Abstract
The size and shape of nanocarriers can affect their fate in vivo, but little is known about the effect of nanocarrier aspect ratio on biodistribution in the setting of cancer imaging and drug delivery. The production of nanoscale anisotropic materials is a technical challenge. A unique biotemplating approach based on of rod-shaped nucleoprotein nanoparticles with predetermined aspect ratios (AR 3.5, 7, and 16.5) is used. These rigid, soft-matter nanoassemblies are derived from tobacco mosaic virus (TMV) components. The role of nanoparticle aspect ratio is investigated, while keeping the surface chemistries constant, using either PEGylated stealth nanoparticles or receptor-targeted RGD-displaying formulations. Aspect ratio has a profound impact on the behavior of the nanoparticles in vivo and in vitro. PEGylated nanorods with the lowest aspect ratio (AR 3.5) achieve the most efficient passive tumorhoming behavior because they can diffuse most easily, whereas RGD-labeled particles with a medium aspect ratio (AR 7) are more efficient at tumor targeting because this requires a balance between infusibility and ligand–receptor interactions. The in vivo behavior of nanoparticles can therefore be tailored to control biodistribution, longevity, and tumor penetration by modulating a single parameter: the aspect ratio of the nanocarrier.
1. Introduction
Nanotechnology can be defined as the development and application of materials that are 1–100 nm in diameter. This scale confers unique properties that are useful and valuable in fields such as chemistry, medicine, materials science, and electronics. One major application area is the use of nanoparticles in medicine, either for imaging or drug delivery. Because nanoparticles are defined only by their size, the concept embraces diverse structures including inorganic particles, polymers, lipid vesicles, and even viruses. Many different nanoparticle formulations are currently undergoing preclinical and clinical testing, and much of the development pipeline focuses on the use of nanoparticles for cancer diagnosis and therapy. This reflects the comparatively recent development of targeted nanoparticles that home in on tumors either to facilitate medical imaging or to concentrate drugs at the tumor site thus avoiding systemic effects.
The desirability of targeted therapy has prompted researchers to investigate how the properties of nanoparticles affect their biodistribution, tissue penetration, uptake, and elimination. Much of this research has focused on surface properties, which can be modified to achieve general objectives such as immune system evasion, or specific objectives such as tumor targeting by decorating the particle with tumor-specific ligands. The precise size and shape of nanoparticles also affect their in vivo behavior, but these properties have received far less attention. Although diverse nanoparticle shapes have been considered,[1,2] the vast majority of platforms currently undergoing testing are spherical or low-aspect-ratio materials. Filomicelles, which are polymeric high-aspect-ratio materials that mimic filamentous viruses, are notable exceptions.[3] The in vivo comparison of filamentous and spherical nanoparticles with similar chemical properties has shown that the high-aspect-ratio filaments have preferable tumor-homing properties because their shape allows them to evade macrophages leading to prolonged circulation,[3,4] and confers enhanced margination properties for improved endothelial targeting.[3,5–10] We have made similar observations studying the filamentous nanomaterials formed by the plant virus potato virus X (PVX), which measures 515 nm × 13 nm and is a highly flexible material. We observed that the PVX filaments accumulate and penetrate tumors due to their elongated and flexible nature.[11,12]
Because few studies have been carried out to determine the impact of nanoparticle shape and size, there is a lack of data considering the design virtues of nanomaterials that differ in terms of their aspect ratio only. Some reports indicate that high-aspect-ratio materials (nanorods and nanofilaments) are taken up more quickly by cells than low-aspect-ratio materials (nanospheres)[13,14] whereas other reports suggest the opposite.[15–18] These discrepancies clearly reflect the lack of comparability between different experimental set ups, e.g., the use of different cell lines, different growth media, varying incubation times, and distinct fixation methods. Furthermore, the direct comparison of nanoparticles with different shapes is only valid if their chemical composition and surface chemistry are stringently controlled to ensure they are equal. For example, gold nanorods and nanospheres may exhibit opposite surface charges due to differences in the method of synthesis and the capping agents employed.[14]
Caution must nevertheless be exercised when considering the data presented above because spherical nanoparticles with diverse sizes and chemical properties have been studied in detail, whereas there are fewer studies involving nanorods and nanofilaments and only a small number of these have considered how the aspect ratio affects biodistribution and targeting. In part, this reflects the fact that synthetic nanospheres are easier to create than nanorods and nanofilaments. In fact, tailoring nanomaterials in two dimensions on the nanoscale to create anisotropic materials of defined aspect ratios while keeping the surface chemistry consistent remains technologically challenging using synthetic chemistry. To overcome this barrier, we used a unique biotemplated self-assembly approach to produce nucleoprotein-based nanorods formed by tobacco mosaic virus (TMV) protein and different RNA scaffolds.
TMV was the first virus to be identified and remains one of the best-characterized viruses in terms of genetics, molecular biology, and structural biology. It has served as a model system in virology and facilitated the development of key structural biology techniques.[19] More recently, it has become a key model of bioinspired materials in the field of nanotechnology, where it has been developed, e.g., as a scaffold for light harvesting,[20] sensors, and battery electrodes,[21] as well as semiconducting transistor materials.[22] Virus-like particles in predetermined distinct length classes, generated using engineered TMV components in vitro, served as beneficial additives in technically applied ferrofluids, regulating both magnetoviscosity and shear stability according to the particles’ aspect ratio.[23] In a medical context, TMV has been developed as a vaccination platform,[24] a tissue-engineering scaffold,[25] and molecular contrast agent for bioimaging applications.[26]
The native TMV capsid comprises 2130 identical copies of the virus coat protein (CP), which self-assemble with the single-stranded RNA genome to form a 300 × 18 nm nanotube with a 4-nm longitudinal lumen, thereby integrating the RNA within a CP helix (see also Figure 1A). TMV was first identified in tobacco but it infects many different plant species in at least 30 families.[27,28] The virus can be propagated easily, which means that large amounts of the virus can be effectively “farmed” with little expense, providing a significant economic advantage over synthetic nanomaterials, which are more expensive to produce, especially in large quantities. The structure of TMV is known to atomic resolution, and protocols for genetic engineering[29–32] and chemical bioconjugation[33] are well-established, allowing the surface to be functionalized in a precise and predictable manner to facilitate the conjugation of imaging probes, targeting ligands, and drugs. For example, we recently achieved dual-modal optical-magnetic resonance imaging of atherosclerotic plaques in a mouse model using a TMV-based contrast agent targeted to vascular cell adhesion molecule (VCAM)-1.[26] Our novel contrast agent was more sensitive than commercially-available products such as Magnevist, reflecting the combined impact of the more concentrated distribution of imaging molecules (chelated Gd(DOTA)) on each nanoparticle, the specific targeting of the nanoparticle to the disease tissue, and the elongated shape of the particle, which promoted margination and endothelial targeting.[26]
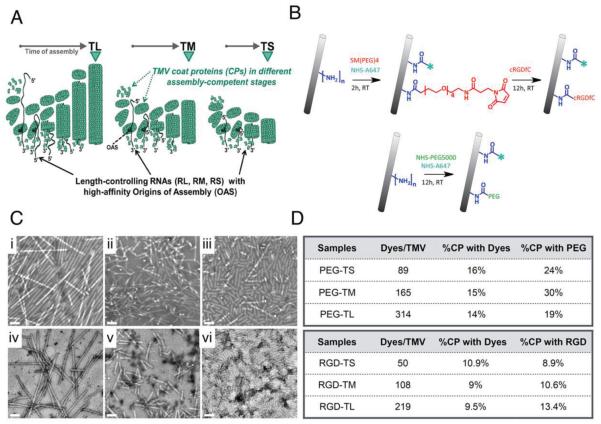
Figure 1.
