12 crystallographic structures of T. thermophilus HB27 multicopper oxidase in holo, apo and Hg-bound forms and with different absorbed X-ray doses are reported, revealing the first structural evidence for the proton-relay mechanism in the X-ray-induced reduction of O2 to 2H2O at the trinuclear copper cluster of the enzyme. Different O2-reduction states and a total depletion of T2Cu at radiation doses higher than 0.2 MGy were also observed.
Keywords: copper depletion, dioxygen reduction, laccase, multicopper oxidase, proton-relay mechanism, radiation damage, X-ray-induced reduction
Abstract
During X-ray data collection from a multicopper oxidase (MCO) crystal, electrons and protons are mainly released into the system by the radiolysis of water molecules, leading to the X-ray-induced reduction of O2 to 2H2O at the trinuclear copper cluster (TNC) of the enzyme. In this work, 12 crystallographic structures of Thermus thermophilus HB27 multicopper oxidase (Tth-MCO) in holo, apo and Hg-bound forms and with different X-ray absorbed doses have been determined. In holo Tth-MCO structures with four Cu atoms, the proton-donor residue Glu451 involved in O2 reduction was found in a double conformation: Glu451a (∼7 Å from the TNC) and Glu451b (∼4.5 Å from the TNC). A positive peak of electron density above 3.5σ in an F o − F c map for Glu451a O∊2 indicates the presence of a carboxyl functional group at the side chain, while its significant absence in Glu451b strongly suggests a carboxylate functional group. In contrast, for apo Tth-MCO and in Hg-bound structures neither the positive peak nor double conformations were observed. Together, these observations provide the first structural evidence for a proton-relay mechanism in the MCO family and also support previous studies indicating that Asp106 does not provide protons for this mechanism. In addition, eight composite structures (Tth-MCO-C1–8) with different X-ray-absorbed doses allowed the observation of different O2-reduction states, and a total depletion of T2Cu at doses higher than 0.2 MGy showed the high susceptibility of this Cu atom to radiation damage, highlighting the importance of taking radiation effects into account in biochemical interpretations of an MCO structure.
1. Introduction
Multicopper oxidases (MCOs) are a diverse family of metalloenzymes that are widely distributed in all kingdoms, with bacterial and fungal laccases being the most numerous members (Hoegger et al., 2006 ▸; Sharma et al., 2007 ▸). MCOs couple four one-electron oxidations of several substrates to the four-electron reduction of O2 to 2H2O using at least four Cu atoms distributed in two active sites: one type 1 copper (T1Cu) site, where an organic and/or inorganic substrate is oxidized, and a trinuclear copper cluster (TNC), where O2 is bound, activated and reduced to 2H2O (Solomon et al., 2001 ▸). The TNC consists of one type 2 copper (T2Cu) site and a type 3 binuclear T3Cu–T3′Cu cluster (Fig. 1 ▸; Solomon et al., 1996 ▸). T1Cu has a typical electron paramagnetic resonance (EPR) signal and also forms a covalent S—Cu bond with an intense SCys→CuII charge-transfer band at around 610 nm, which provides the distinctive blue colour of MCOs. T2Cu also has a distinctive EPR signal but it does not have any noticeable features in the UV–visible region, while the binuclear T3Cu–T3′Cu cluster does not have an EPR signal but has an absorption band at around 330 nm (Solomon et al., 2008 ▸). The binuclear T3Cu–T3′Cu cluster is coordinated by three His residues on each of the two Cu atoms, T2Cu is coordinated by two His residues and T1Cu is coordinated by two His residues and one Cys residue (Sharma et al., 2007 ▸; Zhukhlistova et al., 2008 ▸).
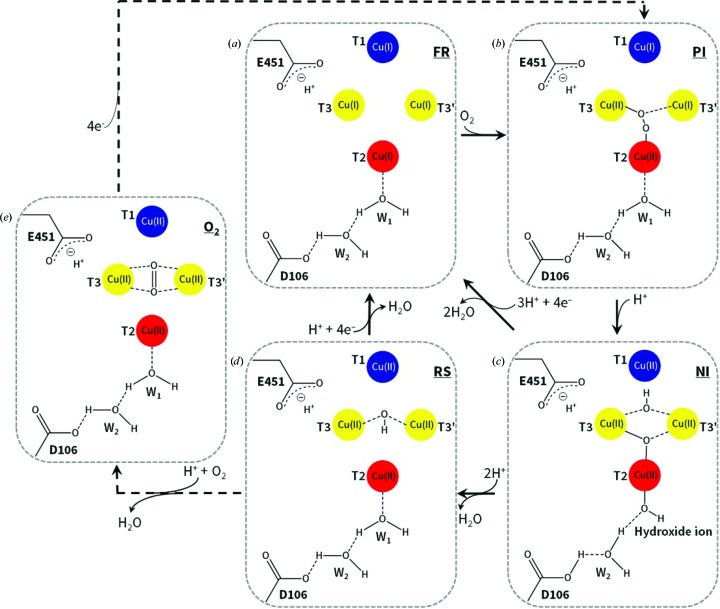
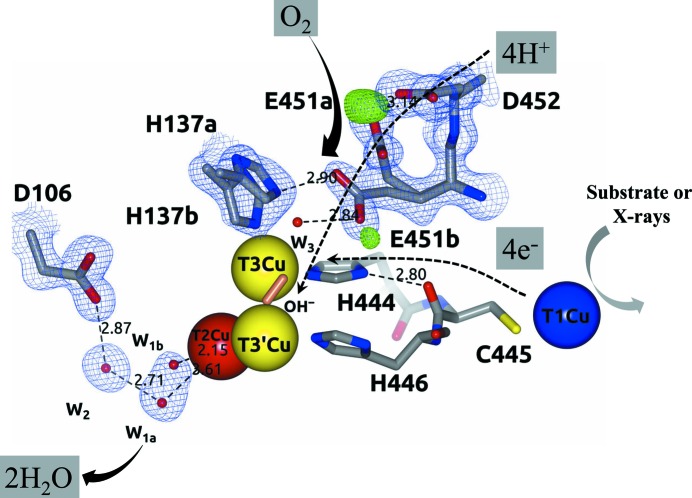
Figure 1.
Schematic representation of the catalytic mechanism of O2 reduction to 2H2O by MCOs. (a) FR state. (b) PI. (c) NI. (d) RS. (e) O2 state. Note that the Glu451/Asp106 residues belong to Tth-MCO. W1 and W2 are water molecules. The same colour code for the four Cu atoms is used in all other figures. Adapted from Bento et al. (2005 ▸), Quintanar et al. (2005 ▸) and Augustine et al. (2010 ▸).
Four protons and four electrons generated by T1Cu from four substrate molecules are required for the reduction of O2 at the TNC (Solomon et al., 2008 ▸). Particularly, after substrate oxidation occurs at the T1Cu site, the electrons are shuttled over a distance of ∼13 Å through a T1CuII–Cys–His–T3CuII electron-transfer pathway to the TNC (Augustine et al., 2010 ▸). However, O2 reduction at the TNC could also be observed in the crystalline state without substrate oxidation, since electrons and protons are released into the crystal by the radiolysis of water molecules during X-ray data collection (Hakulinen et al., 2006 ▸; Macedo et al., 2009 ▸; Garman, 2010 ▸). Thus, the final crystallographic structure of an MCO is an average model of different Cu oxidation states and X-ray-induced O2-reduction states and intermediates (Hakulinen et al., 2006 ▸; Ferraroni et al., 2012 ▸). Accordingly, the structure of Melanocarpus albomyces laccase at high absorbed X-ray dose was found to be in the resting state (RS; Fig. 1 ▸ d) with the binuclear T3Cu–T3′Cu cluster coupled through a bridging hydroxide (OH−) ligand, while at low absorbed X-ray dose the structure was found with an almost symmetrically coordinated O2 amidst the binuclear T3Cu–T3′Cu cluster (Fig. 1 ▸ e; Hakulinen et al., 2006 ▸). Since an MCO crystal is usually exposed from low to high absorbed radiation dose during a classic X-ray diffraction data acquisition, the latter observations revealed the reduction of O2 in the crystalline state. Remarkably, in order to systematically study such catalytic reactions of metalloenzymes in the course of X-ray measurements, a multicrystal data-collection strategy based on a systematic diffusion of absorbed dose over a set of crystals has been designed (Berglund et al., 2002 ▸). Using this technique of data combination from different crystals and the generation of composite structures with different absorbed X-ray doses, the catalytic mechanism of X-ray-induced reduction of O2 in horseradish peroxidase has been described (Berglund et al., 2002 ▸).
The reaction mechanism for the reduction of O2 to 2H2O at the TNC has been extensively studied and involves two two-electron transfers (Fig. 1 ▸) starting from the fully Cu reduced (FR) enzyme with four catalytic Cu ions in the cuprous state (Fig. 1 ▸ a; Augustine et al., 2010 ▸). The first two-electron reduction from O2 to the peroxide intermediate (PI) is rate-limiting (Figs. 1 ▸ a and 1 ▸ b), while the conversion of the PI to the native intermediate (NI) is extremely fast in the holoenzyme (Figs. 1 ▸ b and 1 ▸ c; Lee, George et al., 2002 ▸; Palmer et al., 2002 ▸; Yoon et al., 2007 ▸; Yoon & Solomon, 2007 ▸). It has previously been demonstrated that protons are not involved in all steps of the reduction of O2 at the TNC (Augustine et al., 2007 ▸). For instance, no protons are involved in the reductive cleavage of the molecular oxygen O=O bond from O2 to give the PI (Figs. 1 ▸ a and 1 ▸ b; Augustine et al., 2007 ▸). However, there is one proton involved in the conversion of the PI to the NI, and therefore in the reductive cleavage of the peroxide −O—O− bond (Figs. 1 ▸ b and 1 ▸ c; Palmer et al., 2002 ▸). Structurally, one O atom of the PI is coordinated to T3Cu and another O atom is coordinated to T2Cu, while in the NI one O atom is in the middle of the TNC and another O atom (OH−) is symmetrically located between the binuclear T3Cu–T3′Cu cluster (Augustine et al., 2010 ▸). Thus, after the two two-electron transfers have taken place, the NI displays four catalytic Cu ions in the cupric state and forms the RS by releasing one H2O molecule (Fig. 1 ▸ d; Yoon et al., 2007 ▸). As mentioned above, the RS presents an OH− ligand bridging the binuclear T3Cu–T3′Cu cluster as well as four catalytic Cu ions in the cupric state (Quintanar et al., 2005 ▸). However, the decay of the NI to the RS is slower than the turnover rate of the enzyme and therefore is not catalytically relevant (Yoon et al., 2007 ▸). In fact, under catalytic conditions the cycle is completed upon reduction of the NI by a total of four electrons and the release of 2H2O molecules by the addition of three protons, regenerating the FR enzyme for the next enzyme cycle without decaying to the RS. Nevertheless, because of the lack of sufficient electrons to form the FR enzyme, the NI necessarily decays to the RS (Augustine et al., 2010 ▸). Indeed, the RS is rather common in a large number of crystallographic structures of MCOs deposited in the PDB (~15 MCO structures), since the catalytic cycle for O2 reduction in the crystalline state using the protons and electrons released by X-rays is rather inefficient (Hakulinen et al., 2006 ▸; Kjaergaard et al., 2012 ▸). Thus, since the FR enzyme is the first state of the catalytic cycle for O2 reduction of MCOs (Augustine et al., 2010 ▸), and reacts immediately with O2 to form the PI, the structural stabilization of the O2 state (Fig. 1 ▸e) observed in the structure of M. albomyces laccase at low absorbed X-ray dose (Hakulinen et al., 2006 ▸) seems to be a process that is exclusive to the crystalline state as a result of the deficiency of electrons to generate the FR enzyme at low doses.
