall biosynthesis since the cell wall is the major structural component of the cell surface. We grew cells in very low doses of fosfomycin, a covalent inhibitor of MurA, the cytoplasmic enzyme that catalyzes the first committed step of PG biosynthesis (Kahan et al., 1974; Marquardt et al., 1994). Concentrations were chosen where the bulk doubling time of cultures was unaffected. Figure 2A-B shows C. crescentus cells that had been grown for several mass doubling times in the presence of different concentrations of fosfomycin. As predicted by the model, the average SA/V decreased in a dose-dependent manner with increasing concentrations of fosfomycin. To achieve this, individual cells became larger on average. Because the mass doubling time of these cells was unchanged, individual cells appear to have delayed division to become longer and also become wider as a way of reducing the amount of surface needed to cover a given amount of volume (i.e. reduced their overall SA/V).
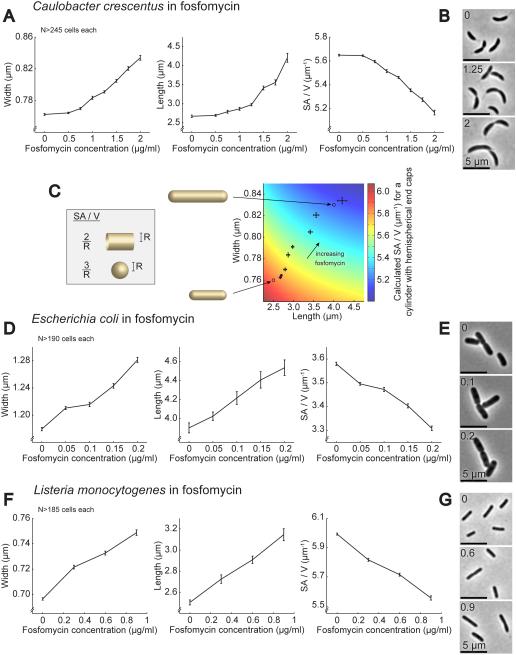
Figure 2. Growth in Sublethal Concentrations of a PG Biosynthesis Inhibitor Causes Diverse Species to Become Larger and Decrease Their SA/V in a Dose-dependent Manner.
A. Average width, length, and SA/V for C. crescentus cells grown for 5 h in the presence of different concentrations of fosfomycin. N>245 cells for each condition. Error bars = SEM.
B. Representative images of C. crescentus cells from three of the fosfomycin concentrations in (A).
C. Calculated SA/V (color scale) for a cylinder with hemispherical end caps of the indicated length and width, superimposed with the average width and length values for C. crescentus cells grown in fosfomycin (same data as (A)). Error bars = SEM. Circles show where the two idealized cells shown to the left would fall on this plot.
D. Average width, length, and SA/V for E. coli cells grown in different concentrations of fosfomycin for 3 h. N>190 cells for each condition. Error bars = SEM.
E. Representative images of E. coli cells from three of the fosfomycin concentrations in (D).
F. Average width, length, and SA/V for L. monocytogenes cells grown in different concentrations of fosfomycin for 4 h. N>185 cells for each condition. Error bars = SEM.
G. Representative images of L. monocytogenes cells from three of the fosfomycin concentrations in (F).
To investigate the relative impact of length and width increases on the final cell SA/V, we calculated the expected SA/V for a cylinder with hemispherical end caps over a range of widths and lengths similar to those measured in fosfomycin and overlaid this with the measured values for width and length (Figure 2C). It is clear that in this regime cell lengthening causes a significant drop in SA/V, even in the absence of cell widening. This effect arises because the SA/V of the hemispherical end caps is higher than the SA/V of the cylindrical cell body (Figure 2C inset). Thus, increases in length reduce the relative contribution of the end caps, lowering the overall SA/V.
Wondering if this change in cell size in response to low levels of cell wall biosynthesis inhibition is widely conserved, we also measured the effects of fosfomycin on another Gram-negative bacterium, Escherichia coli, as well as on Listeria monocytogenes, a Gram-positive pathogen (Figure 2D-G). In both cases SA/V decreased in a dose-dependent manner after growth in fosfomycin for several generations, achieved through increases in both width and length. This result implies that the ability to fluidly modulate width and length in response to PG biosynthesis inhibition is widely conserved, and that the availability of PG precursors plays an integral role in determining cellular dimensions across diverse species of bacteria. Furthermore, previous experiments in the Gram-positive bacterium Bacillus subtilis revealed that depletion of the second enzyme in the PG biosynthesis pathway, MurB, led to an elongated, widened phenotype (Real and Henriques, 2006), and recent CRISPRi knockdowns of other enzymes in the pathway in B. subtilis also produced wide cells (Peters et al. unpublished data), supporting our conclusion that restricting the flux through this pathway is sufficient to cause a drop in SA/V.
In addition to predicting a decrease in the steady-state SA/V, the model also makes specific predictions about the dynamic behavior of α and β upon inhibition of PG precursor synthesis – specifically that β should decrease while α is less strongly affected. To directly measure this, we developed a long-term, single-cell imaging platform based on a previously published method that allowed us to accurately track cell shape and growth dynamics for many generations (Figure 3A-E and Movie S1) (Iyer-Biswas et al., 2014). We calculated α and β by estimating the instantaneous linearized volume and SA growth rates and dividing these by the current volume. We observed that cell width, SA/V, α, and β were all remarkably stable over many hours and generations (Figure 3C-E).
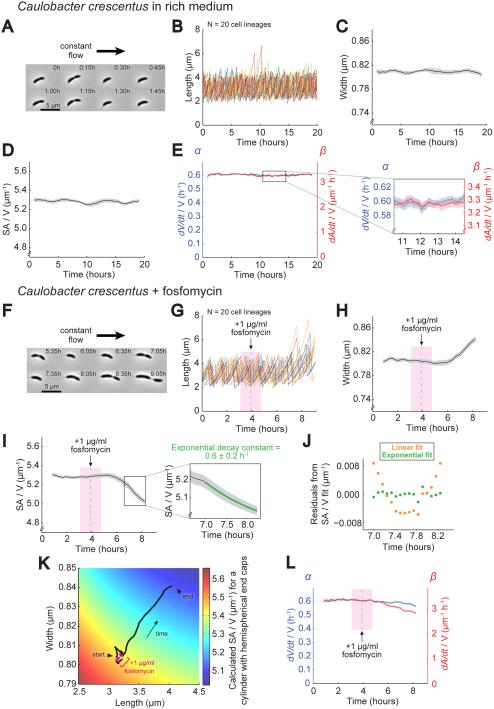
Figure 3. Inhibition of PG Biosynthesis Slows Down SA Growth Relative to Volume Growth, Leading to a Drop in SA/V.