A) Bottom-up assembly of TMV nanoparticles using synthetic RNA templates containing an OAS, and purified coat proteins (mechanistic scheme; proportions not in scale). B) Bioconjugation scheme: lysine 158 on each CP sub-unit is modified with A647 and RGD or PEG using NHS-reactive esters. C) TEM of negatively-stained i) PEG-TL, ii) PEG-TM, iii) PEG-TS, iv) RGD-TL, v) RGD-TM, vi) RGD-TS. Scale bar = 100 nm. D) Quantification of A647 dye and PEG ligands per TMV particle.
Viruses are versatile materials for nanomedical engineering because the structure is genetically encoded and therefore highly tunable, and the CP subunits have evolved to facilitate self-assembly. In the case of TMV, reconstitution experiments using purified CPs with and without RNA templates yield either disk-shaped or rod-shaped structures.[34] During normal infections, TMV capsid assembly initiates at an RNA hairpin loop-forming sequence known as the origin of assembly site (OAS),[35] and the length of the resulting virion depends on the length of the genomic RNA. The wild-type TMV genome is 6395 nucleotides in length, which yields a 300-nm rod. However, the OAS is the only sequence necessary to initiate bidirectional encapsidation of an RNA strand by TMV CP.[36,37] This enables a unique biotemplated approach to produce TMV-based nanorods of defined aspect ratio programmed by the length of the RNA molecule carrying the OAS. Based on this principle, synthetic RNA scaffolds freely dissolved or immobilized at one end, containing single or multiple OAS assembly nucleation sequences, have been combined with purified TMV CPs to produce nanotubes with distinct longitudinal domains,[38] uncommon kinked and branched nanoobjects,[39] and bottom-up-grown flat[40] or star-shaped nanoparticle arrays.[41] We have developed this unique nanoparticle-programming technology to gain novel insights into the effect of aspect ratio on biodistribution in the setting of cancer imaging and drug delivery.
We set out to fabricate TMV-based nanoparticle formulations with constant diameters and equal surfaces, but different aspect ratios as governed by distinct RNA scaffolds, allowing a systematic comparison of biodistribution and tumor-targeting properties. Stealth TMV formulations were produced by covering the particle surface with polyethylene glycol (PEG) to overcome immune surveillance, and targeted TMV formulations were produced by displaying the integrin-targeting cyclic peptide ligand RGD via an intervening PEG spacer. PEG and RGD are commonly used surface modifiers, specifically chosen as tools to provide surface chemistries that could be compared to other nanoparticle platform technologies currently in the development pipeline. Biodistribution and tumor-homing behavior were analyzed by Maestro imaging and fluorescence molecular tomography (FMT) showing that both the aspect ratio and surface chemistry of the particles had distinct effects on in vivo behavior, including tumor localization and cancer cell versus immune cell targeting.
2. Results and Discussion
2.1. Synthesis of TMV Nanoparticles
We prepared three TMV particle formulations varying in length but identical in width by assembling purified TMV CPs with in vitro synthesized RNA molecules of defined lengths, each containing an OAS (Figure 1A, Table 1). Detailed methods are provided in the Supporting Information. This yielded TMV rods with median lengths of ≈300, 130, and 60 nm, corresponding to aspect ratios of 16.5, 7, and 3.5, respectively (Figure S1, Supporting Information). These are described hereafter as TL, TM, and TS for TMV long, medium, and short. In each case, we used a well-characterized TMV CP mutant, which replaces the threonine residue at position 158 with lysine.[38] The amine group of the lysine side chain is displayed on the particle surface to allow functionalization with fluorophores, PEG, and the RGD ligand.
Table 1.
RNA molecules and underlying constructs used to generate TL, TM, and TS TMV-like particles.
| TMV/RNA name | Theoret. particle length [nm] |
RNA length [nts] |
Plasmid name | RNA portions 5′/OAS/3′ [nts]a) |
Nanorod 5′/3′ portions [nm] |
|---|---|---|---|---|---|
| TL/RL | 300 | 6395 | p843pe35TMVr.1 b) | 5472/9/914 | 257/43 |
| TM/RM | 135 | 2884 c) | p843peTMVΔ9353–3391 d) | 2107/9/768 | 99/36 |
| TS/RS | 59 | 1294 c) | pGEM-T/N 13p843pe TMV 5081–6245N35 |
408/9/832 | 20/39 |
OAS: only a 9-nt loop portion is denoted;
infectious plasmid described by Kadri et al. 2011, resulting in genomic TMV RNA inside plants;
after linearization with BsiW1 (for TM) or NdeI (for TS), respectively, and in vitro transcription;
in vitro transcription plasmid described by Azucena, Eber et al. 2012. [42]
Conjugation using the lysine side chain described above should not affect the integrity of the TMV particles. We therefore conjugated TL, TM, and TS with AlexaFluor647 (A647) for imaging, PEG5000 to evade immune surveillance, or the cyclic RGDfC peptide for tumor targeting (Figure 1B). In the latter case, the cysteine side chain of the peptide was used for conjugation via a 500-Da PEG spacer. The reactions and methods are described in detail in the Supporting Information. PEG is a validated stealth polymer with low toxicity and immunogenicity, which reduces serum protein adsorption and thus increases residence time in the circulation, and ensures that less of the nanoparticle formulation is deposited in the liver and spleen.[43–47] RGD targets αvβ3 and αvβ5 integrins, which are overexpressed on tumor cells and the neovasculature in many forms of cancer, and the upregulation of integrins is associated with tumor progression and resistance.[48,49] Integrins are therefore validated targets for molecular imaging and targeted therapy, hence several RGD-based, integrin-specific contrast agents are currently undergoing clinical development.[49–51] PEG and RGD are widely used surface modifiers; we chose these molecules because this should allow us to correlate and translate our findings to other nanoparticulate systems under development.