MCOs have a pair of highly conserved acidic residues in the second coordination sphere of the TNC which are involved in the proton-relay mechanism for O2 reduction mentioned above (Quintanar et al., 2005 ▸; Augustine et al., 2007 ▸; Kataoka et al., 2009 ▸; Bento et al., 2010 ▸; Chen, Durão et al., 2010 ▸); specifically, a Glu residue next to T3Cu or in the middle of the TNC in the entry channel for O2 and an Asp residue close to T2Cu in the exit channel for the water molecules produced. The pair of Glu/Asp residues in CotA from Bacillus subtilis (Bento et al., 2010 ▸; Chen, Durão et al., 2010 ▸), CueO from Escherichia coli (Kataoka et al., 2009 ▸) and Fet3p from Saccharomyces cerevisiae (Quintanar et al., 2005 ▸; Augustine et al., 2007 ▸) are Glu498/Asp116, Glu506/Asp112 and Glu487/Asp94, respectively. As expected, Thermus thermophilus HB27 multicopper oxidase (Tth-MCO; Miyazaki, 2005 ▸; Serrano-Posada et al., 2011 ▸) also possesses these two conserved acidic residues: Glu451/Asp106 (Fig. 1 ▸). It has been strongly suggested that Glu498 in CotA, Glu506 in CueO and Glu487 in Fet3p is the only proton-donor residue that plays a crucial role in the protonation events at the TNC, channelling the four protons required for O2 reduction. In contrast, Asp116 in CotA, Asp112 in CueO and Asp94 in Fet3p is not a proton-donor residue but is important in the decay of the PI to the NI by the stabilization of both an H2O molecule (W1; Fig. 1 ▸) and an OH− ion (Fig. 1 ▸ c), which are coordinated to T2Cu in the PI and the NI, respectively (Quintanar et al., 2005 ▸; Augustine et al., 2007 ▸; Kataoka et al., 2009 ▸; Bento et al., 2010 ▸; Chen, Durão et al., 2010 ▸; Silva, Damas et al., 2012 ▸). In fact, both H2O/OH− species interact in each intermediate with the side chain of the Asp residue through a structural water molecule (W2; Fig. 1 ▸). It has been shown that the absence of these two Glu/Asp residues, as in several mutants of CotA (Chen, Durão et al., 2010 ▸; Silva, Damas et al., 2012 ▸), CueO (Kataoka et al., 2009 ▸) and Fet3p (Quintanar et al., 2005 ▸; Augustine et al., 2007 ▸), results in a severe catalytic impairment of O2 reduction, reinforcing their importance in the proton-relay mechanism. Despite extensive study of the reduction of O2 at the TNC in MCOs, the structural details of the proton-relay mechanism involved in this catalytic reaction have not currently been described.
Tth-MCO is a thermophilic metalloenzyme which exhibits oxidase activity towards laccase substrates such as guaiacol and ABTS, with an optimal temperature of 365 K for ABTS oxidation and a half-time of thermal inactivation of over 14 h at 353 K (Miyazaki, 2005 ▸). In this work, 12 different crystallographic structures of Tth-MCO have been determined in order to study the X-ray-induced catalytic active-site reduction of the enzyme and the role of the Glu451/Asp106 residues in the proton-relay mechanism. Firstly, the Tth-MCO structure at 1.5 Å resolution and three inactive forms of the enzyme at 1.7 Å resolution, apo Tth-MCO, Hg-Tth-MCO and Hg-Tth-MCO-2h, were determined using classic data acquisition from a single crystal. Secondly, eight composite structures (Tth-MCO-C1–8) at 1.8 Å resolution with different absorbed X-ray doses were determined using a multicrystal data-collection strategy. In the latter case, different states of the X-ray-induced reduction of O2 were trapped. The noticeably different behaviour of the Glu451/Asp106 residues in active and inactive forms of Tth-MCO provided structural details of the proton-relay mechanism and indicated that they act together, providing part of the necessary driving force to reduce O2. Moreover, the four catalytic Cu atoms of Tth-MCO revealed uneven radiation damage by absorbed X-rays, where T2Cu is completely depleted at doses of >0.2 MGy.
2. Experimental procedures
All chemical supplies were analytical grade and were purchased from Sigma–Aldrich unless stated otherwise.
2.1. Gene cloning, protein expression, purification and crystallization
Gene cloning, protein expression, protein purification and crystallization of Tth-MCO and apo Tth-MCO from T. thermophilus HB27 were performed as described previously (Serrano-Posada et al., 2011 ▸). The purified recombinant Tth-MCO was the mature form of the enzyme, a 439-residue protein without the first 22 residues (Met1–Ala22), which are part of the signal peptide (Miyazaki, 2005 ▸), and exhibited the typical blue colour of MCOs. Additionally, two inactive Hg-Tth-MCO and Hg-Tth-MCO-2h forms of Tth-MCO with Hg instead of Cu were obtained at 278 K by soaking two apo Tth-MCO crystals with 5 mM mercury(II) chloride for 5 min and 2 h, respectively. For the multicrystal data-collection strategy, eight different Tth-MCO crystals of approximate dimensions ∼0.025 × 0.05 × 0.1 mm were grown at 278 K by the hanging-drop vapour-diffusion method using the microseeding technique (Serrano-Posada et al., 2011 ▸).
2.2. Copper content
The copper content of Tth-MCO and apo Tth-MCO was determined spectrophotometrically by the 2,2′-biquinoline method as described elsewhere (Felsenfeld, 1960 ▸). A solution of 2,2′-biquinoline at 0.5 mg ml−1 in glacial acetic acid was prepared and standardized against five submicromolar solutions of copper(II) sulfate incubated with an excess of ascorbic acid (8 mM). Protein solutions (100 µl at 25 µM) were incubated for 20 min at 383 K for protein denaturation and mixed with 8 mM ascorbic acid for 10 min to reduce native enzyme cupric ions to copper(I). The latter solution was then mixed with 100 µl of the 2,2′-biquinoline solution and the Cu content was determined at λ = 546 nm. Experiments were performed in triplicate and solutions were kept anaerobic by bubbling gaseous N2 through them. All UV–visible spectra were recorded using an Evolution 1000 spectrophotometer (Thermo Scientific) using quartz cells with 1 cm path length.
2.3. X-ray data collection
Data collection was performed on beamline X6A of the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory (BNL), New York, USA using an ADSC Quantum 270 detector. Diffraction data collection for Tth-MCO (12.650 keV, λ = 0.9795 Å and 9.003 keV, λ = 1.3767 Å), apo Tth-MCO (9.019 keV, λ = 1.3747 Å) and Hg-Tth-MCO (14.900 keV, λ = 0.8321 Å) has been described previously (Serrano-Posada et al., 2011 ▸). A data set (12.650 keV, λ = 0.9795 Å) was collected to a resolution of 1.7 Å for an Hg-Tth-MCO-2h crystal; its unit-cell parameters were a = 93.3 Å, b = 110.1 Å, c = 96.2 Å, α = β = γ = 90°. X-ray absorption spectra in fluorescence mode were collected on beamline X6A using a Si(111) channel-cut monochromator (band pass 1.9 × 10-1 eV) and a custom-made fluorescence detector vertically positioned at an angle of 90° relative to the beam axis. Fluorescence spectra were subjected to background subtraction and normalized using the Athena software package (Ravel & Newville, 2005 ▸).
For the multicrystal data-collection strategy, data were collected from eight different Tth-MCO crystals obtained using the microseeding technique under the same conditions: 12.650 keV (λ = 0.9795 Å), crystal-to-detector distance 200 mm, Δφ = 1.0°, exposure time per image 30 s, slit size 100 × 100 µm. Complete diffraction patterns of 96 images each were collected from eight Tth-MCO crystals at different known starting φ angles.
2.4. Data processing and model refinement
Diffraction images of Tth-MCO, apo Tth-MCO, Hg-Tth-MCO and Hg-Tth-MCO-2h were integrated using XDS (Kabsch, 2010 ▸) and scaling was performed with SCALA from the CCP4 suite (Winn et al., 2011 ▸). For the multicrystal data-collection strategy, each of eight individual data sets was divided into sequential blocks of 12 images. These blocks from eight different crystals but having the absorbed dose in common were separately integrated using XDS (Kabsch, 2010 ▸) and then combined using SORTMTZ from the CCP4 suite (Winn et al., 2011 ▸) in order to obtain eight full composite data sets (Tth-MCO-C1–8) corresponding to increasing absorbed doses. Scaling was then performed as described above.
