ABSTRACT
CGGBP1 (CGG triplet repeat-binding protein 1) regulates cell proliferation, stress response, cytokinesis, telomeric integrity and transcription. It could affect these processes by modulating target gene expression under different conditions. Identification of CGGBP1-target genes and their regulation could reveal how a transcription regulator affects such diverse cellular processes. Here we describe the mechanisms of differential gene expression regulation by CGGBP1 in quiescent or growing cells. By studying global gene expression patterns and genome-wide DNA-binding patterns of CGGBP1, we show that a possible mechanism through which it affects the expression of RNA Pol II-transcribed genes in trans depends on Alu RNA. We also show that it regulates Alu transcription in cis by binding to Alu promoter. Our results also indicate that potential phosphorylation of CGGBP1 upon growth stimulation facilitates its nuclear retention, Alu-binding and dislodging of RNA Pol III therefrom. These findings provide insights into how Alu transcription is regulated in response to growth signals.
Keywords: Alu-SINEs; CGGBP1; ChIP-seq; growth signals; RNA Pol III; transcription; tyrosine phosphorylation
Introduction
Repetitive elements comprise more than 50% of the human genome, with the interspersed repeats alone accounting for more than its one third.1,2 About 20% of the genome is occupied by long interspersed nuclear elements (LINEs) and another 13% by short interspersed nuclear elements (SINEs). The L1-LINEs (full-length ones upto 6–8Kb long) and <300 bps long Alu-SINEs constitute the majority of LINEs and SINEs respectively.3-5 The Alu-SINEs originated from head-to-tail fusion of 7SL RNA5 and are the most numerous known transcription-units of a kind.6 Although they were previously thought to be junk DNA, they are now known to exert important functions. Alu-SINEs play vital roles in shaping the human genome (by affecting the genetic content and participating in DNA damage and repair),7,8 epigenome (through functions as boundary elements, nucleosome-positioning sites, CpG methylation and histone modification sites)9-13 and transcriptome (by influencing splicing, mRNA stability and RNA Pol II activity under stress and by acting as RNA Pol II transcription-regulatory regions).14-18 Thus they exert profound influence on human evolution as well as on development and differentiation at the cellular level. Transcription of Alu elements ultimately determines their prevalence in the transcriptome, and through associated retrotransposition, in the genome as well. Understanding the mechanisms of Alu transcription/silencing is thus of primary importance for comprehending the regulation and function of our genome and transcriptome.
Structurally, the Alu promoters belong to the RNA Pol III type 2 category, same as those of the 7SL genes/pseudogenes and cytoplasmic tRNA genes.19 A defining feature of these promoters is the presence of consensus A-box and B-box sequences downstream of the transcription start site (TSS).19 These sequence elements serve as binding sites for various RNA Pol III subunit proteins such as BRF1, TFIIIC (GTF3C1) and POLR3F and regulate the positioning of RNA Pol III at an upstream start site.19,20
Expression of the tRNA genes, as well as other RNA Pol III target genes such as 7SL and 5S rRNA are important for protein synthesis and are particularly important in growth-stimulated cells. Although endowed with similar promoter structures and RNA polymerase requirements, the much larger population of Alu-SINEs (an estimated 1.5 million) give rise to very low levels of Alu RNA,11,12,15,21-23 whereas the much fewer tRNA genes (<1000 genome-wide) give rise to between 15–30% of total RNA,24,25 indicating discriminate recruitment of RNA Pol III at non-Alu promoters.
Although both genetic and epigenetic mechanisms potentially affect Alu RNA production,11-13,15,23,26-31 the precise mechanisms regulating Alu RNA levels remain elusive. Apparently, there are mechanisms, which preferentially direct RNA Pol III to tRNA genes and not at Alu-SINEs. For instance, in growth-stimulated cells, RNA Pol III is catalytically active and used to generate tRNAs whereas Alus are prevented from getting transcribed. Such target-specifying mechanisms likely reflect sequence differences at critical transcription-regulatory regions of Alu-SINE or tRNA promoters. The dissimilarities between the Alu transcription enhancer (ATE) sequence (containing the A-box) of Alu-SINEs and the corresponding region of tRNA genes are more pronounced than those between their transcription-directing elements (containing the B-box)32 (Singh and Westermark, unpublished observation). Although the ATE sequence is known to be an RNA Pol III-enhancing sequence with positive effects on transcription,32 its potential role in Alu-silencing is not reported. Any possible mechanism of Alu-silencing through recruitment of repressor proteins on the A-box, however, should exhibit heat shock-sensitivity as the epigenetically silenced Alu-SINEs are promptly transcribed in response to heat shock stress.18,33 We show here that the ATE sequence serves as a target site for CGGBP1, a repeat-binding transcription regulatory protein, which antagonizes sequestering of RNA Pol III at Alu-SINEs specifically in growth-stimulated cells.
CGGBP1 (CGG triplet repeat-binding protein 1)34 has been implicated in a variety of functions, including rRNA transcription, transcription regulation of FMR1, HSF1, GAS1 and CDKN1A in-cis, and telomere protection.35-40 Depletion of CGGBP1 impacts cell cycle progression and causes G1 or G2 arrest with abscission failure.39,40 Notably, one of the functions of CGGBP1 is immediate transcription regulation in response to heat-shock and its depletion mimics heat-shock in terms of gene expression changes, viz. up-regulation of HSF1 transcription.38 The pan-nuclear staining of CGGBP1, however, suggests that the rarely occurring (CGG)xn sequences could not be the only sites to which CGGBP1 is recruited. Either CGGBP1 binds to a variety of diverse sequences, or has a sequence-independent chromatin binding property, or binds to highly prevalent sequence motifs.
We show here that CGGBP1 is a serum-dependent mediator of serum-induced changes in global gene expression, through enhanced nuclear presence with no detectable preference for CGG repeat-associated genes. By sequencing genomic DNA bound to CGGBP1, we identify that CGGBP1 is bound to the Alu-SINEs preferentially at the ATE region and suppresses Alu transcription in cis. The global effect of CGGBP1 on expression of RNA Pol II-transcribed genes seems to be partially in trans through Alu RNA-dependent inhibition of RNA Pol II. Additionally, our results indicate that in response to growth factor stimulation, potential Y20 phosphorylation of CGGBP1 and subsequent dislodging of RNA Pol III from Alu promoters could be the underlying mechanism.
We thus propose a novel mechanism of growth-associated silencing of Alu-SINEs in which CGGBP1 translates growth factor signals into transcriptional regulation of growth associated gene expression.
Results
Global gene expression changes caused by CGGBP1 depletion are diminished by serum-starvation
CGGBP1 regulates transcription as well as cellular proliferation and growth. Here we explored the possibility that CGGBP1 targets genes differently in growing (serum-stimulated) and quiescent (serum-deprived) cells. Normal human foreskin fibroblasts 1064Sk, stably transduced with lentivirally-expressed CGGBP1 shmiR or control shmiR (efficiency of CGGBP1 knockdown shown in Fig. S1), were mock-stimulated (serum-free medium) or serum-stimulated (10% fetal calf serum) for 12 h after 72 h of serum-deprivation. Global transcriptome profiling in triplicates using Affymetrix arrays showed that the differences in gene expression caused by CGGBP1-depletion are diminished in quiescent cells (Two-way ANOVA; p value cutoff 0.01) (Fig. 1A and B and Table S1). The expression levels of 802 serum-and-CGGBP1-co-regulated genes (Two-way ANOVA P < 0.01) showed extremely high correlation between control and CGGBP1-depleted quiescent samples (r2 = 0.995, Fisher test F = 3.405, Fig. 1B; blue dots). This correlation was lost in serum-stimulated samples due to differential induction/repression of genes between control- or CGGBP1-depleted cells (r2 = 0.444, Fisher test F = 0.804, Fig. 1B; red dots). Supporting this, the hierarchical clustering of samples based on differentially expressed genes showed that the quiescent samples (both control or CGGBP1 shmiR-transduced) closely clustered together, whereas the serum-stimulated samples clustered together but with larger distance between them. (Two-way ANOVA P < .01) (Fig. S2, A to E). This suggested that serum-induced gene expression changes involve serum-dependent transcription-regulatory functions of CGGBP1. Interestingly, among the serum-and-CGGBP1-co-regulated genes, no specific functional category was enriched.
Figure 1.