After purifying the modified TMV particles, the new surface chemistries were verified using a combination of transmission electron microscopy (TEM) (Figure 1C), UV–visible spectroscopy (Figure S2A, Supporting Information), and protein gel electrophoresis (Figure S2B, Supporting Information). These experiments confirmed that the stealth particles (PEG-TMV) and targeted particles (RGD-TMV) were intact and incorporated the anticipated modifications. In the case of PEG-TS, PEG-TM, and PEG-TL, approximately 15% of the CPs on each particle were labeled with A647 and 25% displayed PEG5000. In the case of RGD-TS, RGD-TM, and RGD-TL, approximately 10% of the CPs on each particle were labeled with A647 and 10% displayed the RGD peptide (Figure 1D).
2.2. Tumor-Homing Properties
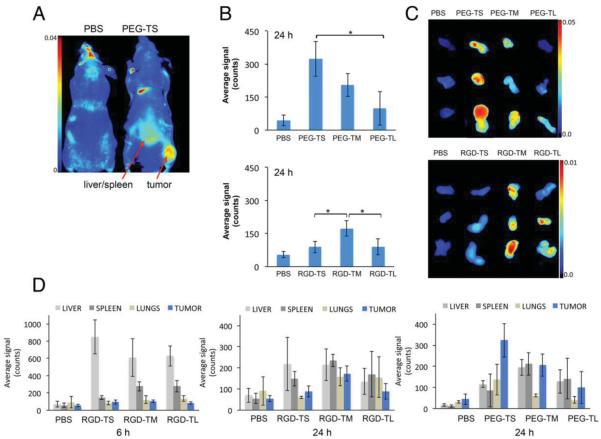
We tested the tumor-homing properties of TMV formulations with different aspect ratios using an established mouse model of colon cancer (NCr nu/nu nude mice with subcutaneous HT-29 xenografts). All animal studies were carried out using IACUC-approved procedures for in vivo imaging, ex vivo imaging, and histology, which are described in detail in the Supporting Information. The nanoparticle formulations were administered intravenously as a single 10 mg protein per kg body weight dose, and the animals were examined using the Maestro imaging system 6 and 24 h post-administration (Figure 2A). The mice were then sacrificed and the tumors, livers, spleens, and lungs removed for ex vivo quantitative tissue analysis. Both the stealth particles and the targeted particles showed aspect ratio-dependent effects; notably, the optimum aspect ratio differed for distinct surface functionalizations. The accumulation of stealth particles in tumors appeared to be aspect ratio-dependent, with the lowest-aspect-ratio particles accumulating at higher concentrations (PEG-TS > PEG-TM > PEG-TL). In contrast, although the accumulation of the targeted particles was also aspect ratio-dependent, it was the 130-nm particles (RGD-TM) that accumulated to the highest levels in tumors (Figure 2B).
Figure 2.
A) Maestro imaging of NCr nu/nu mice with subcutaneous HT-29 tumor xenografts 24 h post-administration of PBS (control) or fluorescently-labeled PEG-TS. B) Tumor accumulation of stealth and targeted TMV formulations 24 h post-administration; quantification is based on ex vivo tissue analysis; C) Maestro images of the above. D) Biodistribution profiles determined by quantitative ex vivo tissue analysis using a Maestro imaging system. For all experiments we used n = 3 animals per formulation; graphs show average fluorescence signals and standard deviations.
Tumor homing by stealth particles is mediated passively by the enhanced permeability and retention (EPR) effect,[52] whereas the RGD-targeted nanoparticles are actively targeted to tumor-associated integrin receptors (the integrin-specificity of the cRGDfK peptides and their conjugates was confirmed by competition binding experiments in vitro; Supporting Information Figure S4, not shown). The differences in tumor-homing properties between the stealth and targeted particles therefore appear to reflect their different targeting mechanisms. The efficiency of tumor accumulation was similar when we compared PEG-TM and RGD-TM, or PEG-TL and RGD-TL (Figure 2B). Others have also shown that targeting ligands only marginally improve the tumor-homing properties of nanoparticles compared to nontargeted controls, but there were differences in the overall biodistribution profiles and the localization of the nano-particles within the tumors,[53] which is consistent with our histological observations described below.
In contrast to the longer particles, there were significant differences in tumor accumulation between the low-aspect-ratio stealth and targeted formulations, with PEG-TS showing more efficient tumor targeting than RGD-TS (Figure 2B). The high-molecular-weight PEG layer in the PEG-TS formulation may provide a more effective stealth effect than the shorter intervening PEG spacer in the RGD-TS formulation, protecting the particles from serum protein adsorption and thus achieving immune evasion and more efficient passive tumor accumulation. This is consistent with reports describing the prolonged circulation of PEGylated liposomal doxorubicin formulations, which promotes EPR-mediated tumor homing.[54,55] Increased passive tumor homing of PEGylated formulations, such as stealth liposomes, can be explained by their increased circulation times. The greater bioavailability increases the likelihood of nanoparticle accumulation mediated by the EPR effect. Because the RGD-TS formulations incorporate shorter PEG brushes compared to the PEG-TS formulation, we suggest that the differences in tumor accumulation result from differences in their pharmacokinetics profiles.