The Tth-MCO structure at 1.5 Å resolution was determined by a combination of the molecular-replacement (MR) and single-wavelength anomalous dispersion (SAD) techniques (Serrano-Posada et al., 2011 ▸) using Phaser in MR-SAD mode (McCoy et al., 2007 ▸). Initial model building was performed automatically using ARP/wARP (Langer et al., 2008 ▸). The structures of the inactive forms of Tth-MCO at 1.7 Å resolution, apo Tth-MCO, Hg-Tth-MCO and Hg-Tth-MCO-2h, as well as the active composite structures Tth-MCO-C1–8 at 1.8 Å resolution, were determined by MR using Phaser (McCoy et al., 2007 ▸). The atomic coordinates of Tth-MCO (PDB entry 2xu9), from which all of the Cu ions, MPD molecules and water molecules had been removed, were used as a structural model for MR trials. All crystallographic structures determined in this work belonged to the C-centred orthorhombic space group C2221 as suggested by POINTLESS (Evans, 2006 ▸). Matthews coefficient calculations suggested that there was one molecule per asymmetric unit in all structures (∼50% solvent content in all structures).
In all cases, refinement was performed using PHENIX (Adams et al., 2010 ▸) until the R work and R free values were lower than 0.19 with satisfactory r.m.s. deviations from ideal bond lengths and bond angles. Refinement was alternated with manual building/refinement in Coot (Emsley et al., 2010 ▸). 5% of the data were randomly chosen and reserved to determine R free. Water molecules were first automatically located using PHENIX and then validated in Coot. Meanwhile, several MPD molecules and metal ions were positioned manually in Coot and then refined using PHENIX. The occupancies of the metal atoms in all structures were adjusted so that their isotropic thermal vibration parameters were refined approximately to the values of the neighbouring atoms in the structure. For the Tth-MCO-C1 composite structure with the lowest absorbed X-ray dose (0.2 MGy), a symmetrical and elongated electron density was found between the binuclear T3Cu–T3′Cu cluster and was assumed to be O2. Refinement then proceeded constraining the O=O distance to the target value of 1.21 Å. TLS refinement for the His95 residue of the composite structures Tth-MCO-C2–8 was performed using PHENIX. Changes in the electron density for the Tth-MCO-C1–8 structures, structural damage at each Cu atom and the decrease of the occupancy values induced by X-ray radiation were observed and analyzed as a function of the absorbed dose.
Model validation was performed using MolProbity (Chen, Arendall et al., 2010 ▸). The graphical representations were made using CCP4mg v.2.7.3 (McNicholas et al., 2011 ▸) and PyMOL (DeLano, 2002 ▸). Data-collection and refinement statistics for the Tth-MCO, apo Tth-MCO, Hg-Tth-MCO and Hg-Tth-MCO-2h structures determined using classic single-crystal data collection are shown in Table 1 ▸. Data-collection and refinement statistics for the composite structures Tth-MCO-C1–8 determined using multicrystal data collection are shown in Table 2 ▸.
Table 1. X-ray data-collection and refinement statistics.
Values in parentheses are for the last resolution shell.
| Tth-MCO | Apo Tth-MCO | Hg-Tth-MCO | Hg-Tth-MCO-2h | |
|---|---|---|---|---|
| Data-collection statistics | ||||
| Space group | C2221 | C2221 | C2221 | C2221 |
| Unit-cell parameters | ||||
| a (Å) | 93.6 | 93.0 | 93.5 | 93.3 |
| b (Å) | 110.3 | 110.1 | 110.2 | 110.1 |
| c (Å) | 96.3 | 96.3 | 96.3 | 96.2 |
| α = β = γ (°) | 90.0 | 90.0 | 90.0 | 90.0 |
| Resolution range (Å) | 23.0–1.50 (1.60–1.50) | 26.0–1.70 (1.80–1.70) | 20.0–1.70 (1.80–1.70) | 28.0–1.70 (1.80–1.70) |
| No. of reflections | 899414 | 435926 | 542926 | 245055 |
| No. of unique reflections | 78896 (10654) | 53818 (7154) | 54841 (8585) | 53273 (7694) |
| Completeness (%) | 94.7 (83.9) | 98.9 (82.1) | 94.6 (91.4) | 97.9 (97.5) |
| R merge (%) | 8.6 (39.5) | 9.0 (45.0) | 13.3 (35.3) | 5.5 (27.9) |
| CC1/2 (%) | 96.7 (96.3) | 96.5 (96.0) | 96.3 (95.8) | 96.2 (95.8) |
| 〈I/σ(I)〉 | 19.4 (4.1) | 18.5 (3.8) | 9.1 (3.7) | 18.4 (5.1) |
| Multiplicity | 11.4 (9.9) | 8.1 (6.6) | 9.9 (10.3) | 4.6 (4.7) |
| Monomers per asymmetric unit | 1 | 1 | 1 | 1 |
| Refinement statistics | ||||
| Resolution range (Å) | 23.0–1.50 | 26.0–1.70 | 20.0–1.70 | 28.0–1.70 |
| R work/R free (%) | 15.5/17.8 | 15.1/18.5 | 15.3/17.8 | 15.2/17.0 |
| No. of atoms | ||||
| Protein | 4259 | 4045 | 3837 | 3799 |
| Ion/ligand | 134 | 128 | 141 | 133 |
| Water | 549 | 525 | 455 | 504 |
| Mean B values (Å2) | ||||
| Protein | 13.7 | 16.5 | 17.2 | 15.8 |
| Ion/ligand | 33.1 | 37.8 | 38.3 | 34.8 |
| Water | 31.5 | 33.2 | 31.5 | 31.7 |
| All atoms | 16.2 | 18.9 | 19.0 | 18.2 |
| Wilson plot | 12.5 | 16.3 | 15.8 | 14.1 |
| R.m.s.d. from ideal stereochemistry | ||||
| Bond lengths (Å) | 0.01 | 0.02 | 0.02 | 0.01 |
| Bond angles (°) | 1.51 | 1.99 | 1.98 | 2.32 |
| Coordinate error (maximum-likelihood-based) | 0.19 | 0.18 | 0.17 | 0.17 |
| Ramachandran plot (%) | ||||
| Most favoured regions | 97.3 | 97.5 | 97.7 | 97.8 |
| Additional allowed regions | 2.4 | 2.2 | 1.9 | 2.2 |
| Disallowed regions | 0.3 | 0.3 | 0.4 | 0.0 |
| PDB code | 2xu9 | 2xuw | 2xvb | 4ai7 |
Table 2. X-ray data-collection and refinement statistics.
Values in parentheses are for the last resolution shell.
| Tth-MCO-C1 | Tth-MCO-C2 | Tth-MCO-C3 | Tth-MCO-C4 | Tth-MCO-C5 | Tth-MCO-C6 | Tth-MCO-C7 | Tth-MCO-C8 | |
|---|---|---|---|---|---|---|---|---|
| Data-collection statistics | ||||||||
| Space group | C2221 | C2221 | C2221 | C2221 | C2221 | C2221 | C2221 | C2221 |
| Unit-cell parameters | ||||||||
| a (Å) | 93.6 | 93.7 | 93.6 | 93.5 | 93.5 | 93.5 | 93.7 | 93.7 |
| b (Å) | 110.3 | 110.3 | 110.3 | 110.2 | 110.2 | 110.2 | 110.4 | 110.4 |
| c (Å) | 96.3 | 96.3 | 96.3 | 96.3 | 96.3 | 96.2 | 96.4 | 96.5 |
| α = β = γ (°) | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 |
| Resolution range (Å) | 30.0–1.80 (1.90–1.80) | 29.0–1.80 (1.90–1.80) | 29.0–1.80 (1.90–1.80) | 28.0–1.80 (1.90–1.80) | 30.0–1.80 (1.90–1.80) | 29.0–1.80 (1.90–1.80) | 29.0–1.80 (1.90–1.80) | 29.0–1.80 (1.90–1.80) |
| No. of reflections | 174732 | 176253 | 151457 | 175957 | 153500 | 173277 | 175181 | 172277 |
| No. of unique reflections | 44803 (6341) | 45193 (6600) | 44546 (6378) | 45117 (6463) | 43857 (6385) | 44430 (6520) | 44918 (6345) | 45336 (6589) |
| Completeness (%) | 96.5 (95.2) | 97.7 (98.7) | 96.9 (95.5) | 97.6 (96.6) | 95.4 (95.5) | 96.7 (97.8) | 97.2 (95.8) | 97.9 (98.4) |
| R merge (%) | 13.0 (28.0) | 27.6 (40.1) | 12.4 (29.6) | 12.6 (30.0) | 13.6 (32.6) | 13.4 (32.9) | 13.1 (32.4) | 13.5 (31.2) |
| CC1/2 (%) | 95.8 (95.4) | 95.1 (94.9) | 95.5 (95.1) | 95.8 (95.4) | 95.5 (95.0) | 95.7 (95.2) | 96.0 (95.4) | 95.4 (94.9) |
| 〈I/σ(I)〉 | 7.6 (4.2) | 6.8 (3.5) | 7.8 (4.2) | 8.1 (4.2) | 7.4 (4.0) | 8.0 (4.1) | 8.1 (4.1) | 7.9 (4.0) |
| Multiplicity | 3.9 (4.0) | 3.9 (3.8) | 3.4 (3.4) | 3.9 (3.9) | 3.5 (3.5) | 3.9 (3.9) | 3.9 (4.0) | 3.8 (3.8) |
| Monomers per asymmetric unit | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Refinement statistics | ||||||||
| Resolution range (Å) | 30.0–1.80 | 29.0–1.80 | 29.0–1.80 | 28.0–1.80 | 30.0–1.80 | 29.0–1.80 | 29.0–1.80 | 29.0–1.80 |
| R work/R free (%) | 14.7/17.4 | 15.4/18.9 | 14.8/17.9 | 14.7/17.2 | 14.8/18.2 | 14.9/17.9 | 14.6/16.8 | 15.0/18.3 |
| No. of atoms | ||||||||
| Protein | 3798 | 3786 | 3770 | 3763 | 3746 | 3790 | 3786 | 3762 |
| Ion/ligand | 134 | 125 | 133 | 133 | 133 | 131 | 133 | 117 |
| Water | 500 | 516 | 509 | 498 | 543 | 512 | 508 | 501 |
| Mean B values (Å2) | ||||||||
| Protein | 12.1 | 12.0 | 12.0 | 11.9 | 11.9 | 12.5 | 13.0 | 12.4 |
| Ion/ligand | 31.8 | 30.6 | 31.3 | 31.3 | 31.3 | 32.0 | 32.5 | 31.1 |
| Water | 28.3 | 28.6 | 28.8 | 28.2 | 29.7 | 30.0 | 31.0 | 29.4 |
| All atoms | 14.6 | 14.5 | 14.6 | 14.3 | 14.7 | 15.1 | 15.7 | 14.8 |
| Wilson plot | 11.2 | 11.3 | 11.2 | 11.2 | 11.1 | 12.2 | 12.9 | 11.9 |
| R.m.s.d. from ideal stereochemistry | ||||||||
| Bond lengths (Å) | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| Bond angles (°) | 1.57 | 1.43 | 1.39 | 1.40 | 1.42 | 1.42 | 1.41 | 1.43 |
| Coordinate error (maximum-likelihood-based) | 0.17 | 0.18 | 0.15 | 0.15 | 0.17 | 0.18 | 0.17 | 0.19 |
| Ramachandran plot (%) | ||||||||
| Most favoured regions | 97.6 | 96.9 | 97.8 | 97.8 | 97.8 | 97.5 | 97.6 | 97.4 |
| Additional allowed regions | 2.2 | 3.1 | 2.2 | 2.2 | 1.8 | 2.1 | 2.0 | 2.2 |
| Disallowed regions | 0.2 | 0.0 | 0.0 | 0.0 | 0.4 | 0.4 | 0.4 | 0.4 |
| PDB code | 2yae | 2yaf | 2yah | 2yam | 2yao | 2yap | 2yaq | 2yar |
2.5. Dose calculation
Calculations of the absorbed dose, in megagrays (MGy), were performed using RADDOSE v.2 (Murray et al., 2004 ▸; Paithankar et al., 2009 ▸). The values of the beam parameters (including the energy, beam profile, size/area and flux) and the crystal properties (unit cell, space group, number of molecules per asymmetric unit, composition, size and thickness) were used in order to calculate the absorbed X-ray dose. An estimate of the X6A beam flux in photons s−1 was obtained using a silicon pin diode to calibrate the beam intensity as described previously (Owen et al., 2009 ▸). The sizes of the eight Tth-MCO crystals for the composite structures Tth-MCO-C1–8 were chosen in order to match the beam size, ensuring a uniform dose for all crystal regions.