A. Representative images of a C. crescentus lineage grown under a constant flow of fresh rich medium (PYE) where the adherent mother cell was retained on the coverslip while daughter cells were swept away after division. See also Movie S1.
B. Plot of cell length for 20 lineages grown as in (A). Images were taken every 5 m. Each color represents a different lineage.
C. Average cell width for lineages from (B) over time bounded by SEM.
D. Average SA/V for lineages from (B) plotted as in (C).
E. Average α and β for lineages from (B) plotted as in (C). Inset shows the magnitude of fluctuations in α and β.
F. Representative images of a C. crescentus lineage grown using the same perfusion setup as in (A). Cells were grown initially in rich medium and then switched to rich medium plus 1 μg/ml fosfomycin. Images shown were taken starting ~1.5 h after addition of drug. See also Movie S2.
G. Plot of cell length for 20 lineages grown as in (F). The gray dashed line marks the addition of fosfomycin, and the bounding pink box denotes the size of the sliding average window. Images were taken every 5 m. Each color represents a different lineage.
H. Average cell width for lineages from (G) plotted as in (C).
I. Average SA/V for lineages from (G) plotted as in (C). The inset shows the region where the SA/V traces were fit with a decaying exponential. See also Figure S1.
J. Residuals from fitting the average SA/V values from the region shown in the inset of (I) with both a linear and an exponential function.
K. Calculated SA/V (color scale) superimposed with the trajectory of average width and length for lineages treated with fosfomycin (same data as (G-H)). The white point marks when drug was first added, and the bounding pink region represents the sliding average window.
L. Average α and β for lineages from (G) calculated as in (E), plotted as in (C).
We next observed the dynamics during fosfomycin treatment. We initially flowed in medium without drug to measure pre-treatment growth rates and cellular dimensions and then switched to medium containing drug and imaged until cells were too large to consistently lie flat in the imaging plane (Figure 3F and Movie S2). As seen in the dose-response experiments (Figure 2A), C. crescentus cells treated with fosfomycin became both longer and wider and SA/V decreased (Figure 3G-J). Plotting the dynamic changes in average length and width on top of predicted SA/V values revealed that both increases in width and length contributed to the drop in SA/V (Figure 3K). Importantly, treatment with fosfomycin caused β to decrease significantly, while α remained steady for most of the experiment, dropping slightly at the end but by a smaller percentage than β (Figure 3L). The model predicts this behavior – that restricting the flux through the PG biosynthesis pathway per unit volume would decrease surface growth (β) to a greater extent than volume growth (α), and thereby significantly decrease SA/V.
After hour 7 of treatment, α and β adopted relatively constant values, providing an opportunity to test the model’s quantitative, non-trivial prediction that in this scenario the SA/V will move toward a new steady state exponentially with decay constant α. Fitting the relevant portion of the single-cell SA/V traces (see Supplemental Experimental Procedures and Figure S1A-D) revealed that these trajectories were well-described by a decaying exponential with decay constant 0.6±0.2 h−1 (for error estimation see Supplemental Experimental Procedures) (Figure 3I inset and 3J). This is remarkably close to the measured value of α over this time frame, 0.56-0.60 h−1 (Figure 3L). The fact that the model was able to predict both the exponential nature of this movement toward a new steady state as well as the specific decay constant provides strong support for the proposed mechanism of SA/V maintenance and size control.
Nutritional Upshift Increases Size and Decreases SA/V
We next wanted to use the “relative rates” model to understand the classic observation that bacteria growing quickly (e.g. in rich medium) are typically larger and have lower SA/V values than slower-growing cells (Schaechter et al., 1958). To do this, we performed nutritional upshift experiments with E. coli cells, which exhibit a quick escape from stationary phase upon introduction of fresh LB medium. We imaged stationary phase cells on a microfluidic device where medium can be exchanged, and tracked a single lineage that remained on the periphery of the microcolony (Figure 4A). As expected, the nutritional upshift was marked by an increase in cell length and width and a decrease in SA/V (Figure 4B-D). Plotting the average width and length on top of the predicted SA/V showed that increases in both width and length contributed to the drop in SA/V (Figure 4E).
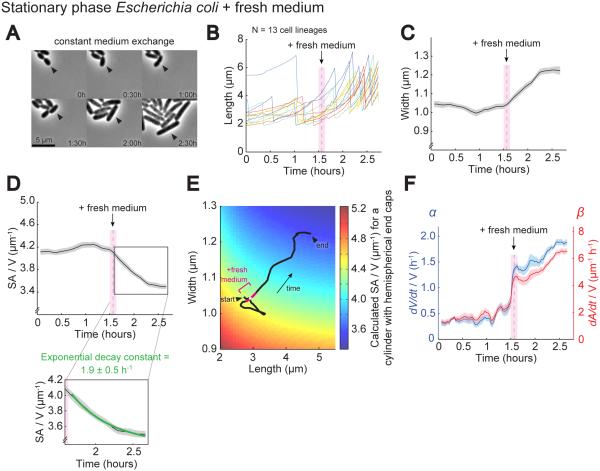
Figure 4. Nutritional Upshift Increases Volume Growth More Than SA Growth, Causing a Reduction in SA/V.
A. Representative images of stationary phase E. coli cells grown in a microfluidic device, initially perfused with spent stationary phase medium, and then switched to fresh LB. Arrowheads mark the daughter cell that remained on the periphery of the microcolony that was tracked over time.
B. Plot of cell length for 13 lineages grown as in (A). The gray dashed line marks when fresh medium was added, and the bounding pink box denotes the size of the sliding average window. Images were taken every 2 m. Each color represents a different lineage.
C. Average cell width for lineages from (B) over time bounded by SEM.
D. Average SA/V for lineages from (B) plotted as in (C). The inset shows the region where the SA/V traces were fit with a decaying exponential.
E. Calculated SA/V (color scale) superimposed with the trajectory of average width and length for lineages undergoing a nutritional upshift (same data as (B-C)). The white point marks when fresh medium was added, and the bounding pink region represents the sliding average window.