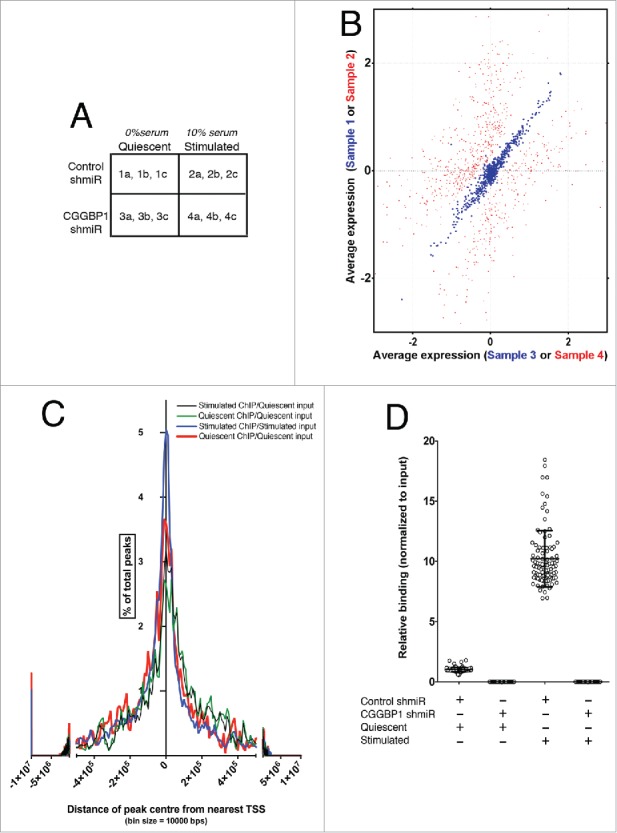
Serum stimulation affects gene expression regulation by and DNA-binding pattern of CGGBP1. (A) A matrix describing the sample treatments and nomenclature. The alphabets “a," “b” and “c” denote technical replicates for each sample derived from a pool of 5 biological and experimental replicates. (B) Correlation plot showing lack of co-variability in expression values of CGGBP1-and-serum co-regulated genes between samples 2 and 4 (red spots, r2 = 0.444, Fisher test F = 0.804) and very high co-variability in expression values of CGGBP1-serum affected genes between samples 1 and 3 (blue spots, r2 = 0.995, Fisher test F = 3.405). Data are from mean expression values from 3 technical replicates of 5 pooled biological and experimental samples. (C) Frequency distribution of CGGBP1-ChIP-seq peaks in relation to the TSS of nearest protein-coding/non-coding genes. Percent of total number of peaks is shown on Y-axis and distance from TSS on X-axis. The summit shows an enrichment of genes around the TSS, although the long units on X-axis means that these distance are still very large. (D) ChIP qPCR showing the specificity of (by using shRNA-knockdown) and changes in the binding (upon serum-stimulation) of CGGBP1 to ChIP-seq peaks. Increase in binding in Stimulated over Quiescent is highly significant (T-test, p = 3.785E-60). Different data points represent one peak randomly chosen from each chromosome (n = 23 × 4 replicates).
Using MEME Suite (Multiple Em for Motif Elicitation) tool,41 a discriminative motif-search on promoter sequences (−1kb from TSS) of the serum-and-CGGBP1 co-regulated genes (using the promoters of 1000 genes which exhibited the least change in expression as background) did not reveal any motifs associated with the deregulated genes.
We next asked how CGGBP1 regulates gene expression in response to serum stimulation. To objectively identify potential growth-associated CGGBP1-binding sites, we performed chromatin immunoprecipitation-sequencing (ChIP-seq) for CGGBP1 under serum-stimulated and quiescent conditions.
CGGBP1 ChIP sequencing reveals a DNA motif switch upon serum stimulation
ChIP was performed to identify DNA bound to CGGBP1 in quiescent or stimulated 1064Sk human foreskin fibroblasts. ChIP was performed in 5 replicates, pooled and sequenced in triplicates. Peaks of CGGBP1 binding were identified, using input controls for each group. The 4 combinations of 2 ChIP and 2 input samples were as follows: Quiescent ChIP/Stimulated input, Stimulated ChIP/Stimulated input, Stimulated ChIP/Quiescent input, and Quiescent ChIP/Quiescent input. The quality control parameters of ChIP-seq, read mapping and peak calling are provided in Table S2. All peaks satisfied a maximum p value of 1.0E-05. The genomic locations and statistical properties of the peaks are shown in the Table S3.
The mean distance between the peaks and nearest genes ranged between 280 Kb in Stimulated ChIP/Quiescent input to 670 Kb in Stimulated ChIP/Stimulated input (Fig. 1C). There was no proximity between the CGGBP1-target genes identified in the microarray-based analysis and the ChIP peaks. These findings indicated a trans regulation of gene expression by CGGBP1.
To identify signature sequences that define CGGBP1-binding sites, we performed a motif search41 on the peak sequences using MEME motif identification tool. We observed that a long sequence motif ([GA]CCTGTA[AG]TCCCAGC[TA][AC]-[CT]T[TC][GA]GGAGGC[TC]GAGGC[AG]G) was highly enriched in Stimulated peaks compared to the Quiescent peaks (referred to as Stimulation-enriched or SE motif). By qPCR on ChIP DNA we confirmed that CGGBP1 was indeed bound to the identified peaks (one SE motif-containing peak randomly selected from each chromosome) and that this binding was strongly and highly significantly enhanced upon serum stimulation (T-test, p = 3.785E-60; Fig. 1D). When performing the same motif search on repeat-masked peak sequences (by using RepeatMasker42), the SE motif was not detected.
These results showed (i) that CGGBP1 preferentially binds to the SE motif in Stimulated cells, and (ii) that the SE motif is associated with a repetitive sequence.
CGGBP1 binding switches from predominantly on L1-LINEs to Alu-SINEs upon serum stimulation
RepeatMasker analysis of peak sequences obtained by using only 100% uniquely mapped reads revealed the presence of interspersed repeats (Table 1 and Table S4). Alu-SINEs and L1-LINEs, the most abundant repeat elements identified in the peaks, showed an increase and decrease, respectively, in Stimulated peaks compared to Quiescent peaks (Table 2). Thus, CGGBP1 emerged as an interspersed repeat-binding protein exhibiting a shift from L1 to Alus upon serum stimulation.
Table 1.
The table shows repeat contents in Quiescent ChIP/Stimulated input, Stimulated ChIP/Stimulated input, Stimulated ChIP/Quiescent input, and Quiescent ChIP/Quiescent input peaks identified by RepeatMasker. The values are percentages of the combined peak lengths for each of the 4 samples separately. It is noticeable that the percentage share of Alus in the Stimulated ChIP samples (second and fourth columns, top row) are between 2 to 4 folds higher than the expected 10% (approximately) content in the genomic sequence. A converse pattern of enrichment of prevalence of LINEs in Quiescent ChIP samples is seen (first and third columns, second row). The percentages of other types of sequences do not show such marked variability. The Alu and LINE values in the dagger-marked rows were subjected to statistical testing as shown in Table 2.
| Quiescent ChIP/Stimulated input | Stimulated ChIP/Stimulated input | Quiescent ChIP/Quiescent input | Stimulated ChIP/Quiescent input | |
|---|---|---|---|---|
| † SINEs | 7.27% | 38.64% | 11.52% | 26.3% |
| † LINEs | 39.04% | 22.87% | 34.1% | 28.69% |
| Satellites | 23.89% | 16.74% | 18.05% | 17.61% |
| Simple repeats | 8.75% | 6.99% | 13.88% | 10.32% |
| LTR elements | 0.91% | 1.07% | 0.71% | 0.98% |
| Not Masked | 19.82% | 12.54% | 21.53% | 15.67% |
| Low Complexity repeats | 0.05% | 0.09% | 0.04% | 0.03% |
| DNA elements | 0.23% | 0.14% | 0.05% | 0.03% |
| Unclassified | 0.04% | 0.76% | 0.12% | 0.32% |
| Small RNA | 0% | 0.16% | 0% | 0.05% |
Table 2.
A 2-tailed Fisher's exact test (99% confidence interval) shows that the increase of Alu content and the decrease of L1 content in Stimulated peaks as compared to Quiescent peaks is highly significant. The “–” marked entries indicate that no tests were performed for comparison between same samples. p values of Fisher's exact test are mentioned for each set of comparison between the 2 ChIP samples using the 2 inputs as denominators.
| Quiescent ChIP/Stimulated input | Stimulated ChIP/Stimulated input | Quiescent ChIP/Quiescent input | Stimulated ChIP/Quiescent input | |
|---|---|---|---|---|
| Quiescent ChIP/Stimulated input | – | <0.0001 **** | 0.303 | 0.0005 *** |
| Stimulated ChIP/Stimulated input | <0.0001 **** | – | <0.0001 **** | 0.1864 |
| Quiescent ChIP/Quiescent input | 0.303 | <0.0001 **** | – | 0.0135 * |
| Stimulated ChIP/Quiescent input | 0.0005 *** | 0.1864 | 0.0135 * | – |
To establish that the identification of Alu elements was not confounded by their sequence homology with the 7SL genes we applied 2 conditions: (i) using only 100% uniquely-mapped reads for peak-calling, and (ii) using >35 bp long reads only (minimum read length required to differentiate 7SL from Alus; Fig. S3A). The mean read length was 52 bps for Stimulated sample and 46 bps for Quiescent sample. We then verified the authenticity of CGGBP1-Alu binding by rigorous computational and experimental analyses.
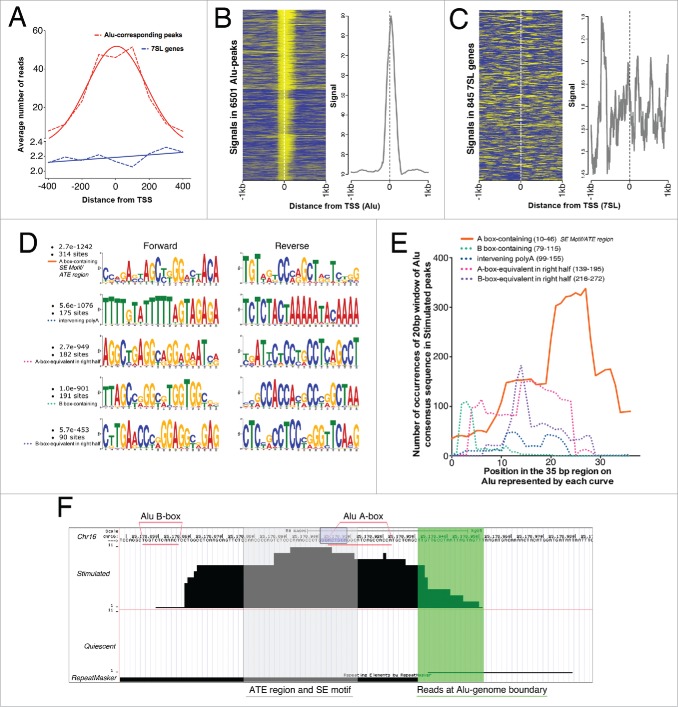
Any carry-over of reads between 7SL and Alus would affect the normal distribution of reads in the peaks. Mapping of reads from −400 to +400 bps from the start sites of Alus showed that (i) the reads were normally distributed in the peaks, (ii) the peaks’ summit was just downstream of the Alu start site, and (iii) the tails of the peaks extended similarly into the Alus as well as out of the Alus into non-repetitive genomic sequences (Fig. 2A). For the 7SL sequences, there was neither any read-buildup within the genes or outside them (Fig. 2A). Since the post-sonication fragment size subjected to ChIP was 150 bps, we plotted the signals43 in units of 150 bps (by collecting the reads mapping to 150 bp segments) from −1kb to +1kb at the Alu-matching regions of Stimulated peaks. Indeed, we observed clustering of signals just downstream of Alu start sites with a 2-tailed presence of signal both into Alus and upstream of Alus into non-repeat regions (Fig. 2B). In contrast, the signals on the 7SL genes were scattered with no clear peak-like buildup and no binomial distribution (Fig. 2C). Further, qPCR on Stimulated CGGBP1-ChIP DNA showed that the relative enrichment for Alus was approximately 2 orders of magnitude higher than that of 7SL genes (Fig. S3B, P < .0001). It was thus confirmed that the Alus are bona-fide CGGBP1-binding sites without any interference from their repetitious nature and sequence similarity with the 7SL genes.