3. Results and discussion
3.1. Crystallographic structures
The classic single-crystal data-acquisition and multicrystal data-collection techniques were employed to determine 12 crystallographic structures of Tth-MCO in holo, apo and Hg-bound states and with different absorbed X-ray doses. The latter structures were determined with increasing doses from Tth-MCO-C1 (0.2 MGy, 12.5% dose) to Tth-MCO-C8 (1.6 MGy, 100% dose). The overall architecture of inactive forms of the enzyme (apo Tth-MCO, Hg-Tth-MCO and Hg-Tth-MCO-2h) is identical to that of Tth-MCO; a calculated r.m.s. deviation of 0.16 Å was obtained after superimposing 439 Cα atoms. The most interesting changes were observed in metal-coordinating residues and Glu451/Asp106 at the TNC, which are involved in the first and second coordination spheres, respectively. The markedly different behaviour of Glu451/Asp106 in the active and inactive forms of Tth-MCO provided structural details of the proton-relay mechanism of O2 reduction. In addition, a methionine-rich β-hairpin motif, which seems to be involved in an extra metal-coordinating site, was also observed in all structures. Different states (the O2 state, RS and FR state) of the X-ray-induced reduction of O2 were trapped in the Tth-MCO-C1, Tth-MCO-C2 and Tth-MCO-C6 structures. The four catalytic Cu atoms exhibited a different radiation-damage behaviour from absorbed X-rays. Accordingly, while T2Cu and T3Cu showed a high susceptibility to radiation damage, both T1Cu and T3′Cu seem to be quite stable to X-rays.
3.1.1. Tth-MCO structure
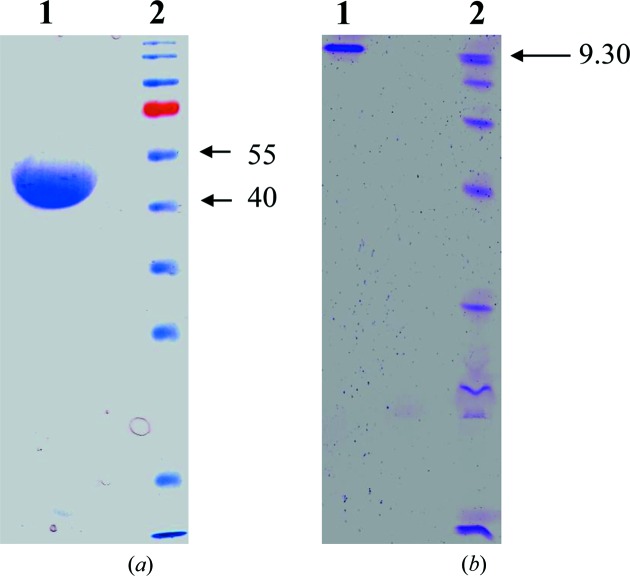
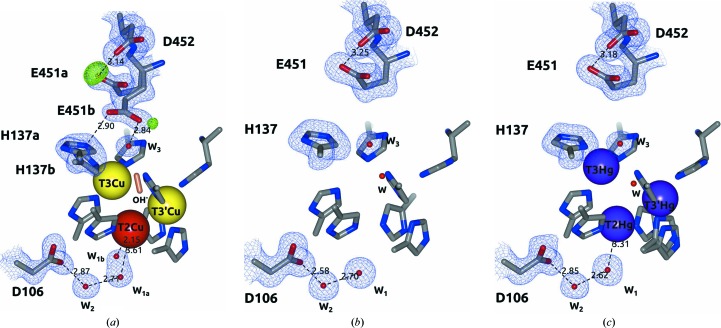
Tth-MCO is a monomeric enzyme (Miyazaki, 2005 ▸) which showed a molecular mass of ∼50 kDa on SDS–PAGE and an isoelectric point (pI) of ∼9.3 on an isoelectric focusing (IEF) gel (Fig. 2 ▸). The overall Tth-MCO fold comprises three sequential cupredoxin globular domains: domain 1, Gly24–Ala169, domain 2, Glu170–Val343; domain 3, Val344–Gly462 (Fig. 3 ▸ a). This fold was first observed in the cupredoxin family, which includes the single-domain blue copper proteins plastocyanin and azurin (Murphy et al., 1997 ▸), and was subsequently detected in two-domain (Komori et al., 2009 ▸) and three-domain (Zhukhlistova et al., 2008 ▸) MCOs. The cupredoxin fold basically consists of an eight-stranded Greek-key β-barrel comprising two β-sheets composed of four strands each arranged in a sandwich conformation (Murphy et al., 1997 ▸). The Tth-MCO fold contains 35 β-strands and five α-helices distributed in the three domains. The four catalytic Cu ions are distributed among two of the three domains in two different active sites (Fig. 3 ▸ a). Thus, while domain 3 contains the T1Cu-coordinating residues His393, His450 and Cys445, the interface between domains 1 and 3 contains TNC-coordinating residues (Figs. 3 ▸ a and 4 ▸ a). T2Cu is coordinated by His95 and His396. The binuclear T3Cu–T3′Cu cluster is coordinated by His137, His398 and His444 on T3Cu and His97, His135 and His446 on T3′Cu (Fig. 4 ▸ a). In contrast, domain 2 is not involved in Cu coordination, but structurally connects domains 1 and 3. A 16-residue methionine-rich β-hairpin motif located over the T1Cu site was found in domain 2 (Fig. 3 ▸). As expected, the Tth-MCO structure was found in the RS with an OH− (occupancy of 0.57) asymmetrically bridging the binuclear T3Cu–T3′Cu cluster (Fig. 4 ▸ a). The TNC geometry corresponds to a scalene triangle in which T3Cu and T3′Cu are 5.01 Å apart, while the T3Cu–T2Cu and T3′Cu–T2Cu distances are 3.65 and 3.95 Å, respectively. This TNC geometry has been previously described for the RS in other MCO structures in which the four Cu atoms are in the oxidized state (Ferraroni et al., 2007 ▸; De la Mora et al., 2012 ▸). Furthermore, Met455 in a double conformation was found flanking T1Cu (Fig. 4 ▸ a). This axial methionine residue has been proposed to be involved in tuning the reduction potential (E°) of T1Cu (Quintanar et al., 2007 ▸) and is frequently found in enzymes with low E° (Zhukhlistova et al., 2008 ▸).
Figure 2.
(a) Coomassie Blue-stained SDS–PAGE (12%) of purified recombinant Tth-MCO. Lane 1, purified Tth-MCO. Lane 2, molecular-weight markers (Fermentas; labelled in kDa). (b) IEF gel. Lane 1, purified Tth-MCO. Lane 2, isoelectric point markers (Pharmacia; pI value labelled).
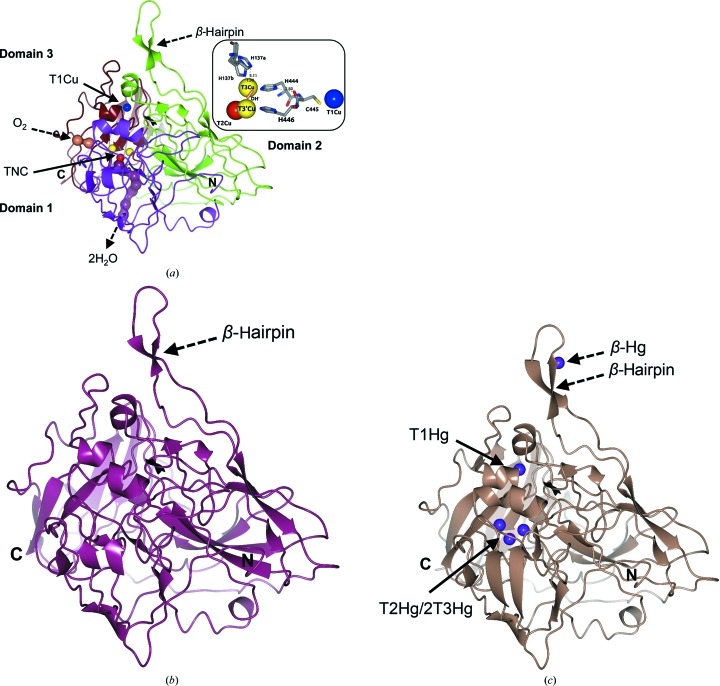
Figure 3.