F. Average α and β for lineages from (B) plotted as in (C).
Interestingly, both volume growth (α) and SA growth (β) increased immediately upon addition of LB, likely due to the influx of raw materials available for metabolism (Figure 4F). In addition to this immediate jump, both α and β continued climbing in the hour after upshift, perhaps due to transcriptional changes (Battesti et al., 2011). The fact that β also increased after the upshift suggests that the rate of SA synthesis per unit volume increased in rich medium, but to a significantly lesser degree than α. Within the framework of the model, it is this mismatch between the percent increase in α and β that leads to the decrease in β/α, and thus lower SA/V, at high growth rates. Additionally, we observed that the movement of SA/V toward a new steady state following nutritional upshift was well-described by a decaying exponential, and fitting the SA/V traces revealed a decay constant of 1.9±0.5 h−1 (Figure 4D inset), very close to the measured value of α during this time, 1.4-1.9 h−1 (Figure 4F). Because the prediction that cells will move toward a new steady state exponentially with decay constant α is based on the assumption that α and β are constant, we used the “relative rates” model to simulate the predicted response of SA/V to continued changes in α and β similar to those observed in Figure 4F. The results (Figure S1E-H) show that as long as the ratio of β/α is relatively constant, as is the case after introduction of fresh medium (Figure S1E), movement of SA/V is still expected to behave like a decaying exponential and have decay constant 1.9 h−1, as we observed. These results provide further quantitative support for the “relative rates” model.
Global Repurposing of Translational Capacity Attenuates SA Growth, Leading to a Decrease in SA/V
In order to further probe the effect of changes in growth rate on SA/V, we used the ribosome inhibitor chloramphenicol, which causes growth rate to decrease approximately linearly with concentration (Scott et al., 2010). We expected that chloramphenicol treatment would slow down mass doubling and thus volume growth (α), but we did not know what effect it would have on the rate of SA synthesis per unit volume (β). Strikingly, we found that decreases in growth rate for C. crescentus, E. coli, and L. monocytogenes in increasing concentrations of chloramphenicol were accompanied by dose-dependent drops in SA/V, achieved through increases in cell width and length (Figure 5A-B and S2). This result goes against the well-established trend that slow-growing cells tend to be small and have high SA/V values. Since we know that the volume growth rate, α, decreased with increasing chloramphenicol, if the model is correct and the ratio of SA growth (β) to volume growth (α) sets SA/V, β must have decreased to an even greater extent than α.
Figure 5. Translational Repurposing Reduces SA Growth More Than Volume Growth, Causing a Reduction in SA/V.
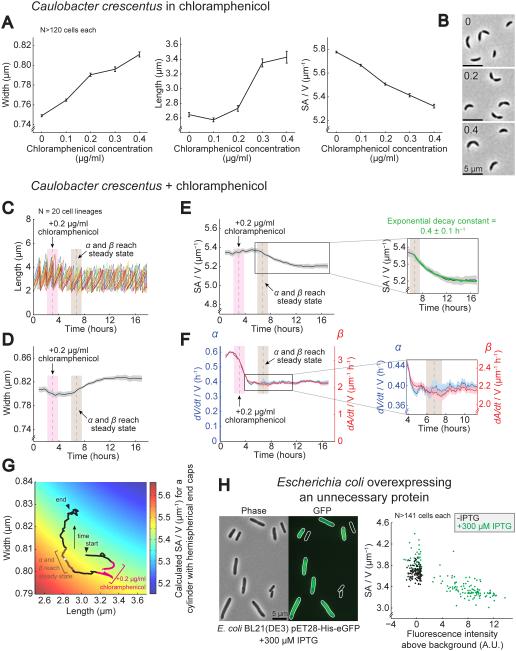
A. Average width, length, and SA/V for C. crescentus cells grown for 5 h in the presence of different concentrations of the ribosome inhibitor chloramphenicol. N>120 cells for each condition. Error bars = SEM. See also Figure S2.
B. Representative images of C. crescentus cells from three of the chloramphenicol concentrations in (A).
C. Plot of cell length for 20 C. crescentus lineages grown under constant flow. Each color represents a different lineage. Cells were grown first in rich medium and then 0.2 μg/ml chloramphenicol was added. The gray dashed line with bounding pink box marks when chloramphenicol was added, and the gray dashed line with bounding brown box marks when α and β reached steady state. Both bounding boxes represent the sliding average window. Images were taken every 5 m.
D. Average cell width for lineages from (C) over time bounded by SEM.
E. Average SA/V for lineages from (C) plotted as in (D). The inset shows the region where the SA/V traces were fit with a decaying exponential.
F. Average α and β for lineages from (C) plotted as in (D).
G. Calculated SA/V (color scale) superimposed with the trajectory of average width and length for lineages treated with chloramphenicol (same data as (C-D)). The white point bounded by pink marks when chloramphenicol was added, and the white point bounded by brown marks when α and β arrived at a steady state. Both bounding regions represent the sliding average window.
H. Representative phase contrast and epifluorescence images of E. coli BL21(DE3) cells containing an IPTG-inducible expression vector encoding His-tagged eGFP grown in the presence of 300 μM IPTG for 3 h, and a scatter plot of fluorescence intensity versus SA/V for the same strain grown with and without IPTG for the same amount of time. N>141 cells for each condition.
To test this prediction experimentally, we directly examined the dynamic behavior of α and β upon chloramphenicol treatment in C. crescentus cells (Figure 5C-G) using the single-cell imaging protocol described above (Figure 3). Immediately after addition of drug (marked in pink), α dropped to a greater extent than β (Figure 5F), and cells became shorter and thinner (Figure 5C, D, G), increasing their SA/V slightly (Figure 5E). This result is expected if volume growth slows to a greater extent than SA growth. However, over the next several hours β continued dropping, until reaching a steady state (marked in brown) that was relatively lower than α (Figure 5F inset). After this point, cell width began increasing steadily and SA/V dropped, consistent with SA growth slowing to a greater extent than volume growth, and yielding the directional change in SA/V that we had observed in static images (Figure 5A-B). We suspect that this gradual drop in β following chloramphenicol introduction is due to decreased expression of the enzymes that synthesize new SA material. It has previously been shown that bacteria upregulate the expression of ribosome-associated proteins in response to translational inhibition, leading to a concomitant, dose-dependent decrease in the expression levels of many other proteins (Scott et al., 2010), and it is possible that the SA biosynthesis enzymes would be subject to this global drop in non-ribosomal protein expression.