Further, the RGD-TS particles carry fewer RGD ligands than the RGD-TM and RGD-TL particles, which may not be sufficient for effective tumor targeting; multivalency and avidity are critical parameters to achieve efficient targeting and integrin interactions.[56] In contrast, the RGD-TM and RGD-TL particles would be expected to bind two or more integrins simultaneously. Furthermore, recent studies have suggested that RGD ligands are useful for the imaging of tumor neovasculature but that the targeting ligand may not increase tumor homing[53] but instead may increase overall tissue distribution,[57] which is also consistent with our findings (Figure 2C). Effective integrin targeting is highly dependent on the ligand density.[58] It would therefore be beneficial to investigate the role of RGD density and PEG spacer length on the efficiency of tumor targeting.
The more efficient passive tumor homing of the low-aspect-ratio stealth formulation may reflect the pore size of the leaky vasculature, which differs among tumor types and the stage of disease.[59] The pore size limits the extravasation of nanocarriers, with smaller nanoparticles having a greater opportunity to exit the circulation and accumulate within the tumor tissue.[60] Nanomaterials with smaller dimensions generally show more efficient tumor penetration.[61] This is consistent with PEG-TS showing higher tumor accumulation and penetration compared to PEG-TM and PEG-TL. Also, the more efficient tumor homing shown by the mid-sized RGD-TM particles suggests that tumor homing involves passive and active mechanisms acting in synergy, with the mid-sized particles showing the most beneficial combination of size and ligand density. In summary, the aspect ratio of nanomaterials affects their tumorhoming properties; aspect ratio can be used as a design parameter (in addition to surface chemistry) to tailor the in vivo profiles of nanoparticle-based contrast agents or drug carriers.
2.3. Biodistribution and Clearance
We measured the distribution of each formulation between the healthy and tumor tissue 6 and 24 h post-administration. We found that there was more of the targeted formulation in the liver, spleen, and lungs compared to the tumors after 6 h, and only residual amounts left in the body after 24 h indicating rapid tissue clearance (Figure 3). Nanoparticles decorated with RGD ligands have previously been shown to accumulate in the organs of the macrophagocytic system, reflecting structural differences in the protein corona compared to stealth nanoparticles.[57] Rapid tissue clearance is consistent with our previous TMV biodistribution studies,[62] and makes TMV particularly attractive for molecular imaging and drug delivery approaches where rapid tissue clearance is necessary.
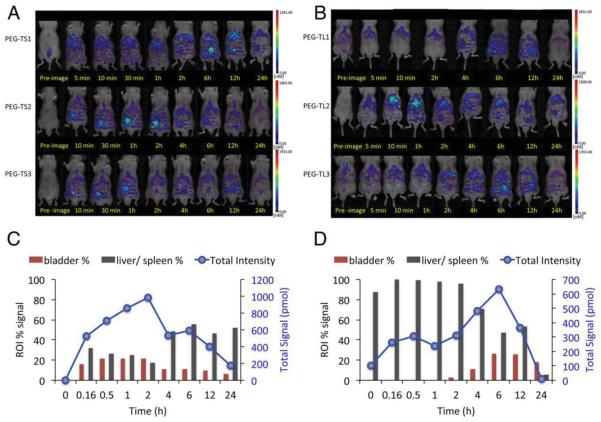
Figure 3.
FMT imaging of NCr nu/nu mice with subcutaneous HT-29 tumor xenografts (n = 3 mice per group) after the intravenous administration of A) PEG-TS or B) PEG-TL. Quantitative analysis showing fluorescence signals from bladder and liver/spleen as a percentage of the total fluorescence signal from fluorescently-labeled C) PEG-TS or D) PEG-TL.
The stealth formulations generally had more favorable biodistribution profiles, with PEG-TS in particular achieving effective partitioning between the tumor and healthy tissues. Approximately 50% of the formulation accumulated in the tumor tissue, while the remainder was distributed in the liver, spleen, and lungs (Figure 2C). The aspect ratio therefore appears to affect partitioning during biodistribution, with higher-aspect-ratio materials promoting molecular targeting and lower-aspect-ratio materials promoting passive uptake by EPR.
The kinetics of biodistribution and clearance were studied by using fluorescence molecular tomography (FMT) to compare the total abundance of PEG-TS and PEG-TL and their partition in the bladder and liver/spleen over time (Figure 3). Peak tissue accumulation occurred after 2 h for PEG-TS and 6 h for PEG-TL, probably reflecting differences in the pharmacokinetic and clearance profiles as a function of aspect ratio. The PEG-TL formulation was expected to persist for longer because it evades the immune system more effectively and deposition in tissues is therefore delayed, as confirmed by our histology and tissue culture experiments (Figure 4). The majority of the total PEG-TL signal was cleared via the liver and spleen, indicating that the reticuloendothelial system (RES) was responsible, whereas the PEG-TS was deposited in the bladder, liver, and spleen, indicating renal filtration, especially at the early time points (Figure 4). The size cut-off for glomerular filtration is less than 20 nm.[63] If TMV particles are aligned in the flow then they can pass through the renal filtration system, but longer TMV rods have a greater likelihood of being trapped while shorter versions pass through the glomerulus.
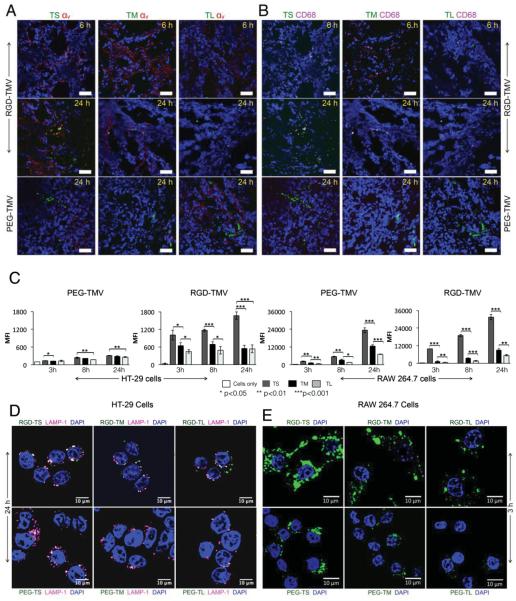
Figure 4.