Crystallographic structures of Tth-MCO in holo, apo and Hg-bound states. (a) High-resolution structure of Tth-MCO showing the cupredoxin-domain organization (domain 1, magenta; domain 2, green; domain 3, brown) with T1Cu (blue sphere) located in domain 3 and the TNC (T2Cu, red sphere; binuclear T3Cu–T3′Cu cluster, yellow spheres) placed at the interface between domains 1 and 3. The molecular oxygen-entrance channel (coral) towards T3Cu and the water-molecule exit channel (dark purple) from T2Cu are represented by water molecules found in the Tth-MCO structure. Right inset, close-up of four catalytic Cu atoms of Tth-MCO, the hydroxide ligand (coral cylinder) bridging the binuclear T3Cu–T3′Cu cluster and the electron-transfer pathway from T1Cu to the TNC. (b, c) Two inactive forms of Tth-MCO: (b) apo Tth-MCO (dark purple) and (c) Hg-Tth-MCO-2h (pale brown) with Hg bound (purple spheres) instead of Cu. It is notable that a fifth Hg-binding site (β-Hg) was observed in (c). Distances are in Å.
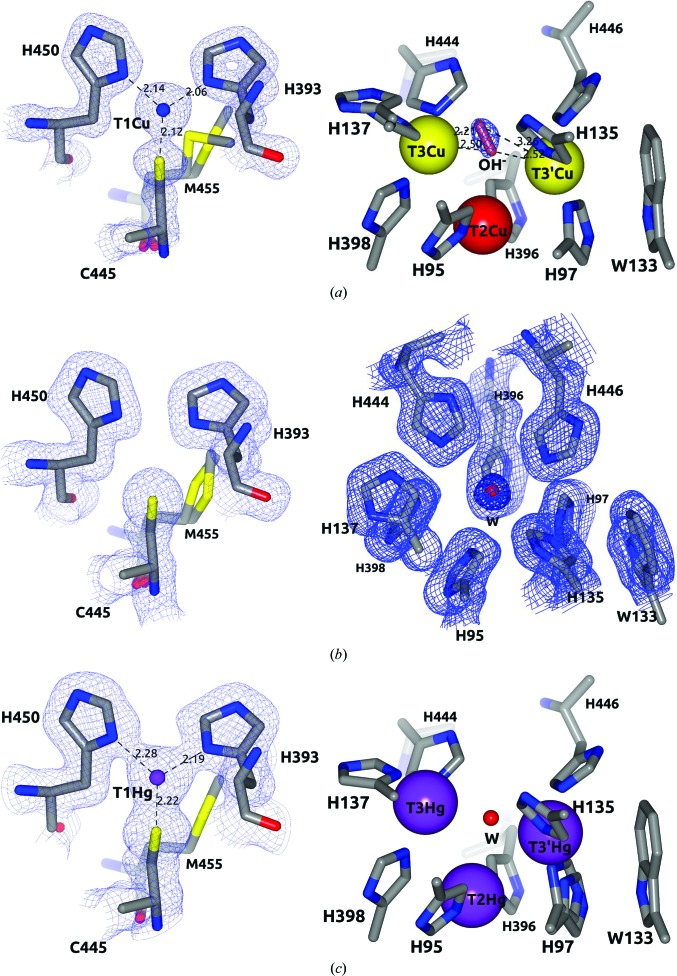
Figure 4.
Geometry of the T1 site and TNC. (a) Tth-MCO. Note the hydroxide ligand (coral cylinder) in the RS asymmetrically bridging the binuclear T3Cu–T3′Cu cluster and Met455 flanking T1Cu. (b) Apo Tth-MCO structure. Note the absence of electron density for T1Cu and the TNC (superior view). A water molecule (red sphere) in the same position as OH− observed in (a) was found in this inactive apo form. (c) Hg-Tth-MCO-2h. Note that the view of TNC is the same that in (a) and a water molecule (red sphere) bridging the T3Cu–T3′Cu cluster was also found in this inactive form with Hg bound. The 2F o − F c electron-density map contoured at the 1σ level is drawn in blue. Metal ions are represented as spheres. Distances are in Å.
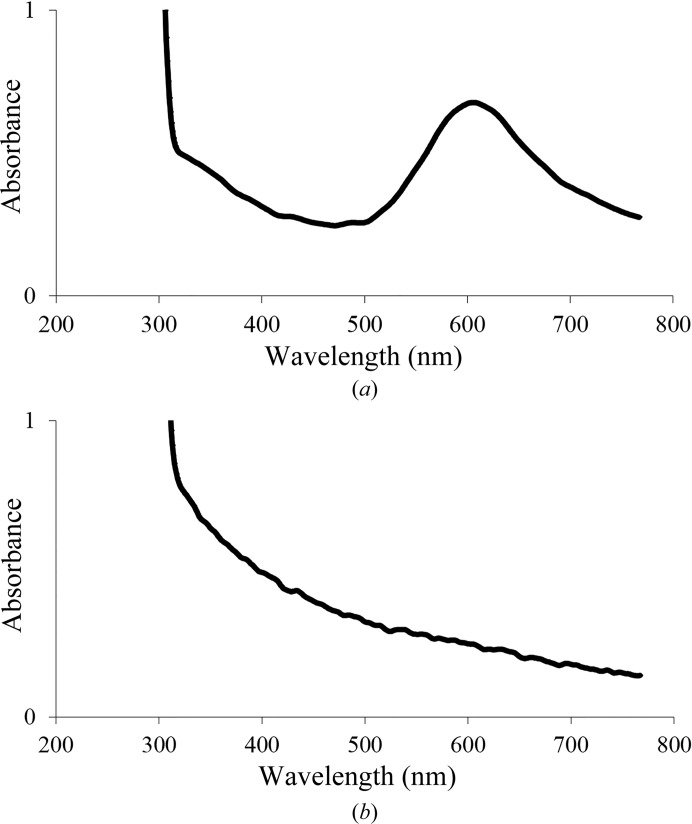
Interestingly, T1Cu and T3′Cu proved to be highly stable to radiation damage, with occupancies of 0.87 and 0.90, respectively. In contrast, T2Cu and T3Cu showed occupancies of 0.13 and 0.43, respectively. The low occupancies of T2Cu and T3Cu in the crystalline state indicate the high susceptibility of both Cu atoms to radiation damage, since in solution the 2,2′-biquinoline method resulted in a ratio of 4.1 ± 0.1 Cu atoms per Tth-MCO molecule. Moreover, a solution of Tth-MCO before crystallization trials also showed typical absorption peaks for both the C445S→T1CuII charge-transfer band at ∼610 nm and the shoulder of the T3CuII←OH−→T3′CuII charge-transfer band at ∼330 nm in UV–visible spectra (Fig. 5 ▸ a). This information suggests that in the crystalline state there are probably several factors that increase the susceptibility of T2Cu and T3Cu to radiation damage. For instance, partial occupancy of T3Cu could be explained by the significant increase from 1.90 to 3.11 Å of the His137 N∊–T3Cu coordination-bond distance (see the inset in Fig. 3 ▸ a). Additionally, as Augustine et al. (2010 ▸) have previously described for Fet3p, the electron-transfer pathway from T1Cu to TNC is conserved in Tth-MCO via T1CuII–Cys445–His444–T3CuII and involves a hydrogen bond of 2.80 Å between the carbonyl group of Cys445 and His444 Nδ (see inset in Fig. 3 ▸ a). Furthermore, unlike T3′Cu, which is flanked by Trp133 in a cation–π interaction (Fig. 4 ▸ a), T3Cu is located in the entry channel for the reduction of O2 (Fig. 3 ▸ a). Therefore, since T3Cu is located in both the entry channel of O2 and at the end of the electron-transfer pathway for the reduction of O2, this Cu atom is probably more affected than T3′Cu by electrons released by X-rays during data collection. On the other hand, although the location of T2Cu in the exit channel for the water molecules produced is structurally important for the catalytic activity of the enzyme (Fig. 3 ▸ a), the partial occupancy of T2Cu suggests that at this position specially designed for the removal of water molecules in MCOs during the reduction of O2, T2Cu could be continually exposed to hydrated electrons released by X-rays.
Figure 5.
Spectra of purified Tth-MCO in 20 mM Tris–HCl pH 8.0. (a) Holo form (450 µM). (b) Apo form (500 µM). Note that although the concentration in (b) is higher than that in (a), the typical absorption peaks at both ∼330 and ∼610 nm were not observed.
3.1.2. Apo Tth-MCO, Hg-Tth-MCO and Hg-Tth-MCO-2h
To assess the significantly different behaviour between the active and inactive forms of Tth-MCO, three inactive structures of the apo and Hg-bound forms were determined. As expected, no electron density was observed for the four catalytic Cu atoms in apo Tth-MCO (Figs. 3 ▸ b and 4 ▸ b), indicating that the overexpression and purification processes did not incorporate Cu atoms into this inactive form of Tth-MCO. In solution, the 2,2′-biquinoline method also resulted in a ratio of 0.4 ± 0.1 Cu atoms per apo Tth-MCO molecule and a UV–visible spectrum revealed the absence of the typical absorption peaks at both ∼610 and ∼330 nm (Fig. 5 ▸ b).