Regardless of what caused the final value of β to drop relative to α, the steady-state ratio of these two held remarkably steady for the following 10 hours (Figure S1I), providing another opportunity to observe the dynamic behavior of SA/V in a regime where α and β were not changing. Over these hours, SA/V moved exponentially toward a new steady state, with decay constant 0.4±0.1 h−1 (Figure 5E inset). This number matches exactly the growth rate, α, over this timeframe, 0.39-0.41 h−1 (Figure 5F). This is yet a third example – in addition to C. crescentus subjected to PG biosynthesis inhibition (Figure 3F-L and Figure S1A-D) and E. coli undergoing nutrient upshift (Figure 4 and Figure S1E-H) – of cells at a different steady-state growth rate, experiencing a fundamentally different type of perturbation, again altering their SA/V in a manner quantitatively predicted by the model (see summary in Figure S1J).
Wondering if the drop in SA/V associated with chloramphenicol treatment was a specific effect of the drug or a general hallmark of global translational repurposing, we chose to also investigate the effect of unnecessary protein overexpression. Similar to chloramphenicol treatment, the gross overexpression of an unnecessary protein in bacteria slows down growth by directing resources away from normal translation, and leads to a global drop in the expression levels of many other proteins (Scott et al., 2010). Inducing overexpression of GFP in an isogenic population of E. coli led to a heterogeneous pattern of induction, where some cells did not contain GFP fluorescence while others displayed high levels of fluorescence (Figure 5H). Strikingly, cells with high fluorescence were also significantly larger and had lower SA/V values than those that were not fluorescent. Similar results have also recently been observed by others (Basan et al., 2015). In addition to chloramphenicol treatment, this provides another example of a situation where α slowed down but did not lead to an increase in SA/V because the associated decrease in β must have been even larger and dominated the final effect on SA/V, further supporting the model that it is the balance between these two rates – rather than their absolute values – that determines SA/V.
Modulation of Cell Length in Response to SA/V Requirements Implies a Connection to Division Timing
In testing the predictions of the “relative rates” model, we found that in addition to changes in cell width, changes in cell length also played a key role in allowing cells to achieve a target SA/V. Because average length is determined by how much cells typically elongate before dividing, this implies a direct connection between SA/V requirements and the triggering of cell division. Indeed, treatment with the cell wall biosynthesis inhibitor fosfomycin led to a dose-dependent increase in cell length in a diverse array of species (Figure 2), suggesting that when surface synthesis is inhibited cells delay division.
Because the timing of cell division has long been thought to be tied to chromosome replication initiation (Cooper and Helmstetter, 1968), we characterized the number of chromosomes present in cells that had delayed division after growth in low levels of fosfomycin. Growing both C. crescentus and E. coli cells with fluorescently labeled chromosomes in fosfomycin (Figure S3), we found that while both species delayed division, fosfomycin-treated C. crescentus cells had on average the same number of chromosomes per cell, likely because compartmentalization of daughter cells is critical for the licensing of chromosome replication in this organism (Domian et al., 1999; Judd et al., 2003), while fosfomycin-treated E. coli cells contained more chromosomes on average, suggesting that replication initiation was not significantly interrupted. These results demonstrate that species with different chromosome replication programs delay division similarly in response to fosfomycin, suggesting that the mechanism underlying this delay is independent of chromosome replication.
Natural Periodicity in SA/V Over the Cell Cycle May Lead to Accumulation of Excess Surface Material Prior to Septation
In considering the implications of the “relative rates” model for division timing, it is critical to understand what is happening to SA/V over an individual cell cycle. So far in our investigation of this model, we have been averaging over multiple cells and cell cycles. On an individual cell basis, however, SA/V must necessarily show some periodicity over the cell cycle due to the alternating processes of elongation and septation, with elongation reducing SA/V by adding low-SA/V cylindrical cell body and septation raising it again by adding high-SA/V end caps (Morgan et al., 2004). Furthermore, because SA/V must vary over the cell cycle, the measured values of α and/or β for an individual cell must also show some cell cycle dependence.
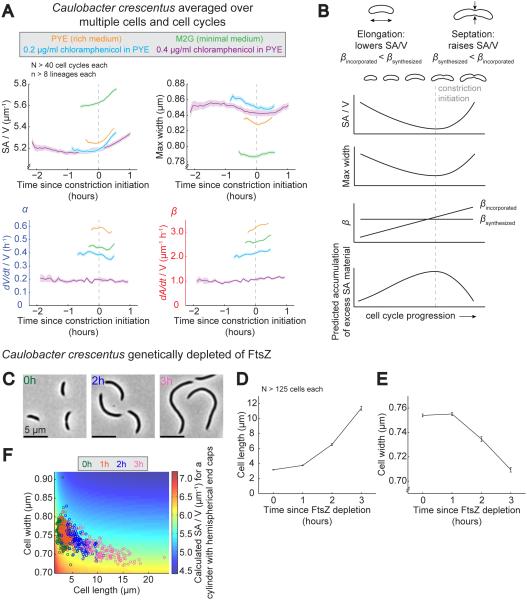
To directly examine these cell cycle dynamics, we subdivided dynamic C. crescentus imaging data from a variety of steady-state growth conditions into individual cell cycles and averaged different shape and growth parameters as a function of time in the cell cycle. In order to facilitate comparison among different growth conditions, we aligned the measured values with respect to the time of constriction initiation (time 0 in Figure 6A). Conditions included rich medium (same data as Figure 3A-E), minimal medium, 0.2 μg/ml chloramphenicol (same data as Figure 5 C-G, but only cell cycles starting after hour 7), and 0.4 μg/ml chloramphenicol. Among these conditions, the length of the cell cycle varied from ~1 h to ~4 h, and α and β each varied over a three-fold range (from 0.2-0.6 h−1 for α and from 1 to 3 μm−1 h−1 for β; Figure 6A). As expected, SA/V displayed cell cycle-dependent variation in every condition, though notably it did not decrease during elongation as would be expected for an elongating cylinder of constant width. Instead, maximum cell width decreased during elongation (Figure 6A), partially offsetting the SA/V-depressing effect of elongation. Additionally, the final value of SA/V in each condition was significantly higher than its initial value because after each division event the smaller swarmer cell was washed away, taking with it a significant amount of the accumulated SA/V and leaving behind the larger, lower SA/V stalked cell.
Figure 6. Excess Surface Material May Accumulate During Cell Elongation.
A. Cell cycle-dependent averages of SA/V, maximum width, α, and β for C. crescentus cells grown in a variety of steady-state conditions. Single-cell lineages from perfusion experiments were subdivided into individual cell cycles, aligned, averaged, and centered about constriction initiation. Shown is the average for each condition over time bounded by the SEM. N>40 cell cycles each. See also Figure S3.