Histological analysis of tumor tissues collected from animals injected with the different TMV formulations. Nuclei were stained with DAPI (shown in blue). TMV was imaged by detecting the conjugated A647 dye (pseudocolored in green). Scale bar = 50 µm. A) Staining of integrin αv shown in red. B) Staining of macrophages shown in pink. C) Cellular uptake of TMV formulations by HT-29 cells (left panel) and RAW264.7 macrophages (right panel). At least 10 000 gated events were recorded. All samples were analyzed in triplicate and averages and standard deviations are shown. MFI = mean fluorescence intensity. D) Confocal microscopy showing the cellular uptake of TMV formulations by HT-29 cells. The endosomal marker LAMP-1 is shown in pink. E) Confocal microscopy showing the cellular uptake of TMV formulations by RAW264.7 macrophages. TMV was imaged by detecting the conjugated A647 dye (pseudocolored in green). Scale bar = 10 µm.
The biodistribution and clearance profiles are consistent with previous reports in which native TMV, PEG-TMV rods, and 50-nm TMV spheres were administered to healthy BALB/c mice.[62] It should be noted that spheres were not considered in the present study, because these materials display different protein domains on their surface—therefore TMV spheres do not compare to TMV rods in terms of their surface properties; and the essence of this study was to evaluate the effect of aspect ratio of otherwise identical anisotropic nanomaterials. Clearance via the liver and spleen was independent of surface chemistry and shape, but there were differences in clearance rate and pharmacokinetic behavior, with the spherical particles clearing more rapidly than their rod-shaped counterparts, and rod-shaped particles persisting for longer because of their more efficient immune system evasion.
2.4. Histological Analysis and Interactions with Cancer and Immune Cells
Histological analysis showed that the stealth and targeted formulations were generally located in proximity to the tumor endothelium, but not necessarily co-localized with the tumor marker CD31 (Figure S3, Supporting Information). Furthermore, only the RGD-targeted formulations were co-localized with integrin αv on the surface of angiogenic tumor endothelium and HT-29 tumor cells (Figure 4A).
The comparison of formulations with different aspect ratios showed that the PEG-TS penetrated further from the vessel wall than the larger versions as anticipated, because diffusion is slower for materials that have a greater molecular weight[64] or size.[65] In an earlier study, 60 × 15 nm gold rods were compared to spherical gold nanoparticles of the same diameter and the rods were found to penetrate further into the tumor, revealing that penetration and diffusion are dependent on particle shape.[6]
The stealth formulations were not co-localized with tumor-associated macrophages (identified by staining with a CD68-specific marker) whereas the RGD-targeted nanoparticles co-localized with macrophages in a size-dependent and time-dependent manner. RGD-TS showed a greater degree of co-localization with CD68 after 6 h than RGD-TM or RGD-TL, consistent with our in vitro cellular uptake data (Figure 4D), whereas macrophage co-localization was not dependent on size after 24 h (Figure 4B). These data indicated that RGD-labeled nanoparticles targeted tumor cells more effectively if they had a higher aspect ratio but were recognized by immune cells more effectively if they had a lower aspect ratio. A recent study suggested that nanoparticle uptake by tumor tissue reflects a combination of extravasation and monocyte delivery, i.e., carbon nanotubes were transported into the tumor tissue by monocytes, and this effect was increased for RGD-functionalized carbon nanotubes.[66] Although immune cell delivery was confirmed for high-aspect-ratio carbon nanotubes, our data indicate that the immune-based delivery strategies can be tailored as a function of surface chemistry and aspect ratio.
We also investigated the behavior of our nanoparticle formulations in the presence of HT-29 cells (a colon carcinoma cell line) and RAW264.7 cells (a mouse macrophage cell line). Time–course interactions were evaluated quantitatively by flow cytometry and qualitatively by confocal microscopy. Our data confirmed that integrin-expressing HT-29 cells were targeted by all three variants of the RGD-TMV formulations, whereas there was only negligible interaction with the stealth PEG-TMV particles (Figure 4C,D). Comparing the different aspect-ratio formulations, we found that the uptake of RGD-TS was most efficient, and there was no statistically significant difference in uptake rates between RGD-TM and RGD-TL. However, whereas RGD-TS and RGD-TM were internalized as shown by their co-localization with the lysosomal marker Lamp-1, RGD-TL was confined to the cell surface.
The cellular uptake of nanoparticles is dependent on the competition between hydrodynamic driving forces and receptor diffusion kinetics. The optimal particle diameter for cellular uptake is 60 nm for spherical materials including icosahedral viruses[67,68] and this explains the less efficient uptake of formulations with a higher aspect ratio. Nanoparticles with RGD ligands undergo receptor-mediated endocytosis via the clathrin or caveolin pathways, the former involving the formation of clathrin-coated pits that produce vesicles 150–200 nm in diameter, and the latter involving the formation of flask-shaped membrane invaginations 50–100 nm in diameter.[69] Therefore, the vesicle size may inhibit the uptake of the 300-nm RGD-TL particles via receptor-mediated endocytosis (Figure 4D). The orientation and contact angle of the nanoparticle compared to the target cell also influence endocytosis[70] and phagocytosis.[4]
The interactions with RAW264.7 cells also provided insight into the combined impact of shape and surface chemistry on the in vivo behavior of TMV nanoparticles. There was a significant difference between PEG-TS and RGD-TS, with the stealth particles showing a much lower susceptibility to phagocytosis over prolonged periods of time (Figure 4C,D). In contrast, there was no significant difference between PEG-TL and RGD-TL. Neither of the particles was particularly susceptible to phagocytosis, and the stealth coating did not appear to confer extra protection (Figure 4C,D).
3. Conclusion
A growing of body of data suggests that nanoparticle shape and size influence their in vivo biodistribution; yet only a small number of anisotropic nanoscale materials have been investigated, and aspect ratio has been largely overlooked as a design parameter. It remains challenging to produce synthetic nanomaterials with a defined aspect ratio and identical surface chemistry, so we used a biotemplating approach to produce stiff nucleoprotein nanorods by the RNA-guided self-assembly of TMV CPs. This bioinspired approach enables the manufacture of TMV-based nanorods with identical surface chemistry and diverse aspect ratios, i.e., the nanorods are equivalent in all properties except their length.
