Matching supply with demand is an enormous challenge not only for manufacturing companies, but also for biological systems. An excess of pathway activity, for example, can lead to tumorigenesis during adulthood, whereas inadequate low activity of the same pathway can cause developmental delays or failures during embryogenesis.
The same principle can be applied to the field of genome engineering. Here, a set of nucleases can be used to introduce specific sequence modifications through the induction of DNA double strand breaks (DSB) at pre-defined genomic loci. The final genome editing outcome depends on the choice between the endogenous, cell-autonomous DNA repair pathways.
In mammalian cells, two distinct pathways ensure repair of DSB, non-homologous end-joining (NHEJ) and homology-directed repair (HDR). While NHEJ does not depend on sequence complementarities to align and ligate DSB, HDR requires intact homologous sequences that usually locate on a sister chromatid or elsewhere in the genome. Importantly, HDR can also use exogenous-derived cDNA sequences (plasmids, DNA oligonucleotides) for DSB repair. This enables site-specific integrations of specific DNA sequences, which is of relevance for therapeutic applications that try to restore gene function by correcting mutations at the endogenous gene locus. Hence, HDR is considered a high-fidelity DNA repair pathway, whereas NHEJ is much more error-prone and can lead to mutations that disrupt gene function. Therefore, the choice of DNA repair pathway after DSB induction has a huge impact on the repair outcome. However, the activities of these two repair pathways differ and largely depend on the cell-cycle phase. The current model is that NHEJ functions throughout the cell cycle, but is most important in G0/G1. In contrast, HDR is only active in S and G2, and is suppressed in G1 due to the suppression of DNA-end resection coupled with a multistep block of BRCA2 to DNA damage sites.1 A recent study refined this activity model further: cells in G1 repair DSBs exclusively by NHEJ. Cells then gradually increase their use of HDR as they progress from G1 to early S. The HDR activity peaks in mid-S and gradually declines as cells move toward late S and G2. In G2, DSBs are almost entirely repaired by NHEJ.2 Consequently, the NHEJ pathway is largely favored over HDR in unsynchronized cell populations. This makes it difficult to introduce well-defined genomic alterations, e.g. tagging of endogenous loci, and there is a huge need to identify strategies that enhance HDR-mediated genome engineering.
With the cell-cycle dependent activity profile of HDR in mind, we thought to match supply with demand. We turned to the RNA-programmable CRISPR-Cas9 system, which is currently the most extensively used genome editing tool due to its ease of use and its targeting flexibility.3 In its simplest form this system only requires the Cas9 protein and a custom-designed single-guide RNA (sgRNA) to introduce DSB and stimulate genome editing. To avoid Cas9-mediated DNA cleavage in G1, which would induce NHEJ repair, we recently engineered a cell-cycle tailored Cas9 fusion protein that is expressed during S/G2/M, but actively degraded via the ubiquitin-proteasome system during G1.4 In 2008, Sakaue-Sawano et al. showed that the first 110 amino acids of human Geminin are sufficient to confer nuclear localization and cell-cycle-dependent proteolysis of fluorescent reporter proteins in G1, which allowed the visualization of spatiotemporal dynamics of cell-cycle progression.5 We adapted this strategy and generated the Cas9-hGem(1/110) protein, whose expression is cell cycle dependent (Fig. 1). This proofed that Cas9 can be controlled in a temporal manner by an endogenous eukaryotic regulatory circuit. Subsequent side-by-side comparisons of Cas9 and Cas9-hGem(1/110) in genome editing assays revealed that the cell-cycle tailored fusion protein significantly increased the rate of HDR at a single-copy reporter gene locus as well as an endogenous (biallelic) non-coding RNA locus by up to 1.87-fold and 1.42-fold, respectively.
Figure 1.