B. Diagram summarizing cell cycle-dependent changes in SA/V, maximum width, and βincorporated, and our prediction for the accumulation of excess SA material given a constant underlying rate of SA material production per unit volume, βsynthesized.
C. Representative images of C. crescentus cells genetically depleted of FtsZ for the amount of time indicated.
D. Average length of C. crescentus cells genetically depleted of FtsZ for 0-3 h. N>125 cells for each condition. Error bars = SEM.
E. Average width of C. crescentus cells genetically depleted of FtsZ. Same cells as (D). Error bars = SEM.
F. Calculated SA/V (color scale) with length and width values for individual cells from populations genetically depleted of FtsZ. Each circle represents an individual cell, and colors correspond to different times since FtsZ depletion. Same cells as (D-E). See also Figure S4.
Associated with cell cycle-dependent variations in SA/V were also variations in the relative rates α and β. While α (volume growth) remained constant for most of the cell cycle, dipping slightly during constriction, β (SA growth) increased steadily as the cell cycle progressed (Figure 6A). This indicates that SA growth was initially slow compared to volume but increased over time, presumably due to cell thinning at first and then as a result of the constriction process itself. This is consistent with previous studies measuring the relative rates of incorporation of radioactivity into the cell wall and newly synthesized proteins over the cell cycle of E. coli: the rate of PG incorporation increased over the cell cycle, particularly at the onset of constriction, while the rate of protein synthesis was constant throughout (Cooper, 1988; Wientjes and Nanninga, 1989; Woldringh et al., 1987).
Though it is clear that the rate of SA incorporation speeds up over the cell cycle, we have no reason to believe that the rate of new SA material synthesis should necessarily vary over the cell cycle. If this is the case and the rate of production of new SA material per unit volume is relatively constant, given the cell cycle dependence of SA incorporation, we predict the following: during elongation, more material is produced than can be incorporated into the lateral wall, leading to an accumulation of excess SA material, while after constriction initiation this excess material is used up in the SA-intensive process of end cap construction (graphically summarized in Figure 6B).
To test the hypothesis that excess SA material accumulates during elongation, we examined C. crescentus cells that were genetically depleted of FtsZ and therefore prevented from dividing (Wang et al., 2001). Our prediction was that in the absence of septation, excess SA material would accumulate in the cell and lead to cell thinning over time. As predicted, after FtsZ depletion cells became elongated and significantly thinner (Figure 6C-E). Interestingly, plotting the length and width of individual cells from each depletion time point on top of the predicted SA/V revealed that, as cells became longer, decreases in width of the magnitude observed were necessary to keep the overall SA/V in a relatively narrow range (Figure 6F) (for a more detailed analysis of SA/V during FtsZ depletion, see Figure S4). These results are consistent with the model: in FtsZ-depleted cells, accumulated excess SA material was not resolved through end cap synthesis, so as cells increased in length, the accumulated material led to cellular thinning and an overall maintenance of the proper SA/V.
Accumulation of a Threshold Amount of Excess Surface Material May Trigger Division, Making Cell Length Responsive to SA/V Requirements
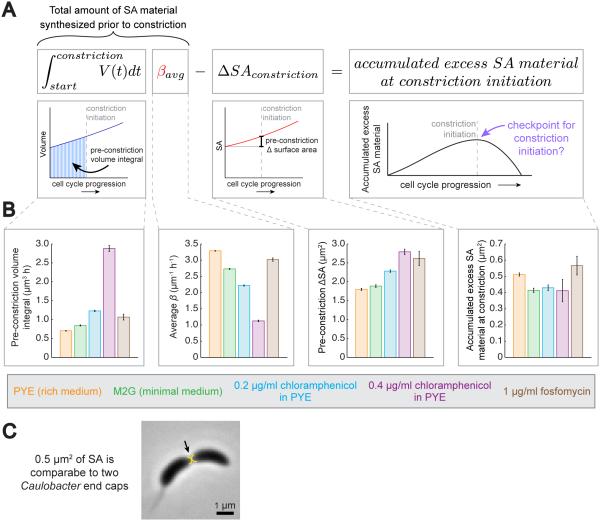
Because cells appear to accumulate excess SA material during elongation (Figure 6), and because this accumulation would necessarily peak at the point of constriction initiation, we wondered if accumulation of a threshold amount of excess SA material could be serving as a trigger for constriction initiation. Such a threshold mechanism would sensitize cell length to the rate of SA material accumulation, and could explain the dose-dependent increases in length observed for diverse species grown in fosfomycin. To investigate this hypothesis, we first demonstrated experimentally that in C. crescentus length control is consistent with the existence of a checkpoint governing the initiation of constriction by showing that the amount of time cells spend elongating prior to the onset of constriction is strongly negatively correlated with their initial size, while the amount of time spent actively constricting is size-independent (Figure S5A). Next, we wanted to estimate how much excess SA material had accumulated in individual cells prior to constriction initiation using measurable growth parameters, hypothesizing that this threshold amount might be similar across different growth conditions. To do this, we developed a framework (Figure 7A) based on the central hypothesis of the “relative rates” model – that SA material is produced at a rate proportional to volume, scaled by β. Thus, by multiplying β by the integral of volume over time for an individual cell from birth until constriction initiation (pre-constriction volume integral), we are able to estimate the amount of SA material produced in that cell prior to constriction. From this, we subtract the SA already incorporated into the lateral wall during elongation (pre-constriction ΔSA). This gives an estimate of the excess SA material present at constriction initiation.
Figure 7. Accumulation of a Threshold Amount of Excess SA Material Could Trigger Constriction Initiation.
A. Mathematical framework for estimating the amount of excess SA material accumulation prior to constriction. βaverage is used to approximate the underlying rate of SA production per unit volume. See also Figures S5 and S6.
B. Bar graphs showing the average values for the measured quantities used to estimate the amount of excess SA material accumulated prior to constriction initiation according to the framework in (A), as well as average values for the estimated accumulated material at constriction. N>19 cell cycles per bar. Error bars = SEM.
C. Representative image of a dividing C. crescentus cell overlaid with two semi-circles representing hemispheres whose SA adds up to 0.5 μm2.