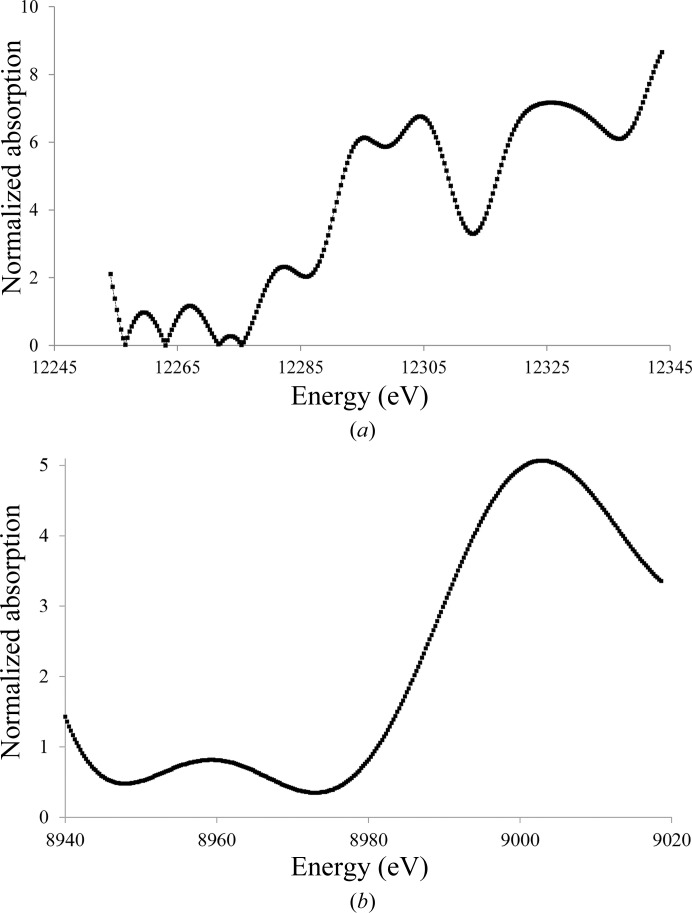
Two inactive derivatives, Hg-Tth-MCO and Hg-Tth-MCO-2h, were prepared with the spectroscopically silent and redox-inactive Hg (Palmer et al., 2001 ▸) instead of Cu. Remarkably, the coordination of Hg atoms with partial occupancies in both the inactive Hg-Tth-MCO and Hg-Tth-MCO-2h (Fig. 3 ▸ c) structures, obtained with different soaking times from two different apo Tth-MCO crystals, occurred in the four vacant Cu sites. Together with this observation, a fifth Hg-binding (β-Hg) site was also found in the methionine-rich β-hairpin motif (Fig. 3 ▸ c). Independently, X-ray fluorescence (XRF) spectra were collected from an Hg-Tth-MCO-2h crystal before data collection. The L III absorption edges of Hg-Tth-MCO-2h were observed at 12.295, 12.304 and 12.325 keV in the fluorescence spectrum (Fig. 6 ▸ a), correlating with the L III absorption edges experimentally determined by Ji et al. (2001 ▸). In addition, a single characteristic peak of copper(II) was always observed at 8.999 keV in the XRF spectrum from Tth-MCO crystals before data collection (Fig. 6 ▸ b). A water molecule bridging the binuclear T3Cu–T3′Cu site was also found in all inactive structures (Figs. 4 ▸ b and 4 ▸ c), indicating the structural relevance of the O2-reduction site at the TNC, in which different states and intermediates of the catalytic cycle are located.
Figure 6.
X-ray absorption spectra of Tth-MCO crystals in the holo form and with Hg bound before X-ray data collection. (a) Hg-Tth-MCO-2h single crystal showing the L III absorption edges of protein-bound Hg. (b) Tth-MCO single crystal showing the Cu K absorption edge of the four native copper(II) atoms of the enzyme.
3.1.3. Composite structures Tth-MCO-C1–8 obtained with different absorbed X-ray doses
To identify the states and/or the intermediates in the X-ray-induced O2 reduction, eight composite structures corresponding to increasing absorbed dose levels from Tth-MCO-C1 (0.2 MGy, 12.5% dose) to Tth-MCO-C8 (1.6 MGy, 100% dose) were determined using the multicrystal data-collection technique.
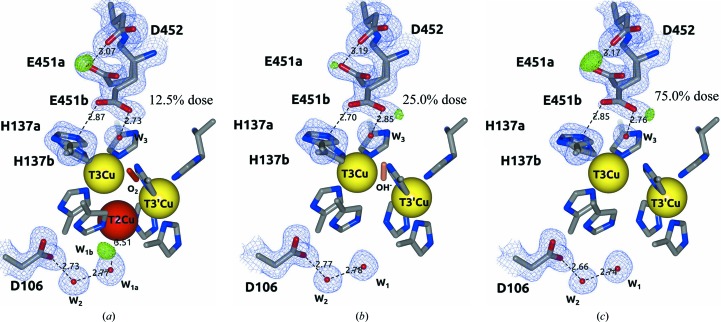
Interestingly, the Tth-MCO-C1 structure at a low absorbed dose was found with an O2 molecule (occupancy of 0.4) almost symmetrically coordinated amongst the binuclear T3Cu–T3′Cu cluster (Fig. 7 ▸ a). This state is structurally similar to the previously described O2 state for low absorbed dose laccase from M. albomyces (PDB entry 2ih8; Hakulinen et al., 2006 ▸) and CotA (PDB entry 1w6l; Bento et al., 2005 ▸). The finding of the O2 state in Tth-MCO-C1 suggests that the electron population released in the crystal by the X-ray radiolysis of water molecules is relatively deficient at doses of ≤0.2 MGy. In agreement with this observation, the stabilization of O2 also suggests that four Cu atoms are oxidized (Fig. 1 ▸ e) and that the FR state has not yet been formed.
Figure 7.
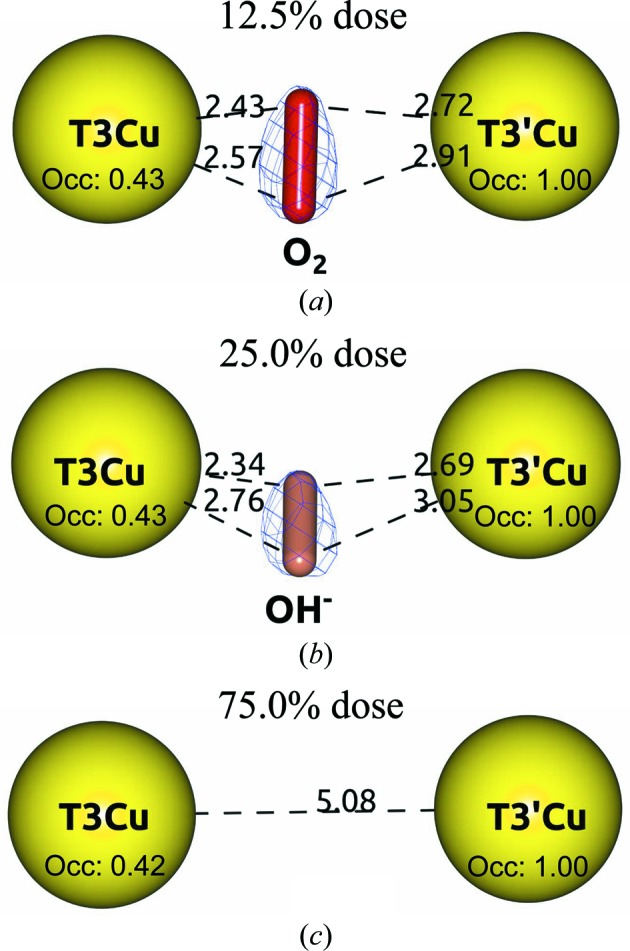
Different states of O2 reduction found at the TNC of the Tth-MCO composite structures. (a) Tth-MCO-C1 in the O2 state. (b) Tth-MCO-C2 in the RS. (c) Tth-MCO-C6 in the FR state. Note that the O2 (red cylinder) is bridging the binuclear T3Cu–T3′Cu cluster at a dose of 0.2 MGy (12.5% dose) in (a), as well as the different occupancy values related to the radiation damage of both Cu atoms. The 2F o − F c electron-density map contoured at the 1σ level is drawn in blue. Metal ions are represented as spheres. Distances are in Å.
Notably, the Tth-MCO-C2 structure at 25.0% dose was found to have an OH− ion (occupancy of 0.7) coordinated between the binuclear T3Cu–T3′Cu cluster (Fig. 7 ▸ b), indicating that the population of protons and electrons in the crystal significantly increased from Tth-MCO-C1 to generate the RS in Tth-MCO-C2. This is particularly interesting with regard to the experiments on Fet3p (Augustine et al., 2010 ▸), since in the path from the Tth-MCO-C1 to the Tth-MCO-C2 structures the system progresses through the following states and intermediates: O2 state (Tth-MCO-C1)→PI→NI→RS (Tth-MCO-C2; Fig. 1 ▸). However, whilst it is well known that the FR state is the first state of the catalytic cycle for the reduction of O2 by MCOs (Augustine et al., 2010 ▸) and that it reacts immediately with O2 to form the PI, the structural stabilization of the O2 state in the Tth-MCO-C1 structure seems to be a process that is exclusive to the crystalline state as the result of a deficiency of electrons to generate the FR state at doses of ≤0.2 MGy. As also mentioned above, in the absence of sufficient electrons to form the FR state in solution, MCOs are usually observed in the RS (Augustine et al., 2010 ▸). Consequently, the formation of the O2 state in Tth-MCO-C1 remains unclear, and in fact even the nature of the RS of MCOs is a question that still remains to be answered (Bento et al., 2010 ▸). Unsurprisingly, the PI and NI cannot be trapped under the conditions of these crystallographic experiments on Tth-MCO owing to their rate of decay and the intrinsic nature of the intermediates.
The RS was always observed in the Tth-MCO-C3 (37.5% dose) to Tth-MCO-C5 (62.5% dose) structures, indicating that the FR state is not stable enough to be trapped, regardless of the increasing levels of absorbed dose. However, when the dose reached 75.0% in the Tth-MCO-C6 structure, no state or intermediate was found at the TNC; a distance of 5.08 Å between the binuclear T3Cu–T3′Cu cluster was also observed (Fig. 7 ▸ c). This observation indicates that the FR state seems to be trapped at 75.0% dose, because in accordance with the mechanism of O2 reduction at the TNC (Fig. 1 ▸), the RS (Tth-MCO-C5) should generate the FR state (Tth-MCO-C6) for the next enzyme cycle (Quintanar et al., 2005 ▸). Naturally, the structural stabilization of the O2 state observed in Tth-MCO-C1 cannot be obtained again considering that the population of protons and electrons is significant at this dose. In addition, the FR state is the only state in the reduction of the O2 in MCOs that presents a binuclear T3Cu–T3′Cu cluster distance of >5 Å (Augustine et al., 2010 ▸). Remarkably, the FR state in Tth-MCO-C6 is also structurally similar to the previously reported FR state for CotA (PDB entry 2bhf; Bento et al., 2005 ▸), which displayed a binuclear T3Cu–T3′Cu cluster distance of 5.10 Å. Finally, according to the mechanism of O2 reduction at the TNC (Fig. 1 ▸), the FR state should generate the RS if the system has progressed as follows: FR (Tth-MCO-C6)→PI→NI→RS (Tth-MCO-C7). In fact, the last two composite structures Tth-MCO-C7 (87.5% dose) and Tth-MCO-C8 (1.6 MGy, 100% dose) were found in the RS.