We used a combination of in vivo imaging, ex vivo imaging, and histology to compare the tumor-homing properties and cellular interactions of stealth and RGD-targeted rod-shaped nanoparticle species of 18 nm core diameter, each prepared as three different formulations with distinct aspect ratios. We found that aspect ratio had a profound impact on the behavior of the particles in vivo and in vitro. Particles concealed from the immune system by PEGylation remained longer in the circulation and accumulated in tumors passively via the EPR system, with the lowest-aspect-ratio nanoparticles outperforming the higher-aspect-ratio particles in terms of tumor accumulation. In contrast, particles that were decorated with the targeting ligand RGD were recognized by immune cells and cleared more rapidly, but were able to interact specifically with the endothelium of the vasculature. Here, the mid-sized aspect ratio particles demonstrated the highest tumor accumulation, reflecting their enhanced immune evasion compared to the lower-aspect-ratio nanoparticles, but enhanced cell targeting and uptake kinetics compared to the higher-aspect-ratio formulation.
Specifically, we found that PEGylated particles with the lowest aspect ratio (3.5) showed enhanced passive tumorhoming behavior because they were more likely to diffuse through the endothelium and into the tumor. In contrast, RGD-labeled particles with the lowest aspect ratio were less efficient at tumor targeting because they were easily removed by macrophages and did not carry enough ligands to bind effectively to target integrins. RGD-labeled particles with the highest aspect ratio (16.5) were unable to pass through the epithelium (and in vitro they were trapped at the cell membrane because they were too large for internalization by endocytosis or phagocytosis). Therefore, it was the medium-sized variant (130 × 18 nm, aspect ratio: 7) that showed the most efficient targeting based on a combination of efficient target binding and efficient diffusion.
Our studies therefore show that the in vivo behavior of elongated nanoparticles can be exquisitely tailored to control biodistribution, longevity, and tumor penetration by modulating the aspect ratio using a biotemplated bioengineering design.
Supplementary Material
Acknowledgements
The authors are very grateful to Diether Gotthardt and Sigi Kober for gardening and technical assistance, and especially to Fania Geiger and Holger Jeske for their continuous support of the study. This work was supported by a grant from the National Science Foundation, NSF CHE-1306447 to N.F.S., Mt. Sinai foundation to N.F.S., and subsidiary funding by the Baden-Wuerttemberg-Stiftung (Research Network “Functional Nanostructures”), and the Deutsche Forschungsgesllschaft (DFG; SPP 1569) to C.W. S.S. performed chemical bioconjugation, tissue culture, histology, tumor homing, biodistribution, and imaging studies. F.J.E. established in vitro transcription plasmid clones for RNA templates, optimized all relevant assembly procedures, conducted detailed analyses of TMV-like particles by TEM and molecular techniques, and instructed N.S. A.S.N. and N.A.D. performed tissue culture and histology experiments. N.S. performed TMV building block preparation, bottom-up assembly procedures, and product characterization. A.M.W. performed TEM studies. S.E. performed TMV propagation and isolation, and supported and guided work of N.S. N.F.S. designed the study and directed its implementation, specifically particle design and preclinical studies; C.W. led the bottom-up assembly approach. N.F.S., C.W., R.T., and S.S. analyzed and interpreted data. N.F.S., C.W., F.J.E., R.T., and S.S. prepared the manuscript; all authors edited the manuscript.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
The authors declare that they have no competing interests that might be perceived to influence the results and/or discussion reported in this article.
Contributor Information
Sourabh Shukla, Department of Biomedical Engineering, Case Western Reserve University Schools of Medicine and Engineering, Cleveland, OH 44106, USA.
Fabian J. Eber, Department of Molecular Biology and Plant Virology, Institute of Biomaterials and Biomolecular Systems, University of Stuttgart, 70569, Stuttgart, Germany
Adithy S. Nagarajan, Department of Biomedical Engineering, Case Western Reserve University Schools of Medicine and Engineering, Cleveland, OH 44106, USA
Nicholas A. DiFranco, Department of Biomedical Engineering, Case Western Reserve University Schools of Medicine and Engineering, Cleveland, OH 44106, USA
Nora Schmidt, Department of Molecular Biology and Plant Virology, Institute of Biomaterials and Biomolecular Systems, University of Stuttgart, 70569, Stuttgart, Germany.
Amy M. Wen, Department of Biomedical Engineering, Case Western Reserve University Schools of Medicine and Engineering, Cleveland, OH 44106, USA
Sabine Eiben, Department of Molecular Biology and Plant Virology, Institute of Biomaterials and Biomolecular Systems, University of Stuttgart, 70569, Stuttgart, Germany.
Richard M. Twyman, TRM Ltd, PO Box 93, York YO43 3WE, UK
Prof. Christina Wege, Department of Molecular Biology and Plant Virology, Institute of Biomaterials and Biomolecular Systems, University of Stuttgart, 70569, Stuttgart, Germany.
Prof. Nicole F. Steinmetz, Department of Biomedical Engineering, Case Western Reserve University Schools of Medicine and Engineering, Cleveland, OH 44106, USA; Department of Radiology, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA; Department of Materials Science and Engineering, Case Western Reserve University School of Engineering, Cleveland, OH 44106, USA; Department of Macromolecular Science and Engineering, Case Western Reserve University School Engineering, Cleveland, OH 44106, USA.