One prediction we can make from this framework is that cells with slightly higher values of β will have smaller pre-constriction volume integrals – a correlation we observe in our C. crescentus steady-state data (Figure S5B). Additionally, both simulations (Figure S6) and analytical modeling (see Supplemental Experimental Procedures) show that division according to this scheme – upon accumulation of a threshold amount of excess SA material – is sufficient to produce the “adder” mode of growth that has recently been described for multiple species in steady-state conditions where cells add a constant amount of volume during each cell cycle regardless of initial size (reviewed in Jun and Taheri-Araghi, 2015). Intuitively, a SA accumulation threshold for division produces an “adder” growth pattern because cells with an initially smaller volume would have to grow for longer before accumulating the same threshold amount as larger cells.
Furthermore, this framework allows us to directly estimate the average amount of excess SA material that had accumulated prior to constriction initiation in C. crescentus cells grown at a variety of steady-state conditions to determine if this “threshold” amount appears constant. We used the same conditions and cell cycle data from Figure 6A as well as fosfomycin-treated cells (same data as Figure 3G-K but only cell cycles starting after hour 6). The measured parameters that went into our estimate of accumulated excess SA material at constriction are shown in Figure 7B. There was a significant spread among the average values of the pre-constriction volume integral, average β, and the pre-constriction ΔSA. However, performing the calculation outlined in Figure 7A for each cell cycle in every condition revealed that these large differences were abolished, and in every condition the amount of excess material predicted to be present at constriction initiation was quite comparable, supporting a threshold mechanism of division (Figure 7B).
Finally, because our calculated estimates for how much SA material had accumulated before constriction initiation is in actual units of μm2, we can estimate how large this amount is compared to the total SA of a C. crescentus cell. Because most of the accumulated material is expected to become new polar end caps, we calculated the diameter of two hemispheres that together represent the average amount of SA that had accumulated in cells grown in rich medium (0.5 μm2). Figure 7C shows the size of these hemispheres relative to a typical dividing cell, revealing that they are approximately the size of two C. crescentus poles. This suggests that the amount of SA material required to trigger constriction roughly corresponds to the amount needed to build two new end caps, and hints at a robust strategy whereby cells delay the onset of septation until they have accumulated sufficient surface material to carry this surface-intensive process through to completion.
Discussion
Though much work has been done to elucidate the origin of size control in bacteria, a unified understanding of the connections between length, width, and growth rate has been lacking. The “relative rates” model of SA/V determination synthesizes many disparate observations into just such a unified theory. By assuming exponential volume growth and postulating a volume-determined rate of surface growth, we were able to quantitatively predict the trajectory of SA/V for distantly related species undergoing a variety of perturbations. In every case, the ratio between the rates of SA and volume growth was the key determinant of cell size, rather than the specific values of these rates themselves. In this way, cells are able to achieve a steady-state SA/V over time without requiring a SA/V sensing system. Interestingly, in the 1970’s several papers suggested (Previc, 1970; Pritchard, 1974) that increases in cell size upon nutritional upshift could be the result of the measured mismatch (Sud and Schaechter, 1964) between the increases in volume and wall synthesis following upshift, a suggestion much in line with our current thinking. More broadly, homeostatic mechanisms similar to the “relative rates” model could be applicable beyond SA synthesis, and potentially beyond bacteria, to describe other processes where the rate of some output scales with volume, similar to mechanisms that have been proposed for eukaryotic systems determining the size and shape of cells and organelles (Chan and Marshall, 2012; Levy and Heald, 2012).
We also demonstrated that inhibiting PG biosynthesis is sufficient to alter the scaling between volume and SA growth rate, supporting the hypothesis that this scaling arises from production of new SA material in the cytoplasm limiting the rate of SA growth. While we have not demonstrated that PG precursor availability is always the rate-limiting factor setting the rate of SA expansion, this is at least the case during fosfomycin treatment, and could be generally true in other contexts as well. Using the flux through the PG biosynthetic pathway as a general determinant of SA growth rate would allow cells to finely tune their SA/V by modulating the flux through this pathway – a pathway particularly amenable to regulation because after the first committed step intermediates are no longer shared with other pathways. Alternatively, membrane biogenesis could be hypothesized to participate in setting the rate of SA growth as well. However, inhibition of fatty acid biosynthesis appears to have dramatic effects on central carbon metabolism, and inhibition of this pathway also reduces volume growth and results in small, slow-growing cells rather than causing cells to reduce only SA growth (Yao et al., 2012).
In response to different perturbations, we observed diverse bacterial species fluidly modulate their width in order to achieve a SA/V consistent with the “relative rates” model. It remains unclear how SA material abundance could cause cells to become wider or thinner. One possibility is that mismatches between volume and SA growth could produce changes in turgor pressure: if volume grows faster than SA, there will be high turgor pressure inside the cell, leading to increases in width, and if SA grows faster than volume, the turgor pressure will be low, leading to decreases in width. However, even if this is true, it is unclear how turgor pressure influences cell width. Perhaps the sidewall insertion machinery or MreB behave differently at different turgor pressures or depending on the abundance of substrate. This could give rise to differences in insertion site placement, PG crosslinking, hydrolytic activity, glycan strand length, strand orientation, and/or insertional stretching. In the future, unraveling the relative contributions of these and other properties will be key to understanding the molecular basis of width changes.
In addition to width, cells also modulated their length in response to SA/V requirements. To address this, we proposed and provided support for a model of division where cells delay division until a threshold amount of excess SA material has been accumulated. Such a checkpoint would presumably be one of several that cells must pass in order to divide. Notably, constriction initiation happens well after formation of the FtsZ ring, and factors that disrupt FtsZ ring formation – such as failed nucleoid segregation, stress response proteins, or nutrient sensing proteins (reviewed in Adams and Errington, 2009) – cause cells to dramatically delay division. This implies that FtsZ ring formation is upstream of any SA material checkpoint. There are several late-arriving divisome proteins (Aarsman et al., 2005; Gamba et al., 2009; Goley et al., 2011) that could be responsible for sensing an accumulated amount of SA material and triggering constriction. Because the cell wall is the structural determinant of the cell surface, and because we observe dose-dependent increases in cell length upon fosfomycin treatment, we propose that such a sensor is most likely measuring the accumulated amount of PG precursors. Future studies of PG precursor levels over the cell cycle as well as biochemical investigation of candidate sensors will be critical for determining if and how such a threshold functions in cells.