Accordingly, the composite structures Tth-MCO-C1–8 support previous observations indicating that the final crystallographic structure of an MCO obtained using classic single-crystal data acquisition such as Tth-MCO (Fig. 3 ▸ a) is an average model of different Cu oxidation states and X-ray-induced O2-reduction states and intermediates. As also mentioned above, Tth-MCO was found in the RS (Figs. 3 ▸ a and 4 ▸ a); this is the statistically relevant state in a full data set of this enzyme, as the composite structures Tth-MCO-C1–8 have shown.
3.2. Methionine-rich β-hairpin motif
The MCOs found in many bacterial copper-detoxification systems have both methionine-rich structural motifs and copper(I) oxidase activity (Cha & Cooksey, 1991 ▸; Lee, Grass et al., 2002 ▸; Fernandes et al., 2007 ▸). Interestingly, a 16-residue methionine-rich β-hairpin motif (Ala292–Gln307; Fig. 3 ▸) over the T1Cu site was observed in all of the crystallographic structures determined in this work. This structural motif contains six methionines, Met293, Met295, Met296, Met298, Met301 and Met305, of the 13 in total in the primary sequence of the enzyme. In addition, the β-Hg (Figs. 3 ▸ c and 8 ▸ a) observed in both proteins with bound Hg is coordinated by four residues of the β-hairpin motif: Met296, Met301, Met305 and His303. However, low occupancies of 0.10 (B value of 83.4 Å2) in Hg-Tth-MCO and 0.20 (B value of 58.5 Å2) in Hg-Tth-MCO-2h were found for this extra Hg atom. This observation corresponds to the structural dynamics of the local environment of the β-Hg, which displayed average B values of 77.2 and 68.2 Å2 for Hg-Tth-MCO and Hg-Tth-MCO-2h, respectively. No electron density was observed at the β-Hg site for Tth-MCO and apo Tth-MCO.
Figure 8.
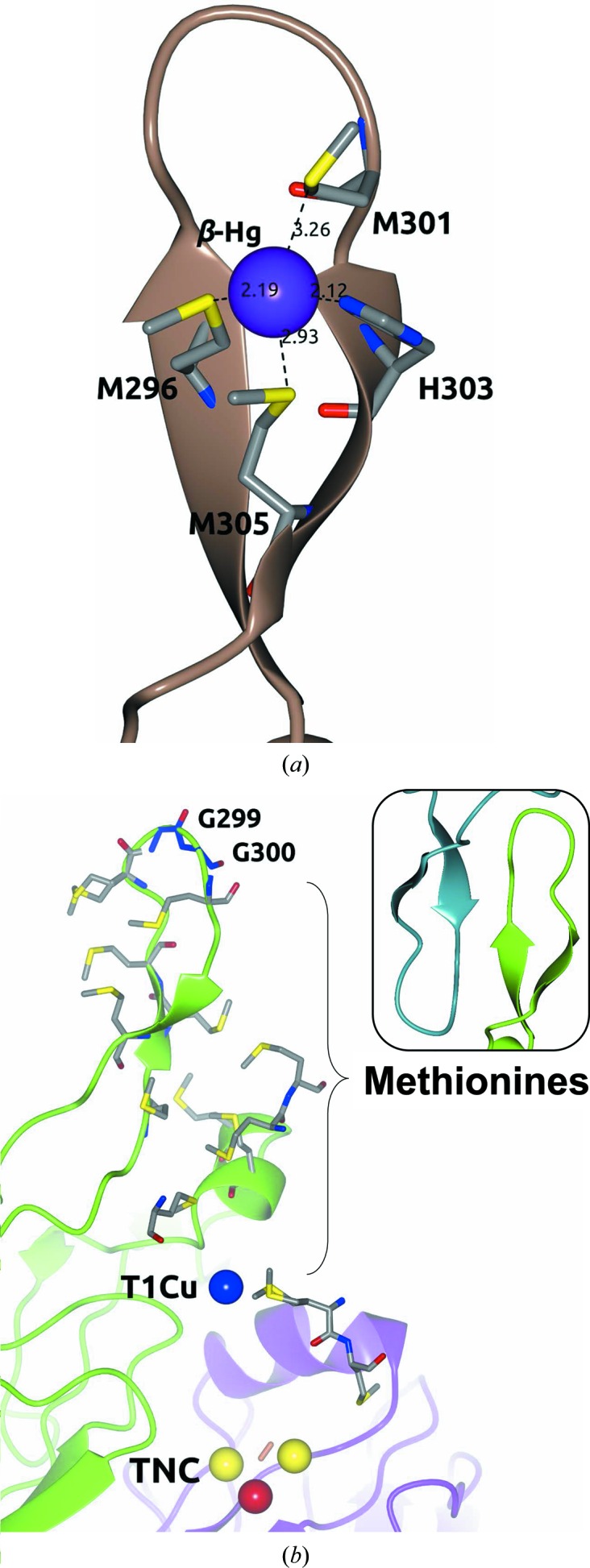
Methionine-rich β-hairpin motif over the T1Cu site. (a) β-Hg coordinated in the β-hairpin motif of Hg-Tth-MCO-2h. (b) Methionine distribution between the top (Gly299-Gly300; blue cylinders) of the β-hairpin motif and T1Cu of Tth-MCO. Right inset: the β-hairpin motif (green) of Tth-MCO stabilized by the same motif (dark cyan) of the crystallographic neighbour. Metal ions are represented as spheres. Distances are in Å.
Similar structural motifs over the T1Cu site have been described previously in other MCOs such as McoP from Pyrobaculum aerophilum (PDB entry 3aw5; Sakuraba et al., 2011 ▸), McoC from Campylobacter jejuni (PDB entry 3zx1; Silva, Durão et al., 2012 ▸) and CueO (PDB entry 1kv7; Roberts et al., 2002 ▸), but the methionine-rich β-hairpin motif is unique to Tth-MCO (Fig. 3 ▸). Molecular-dynamics simulations have shown that this Tth-MCO motif experiences conformational changes which enable exposure of the T1Cu site for substrate oxidation (Bello et al., 2012 ▸), regardless of its stabilization by the same motif in a crystallographic neighbour (see the inset in Fig. 8 ▸ b).
It has also previously been shown that the methionine-rich CueO motif is involved in several copper(I)-coordinating sites apart from the four catalytic Cu atoms, which increase the metal oxidase activity to oxidize copper(I) to the less toxic copper(II) (Roberts et al., 2003 ▸; Singh et al., 2011 ▸). Indeed, the removal of the extra copper(I)-binding sites through the mutation of copper(I)-coordinating residues leads to a catalytic impairment of copper(I) oxidation (Singh et al., 2011 ▸). Moreover, deletion of the methionine-rich motif decreased the copper(I) oxidase activity and increased the organic substrate oxidase activity through increased access to T1Cu (Kataoka et al., 2007 ▸). Nevertheless, the mechanism of copper(I) oxidation is still unknown, but it has been suggested that it involves an extended electron-transfer pathway from the extra copper(I)-binding sites to T1Cu and then to the TNC (Singh et al., 2011 ▸).
Consequently, it appears to be very probable that the methionine-rich β-hairpin motif of Tth-MCO is involved in extra copper(I)-binding sites. Nevertheless, electron transfer from the extra copper(I)-binding sites to T1Cu of Tth-MCO seems to be difficult, since there is a long distance of ∼23 Å from a hypothetical copper(I) placed in the same position as β-Hg to T1Cu. Notably, Tth-MCO possesses 13 methionines in the primary sequence, which are all located between the top (Gly299-Gly300) of the β-hairpin motif and T1Cu (Fig. 8 ▸ b), suggesting that several positions for extra copper(I)-binding sites, differing from the β-Hg site, may be present in the enzyme. As mentioned above, Tth-MCO exhibits laccase activity towards organic substrates such as guaiacol and ABTS (Miyazaki, 2005 ▸). However, this information suggests that Tth-MCO also displays important structural features over the T1Cu site that could be related to its oxidase activity towards inorganic substrates, although there is no experimental evidence to support this view.
3.3. Proton-relay mechanism for the X-ray-induced reduction of O2 to 2H2O
To assess the role in the crystalline state of the highly conserved pair of acidic residues Glu451/Asp106 involved in the proton-relay mechanism for the X-ray-induced reduction of O2, and in particular their behaviour in active and inactive forms of Tth-MCO, several crystallographic structures of the enzyme in holo, apo and Hg-bound forms and with different X-ray absorbed doses were analyzed.
3.3.1. Tth-MCO, apo Tth-MCO and Hg-Tth-MCO-2h
In active holo Tth-MCO (Fig. 3 ▸ a), which contains the four catalytic Cu atoms, a double conformation Glu451a (∼7 Å to the TNC; occupancy of 0.56) and Glu451b (∼4.5 Å to the TNC; occupancy of 0.44) of the proton-donor residue Glu451 located in the entry channel of O2 was observed (Fig. 9 ▸ a). A positive peak of electron density above 3.5σ in an F o − F c map for Glu451a O∊2 strongly suggests a carboxyl functional group at the side chain, while its significant absence at the side chain of Glu451b suggests a carboxylate functional group. This observation indicates that in a proton-relay mechanism the Glu451a conformation seems to accept the protons for O2 reduction, while the Glu451b conformation releases the protons to the TNC in a manner that appears to take place as follows: Glu451a (RCOOH)→Glu451b (RCOO− + H+)→TNC (Fig. 9 ▸ a). In addition, it has previously been shown that the most likely proton-relay path from Glu451b to the TNC is through a structural water molecule (W3; Fig. 9 ▸ a; Bento et al., 2005 ▸, 2010 ▸; Augustine et al., 2007 ▸; Kataoka et al., 2009 ▸). According to the latter, structural data for Tth-MCO also suggest that apart from W3, which is at a distance of 2.84 Å from Glu451b O∊2, a second path with a distance of 2.90 Å between Glu451b O∊1 and His137a N∊ is also plausible (Fig. 9 ▸ a).
Figure 9.
Proton-relay mechanism for the reduction of O2 to 2H2O at the TNC of active and inactive forms of Tth-MCO. Note the presence of a double conformation of Glu541 in the active form of Tth-MCO (a) and its absence in the inactive forms apo Tth-MCO (b) and Hg-Tth-MCO-2h (c). The 2F o − F c (blue) and F o − F c (green) electron-density maps are contoured at the 1σ and 3.5σ levels, respectively. Metal ions and water molecules are represented as spheres. Distances are in Å.