Figure 2.
CGGBP1 shows enhanced binding to a specific subsequence of Alu-SINEs upon serum stimulation. (A) Distribution of reads in Alu-matching Stimulated peaks shows a binomial distribution with the reads piling up to create a consensus summit just downstream of Alu TSS (red curve). The tails of the reads-distribution curve extend into Alus and non-Alu regions showing the robustness of peak calling and general peak structure of Alu-matching Stimulated peaks. Similar analysis for 7SL (blue curve) showed much weaker read density compared to Alus and no normal distribution of the reads thereby supporting the absence of 7SL genes in Stimulated peaks. X-axis shows distance from TSS and Y axis shows number of reads per 6196 Alu-matching peaks and 845 7SL genes/pseudogenes. Broken lines connect exact values and the solid lines are the best Gaussian fit curves. (B) Distribution of signals (yellow) derived by mapping reads in unit sequences of 150 bps (mean length of sonicated DNA subjected to ChIP) in the regions −1kb to +1kb from Stimulated peak-matching Alu start sites shows clustering of signals both upstream and downstream of Alu TSS with summit just downstream of TSS. (C) Same analysis (as shown in (B) for Alu-matching peaks) for 7SL genes show scattered reads with no clear clustering pattern. The differences in the Y-axis values of graphs in (B) and (C) show the high enrichment around Alu TSS but no enrichment around 7SL TSS. Eleven Alu-peaks and 4 7SL values were removed as outliers from the analysis. Similar binding pattern of CGGBP1 on Alu and absence of specific binding on 7SL genes was observed in Quiescent sample also (not shown). (D) Counts and p values of 20 bp Alu sub-sequences in Stimulated peaks (performed on Alu-matching peaks only using MEME suite); the maximum occurrence and highest probability were both associated with the topmost sequence that matches to the most proximal A-box-containing region of Alus corresponding to SE motif and ATE region. (E) Intra-Alu distribution of CGGBP1-binding sequences as occurring in Stimulated peaks derived by counting exact matches between Alu consensus sequence and Stimulated peaks (iterative 100% match of a 20 bp window with one base shift per iteration, using MS Excel). Number of counts on Y-axis and location on Alu on X-axis is shown. The solid orange curve in (E)corresponds to the ATE region/SE motif and has significantly higher occurrence than other regions plotted in broken lines (chi square test, P < .01). (F) A representative Alu-matching Stimulated-specific peak shown with different features highlighted (Region = Chr16: 25,178,839-25,179,005; Alu repeat detected by RepeatMasker = black bar below; Region of reads mapping to non Alu region of the Alu-genome junction = highlighted in green; Region of reads corresponding to peak summit, ATE region/SE motif = shaded in gray; most commonly occurring 8bp motif GGAYTACA = purple box; A-box and B-boxes = underlined with pink; chromosomal coordinates and scale = mentioned on top).
The above analysis, however, indicated an intra-Alu heterogeneity in CGGBP1 binding with maximal binding near the Alu start sites. To identify this further, the Alu-matching Stimulated peak sequences were subjected to a search for most-commonly occurring Alu sub-sequence using a motif size of 20 bps (Fig. 2D). The A-box and adjacent downstream region C[CG][AC]GA[GC][TC]AGC[TC][GT]GG-[AC][CT][TA]ACA (matching with the SE motif and ATE sequence) was found to be the most commonly occurring Alu-sub-sequence with 314 hits in the Stimulated dataset (MEME p = 2.7E-1242) (Fig. 2D, topmost motif). The maximum enrichment of this subset of Alu sequence in Stimulated peaks was recapitulated by measuring the frequency of occurrence of the canonical Alu consensus sequence (using a window size of 20 bases and sliding the window by 1 base per iteration) in Stimulated peaks. Thus the occurrence of ATE sequence at location +10 to +46 bps from Alu start site was significantly higher than the other sequences of the Alus: locations +79 to +115, +99 to +155, +139 to +195 and +216 to +272 (ANOVA P < .0001, Fig. 2E). A representative peak, with different landmarks highlighted, is shown in Figure 2F.
We then wanted to know how CGGBP1-ATE binding was relevant for global gene expression regulation by CGGBP1 as observed in the microarray experiments.
CGGBP1 regulates Alu RNA levels
First we asked if CGGBP1-ATE binding had consequences on Alu RNA levels. To quantify Alu RNA, we avoided any hybridization-based approach because the probes described so far do not discriminate between Alu RNA and 7SL transcripts. Instead we employed a recently described qRT-PCR-based approach with minor modifications.44 The Alu-reverse primer (Alu-rev-RA) sequence was extended by 3 bases into a region of sequence dissimilarity between 7SL and Alu sequences (Fig. 3A) and PCR conditions were optimized to ensure that the Alu-specific primers amplified specifically from the Alu template and not from the 7SL template (Fig. 3B).
Figure 3.
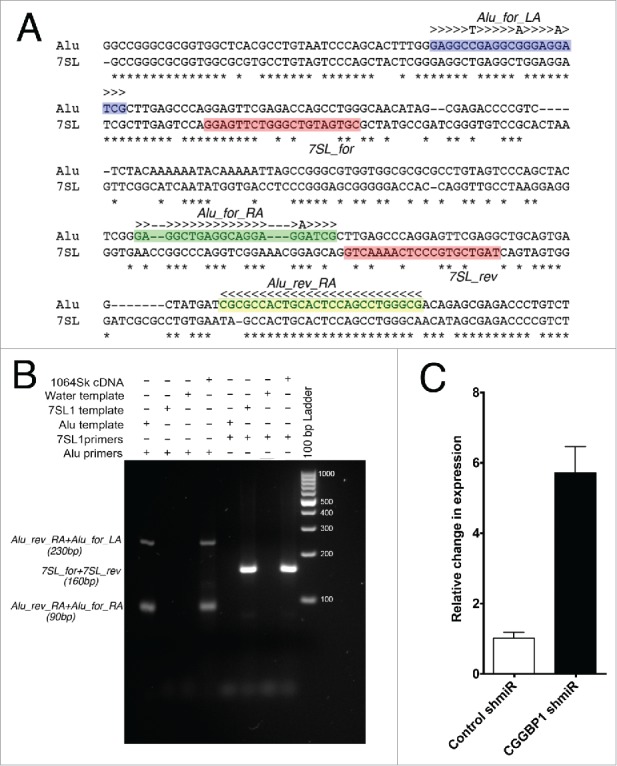
Alu and 7SL PCR primers, test of specificity and quantitation of Alu RNA. (A) Location of primers: Alu_rev_RA (yellow shade) will amplify a 90 bp fragment with Alu_for_RA (Alu forward primer annealing in right arm, green shade) and a 230 bp fragment with Alu_for_LA (Alu forward primer annealing in left arm, blue shade). The 2 7SL primers (7SL_for and 7SL_rev; pink shade) will amplify a 160 bp fragment. Poor sequence complementarity between 7SL primers and Alu consensus sequence ensures no Alu amplification by 7SL primers. Alu_rev_RA primer has been extended by 3 bases at the 3′ end (rest of the sequence the same as described by Marullo et al., 2010) to generate terminal mismatch between Alu_rev_RA primer and 7SL template. This prevent cross amplification of 7SL by Alu primers. (B) By using cDNA as a general template, purified 90 and 160 bps Alu fragments as Alu template and annealed oligos of 7SL1 sequence as 7SL-specific template (see methods for sequence), the specificity of the primers was confirmed. (C) qPCRs show that CGGBP1 depletion by CGGBP1 shmiR induced Alu RNA levels as compared to control shmiR (P < .0001; n = 3). Y-axis values are obtained from subtraction of Ct values of an all-sample-mix as an internal standard from the Ct values of each sample and the difference subjected to negative power of base 2. The values were then normalized to set control (Stimulated) to a mean of 1.
qRT-PCR showed that upon CGGBP1-depletion, there was indeed an increase in Alu RNA levels (Fig. 3C) suggesting that CGGBP1-ATE binding suppresses Alu transcription. In line with our findings that CGGBP1 attains increased Alu binding upon serum deprivation, we also found that Alu RNA levels were increased in Quiescent cells as compared to Stimulated cells (Fig. S4). Together these findings indicated a serum-dependent Alu inhibition by CGGBP1. It should, however, be noted that the Alu RNA detected by this method is derived from RNA Pol III as well as RNA Pol II. As will be discussed below, the increase in Alu RNA in CGGBP1-depleted cells is probably caused by an increase in Pol III regulated transcripts because Pol II is inhibited under the conditions of Alu RNA increase.