References
- [1].Caldorera-Moore M, Guimard N, Shi L, Roy K. Exp. Opin. Drug Delivery. 2010;7:479. doi: 10.1517/17425240903579971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Daum N, Tscheka C, Neumeyer A, Schneider M. Wiley Interdisciplinary Rev. Nanomed. Nanobiotechnol. 2012;4:52. doi: 10.1002/wnan.165. [DOI] [PubMed] [Google Scholar]
- [3].Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Nat. Nanotechnol. 2007;2:249. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Champion JA, Mitragotri S. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4930. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cai S, Vijayan K, Cheng D, Lima EM, Discher DE. Pharm. Res. 2007;24:2099. doi: 10.1007/s11095-007-9335-z. [DOI] [PubMed] [Google Scholar]
- [6].Chauhan VP, Popovic Z, Chen O, Cui J, Fukumura D, Bawendi MG, Jain RK. Angew. Chem. Int. Ed. 2011;50:11417. doi: 10.1002/anie.201104449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Christian DA, Cai S, Garbuzenko OB, Harada T, Zajac AL, Minko T, Discher DE. Mol. Pharm. 2009;6:1343. doi: 10.1021/mp900022m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Decuzzi P, Godin B, Tanaka T, Lee S-Y, Chiappini C, Liu X, Ferrari M. J. Controlled Release. 2010;141:320. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- [9].Gentile F, Chiappini C, Fine D, Bhavane RC, Peluccio MS, Cheng MM, Liu X, Ferrari M, Decuzzi P. J. Biomech. 2008;41:2312. doi: 10.1016/j.jbiomech.2008.03.021. [DOI] [PubMed] [Google Scholar]
- [10].Lee S-Y, Ferrari M, Decuzzi P. Nanotechnology. 2009;20:495101. doi: 10.1088/0957-4484/20/49/495101. [DOI] [PubMed] [Google Scholar]
- [11].Shukla S, Ablack AL, Wen AM, Lee KL, Lewis JD, Steinmetz NF. Mol. Pharm. 2013;10:33. doi: 10.1021/mp300240m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shukla S, Wen AM, Ayat NR, Commandeur U, Gopalkrishnan R, Broome AM, Lozada KW, Keri RA, Steinmetz NF. Nanomedicine. 2014;9:221. doi: 10.2217/nnm.13.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11613. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Arnida, Malugin A, Ghandehari H. J. Appl. Toxicol. 2010;30:212. doi: 10.1002/jat.1486. [DOI] [PubMed] [Google Scholar]
- [15].Nan A, Bai X, Son SJ, Lee SB, Ghandehari H. Nano Lett. 2008;8:2150. doi: 10.1021/nl0802741. [DOI] [PubMed] [Google Scholar]
- [16].Chithrani BD, Chan WCW. Nano Lett. 2007;7:1542. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- [17].Chithrani BD, Ghazani AA, Chan WCW. Nano Lett. 2006;6:662. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- [18].Schaeublin NM, Braydich-Stolle LK, Maurer EI, Park K, MacCuspie RI, Afrooz AR, Vaia RA, Saleh NB, Hussain SM. Langmuir. 2012;28:3248. doi: 10.1021/la204081m. [DOI] [PubMed] [Google Scholar]
- [19].Scholthof KB. Annu. Rev. Phytopathol. 2004;42:13. doi: 10.1146/annurev.phyto.42.040803.140322. [DOI] [PubMed] [Google Scholar]
- [20].Miller RA, Presley AD, Francis MB. J. Am. Chem. Soc. 2007;129:3104. doi: 10.1021/ja063887t. [DOI] [PubMed] [Google Scholar]
- [21].Fan XZ, Pomerantseva E, Gnerlicha M, Brown A, Gerasopoulos K, McCarthy M, Culver J, Ghodssi R. J. Vaccum Sci., Technol. A. 2013;31:050815. [Google Scholar]
- [22].Atanasova P, Rothenstein D, Schneider JJ, Hoffmann RC, Dilfer S, Eiben S, Wege C, Jeske H, Bill J. Adv. Mater. 2011;23:4918. doi: 10.1002/adma.201102900. [DOI] [PubMed] [Google Scholar]
- [23].Wu Z, Mueller A, Degenhard S, Ruff SE, Geiger F, Bittner AM, Wege C, Krill CE., 3rd ACS Nano. 2010;4:4531. doi: 10.1021/nn100645e. [DOI] [PubMed] [Google Scholar]
- [24].Plummer EM, Manchester M. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010;3:174. doi: 10.1002/wnan.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Luckanagul JA, Lee LA, You S, Yang X, Wang Q. J. Biomed. Mater. Res. A. 2014 doi: 10.1002/jbm.a.35227. DOI: 10.1002/jbm.a.35227. [DOI] [PubMed] [Google Scholar]
- [26].Bruckman M, Jiang K, Simpson EJ, Randolph L, Luyt LG, Yu X, Steinmetz NF. Nano Lett. 2014 doi: 10.1021/nl404816m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zaitlin M, Palukaitis P. Annu. Rev. Phytopathol. 2000;38:117. doi: 10.1146/annurev.phyto.38.1.117. [DOI] [PubMed] [Google Scholar]
- [28].Adams MJ. In: Ninth Report of the International Committee on Taxonomy of Viruses. King AMQ, Lefkowitz E, Adams MJ, Carstens EB, editors. Elsevier/Academic Press; Amsterdam, Boston, Heidelberg, London, New York, Oxford, Paris, San Diego, San Francisco, Singapore, Sydney, Tokyo: 2012. p. 1139. [Google Scholar]
- [29].Fitchen J, Beachy RN, Hein MB. Vaccine. 1995;13:1051. doi: 10.1016/0264-410x(95)00075-c. [DOI] [PubMed] [Google Scholar]
- [30].Fujiyama K, Saejung W, Yanagihara I, Nakado J, Misaki R, Honda T, Watanabe Y, Seki T. J. Biosci. Bioeng. 2006;101:398. doi: 10.1263/jbb.101.398. [DOI] [PubMed] [Google Scholar]
- [31].Koo M, Bendahmane M, Lettieri GA, Paoletti AD, Lane TE, Fitchen JH, Buchmeier MJ, Beachy RN. Proc. Natl. Acad. Sci. U.S.A. 1999;96:7774. doi: 10.1073/pnas.96.14.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Smith ML, Fitzmaurice WP, Turpen TH, Palmer KE. Curr. Top Microbiol. Immunol. 2009;332:13. doi: 10.1007/978-3-540-70868-1_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bruckman MA, Steinmetz NF. Methods Mol. Biol. 2014;1108:173. doi: 10.1007/978-1-62703-751-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fraenkel-Conrat H, Williams RC. Proc. Natl. Acad. Sci. U.S.A. 1955;41:690. doi: 10.1073/pnas.41.10.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zimmern D, Butler PJ. Cell. 1977;11:455. doi: 10.1016/0092-8674(77)90064-2. [DOI] [PubMed] [Google Scholar]