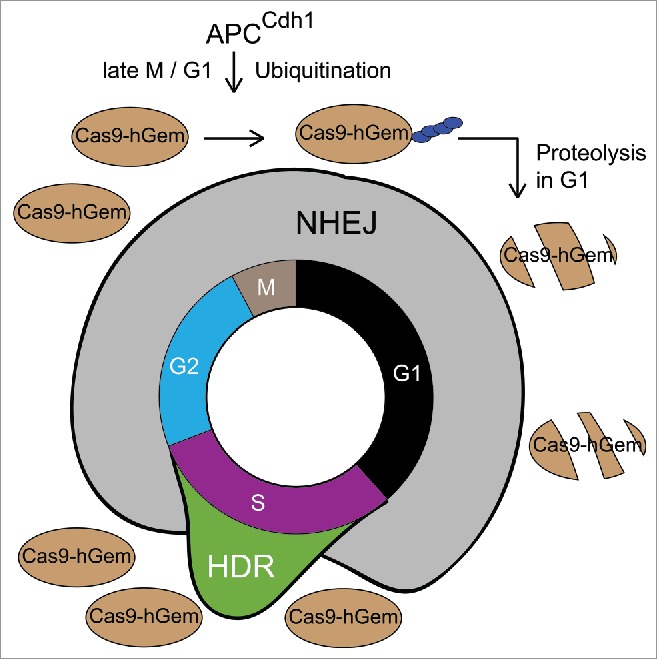
A cell-cycle tailored genome editing system to enhance HDR. The NHEJ pathway dominates DSB repair in G1 and G2 whereas HDR activity peaks in mid-S phase. The Cas9-hGem(1/110) fusion protein is subjected to proteolytic destruction during G1, but its expression is restored in S/G2/M. This partially synchronizes genome editing with HDR activity and enhances rates of the latter.
Importantly, the steady-state protein expression level of Cas9-hGem(1/110) was on average half of the level of Cas9. This is expected for an S/G2/M-specific protein and supports the idea of matching supply with demand to increase HDR. Of note, the reduced expression level of Cas9-hGem(1/110) had no detectable impact on NHEJ-mediated repair, as we observed the same amount of insertions and deletions at the endogenous non-coding RNA locus when we delivered the protein and sgRNA only, but omitted the repair donor plasmid. This might be explained by the refined activity profiles of NHEJ and HDR.2 While we did not investigate off-target effects, a previous study could show that reduced dosages of Cas9 and sgRNA could reduce off-target modifications.6 However, another study found that reducing expression levels of sgRNA and Cas9 in cells is not likely to provide a simple solution for reducing off-target effects.7 Hence, it will be interesting to investigate the off-target effects of the newly developed Cas9-hGem(1/110) protein in the future to see if the timing of expression has an impact on off-target effects.
Taken together, we propose that high-resolution expression strategies that tailor Cas9 or alternative genome-engineering proteins to the HDR activity profile of a cell will improve HDR rates without the need for sophisticated manipulation of cells by drugs or additional gene targeting reagents that could cause unpredictable effects on cell viability and/or genome integrity.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Orthwein A, et al.. Nature 2015; 528:422-26; PMID:26649820; http://dx.doi.org/ 10.1038/nature16142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Karanam K, et al.. Mol Cell 2012; 47:320-9; PMID:22841003; http://dx.doi.org/ 10.1016/j.molcel.2012.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Doudna JA, Charpentier E. Science 2014; 346:1258096; PMID:25430774; http://dx.doi.org/ 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- [4].Gutschner T, et al.. Cell Rep 2016; 14:1555-66; PMID:26854237; http://dx.doi.org/ 10.1016/j.celrep.2016.01.019 [DOI] [PubMed] [Google Scholar]
- [5].Sakaue-Sawano A, et al.. Cell 2008; 132:487-98; PMID:18267078; http://dx.doi.org/ 10.1016/j.cell.2007.12.033 [DOI] [PubMed] [Google Scholar]
- [6].Hsu PD, et al.. Nat Biotechnol 2013; 31:827-32; PMID:23873081; http://dx.doi.org/ 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fu Y, et al.. Nat Biotechnol 2013; 31:822-26; PMID:23792628; http://dx.doi.org/ 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]