Increase in Alu RNA causes a decrease in RNA Pol II activity and thereby affects transcription of genes of all functional categories except those required for heat shock response.18 This mechanism could explain the generalized gene expression changes upon CGGBP1-depletion without enrichment of any specific functional category.
Alu RNA induction by CGGBP1 depletion inhibits RNA Pol II activity
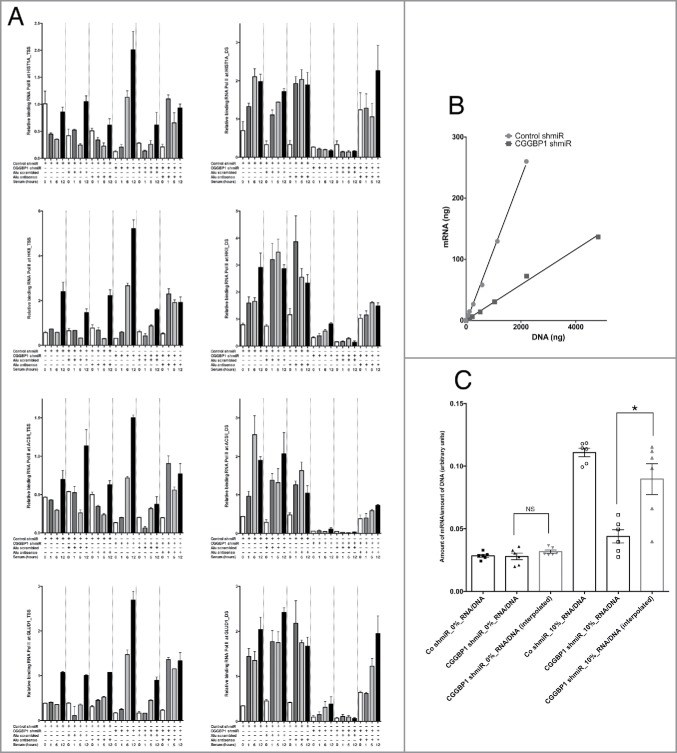
To ascertain that the increase in Alu RNA levels upon CGGBP1-depletion plays a role in RNA Pol II regulation, we employed a previously described experimental strategy.18 For HSP70 and U2 RNA genes, CGGBP1 depletion or antisense oligonucleotides against Alus in combination with serum stimulation or starvation did not bring any consistent differences in RNA Pol II occupancy at the TSS or at the downstream (DS) regions (Fig. S5). These genes were chosen as Alu-independent RNA Pol II-transcribed genes since Alu RNA impedes RNA Pol II function only at protein-coding genes not required for heat shock response.18 At the TSS of the protein-coding genes (not involved in heat shock response) HIST1A, HKII, ACSII and GLUD1, RNA Pol II showed a consistent occupancy, with small increases seen upon serum-stimulation, which was not affected by antisense oligonucleotides against Alus (Fig. 4A). At the DS region of these genes however, CGGBP1-depletion negatively affected RNA Pol II recruitment, which could be rescued by Alu-antisense oligonucleotides (Fig. 4A). The relative increase in RNA Pol II recruitment after 12 h serum stimulation in Alu antisense-treated sample as compared to Alu scrambled-treated sample is significant for all genes tested (Fig. 4A, T-test P < .05).
Figure 4.
Effect of Alu induction (upon CGGBP1-depletion) on RNA Pol II activity. (A) Measurement of RNA Pol II occupancy on TSS or DS regions of HIST1A, HKII, ACSII and GLUD1 showed that serum stimulation had a consistent enhancing effect on RNA Pol II occupancy at the DS regions. Unlike at the TSS (left panel), at the DS regions (right panel), CGGBP1 depletion reduced the RNA Pol II occupancy. At the DS regions, the RNA Pol II occupancy was not affected by Alu antisense treatment without CGGBP1 depletion, whereas in CGGBP1 depleted samples, a concomitant RNA interference against Alu rescued the RNA Pol II occupancy. Serum treatment also enhanced the RNA Pol II occupancy at the DS regions and at 2 (HIST1A and ACSII) of the 4 housekeeping genes tested, this effect of serum was vanished by depleting CGGBP1 but could be rescued by combining CGGBP1 depletion with Alu antisense. Input-normalized ddCt values are plotted on Y-axis (n = 4 for each bar; for serum 12h data-points all comparisons between Alu scrambled and Alu antisense satisfy T-test with P < .05). (B) Measurement of correlation between mRNA productions per unit amounts of genomic DNA (different amounts derived from different densities of cells harvested). The slopes of correlation curves show lower mRNA production per unit amount of DNA in CGGBP1-depleted cells (Fishers exact test P < .0001). These curves are for cells cultured in 10% serum. (C) Histograms of mRNA/DNA ratios from control or CGGBP1-depleted cells serum-deprived or stimulated with serum for 48h. Interpolations to derive expected mRNA amounts in CGGBP1-depleted cells based on the mRNA/DNA ratios of control cells show no significant reduction under serum deprivation (left panel; Wilcoxon's test p = >0.999, labeled NS), whereas in presence of serum, the actual value of the mRNA production is significantly lower than the expected value of mRNA production (right panel; Wilcoxon's test, p = 0.0313, labeled *).
With these indications that CGGBP1 depletion caused RNA Pol II inhibition, we quantified the amount of polyA-RNA (mRNA), and normalized it to genomic DNA produced from a particular amount of cells, in control-shmiR or CGGBP1-shmiR transduced cells under quiescent or serum-stimulated conditions. Indeed, the amount of mRNA obtained per unit amount of genomic DNA was reduced to approximately half the levels in CGGBP1-depleted cells as compared to control cells, showing that CGGBP1-depletion negatively impacts the amount of mRNA available for protein production (Fisher test F < 0.0001, Fig. 4B and C). A likely interpretation of this result is that the lack of suppression of Alu-SINEs in CGGBP1-depleted serum-stimulated cells results in Alu-mediated inhibition of RNA Pol II activity and reduced mRNA production. Through Alu RNA, CGGBP1 thus appeared to regulate global gene expression in trans although there could be other mechanisms involved too. As RNA Pol II was inhibited by CGGBP1 depletion in an Alu RNA-dependent manner and since RNA Pol III is required for Alu RNA production, we next wanted to find out the mechanisms of CGGBP1-ATE binding and their consequences on RNA Pol III activity.
Possible mechanisms of Alu regulation by CGGBP1: phosphorylation associated with Y20 CGGBP1 antagonizes RNA Pol III at ATE
We explored various mechanisms through which CGGBP1 might regulate Alu expression in a serum-dependent manner. A Eukaryotic Linear Motif prediction for functional sites in proteins45 showed that the post-translational modification site predicted with maximum probability on CGGBP1 was Y20 (ELM prediction p = 2.454e-04) as a phosphorylable residue. The 2 other tyrosine residues Y155 and Y150 were predicted as potential modification sites at relatively lower ELM prediction probabilities with p values of 4.787e-04 and 3.296e-03 respectively. The Y20 flanking region on CGGBP1 also showed structure-based sequence similarity46 with the Y364 region of Ewing's Sarcoma protein (EWS), which exhibits Y364 phosphorylation-dependent nuclear localization47 (Fig. S6). The expression of endogenous CGGBP1 in the nuclei decreased and became diffusely pan-cellular upon serum starvation for 48 h (Fig. S7). With the premise that growth factors in serum can induce tyrosine phosphorylation, we wanted to measured phospho-tyrosine levels on CGGBP1 using lentiviral overexpression system for V5-tagged WT, or tyrosine residue mutant forms of CGGBP1. For this, we first generated tyrosine to phenylalanine mutations of 2 tyrosine residues with highest modification probabilities, Y20 and Y155. Tyrosine phosphorylation levels were studied in 1064Sk cells overexpressing WT, Y20F or Y155F CGGBP1 and subjected to stimulation with various growth factors (Fig. 5A). The pY signal on WT CGGBP1 seen upon IP:V5-Blot:pY was the lowest in Quiescent cells, unaffected by stimulation with SCF and insulin (not shown) or FGF, increased by EGF strongly and PDGFB mildly (Fig. 5A). While the EGF-induced phosphorylation was reduced by Y155F and Y20F mutations both, PDGFB-induced phosphorylation was sensitive only to Y20F mutation (Fig. 5A).
Figure 5.