- [36].Lebeurier G, Nicolaieff A, Richards KE. Proc. Natl. Acad. Sci. U.S.A. 1977;74:149. doi: 10.1073/pnas.74.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Otsuki Y, Takebe I, Ohno T, Fukuda M, Okada Y. Proc. Natl. Acad. Sci. U.S.A. 1977;74:1913. doi: 10.1073/pnas.74.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Geiger FC, Eber FJ, Eiben S, Mueller A, Jeske H, Spatz JP, Wege C. Nanoscale. 2013;5:3808. doi: 10.1039/c3nr33724c. [DOI] [PubMed] [Google Scholar]
- [39].Eber FJ, Eiben S, Jeske H, Wege C. Nanoscale. 2015;7:344. doi: 10.1039/c4nr05434b. [DOI] [PubMed] [Google Scholar]
- [40].Mueller A, Eber FJ, Azucena C, Petershans A, Bittner AM, Gliemann H, Jeske H, Wege C. ACS Nano. 2011;5:4512. doi: 10.1021/nn103557s. [DOI] [PubMed] [Google Scholar]
- [41].Eber FJ, Eiben S, Jeske H, Wege C. Angew. Chem. Int. Ed. 2013;52:7203. doi: 10.1002/anie.201300834. [DOI] [PubMed] [Google Scholar]
- [42].Azucena C, Eber FJ, Trouillet V, Hirtz M, Heissler S, Franzreb M, Fuchs H, Wege C, Gliemann H. Langmuir. 2012;28:14867. doi: 10.1021/la302774h. [DOI] [PubMed] [Google Scholar]
- [43].Harris JM, Chess RB. Nat. Rev. Drug Discovery. 2003;2:214. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- [44].Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanomedicine. 2011;6:715. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Roberts MJ, Bentley MD, Harris JM. Adv. Drug Delivery Rev. 2002;54:459. doi: 10.1016/s0169-409x(02)00022-4. [DOI] [PubMed] [Google Scholar]
- [46].Veronese FM, Pasut G. Drug Discovery Today. 2005;10:1451. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- [47].Wattendorf U, Merkle HP. J. Pharm. Sci. 2008;97:4655. doi: 10.1002/jps.21350. [DOI] [PubMed] [Google Scholar]
- [48].Brooks PC, Montgomery AM, Rosenfeld M, Reisfeld RA, Hu T, Klier G, Cheresh DA. Cell. 1994;79:1157. doi: 10.1016/0092-8674(94)90007-8. [DOI] [PubMed] [Google Scholar]
- [49].Sutherland M, Gordon A, Shnyder SD, Patterson LH, Sheldrake HM. Cancers. 2012;4:1106. doi: 10.3390/cancers4041106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Danhier F, Breton Le A., Preat V. Mol. Pharm. 2012;9:2961. doi: 10.1021/mp3002733. [DOI] [PubMed] [Google Scholar]
- [51].Kenny LM, Coombes RC, Oulie I, Contractor KB, Miller M, Spinks TJ, McParland B, Cohen PS, Hui AM, Palmieri C, Osman S, Glaser M, Turton D, Al-Nahhas A, Aboagye EO. J. Nucl. Med. 2008;49:879. doi: 10.2967/jnumed.107.049452. [DOI] [PubMed] [Google Scholar]
- [52].Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. J. Controlled Release. 2000;65:271. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- [53].Huang X, Peng X, Wang Y, Wang Y, Shin DM, Sayed El-M. A., Nie S. ACS Nano. 2010;4:5887. doi: 10.1021/nn102055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Papahadjopoulos D, Allen TM, Gabizon A, Mayhew E, Matthay K, Huang SK, Lee KD, Woodle MC, Lasic DD, Redemann C. Proc. Natl. Acad. Sci. U.S.A. 1991;88:11460. doi: 10.1073/pnas.88.24.11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nagayasu A, Uchiyama K, Kiwada H. Adv. Drug Delivery Rev. 1999;40:75. doi: 10.1016/s0169-409x(99)00041-1. [DOI] [PubMed] [Google Scholar]
- [56].Arnold M, Cavalcanti-Adam EA, Glass R, Blummel J, Eck W, Kantlehner M, Kessler H, Spatz JP. ChemPhysChem. 2004;5:383. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- [57].Shuhendler AJ, Prasad P, Leung M, Rauth AM, Dacosta RS, Wu XY. Adv. Healthcare Mater. 2012;1:600. doi: 10.1002/adhm.201200006. [DOI] [PubMed] [Google Scholar]
- [58].Rangger C, Helbok A, von Guggenberg E, Sosabowski J, Radolf T, Prassl R, Andreae F, Thurner GC, Haubner R, Decristoforo C. Int. J. Nanomed. 2012;7:5889. doi: 10.2147/IJN.S36847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Matsumura Y, Maeda H. Cancer Res. 1986;46:6387. [PubMed] [Google Scholar]
- [60].Prabhakar U, Maeda H, Jain RK, Muraca Sevick-E. M., Zamboni W, Farokhzad OC, Barry ST, Gabizon A, Grodzinski P, Blakey DC. Cancer Res. 2013;73:2412. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Cabral H, Matsumoto Y, Mizuno K, Chen Q, Murakami M, Kimura M, Terada Y, Kano MR, Miyazono K, Uesaka M, Nishiyama N, Kataoka K. Nat. Nanotechnol. 2011;6:815. doi: 10.1038/nnano.2011.166. [DOI] [PubMed] [Google Scholar]
- [62].Bruckman MA, Randolph LN, Vanmeter A, Hern S, Shoffstall AJ, Taurog RE, Steinmetz NF. Virology. 2014;449:163. doi: 10.1016/j.virol.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kostarelos K. Nat. Mater. 2010;9:793. doi: 10.1038/nmat2871. [DOI] [PubMed] [Google Scholar]
- [64].Thurber GM, Schmidt MM, Wittrup KD. Adv Drug Delivery Rev. 2008;60:1421. doi: 10.1016/j.addr.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WC. Nano Lett. 2009;9:1909. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- [66].Smith BR, Ghosn EE, Rallapalli H, Prescher JA, Larson T, Herzenberg LA, Gambhir SS. Nat. Nanotechnol. 2014;9:481. doi: 10.1038/nnano.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gao H, Shi W, Freund LB. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9469. doi: 10.1073/pnas.0503879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Jiang W, Kim BYS, Rutka JT, Chan WCW. Nat. Nanotechnol. 2008;3:145. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- [69].Hillaireau H, Couvreur P. Cell Mol. Life Sci. 2009;66:2873. doi: 10.1007/s00018-009-0053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Herd H, Daum N, Jones AT, Huwer H, Ghandehari H, Lehr CM. ACS Nano. 2013;7:1961. doi: 10.1021/nn304439f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.