Because multiple checkpoints must be passed for cells to complete division, the one that is rate limiting could change based on the physiological state of the cell, potentially giving rise to different growth patterns. For fast-growing cells at steady state, it was recently demonstrated that an “adder” mechanism is at play (reviewed in Jun and Taheri-Araghi, 2015). Our simulations revealed that dividing after a target amount of excess SA material has been accumulated produces just such a growth pattern. Indeed, we found that to achieve an “adder” mechanism within the context of our simulations, the “trigger” molecule must be produced at a rate proportional to volume, a characteristic that we have shown to be the case for SA material, since this mode of synthesis is the central hypothesis underlying the “relative rates” model. Furthermore, the accumulated amount must be reset at the start of every cell cycle, a requirement that would be robustly achieved by using the accumulated SA material to synthesize two new end caps. This type of general model has been previously proposed without a specific molecular mechanism (Fantes et al., 1975; Sompayrac and Maaloe, 1973). These facts, coupled with evidence presented above, strongly suggest that SA material accumulation could underlie the observed “adder” pattern of bacterial growth at steady state, as well as the modulation of cell length in different physiological conditions in order to meet the SA/V requirements of the “relative rates” model.
Experimental Procedures
Growth Conditions
All bacterial cultures were inoculated from frozen stocks (C. crescentus and E. coli) or from a plate (L. monocytogenes), grown overnight, diluted into fresh medium in the morning, and grown according to standard conditions. See Supplemental Experimental Procedures for details.
Imaging Conditions
For dose-response experiments, static images were acquired by imaging cells on a 1% agarose pad made using the medium cells had been grown in using an upright fluorescence microscope (Zeiss, Thornwood, NY, USA) equipped with a Plan-Apo 1.4 100x phase objective, conventional epifluorescence filter set, and 1024 × 1024 back illuminated EM-CCD camera (Andor, South Windsor, CT, USA), producing an effective pixel size of 66 nm. Dynamic experiments were imaged using a Nikon Eclipse TiE enclosed in a temperature controlled chamber (Haison Technology, Taipei, Taiwan) set to 30°C in the case of C. crescentus and 37°C in the case of E. coli using a Plan-Apo 1.45 100x phase objective and 1024 × 1024 back illuminated EM-CCD camera (Andor, South Windsor, CT, USA), producing an effective pixel size of 86 nm. E. coli nutritional upshift experiments were performed using an ONIX Microfluidic Perfusion Platform (CellASIC, Hayward, CA, USA). C. crescentus perfusion experiments were performed using a low profile RC-31 flow chamber (Warner, Hamden, CT, USA). See Supplemental Experimental Procedures for further details.
Cell Segmentation, Shape, α, and β Determination
All image analysis was performed using MicrobeTracker (Sliusarenko et al., 2011) to find cell outlines and divide cells into 1-pixel-thick segments perpendicular to their long axes. The rate of α for each time point, t, was calculated by finding the difference in volume between the following (t+1) and previous (t−1) frames, ensuring that this difference was not large and negative meaning a division event had occurred, and dividing this difference by the volume at the current time. β was calculated similarly, except the difference in SA was divided by the current volume. See Supplemental Experimental Procedures and Figure S7 for further details.
Supplementary Material
Acknowledgments
We thank members of the Theriot lab as well as K.C. Huang and L. Shapiro and their labs for helpful suggestions, particularly E. Rojas, X. Zhou, and M. Footer. We thank S. Crosson, L. Shapiro, D. Portnoy, J. Dawson, and S. Austin for strains and C. Gross and J. Peters for sharing unpublished data. Helpful comments on the manuscript were provided by A. Colavin, N. Dye, A. Kennard, and E. Rojas. This work was supported by an NSF Graduate Research Fellowship to L.K.H., NIH R01 AI067712, NIH R01 AI036929, the Stanford Center for Systems Biology (P50-GM107615), and HHMI.
Footnotes
Author Contributions
L.K.H. and J.A.T. conceived of the project and wrote the paper. L.K.H. performed all experiments and data analysis.
References
- Aarsman MEG, Piette A, Fraipont C, Vinkenvleugel TMF, Nguyen-Distèche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Amir A. Cell Size Regulation in Bacteria. Phys. Rev. Lett. 2014;112 [Google Scholar]
- Basan M, Zhu M, Dai X, Warren M, Sévin D, Wang Y-P, Hwa T. Inflating bacterial cells by increased protein synthesis. Mol. Syst. Biol. 2015;11 doi: 10.15252/msb.20156178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-Mediated General Stress Response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Surovtsev IV, Kato S, Paintdakhi A, Beltran B, Ebmeier SE, Jacobs-Wagner C. A Constant Size Extension Drives Bacterial Cell Size Homeostasis. Cell. 2014;159:1433–1446. doi: 10.1016/j.cell.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y-HM, Marshall WF. How Cells Know the Size of Their Organelles. Science. 2012;337:1186–1189. doi: 10.1126/science.1223539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastanet A, Carballido-Lopez R. The actin-like MreB proteins in Bacillus subtilis: a new turn. Front. Biosci. Sch. Ed. 2012;4:1582–1606. doi: 10.2741/s354. [DOI] [PubMed] [Google Scholar]
- Cooper S. Rate and topography of cell wall synthesis during the division cycle of Salmonella typhimurium. J. Bacteriol. 1988;170:422–430. doi: 10.1128/jb.170.1.422-430.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli. Br. J. Mol. Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- Deforet M, van Ditmarsch D, Xavier JB. Cell-Size Homeostasis and the Incremental Rule in a Bacterial Pathogen. Biophys. J. 2015;109:521–528. doi: 10.1016/j.bpj.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Reisenauer A, Shapiro L. Feedback control of a master bacterial cell-cycle regulator. Proc. Natl. Acad. Sci. 1999;96:6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Fisher IC, Shapiro L, Theriot JA. Mutations in the nucleotide binding pocket of MreB can alter cell curvature and polar morphology in Caulobacter. Mol. Microbiol. 2011;81:368–394. doi: 10.1111/j.1365-2958.2011.07698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes PA, Grant WD, Pritchard RH, Sudbery PE, Wheals AE. The regulation of cell size and the control of mitosis. J. Theor. Biol. 1975;50:213–244. doi: 10.1016/0022-5193(75)90034-x. [DOI] [PubMed] [Google Scholar]