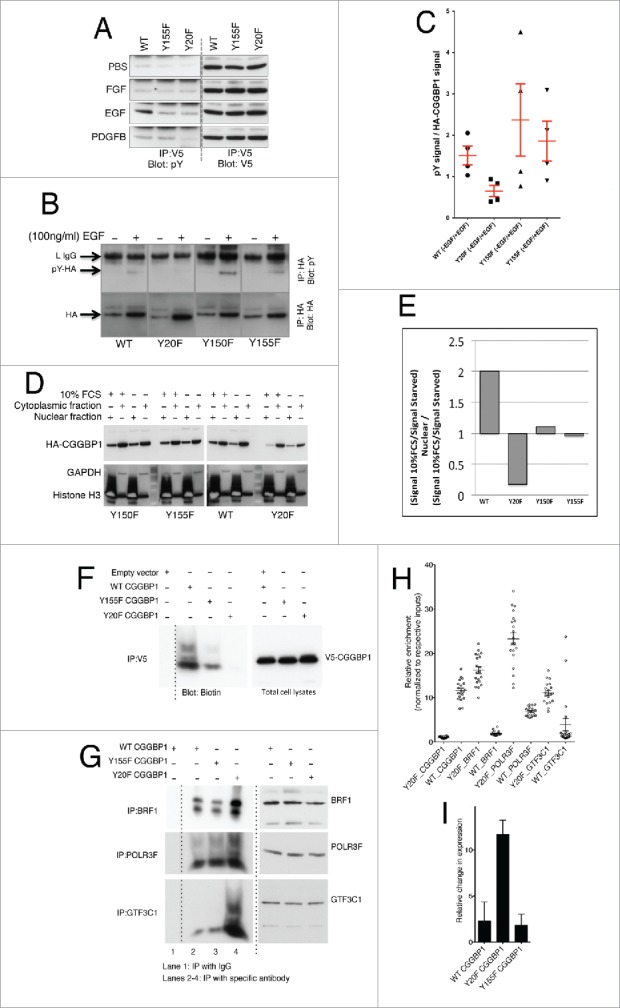
Growth stimulation induces tyrosine phosphorylation and nuclear localization of CGGBP1 that antagonizes RNA Pol III recruitment at ATE DNA. (A) 1064Sk cells transduced witAh V5-tagged WT, Y20F or Y155F CGGBP1 lentiviruses subjected to IP: V5-blot: pY after stimulation with indicated growth factors. Y20 seems to be a common phosphorylation site upon EGF and PDGFB stimulation. In all panels the upper band correspond to the light IgG chain (25 KDa approximately) and the lower band correspond to CGGBP1 (20 KDa approximately). The specificity of the IPs have been determined (not shown). (B) Measurement of the effect of EGF stimulation on tyrosine phosphorylation levels in HEK293T cells showed that Y20F mutant CGGBP1 does not respond to EGF-induced tyrosine phosphorylation. HA-tagged CGGBP1 was immunoprecipitated and blotted for pY, stripped and rebottled for HA. The HA and pY bands correspond to 20 KDa. pY signal at other molecular weights, which was stronger in EGF-treated samples, occurring due to pY-containing proteins co-immunoprecipitated with HA-CGGBP1, was ignored. (C) The net change in phosphorylation was calculated as a ratio of pY signal to total HA signal from 4 observations (WT versus Y20F; T-test p = 0.0181). (D) Consistent with immunofluorescence findings in 1064Sk cells, nuclear-cytoplasmic fractionation experiments in HEK293T cells showed that the ratio of nuclear to cytoplasmic distribution of WT CGGBP1 was increased whereas Y20F CGGBP1 was decreased upon serum stimulation; Y150F and Y155F mutations had no effect on the nuclear-cytoplasmic distribution upon serum stimulation. (E) The change in nuclear/cytoplasmic distribution upon serum stimulation was quantified using NIH ImageJ. The values are calculated using the formula ((Signal 10%FCS / Signal Quiescent) Nuclear) / ((Signal 10%FCS / Signal Quiescent) Cytoplasmic). (F) Y20 CGGBP1 exhibits strongly reduced binding to ATE DNA in in vitro DNA IPs. Equal expression of CGGBP1 and specificity of IPs have been confirmed (not shown). (G) Binding of BRF1, POLR3F and GTF3C1 were increased strongly by Y20F CGGBP1 as compared to WT CGGBP1 in in vitro DNA IPs using ATE dsDNA. For assays shown in (F) and (G), the only DNA incubated with lysates is the ATE DNA oligo pair. The smeary migration pattern on non-denaturing gel shows the mobility shift associated with interactions. (H) ChIP qPCR shows that in vivo also the binding of CGGBP1 is decreased (T-test, P < .01) but binding of BRF1, POLR3F and GTF3C1 is increased by Y20F CGGBP1 as compared to WT CGGBP1 (T-test, P < .01). (I) qPCRs show that overexpression of Y20F CGGBP1-induced Alu RNA levels compared to WT or Y155F CGGBP1-overexpression (T-test P < .0001). CGGBP1 depletion by CGGBP1 shmiR also induced Alu RNA levels compared to control shmiR (P < .0001).
Immunodetection of V5-CGGBP1 showed that WT-V5 and Y155F-V5 CGGBP1 were present strongly in the nuclei with weaker but specific staining also detected in the cytoplasm (Fig. S8). Y20F CGGBP1 showed a clear exclusion from the nuclei, with approximately 5% of cells having a widely cytoplasmic staining whereas the majority of cells displayed accumulation of CGGBP1 in a perinuclear ring with very weak presence in nuclei (Fig. S8). Tagging Y20F CGGBP1 with a constitutive nuclear localization signal from SV40 T-antigen failed to generate a clear nuclear localization (Fig. S8), whereas V5-Y20ex CGGBP1 (CGGBP1 with Y20 as the only tyrosine residue; Y150 and 155 mutated to F) yielded a WT CGGBP1-like nuclear localization (Fig. S8). To verify these findings, we studied EGF-induced tyrosine phosphorylation and its effect on CGGBP1 nuclear localization in transiently transfected HEK293T cells also. In these experiments we included all tyrosine-to-phenylalanine mutations of CGGBP1 (Y20F, Y150F and Y155F). EGF-induced phosphorylation was ablated by Y20F mutation (Fig. 5B and C). Nuclear-cytoplasmic fractionation assays showed that the nuclear to cytoplasmic ratio of CGGBP1 levels was drastically reduced upon Y20F mutation in serum-stimulated HEK293T cells (Fig. 5D and E) showing that the Y20 phosphorylation of CGGBP1 affects its nuclear presence. DNA-protein immunoprecipitations (DNA-IPs) performed using ds-oligonucleotides representing the ATE region of Alu-SINEs and using lysates from WT, Y20F or Y155F CGGBP1-transduced 1064Sk cells, showed that CGGBP1-ATE binding was diminished by Y20F mutation (Fig. 5F). This finding suggests that the potential Y20 phosphorylation of CGGBP1 aids in its binding to ATE and nuclear retention. DNA-IPs using the known RNA Pol III constituents BRF1, POLR3F and GTF3C1, showed that their association with the ATE DNA was inversely correlated with potential Y20 phosphorylation of CGGBP1 (Fig. 5G). ChIP assays showed that in vivo also WT CGGBP1 was bound to ATE-flanking region significantly more than Y20F CGGBP1 (T-test P < .01, Fig. 5H). Conversely, the binding of BRF1, POLR3F and GTF3C1 to ATE-flanking region was increased by Y20F mutation (Fig. 5H). POLR3F ChIP and qPCRs on candidate tRNA genes showed that Y20F CGGBP1 overexpression either did not affect or decreased POLR3F recruitment on tRNA genes (Fig. S9, P < .02).
In agreement with these observations, we found that Alu RNA levels were increased upon Y20F CGGBP1-overexpression (Fig. 5I, P < .0001). These results collectively indicate that a potential Y20 phosphorylation upon growth stimulation facilitates CGGBP1-ATE binding and counteracts recruitment of RNA Pol III components specifically at Alu promoters.
Discussion
We have found that CGGBP1 affects global gene expression through regulation in cis of Alu RNA levels, which in turn affects RNA Pol II. We also provide some mechanistic insight into possible mechanisms of Alu regulation by CGGBP1. Mutually exclusive binding patterns of RNA Pol III components and CGGBP1 (that is potentially phosphorylated at Y20) indicate that CGGBP1 is an Alu-specific RNA Pol III regulator. These results advance our knowledge about how Alu transcription is regulated and pave the way for investigations into the possible functions of CGGBP1 as a regulator of RNA Pol III, like MAF1, the only previously known RNA Pol III regulator.26
The absence of enrichment of specific functional categories and no conserved DNA sequence motifs in the promoters of the genes deregulated by CGGBP1 depletion, is in accordance with the finding that the global mRNA production was lowered as a consequence of RNA Pol II inhibition by Alu RNA. Transcriptome-wide techniques such as microarrays inherently fail to report such large-scale alterations in mRNA levels as they rely on equality of RNA from different samples and statistical normalization of signals. To this end, the calculation of mRNA per unit amount of DNA is a useful means to get a relative measure of global mRNA production. Because we have measured the mRNA/DNA ratio from multiple samples with different cell densities, the slopes of the curves are statistically reliable for a relative quantitation. However, for objective measurements of rate of mRNA productions in real-time, innovative new methods need to be devised. Nevertheless, these results highlight the need of discriminating between global gene expression retardation and target-specific gene expression change as most conventional assays are based on the assumption that the amount of total mRNA produced per cell (refection of a unit amount of DNA) remains unchanged and the changes are only for some specific target genes.
We performed ChIP-seq to identify DNA-binding sites of CGGBP1 that might reveal its direct transcription-targets. The large distances between ChIP-seq peaks and nearest promoters suggest no cis-regulation of gene expression by CGGBP1 binding to Alus. However, any cis-regulatory CGGBP1-binding sites could evade detection in ChIP-seq due to their repetitive nature or low enrichment as compared to interspersed repeats. The inhibition of RNA Pol II upon CGGBP1-depletion would result in altered levels of various transcription-regulatory factors thereby secondarily affecting gene expression. Together, these will bring about a complex and broad change in the transcriptome as seen in our microarray results. Despite the evidence we provide for RNA Pol II inhibition by Alu RNA increase due to loss of CGGBP1 function, it is likely that this is only one of many known and possible mechanisms. For example, we do not provide any evidence that CGGBP1 does not affect RNA Pol II directly by bypassing Alus. Such a mechanism would be possible if RNA Pol II components interact with CGGBP1; a possibility not ruled out based on the presented results. Also, the cell cycle arrest seen upon loss of CGGBP1 function could affect RNA Pol II indirectly, without involving Alu RNA. It is important to note that basal RNA Pol II activity is indeed regulated in a cell cycle dependent manner and Alus RNA has not been implicated in this process yet.48
It is interesting that the increase in DNA binding of CGGBP1 upon growth-stimulation is not universal and unspecific but restricted mainly to Alu-SINEs. Conversely, under conditions of starvation, CGGBP1-bound mostly to L1-LINEs. It thus seems that the affinity of CGGBP1 for LINEs is independent of growth signals, although it could have a general effect on CGGBP1-DNA binding. It is likely that some additional target sequence-specifying mechanisms cooperate with growth-stimulation to determine CGGBP1-DNA binding pattern at LINEs and other target sequences. It is interesting that the LINEs and SINEs have overtly mutually exclusive location patterns: LINEs in the G/C-poor G-bands and SINEs in the G/C-rich R-bands.49 The differential CGGBP1-binding patterns on on Alus and L1 could depend on the differences in G/C and A/T richness of the genomic regions in which they are located. The sequences of the exact target sites will also determine the local chromatin context that could affect CGGBP1-binding.
