Protein synthesis is a central biological process in which all proteins needed for the life of the organism are produced. Its base is the ribosome, a complex system with several RNA and many protein molecules and many functional steps.1 Deletion of some component may in some cases lead to marginal effects but in other cases a catastrophe. Thus the large RNA molecules are absolutely essential, but the smallest ribosomal RNA, the 5S RNA, is absent in mitochondrial ribosomes. Some proteins have been deleted without severe consequences, but some are crucial for well functioning ribosomes.
The protein L11 is found in ribosomes from all species and its universal occurrence has therefore led to the addition of the letter u before its name, uL11.2 uL11 has received significant attention for a long time. Now, when ribosomal structures are known from many species, we know that uL11 has 2 lobes or domains connected by a linker. Its location in the large (60S) subunit is at the GTPase activating center (GAC), which also is composed of many components. Their individual roles are gradually emerging. uL11 is next to a structure called the stalk composed of different proteins in bacteria (bL12) and eukaryotes (P1/P2). Their roles are not fully established, but they have important interactions with all ribosomal GTPases. Protein uL10 (formerly P0 in eukaryotes) forms the base of the stalk and is a neighbor of uL11.
The role of uL11 has not been fully characterized, but in a recent paper Wawiórka et al.3 have studied yeast ribosomes where both genes producing uL11 were shut off to analyze the deficient ribosomes in vivo. A prime observation is that yeast cells survive without uL11, but the translation efficiency was reduced by 70%. The further analysis showed that a multitude of functional steps are affected. Starting from the assembly of ribosomes or its biogenesis uL11 is important also in these early steps.
The biogenesis of eukaryotic ribosomes is a complex process. Part of it concerns the ribosomal RNAs that are processed stepwise from a single transcript. The maturation of the 40S and 60S subunits starts in the nucleolus and is completed in the cytoplasm. Several proteins with chaperone-like activities participate to assist in the maturation of ribosomal subunits. In many of these steps the GAC has an important role. The initial steps in producing the correct lengths, 25S, 18S and 5.8S, of the ribosomal RNAs (rRNAs) are not affected by the lack of uL11. However, the 60S subunits do not mature properly since the rRNAs are degraded prematurely. How does the defective GAC prevent 60S maturation?
One of the last steps in maturation of 60S subunits is the release of the anti-association factor Tif6 from the subunit. This is done by Efl1.4 Efl1 is a paralogue of eEF2, the elongation factor that in complex with GTP primarily translocates peptidyl-tRNA from the A-site to the P-site in order to leave room for a new aminoacyl-tRNA to bind to the A-site. Tif6 (also called eIF6) is a eukaryotic initiation factor preventing premature association between large and small subunits. From the series of experiments by Wawiórka et al.3 it is clear that Tif6 accumulates bound to 60S subunits preventing the full maturation of the subunits when uL11 is deleted. The same effect is also observed when Efl1 is deleted. Thus, the proper release of Tif6 requires both Efl1 and uL11 (Fig. 1). The joining of the 2 subunits depends on the GTPase eIF5B and is also affected by lack of uL11 in the GAC.
Figure 1.
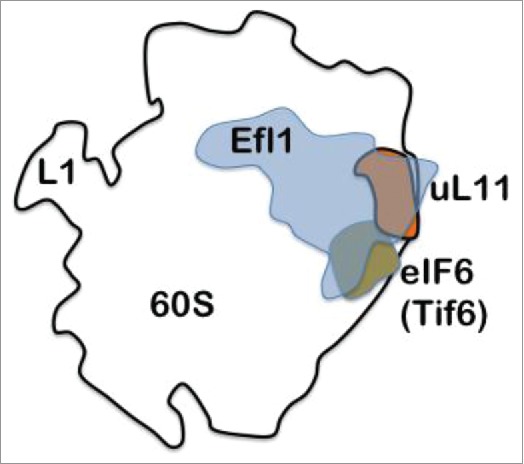
Tif6 or eIF6 prevents the association of the small and large subunits. Its location is near ribosomal protein uL11. Efl1, which depends on the presence of uL11 for its function, is required to release Tif6 from he large subunit. Without uL11 the removal of Tif6 fails.
The elongation rate of uL11-deprived ribosomes is reduced by 30% from wild-type rate. The two factors interacting with GAC during elongation are eEF1A and eEF2. eEF1A in complex with GTP carries aminoacyl-tRNA into the ribosome for the decoding process and subsequent peptidyl transfer. The release of eEF1A is much accelerated by GTP hydrolysis, which depends on a fully functional GAC. Likewise GAC is needed for eEF2 translocation. Evidently the lack of uL11 affects several activities that require GTP hydrolysis. uL11 is obviously a central player in translation! Further analysis is needed to fully understand the process and the detailed role of uL11. In this process mutational analysis and structural investigations will be useful.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Liljas A and Ehrenberg M. 2013. Structural aspects of protein synthesis. 2nd edition World Scientific, Singapore: ISBN 978-9814313209. [Google Scholar]
- [2].Ban N, et al.. Curr. Op. Struct. Biol. 2014; 24:1-5 http://dx.doi.org/ 10.1016/j.sbi.2014.01.002. [DOI] [Google Scholar]
- [3].Wawiórka et al.. Cell Cycle 2016; 15:1060-72; http://dx.doi.org/ 10.1080/15384101.2016.1154245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Weis F, et al. Nature Struct Mol Biol 2015: 22:914-919; PMID: 26479198; http;//dx.doi.org/ 10.1038/nsmb.3112 [DOI] [PMC free article] [PubMed] [Google Scholar]