- Gamba P, Veening J-W, Saunders NJ, Hamoen LW, Daniel RA. Two-Step Assembly Dynamics of the Bacillus subtilis Divisome. J. Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gánti T. Organization of chemical reactions into dividing and metabolizing units: the chemotons. Biosystems. 1975;7:15–21. doi: 10.1016/0303-2647(75)90038-6. [DOI] [PubMed] [Google Scholar]
- Godin M, Delgado FF, Son S, Grover WH, Bryan AK, Tzur A, Jorgensen P, Payer K, Grossman AD, Kirschner MW, et al. Using buoyant mass to measure the growth of single cells. Nat. Methods. 2010;7:387–390. doi: 10.1038/nmeth.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Yeh Y-C, Hong S-H, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. Assembly of the Caulobacter cell division machine. Mol. Microbiol. 2011;80:1680–1698. doi: 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LK, Dye NA, Theriot JA. A Caulobacter MreB mutant with irregular cell shape exhibits compensatory widening to maintain a preferred surface area to volume ratio. Mol. Microbiol. 2014;94:988–1005. doi: 10.1111/mmi.12811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill NS, Buske PJ, Shi Y, Levin PA. A Moonlighting Enzyme Links Escherichia coli Cell Size with Central Metabolism. PLoS Genet. 2013;9:e1003663. doi: 10.1371/journal.pgen.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Biswas S, Wright CS, Henry JT, Lo K, Burov S, Lin Y, Crooks GE, Crosson S, Dinner AR, Scherer NF. Scaling laws governing stochastic growth and division of single bacterial cells. Proc. Natl. Acad. Sci. 2014;111:15912–15917. doi: 10.1073/pnas.1403232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd EM, Ryan KR, Moerner WE, Shapiro L, McAdams HH. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8235–8240. doi: 10.1073/pnas.1433105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Taheri-Araghi S. Cell-size maintenance: universal strategy revealed. Trends Microbiol. 2015;23:4–6. doi: 10.1016/j.tim.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Kahan FM, Kahan JS, Cassidy PJ, Kropp H. The mechanism of action of fosfomycin (phosphonomycin) Ann. N. Y. Acad. Sci. 1974;235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- Kruse T, Møller-Jensen J, Løbner-Olesen A, Gerdes K. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R. Mechanisms of Intracellular Scaling. Annu. Rev. Cell Dev. Biol. 2012;28:113–135. doi: 10.1146/annurev-cellbio-092910-154158. [DOI] [PubMed] [Google Scholar]
- Marquardt JL, Brown ED, Lane WS, Haley TM, Ichikawa Y, Wong CH, Walsh CT. Kinetics, stoichiometry, and identification of the reactive thiolate in the inactivation of UDP-GlcNAc enolpyruvoyl transferase by the antibiotic fosfomycin. Biochemistry (Mosc.) 1994;33:10646–10651. doi: 10.1021/bi00201a011. [DOI] [PubMed] [Google Scholar]
- Monds RD, Lee TK, Colavin A, Ursell T, Quan S, Cooper TF, Huang KC. Systematic Perturbation of Cytoskeletal Function Reveals a Linear Scaling Relationship between Cell Geometry and Fitness. Cell Rep. 2014;9:1528–1537. doi: 10.1016/j.celrep.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J. The Growth of Bacterial Cultures. Annu. Rev. Microbiol. 1949;3:371–394. [Google Scholar]
- Morgan JJ, Surovtsev IV, Lindahl PA. A framework for whole-cell mathematical modeling. J. Theor. Biol. 2004;231:581–596. doi: 10.1016/j.jtbi.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Previc EP. Biochemical determination of bacterial morphology and the geometry of cell division. J. Theor. Biol. 1970;27:471–497. doi: 10.1016/s0022-5193(70)80010-8. [DOI] [PubMed] [Google Scholar]
- Pritchard RH. Review lecture on the growth and form of a bacterial cell. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1974;267:303–336. doi: 10.1098/rstb.1974.0003. [DOI] [PubMed] [Google Scholar]
- Real G, Henriques AO. Localization of the Bacillus subtilis murB Gene within the dcw Cluster Is Important for Growth and Sporulation. J. Bacteriol. 2006;188:1721–1732. doi: 10.1128/JB.188.5.1721-1732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaechter M, Maaløe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J. Gen. Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Schaechter M, Williamson JP, Hood JR, Koch AL. Growth, Cell and Nuclear Divisions in some Bacteria. J. Gen. Microbiol. 1962;29:421–434. doi: 10.1099/00221287-29-3-421. [DOI] [PubMed] [Google Scholar]
- Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sompayrac L, Maaloe O. Autorepressor model for control of DNA replication. Nature. New Biol. 1973;241:133–135. doi: 10.1038/newbio241133a0. [DOI] [PubMed] [Google Scholar]
- Sud IJ, Schaechter M. Dependence of the Content of Cell Envelopes on the Growth Rate of Bacillus Megaterium. J. Bacteriol. 1964;88:1612–1617. doi: 10.1128/jb.88.6.1612-1617.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, Jun S. Cell-Size Control and Homeostasis in Bacteria. Curr. Biol. 2015;25:385–391. doi: 10.1016/j.cub.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanouchi Y, Pai A, Park H, Huang S, Stamatov R, Buchler NE, You L. A noisy linear map underlies oscillations in cell size and gene expression in bacteria. Nature. 2015;523:357–360. doi: 10.1038/nature14562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2011;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmer B, Heinemann M. Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PloS One. 2011;6:e23126. doi: 10.1371/journal.pone.0023126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Robert L, Pelletier J, Dang WL, Taddei F, Wright A, Jun S. Robust Growth of Escherichia coli. Curr. Biol. 2010;20:1099–1103. doi: 10.1016/j.cub.2010.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jones BD, Brun YV. A set of ftsZ mutants blocked at different stages of cell division in Caulobacter. Mol. Microbiol. 2001;40:347–360. doi: 10.1046/j.1365-2958.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- Weart RB, Lee AH, Chien A-C, Haeusser DP, Hill NS, Levin PA. A Metabolic Sensor Governing Cell Size in Bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wientjes FB, Nanninga N. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J. Bacteriol. 1989;171:3412–3419. doi: 10.1128/jb.171.6.3412-3419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh CL, Huls P, Pas E, Brakenhoff GJ, Nanninga N. Topography of Peptidoglycan Synthesis during Elongation and Polar Cap Formation in a Cell Division Mutant of Escherichia coli MC4100. Microbiology. 1987;133:575–586. [Google Scholar]
- Yao Z, Davis RM, Kishony R, Kahne D, Ruiz N. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc. Natl. Acad. Sci. 2012;109:E2561–E2568. doi: 10.1073/pnas.1209742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. Bacterial shape: two-dimensional questions and possibilities. Annu. Rev. Microbiol. 2010;64:223–240. doi: 10.1146/annurev.micro.112408.134102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.