The ability of CGGBP1 to achieve a proper nuclear presence and bind to Alu-SINEs was strongly dependent on intactness of Y20, which potentially gets phosphorylated in response to serum and growth factors. Some other transcription regulatory proteins have also been shown to undergo nuclear translocation upon tyrosine phosphorylation, such as HNF4A, STAT5, EWS.47,50-55 Our results also show that, although there is an enhanced nuclear presence of CGGBP1 upon serum stimulation, starvation and a potential loss of Y20 phosphorylation do not abrogate nuclear presence of CGGBP1 completely. These results together suggest that the enhanced nuclear levels of CGGBP1 in serum-stimulated cells could be due to a stronger association with target DNA and possibly increased nuclear retention, caused by Y20 phosphorylation. The levels of Y20F CGGBP1 that remain in the nucleus argue that Y20 is not a master regulator of CGGBP1 nuclear localization. There is a dynamic state of CGGBP1 distribution in the cells, which in presence of Y20 CGGBP1 and growth signals favors nuclear presence more than cytoplasmic whereas upon Y20F mutation or lack of growth signals favors the extra-nuclear or peri-nuclear accumulation. What mechanism senses and restricts Y20F CGGBP1 to the peri-nuclear ring remains unknown.
Many important functions of Alu-SINEs, such as Alu-retrotransposition and the inhibition of RNA Pol II upon heat shock by Alu RNA rely on Alu RNA production. Although the functions of Alu-SINEs have been investigated in details, the regulation of Alu-expression has not been clarified.11-13,15,21,23,29-31 The type 2 promoters of Alus are binding sites for the ubiquitously available RNA Pol III subunits involved in the housekeeping transcription of tRNA and 7SL genes. Some mechanisms must discriminatively keep Alu transcription off while tRNA and 7SL genes are transcribed. Our results suggest that CGGBP1 in response to growth factors suppresses Alu promoters specifically, while sparing the tRNA and 7SL genes. The measurement of Alu RNA by us utilizes a well-controlled PCR-based approach that can be used widely to distinguish Alu and 7SL RNA. By using any approach reported so far, the Alu transcripts originating from RNA Pol III-transcribed Alu units as well as from Alu units embedded within RNA Pol II-transcribed mRNA-coding genes cannot be differentiated. However, the conclusion that the increase in Alu RNA upon CGGBP1-depletion is due to enhanced RNA Pol III activity at Alu promoters is logical because upon CGGBP1 depletion RNA Pol II becomes inhibited whereas RNA Pol III exhibits enhanced binding at ATE DNA. Although additional mechanisms may affect CGGBP1-Alu axis, but in the context of the results reported here, a potentiation of RNA Pol III at Alu promoters seems to be the strongest reason. Interestingly, the mutually exclusive binding of CGGBP1 or RNA Pol III components to Alu promoters suggests that binding of CGGBP1 could cause steric hindrance to BRF1, POLR3F and GTF3C1 access to Alu promoter by either physically occupying the ATE region or changing the chromatin configuration there. For conclusive mechanisms, targeted studies on RNA Pol III regulation by CGGBP1 and partner proteins need to be carried out. The differential effects of CGGBP1 on tRNA and Alu promoters could be due to minor sequence differences or major differences in their epigenetic states. Interestingly, silencing of Alu elements by CGGBP1 occurs through regulatory elements, including the ATE region, which are so far only known as positive regulators of transcription.32 The ATE region thus emerges as a bimodal regulator of Alu transcription: repressor when bound by CGGBP1, and activator when not bound by CGGBP1.
CGGBP1 is an amniote-specific protein (NCBI HomoloGene) and seem to have acquired specialized retrotransposon-regulatory functions. It is an interesting coincidence then that CGGBP1 appears to have originated from the DNA-binding domain of a DNA transposase belonging to the Charlie group of hAT transposases (Arian Smit, unpublished results), which has acquired the function of silencing the SINEs in our genome.
These results for the first time describe a mechanism through which Alu-SINEs transcription is regulated and describe a sequence and context-specific regulator of RNA Pol III activity. These results enhance our understanding about how our cells deal with the widespread Alu elements selectively with a positive outcome for cellular growth.
Methods
Cell culture, expression constructs and lentiviral transduction
1064Sk normal human foreskin fibroblasts, passage 8–25, were cultured in MEM (Sigma) supplemented with 10% fetal bovine serum and 1% Glutamine (Sigma). HEK293T cells were cultured in DMEM supplemented the same way. WT HA-tagged CGGBP1 expression plasmids were used as described earlier.40 Point mutations Y20F, Y150F and Y155F were created by using QuickChangeII site directed mutagenesis kit (Agilent, Stratagene). N-terminal V5 epitope tagged WT, Y20F or Y155F CGGBP1 synthetic or PCR-generated inserts were cloned into pLenti-V5-dest into attL1 and attL2 cloning sites (GeneArt). The clones were transformed into and expanded using STBL3 competent E. coli (Invitrogen). Pooled midipreps (Qiagen) from single clones were used for transfection/transduction. The multiplicity of infection was approximately 3–4. Cells were freshly transduced for 96h before harvesting them for RNA extraction, nuclear fraction preparation, growth factor simulation experiments, or lysis for westerns or immunoprecipitations. Control or CGGBP1-shmiR lentiviruses (targeting 3 different regions in CGGBP1 ORF) were obtained from ThermoScientific, mixed in equal proportions and used for transductions. For stable transductions, cells were selected in 5 ug/ml Puromycin. Transfections were performed using standard Fugene (Promega) protocol. 1:5000 diluted 10 mg/ml Polybrene was used for transducing fibroblasts.
RNA extraction, affymetrix microarray assays and qRT-PCRs
RNAs were extracted using Trizol (Invitrogen) protocol. Either total cells or purified nuclear fractions were used for RNA extraction. Total RNA were subjected to GeneTitan 1.0 ST Affymetrix microarray hybridizations using standard protocols at Stockholm BEA Affymetrix facility. Data were analyzed by GeneSpring V12.6.1 using specified ANOVA settings. For qPCRs, cDNA was made out of total RNA using Vilo reverse transcription kit (Invitrogen) as recommended. qRT-PCRs were performed using cDNA template using 2× SYBR-Green PCR mix from Fermentas or BioRad on Stratagene MX3005 machines. Separate thresholds were applied to each primer pair for Ct value calculation. ddCt calculations were performed on quadruplicate Ct values from different runs for relative quantitation. For Alu expression analysis, a standard curve was prepared from known amounts of Alu PCR products as template (1 fg to 10000 fg) and Ct values obtained from samples were interpolated. The differences in Ct values of treatment versus controls were calculated and subjected to negative power of base 2. The values obtained were used to calculate fold changes. For Alu vs. 7SL enrichment, differences in 7SL and Alu Ct values relative to the common input were calculated and subjected to same analysis as described for relative quantitation using standard curve.
The primers for Alu-specific PCR were used from a previous work44 with slight modification. The reverse primer was extended by 3 bases at 3′ end to generate mismatch with 7SL sequences and ensure Alu specificity. The correctness of fragment sized obtained through this PCR, location and sequences of primers and verification of PCR specificity to ensure no cross amplification of 7SL and Alu template from primer cross matches is shown in Figure 3A and B. The Alu-specific template was obtained by purifying the 2 bands from gel together. The 7SL-specific template was obtained by a mixture of 10fmoles of both of the following oligos in the PCR as template: 5′-GGAGTTCTGGGCTGTAGTGCGCTATGCCGA TCGGG-TGTCCGCACTAAGTTCGGCATCAATATGGTGACCTC-CCGGGAGCGGGGGACCACCAGGTTGCCTAAGGAGG-GGTGAACCGG-3′ and 5′-TGCCCA GGCTGGAGTGCA-GTGGCTATTCACAGGCGCGATCCCACTACTGATCAGCACGGGAGTTTTGACCTGCTCCGTTTCCGACCTGG-GCCGGTTCACCCCTCCTTAGGCAACCTGG-3′.
ChIP: sequencing and qPCRs
ChIP was performed using Upstate protocol with certain modifications, as described before.38 The sonication was done at maximum setting, of 30 sec sonication with intervening icing of 30 sec, for 45 min to achieve a mean fragment size of 150 bp (<200 bps). For ChIP sequencing, the diluted lysates were preincubated for 2 h in a cocktail of non-specific antibodies/IgG (described in antibody list) followed by clearing using protein-G-sepharose beads (GE Life Sciences). The cleared diluted lysates were then subjected to specific immunoprecipitation using CGGBP1 antibody cocktail (described in antibody list). ChIP DNA obtained from approximately 100 million cells were pooled, phenol-chloroform purified, precipitated and resuspended to final concentration of 2–4 ng/μl concentration. ChIP DNA QC was performed using BioAnalyzer and using standard protocols sequenced using SOLiD and IonProton platforms, keeping the minimum read size >35 bases (mean read lengths 46–52 bases). ChIP were performed in 5 replicates, pooled and sequenced in triplicates. Raw sequence data was mapped to human genome (hg19) using LifeScope 2.5 software (Life Technologies) following manufacturer's best practice guidelines and maximum stringency for unique mapping. Only uniquely mapped reads were retained in the resulting BAM file. Peak calling was done using MACS software at default settings for statistical significance (p value ≤ 1E-05) using UPPMAX at Uppsala University Next Generation Sequencing Analysis and Data Storage facility. The peaks thus obtained were analyzed using various bioinformatics tools as necessary. For discriminative motif searches, where one set of sequences was used as a negative data set, a background Markov Model file was generated for the negative file.
The distribution of reads in peaks was calculated in bins of 200 bps (−500 to −300 till +300 to +500) and the results were plotted with center of bins as locations from Alu start site. Intersections of different genomic coordinates for the purpose was done using BEDTools.56 For signal mapping, footprints of the CGGBP1 binding around the peaks were created using the SICTIN software and custom R-scripts in 150 bp segments ranging from −1kb to +1kb from Alu start and K-means clustering was performed to find out the major patterns of read pileup in and around peaks using a previously reported method.43
The ChIP seq data is uploaded at NCBI GEO (ID: GSE53571, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53571). The quiescent sample has been referred to as starved sample.
Nuclear-cytoplasmic fractionation
Nuclear-cytoplasmic fractions were prepared using the REAP method described elsewhere.57
Immunofluorescence and western blots
Immunofluorescence and protein gel blots were performed as described previously.38
DNA-immunoprecipitations
DNA immunoprecipitations were performed as described earlier.35,58 10 pmoles of double stranded ATE DNA (complementary strands synthesized, annealed, digested using ExoI and purified again using Qiagen columns) was incubated with indicated cell lysates prepared using lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 1% aprotinin, and 1 mM phenylmethylsulfonyl fluoride) on ice for 2 h in presence of 1 ug/ml Poly (dI.dC). Immunoprecipitations were performed using indicated antibodies. The ATE DNA attached to the precipitated proteins was eluted by heating (95°C, 5 min). Eluates were run on Novex native DR TBE gels and detected using streptavidin-HRP Nucleic Acid detection system (Pierce). Aliquots of lysates used for DNA immunoprecipitations were also subjected to western blot for the immunoprecipitated protein to serve as controls as the amount of protein in input. The sequence of the forward strand of the ds ATE DNA was BIO-CTG CCT CAG CCT CCC GAG TAG CTG GGA CTA CAG GC.
Primers and DNA Oligos
The DNA oligos used in this study are described in supplementary file (Table S5). For specificity and PCR feasibility, primers had to be chosen in <100 bp vicinity of the peak sequences. One peak from each chromosome was chosen randomly for confirmation. For RNA Pol II ChIP-qPCRs, primers were used as described earlier.18
Genomic DNA extraction, mRNA purification and mRNA/DNA ratio calculation
Cells were seeded in different densities of 0.1, 0.2, 0.5, 1.0, 2.0 and 5.0 (in millions), control or CGGBP1 shmiR transduced after 24 h and after another 24 h either subjected to either 72 h serum deprivation or growth in presence of 10% serum. Cells were washed 2 times with 10 ml ice cold DEPC-treated PBS and resuspended in 1 ml of the same. 100 microliters were used for genomic DNA extraction using Qiagen Genomic DNA extraction kit and the remaining used for mRNA extraction using Qiagen PolyA RNA extraction kit. The DNA and mRNA thus extracted were quantified using NanoDrop. Paired values of mRNA and corresponding DNA concentrations were used to derive a correlation plot of mRNA quantities to DNA quantities obtained. Significance of difference between them was calculated using Fisher's test in GraphPad Prism. The control shmiR slope was used to interpolate the mean expected value of mRNA for CGGBP1 shmiR sample. The observed and expected values of CGGBP1 shmiR were analyzed by Wilcoxon's test in GraphPad Prism.
Statistics and graphs
Graphs were generated using MS Excel or GraphPad Prism 6. All statistical tests were performed using inbuilt tools in GraphPad Prism. Statistics from RepeatMasker, DREME/MEME suites, GeneSpring, SICTIN and R-scripts were reported as in the output.
Antibodies
Following antibodies were used in this study: CGGBP1 western (Proteintech), CGGBP1 ChIP (cocktail of Proteintech 10716-1-AP, Abcam ab126095 and ab56412, Santacruz sc-102434 and sc-102433), IgG ChIP preincubation (goat serum (Gibco), HA (Abcam), FLAG (M2 Sigma), mouse IgG (DAKO), HA tag (Santacruz), V5 tag (Santacruz), Histone H3 (Abcam), GAPDH (Sigma), pY (Santacruz), BRF1 (Santacruz sc-17465), POLR3F (Santacruz sc-32125), GTF3C1 (Santacruz sc-22571), POL2 (Abcam ab817).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We acknowledge the freely available software tools MEME Suit, PRALINE, ELM, DAVID and GALAXY bioinformatics tool cluster. Uppsala Genome Center is acknowledged for ChIP sequencing. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under project “b2013008." BEA facility at Karolinska Institute is acknowledged for Affymetrix Microarray expression analysis.
Author contributions
Performed experiments: PA and US, Analyzed data and helped writing manuscript: All authors; Wrote manuscript: US, PA and BW; Shared unpublished data: AS.
Funding
This work was supported by funds from Swedish Cancer Society and Swedish Research Council.
References
- [1]. Strachan T RA. Human Molecular Genetics. New York: Wiley-Liss, 1999:255-95 [Google Scholar]
- [2]. Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev 1996; 6:743-8; PMID:8994846; http://dx.doi.org/ 10.1016/S0959-437X(96)80030-X [DOI] [PubMed] [Google Scholar]
- [3]. Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009; 10:691-703; PMID:19763152; http://dx.doi.org/ 10.1038/nrg2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Singer MF. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell 1982; 28:433-4; PMID:6280868; http://dx.doi.org/ 10.1016/0092-8674(82)90194-5 [DOI] [PubMed] [Google Scholar]
- [5]. Ullu E, Tschudi C. Alu sequences are processed 7SL RNA genes. Nature 1984; 312:171-2; PMID:6209580; http://dx.doi.org/ 10.1038/312171a0 [DOI] [PubMed] [Google Scholar]
- [6]. Burns KH, Boeke JD. Human transposon tectonics. Cell 2012; 149:740-52; PMID:22579280; http://dx.doi.org/ 10.1016/j.cell.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Wang J, Geesman GJ, Hostikka SL, Atallah M, Blackwell B, Lee E, Cook PJ, Pasaniuc B, Shariat G, Halperin E, et al. Inhibition of activated pericentromeric SINEAlu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle 2011; 10:3016-30; PMID:21862875; http://dx.doi.org/ 10.4161/cc.10.17.17543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Schmitz J. SINEs as driving forces in genome evolution. Genome Dyn 2012; 7:92-107; PMID:22759815; http://dx.doi.org/ 10.1159/000337117 [DOI] [PubMed] [Google Scholar]
- [9]. Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell 2006; 125:859-72; PMID:16751097; http://dx.doi.org/ 10.1016/j.cell.2006.04.028 [DOI] [PubMed] [Google Scholar]
- [10]. Lunyak VV, Atallah M. Genomic relationship between SINE retrotransposons, Pol III-Pol II transcription, and chromatin organization: the journey from junk to jewel. Biochem Cell Biol 2011; 89:495-504; PMID:21916613; http://dx.doi.org/ 10.1139/o11-046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Hellmann-Blumberg U, Hintz MF, Gatewood JM, Schmid CW. Developmental differences in methylation of human Alu repeats. Mol Cell Biol 1993; 13:4523-30; PMID:8336699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Kochanek S, Renz D, Doerfler W. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J 1993; 12:1141-51; PMID:8384552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Kondo Y, Issa JP. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J Biol Chem 2003; 278:27658-62; PMID:12724318; http://dx.doi.org/ 10.1074/jbc.M304072200 [DOI] [PubMed] [Google Scholar]
- [14]. Englander EW, Howard BH. Nucleosome positioning by human Alu elements in chromatin. J Biol Chem 1995; 270:10091-6; PMID:7730313; http://dx.doi.org/ 10.1074/jbc.270.17.10091 [DOI] [PubMed] [Google Scholar]
- [15]. Englander EW, Wolffe AP, Howard BH. Nucleosome interactions with a human Alu element. Transcriptional repression and effects of template methylation. J Biol Chem 1993; 268:19565-73; PMID:8366099 [PubMed] [Google Scholar]
- [16]. Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res 2004; 14:1719-25; PMID:15342557; http://dx.doi.org/ 10.1101/gr.2855504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 2003; 300:1288-91; PMID:12764196; http://dx.doi.org/ 10.1126/science.1082588 [DOI] [PubMed] [Google Scholar]
- [18]. Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 2008; 29:499-509; PMID:18313387; http://dx.doi.org/ 10.1016/j.molcel.2007.12.013 [DOI] [PubMed] [Google Scholar]
- [19]. Dumay-Odelot H, Durrieu-Gaillard S, Da Silva D, Roeder RG, Teichmann M. Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle 2010; 9:3687-99; PMID:20890107; http://dx.doi.org/ 10.4161/cc.9.18.13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Oler AJ, Traina-Dorge S, Derbes RS, Canella D, Cairns BR, Roy-Engel AM. Alu expression in human cell lines and their retrotranspositional potential. Mob DNA 2012; 3:11; PMID:22716230; http://dx.doi.org/ 10.1186/1759-8753-3-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Ichiyanagi K, Li Y, Watanabe T, Ichiyanagi T, Fukuda K, Kitayama J, Yamamoto Y, Kuramochi-Miyagawa S, Nakano T, Yabuta Y, et al. Locus- and domain-dependent control of DNA methylation at mouse B1 retrotransposons during male germ cell development. Genome Res 2011; 21:2058-66; PMID:22042642; http://dx.doi.org/ 10.1101/gr.123679.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet 2007; 23:183-91; PMID:17331616; http://dx.doi.org/ 10.1016/j.tig.2007.02.006 [DOI] [PubMed] [Google Scholar]
- [23]. Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 2004; 431:96-9; http://dx.doi.org/ 10.1038/nature02886 [DOI] [PubMed] [Google Scholar]
- [24]. Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res 2009; 37:7268-80; PMID:19783824; http://dx.doi.org/ 10.1093/nar/gkp787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipursky SL, Darnell J. Molecular Cell Biology. New York: W. H. Freeman and Company, 2000; p. 525. [Google Scholar]
- [26]. Ciesla M, Boguta M. Regulation of RNA polymerase III transcription by Maf1 protein. Acta Biochim Pol 2008; 55:215-25; PMID:18560610 [PubMed] [Google Scholar]
- [27]. Goodfellow SJ, Graham EL, Kantidakis T, Marshall L, Coppins BA, Oficjalska-Pham D, Gérard M, Lefebvre O, White RJ. Regulation of RNA polymerase III transcription by Maf1 in mammalian cells. J Mol Biol 2008; 378:481-91; PMID:18377933; http://dx.doi.org/ 10.1016/j.jmb.2008.02.060 [DOI] [PubMed] [Google Scholar]
- [28]. Ichiyanagi K. Epigenetic regulation of transcription and possible functions of mammalian short interspersed elements, SINEs. Genes Genet Syst 2013; 88:19-29; PMID:23676707 [DOI] [PubMed] [Google Scholar]
- [29]. Macia A, Munoz-Lopez M, Cortes JL, Hastings RK, Morell S, Lucena-Aguilar G, Marchal JA, Badge RM, Garcia-Perez JL. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol Cell Biol 2011; 31:300-16; PMID:21041477; http://dx.doi.org/ 10.1128/MCB.00561-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 2010; 17:620-8; PMID:20418882; http://dx.doi.org/ 10.1038/nsmb.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Xie H, Wang M, Bonaldo Mde F, Smith C, Rajaram V, Goldman S, Tomita T, Soares MB. High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum. Nucleic Acids Res 2009; 37:4331-40; PMID:19458156; http://dx.doi.org/ 10.1093/nar/gkp393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Perez-Stable C, Ayres TM, Shen CK. Distinctive sequence organization and functional programming of an Alu repeat promoter. Proc Nat Acad Sci U S A 1984; 81:5291-5; PMID:6089189; http://dx.doi.org/ 10.1073/pnas.81.17.5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Kim C, Rubin CM, Schmid CW. Genome-wide chromatin remodeling modulates the Alu heat shock response. Gene 2001; 276:127-33; PMID:11591479; http://dx.doi.org/ 10.1016/S0378-1119(01)00639-4 [DOI] [PubMed] [Google Scholar]
- [34]. Deissler H, Wilm M, Genc B, Schmitz B, Ternes T, Naumann F, Mann M, Doerfler W. Rapid protein sequencing by tandem mass spectrometry and cDNA cloning of p20-CGGBP. A novel protein that binds to the unstable triplet repeat 5′-d(CGG)n-3′ in the human FMR1 gene. J Biol Chem 1997; 272:16761-8; PMID:9201980; http://dx.doi.org/ 10.1074/jbc.272.27.16761 [DOI] [PubMed] [Google Scholar]
- [35]. Singh U, Maturi V, Jones RE, Paulsson Y, Baird DM, Westermark B. CGGBP1 phosphorylation constitutes a telomere-protection signal. Cell Cycle 2013; 13:96-105; PMID:24196442; http://dx.doi.org/ 10.4161/cc.26813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Muller-Hartmann H, Deissler H, Naumann F, Schmitz B, Schroer J, Doerfler W. The human 20-kDa 5′-(CGG)(n)-3′-binding protein is targeted to the nucleus and\ affects the activity of the FMR1 promoter. J Biol Chem 2000; 275:6447-52; PMID:10692448; http://dx.doi.org/ 10.1074/jbc.275.9.6447 [DOI] [PubMed] [Google Scholar]
- [37]. Naumann F, Remus R, Schmitz B, Doerfler W. Gene structure and expression of the 5′-(CGG)(n)-3′-binding protein (CGGBP1). Genomics 2004; 83:106-18; PMID:14667814; http://dx.doi.org/ 10.1016/S0888-7543(03)00212-X [DOI] [PubMed] [Google Scholar]
- [38]. Singh U, Bongcam-Rudloff E, Westermark B. A DNA sequence directed mutual transcription regulation of HSF1 and NFIX involves\ novel heat sensitive protein interactions. PLoS One 2009; 4:e5050; PMID:19337383; http://dx.doi.org/ 10.1371/journal.pone.0005050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Singh U, Roswall P, Uhrbom L, Westermark B. CGGBP1 regulates cell cycle in cancer cells. BMC Mol Biol 2011; 12:28; PMID:21733196; http://dx.doi.org/ 10.1186/1471-2199-12-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Singh U, Westermark B. CGGBP1 is a nuclear and midbody protein regulating abscission. Exp Cell Res 2011; 317:143-50; PMID:20832400; http://dx.doi.org/ 10.1016/j.yexcr.2010.08.019 [DOI] [PubMed] [Google Scholar]
- [41]. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 2009; 37:W202-8; PMID:19458158; http://dx.doi.org/ 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Tempel S. Using and understanding RepeatMasker. Methods Mol Biol 2012; 859:29-51; PMID:22367864; http://dx.doi.org/ 10.1007/978-1-61779-603-6_2 [DOI] [PubMed] [Google Scholar]
- [43]. Enroth S, Andersson R, Wadelius C, Komorowski J. SICTIN: Rapid footprinting of massively parallel sequencing data. BioData Min 2010; 3:4; PMID:20707885; http://dx.doi.org/ 10.1186/1756-0381-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Marullo M, Zuccato C, Mariotti C, Lahiri N, Tabrizi SJ, Di Donato S, Cattaneo E. Expressed Alu repeats as a novel, reliable tool for normalization of real-time quantitative RT-PCR data. Genome Biol 2010; 11:R9; PMID:20109193; http://dx.doi.org/ 10.1186/gb-2010-11-1-r9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A, et al. ELM–the database of eukaryotic linear motifs. Nucleic Acids Res 2012; 40:D242-51; PMID:22110040; http://dx.doi.org/ 10.1093/nar/gkr1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Simossis VA, Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res 2005; 33:W289-94; PMID:15980472; http://dx.doi.org/ 10.1093/nar/gki390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Leemann-Zakaryan RP, Pahlich S, Grossenbacher D, Gehring H. Tyrosine phosphorylation in the C-terminal nuclear localization and retention signal (C-NLS) of the EWS protein. Sarcoma 2011; 2011:218483; PMID:21647358; http://dx.doi.org/ 10.1155/2011/218483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48]. Yonaha M, Chibazakura T, Kitajima S, Yasukochi Y. Cell cycle-dependent regulation of RNA polymerase II basal transcription activity. Nucleic Acids Res 1995; 23:4050-4; PMID:7479063; http://dx.doi.org/ 10.1093/nar/23.20.4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. Korenberg JR, Rykowski MC. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell 1988; 53:391-400; PMID:3365767; http://dx.doi.org/ 10.1016/0092-8674(88)90159-6 [DOI] [PubMed] [Google Scholar]
- [50]. Chellappa K, Jankova L, Schnabl JM, Pan S, Brelivet Y, Fung CL, Chan C, Dent OF, Clarke SJ, Robertson GR, et al. Src tyrosine kinase phosphorylation of nuclear receptor HNF4alpha correlates with isoform-specific loss of HNF4alpha in human colon cancer. Proc Natl Acad Sci U S A 2012; 109:2302-7; PMID:22308320; http://dx.doi.org/ 10.1073/pnas.1106799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Cotarla I, Ren S, Zhang Y, Gehan E, Singh B, Furth PA. Stat5a is tyrosine phosphorylated and nuclear localized in a high proportion of human breast cancers. Int J Cancer 2004; 108:665-71; PMID:14696092; http://dx.doi.org/ 10.1002/ijc.11619 [DOI] [PubMed] [Google Scholar]
- [52]. Dey-Guha I, Malik N, Lesourne R, Love PE, Westphal H. Tyrosine phosphorylation controls nuclear localization and transcriptional activity of Ssdp1 in mammalian cells. J Cell Biochem 2008; 103:1856-65; PMID:18080319; http://dx.doi.org/ 10.1002/jcb.21576 [DOI] [PubMed] [Google Scholar]
- [53]. Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J 1994; 13:4361-9; PMID:7925280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Humphries MJ, Ohm AM, Schaack J, Adwan TS, Reyland ME. Tyrosine phosphorylation regulates nuclear translocation of PKCdelta. Oncogene 2008; 27:3045-53; PMID:18059334; http://dx.doi.org/ 10.1038/sj.onc.1210967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55]. Madeo F, Schlauer J, Zischka H, Mecke D, Frohlich KU. Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Mol Biol Cell 1998; 9:131-41; PMID:9436996; http://dx.doi.org/ 10.1091/mbc.9.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56]. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010; 26:841-2; PMID:20110278; http://dx.doi.org/ 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Suzuki K, Bose P, Leong-Quong RY, Fujita DJ, Riabowol K. REAP: a two minute cell fractionation method. BMC Res Notes 2010; 3:294; PMID:21067583; http://dx.doi.org/ 10.1186/1756-0500-3-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58]. Nishihara A, Hanai J, Imamura T, Miyazono K, Kawabata M. E1A inhibits transforming growth factor-beta signaling through binding to Smad proteins. J Biol Chem 1999; 274:28716-23; PMID:10497242; http://dx.doi.org/ 10.1074/jbc.274.40.28716 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.