A theoretical calculation has shown that Glu451 presents an unusually high pK a of 9.9 (Bello et al., 2012 ▸), which is probably the result of interactions with other carboxylic groups at the TNC (Harris & Turner, 2002 ▸; Castañeda et al., 2009 ▸). Accordingly, the perturbed pK a value of Glu451 seems to be governed by its interaction with the Asp452 residue (pK a = 4.40, as calculated using the PROPKA server; Olsson et al., 2011 ▸), which is at a hydrogen-bonding distance of 3.14 Å from Glu451a O∊2 (Fig. 9 ▸ a). Remarkably, the Asp452 residue is also conserved in other extensively studied bacterial MCOs such as CueO (Asp507; PDB entry 1kv7; Roberts et al., 2002 ▸) and CotA (Asp499; PDB entry 1gsk; Enguita et al., 2003 ▸), but in the latter cases the double conformation of the proton-donor residue was not observed. On the other hand, the structural factors that govern the unusual pK a value of the proton-donor residues of MCOs from fungi (Taylor et al., 2005 ▸; De la Mora et al., 2012 ▸) are not obvious from inspection of their crystal structures. For instance, the proton-donor residue Glu487 of Fet3p is in the middle of the TNC, and although there are several acidic residues around this residue none of them seems to explain the unusual pK a value of Glu487 (PDB entry 1zpu; Taylor et al., 2005 ▸). However, when a double conformation of Glu487 is drawn in Coot (Emsley et al., 2010 ▸), interactions with several neighbouring carboxylic groups occur, but in particular with the Glu134 residue, which would be at a hydrogen-bonding distance from Glu487. As expected, a theoretical calculation using the PROPKA server also showed that Asp106 (pK a = 1.13) is not a proton-donor residue in the proton-relay mechanism of Tth-MCO (Fig. 9 ▸ a) but is in fact important for the decay of the PI to the NI (Figs. 1 ▸ b and 1 ▸ c; Quintanar et al., 2005 ▸).
As mentioned above, the decay rate of both the PI and the NI restricts their structural characterization as single species, making them difficult to trap in the crystalline state. Furthermore, apart from the rest states and intermediates in O2 reduction, which present a H2O molecule coordinated to T2Cu (Bento et al., 2005 ▸), the NI is the only species that displays an OH− coordinated to T2Cu (Fig. 1 ▸; Yoon et al., 2007 ▸). As also mentioned above, it has previously been shown that these H2O/OH− species coordinated to T2Cu are always stabilized by the side chain of a highly conserved acidic residue (Asp106 in Tth-MCO) through a structural H2O molecule (W2; Fig. 9 ▸ a; Quintanar et al., 2005 ▸). Interestingly, when an H2O molecule (W1a; Fig. 9 ▸ a) at a T2Cu–H2O coordination bond distance of 3.61 Å was modelled in Tth-MCO, a positive peak of electron density above 3.5σ in a F o − F c map was observed close to T2Cu. The latter case was explained when a second H2O molecule (W1b; Fig. 9 ▸ a) with a T2Cu–OH− coordination-bond distance of 2.15 Å was modelled in a positive peak of electron density. Since an MCO structure determined by classic single-crystal data collection is an average model of different O2-reduction states and intermediates, this information suggests that populations of H2O/OH− species coordinated to T2Cu, and stabilized by Asp106 through W2, are found in the Tth-MCO structure (Fig. 1 ▸). Nevertheless, we deduce that the RS is the statistically relevant state for the Tth-MCO structure (Figs. 3 ▸ a and 4 ▸ a), because apart from H2O/OH− species coordinated to T2Cu, an OH− ion bridging the binuclear T3Cu–T3′Cu cluster was also observed.
To demonstrate that the proton-relay mechanism is not preserved in the inactive structures of Tth-MCO, three forms of the enzyme in the apo form and with Hg bound were analyzed (Figs. 3 ▸ b and 3 ▸ c). In fact, only one conformation of the proton-donor residue Glu451 was observed at ∼7 Å to the TNC (Figs. 9 ▸ b and 9 ▸ c), in the equivalent position to Glu451a in active Tth-MCO. Notably, no electron density was found in an F o − F c map for Glu451, suggesting that Glu451 is deprotonated and the proton-relay mechanism is not undertaken in these inactive apo and Hg-bound forms. Moreover, a single H2O molecule (W1; Figs. 9 ▸ b and 9 ▸ c) at the same position as W1a was observed in Tth-MCO (Fig. 9 ▸ a), indicating that Asp106 does not play any role in −O—O− bond breakage in these inactive forms of Tth-MCO (Figs. 1 ▸ b and 1 ▸ c; Quintanar et al., 2005 ▸).
3.3.2. Composite structures Tth-MCO-C1–8 with different absorbed X-ray doses
The proton-relay mechanism for the reduction of O2 was also observed in the active composite structures Tth-MCO-C1–8 (Fig. 10 ▸). A positive peak of electron density in an F o − F c map at a dose of 0.2 MGy again suggests a carboxyl functional group for Glu451a O∊2 of Tth-MCO-C1 (Fig. 10 ▸ a). Furthermore, the Glu451b O∊2–H137a N∊ and Glu451b O∊1–W3 distances of 2.87 and 2.73 Å, respectively, do not show a preferred proton-relay path from Glu451b to TNC at this dose (Fig. 10 ▸ a).
Figure 10.
Proton-relay mechanism for the reduction of O2 to 2H2O at the TNC of active Tth-MCO composite structures. (a) Tth-MCO-C1 (0.2 MGy; 12.5%). (b) Tth-MCO-C2 (25.0%). (c) Tth-MCO-C6 (75.0%). The 2F o − F c (blue) and F o − F c (green) electron-density maps are contoured at the 1σ and 3.5σ levels, respectively. Metal ions and water molecules are represented as spheres. Distances are in Å.
As mentioned above, protons are not involved in formation of the PI (Augustine et al., 2007 ▸), and the population of electrons released by the radiolysis of water molecules in the crystals of Tth-MCO is relatively deficient at doses of ≤0.2 MGy. In consequence, the O2 state at the TNC of the Tth-MCO-C1 structure was stable enough to be trapped (Figs. 7 ▸ a and 10 ▸ a), despite a double conformation of Glu451 being found in the proton-relay mechanism. Remarkably, the Glu451b O∊2–His137a N∊ distance was reduced from 2.87 Å in Tth-MCO-C1 to 2.70 Å in Tth-MCO-C2, while the Glu451b O∊1–W3 distance increased from 2.73 Å in Tth-MCO-C1 to 2.85 Å in Tth-MCO-C2 (Figs. 10 ▸ a and 10 ▸ b). As also mentioned above, protons and electrons are involved from the O2 state (Tth-MCO-C1) to the RS (Tth-MCO-C2), so we deduce that the changes in the Glu451b O∊2–His137a N∊ and Glu451b O∊1–W3 distances are significantly more informative if we take into account that these two composite structures provide dynamic information for X-ray-induced reduction of O2 (Figs. 10 ▸ a and 10 ▸ b). Additionally, the proton-relay path from Glu451b to the TNC, in which the Glu451b O∊2–H137a N∊ distance is less than that between Glu451b O∊1 and W3, was also observed in five of the composite structures (Tth-MCO-C2–5 and Tth-MCO-C8). This information suggests that the preferred proton-relay path in Tth-MCO from Glu451b to TNC seems to be as follows: Glu451b→His137a→His137b→TNC. However, the latter remains unclear, since the changes in the distances are close to the coordinate error (Table 2 ▸) and should be investigated in the future.
On the other hand, the significant absence of a positive peak of electron density in an F o − F c map for Glu451a O∊2 in Tth-MCO-C2 (Fig. 10 ▸ b) also suggests that a significant part of the population of protons were probably transferred to the TNC to reduce O2 in Tth-MCO-C2 as follows: O2 state (Tth-MCO-C1)→PI→NI→RS (Tth-MCO-C2). It is worth noting that except for Tth-MCO-C2 (Fig. 10 ▸ b), a positive peak of electron density in an F o − F c map for Glu451a O∊2 was clearly observed in all composite structures. In fact, Tth-MCO-C2 was the only structure determined in this work that seems to exhibit both conformations of the Glu451 residue with a carboxylate functional group.
Interestingly, a positive peak of electron density in an F o − F c map corresponding to W1b was also found close to W1a in Tth-MCO-C1 (Fig. 10 ▸ a). In agreement with the information mentioned above for Tth-MCO (Fig. 9 ▸ a), W1a is simultaneously coordinated to T2Cu and stabilized by Asp106 through W2 in Tth-MCO-C1 (Fig. 10 ▸ a). Together, this information confirms the above results around the T2Cu site for Tth-MCO (Fig. 9 ▸ a), suggesting that populations of H2O/OH− species coordinated to T2Cu are also present at 0.2 MGy (Fig. 10 ▸ a) owing to the decay of the PI to the NI in every catalytic cycle during X-ray data collection. Naturally, only W1a was found in the Tth-MCO-C2–8 structures owing to the depletion of T2Cu at doses of >0.2 MGy (Fig. 10 ▸ b). Moreover, an anisotropic displacement of His95 in the Tth-MCO-C2–8 structures was also observed, with a concomitant decrease in the His95 N∊—His396 N∊ distance from 3.28 Å in Tth-MCO-C1 to 3.03 Å in Tth-MCO-C8.
Together, the above results revealed structural details of both the T1CuII–Cys445–His444–T3CuII electron-transfer pathway from T1Cu to the TNC and the proton-relay mechanism, which act together as a part of the driving force for X-ray-driven catalytic conversion of O2 to 2H2O (Fig. 11 ▸).
Figure 11.
Proton-relay mechanism and electron-transfer pathway from T1Cu to TNC for the X-ray-driven catalytic conversion of O2 to 2H2O, taking the active Tth-MCO structure as a reference. The 2F o − F c (blue) and F o − F c (green) electron-density maps are contoured at the 1σ and 3.5σ levels, respectively. Metal ions and water molecules are represented as spheres. Distances are in Å.
3.4. Effects of absorbed radiation dose on the Cu centres of the composite structures Tth-MCO-C1–8
Specific structural damage to the four catalytic Cu atoms and/or the decrease in the occupancy values induced by X-ray radiation were analyzed as a function of absorbed dose in the composite structures Tth-MCO-C1–8. Radiation damage to T1Cu and structural changes in its local environment in the eight composite structures were negligible, with a minimal change in occupancy for this Cu atom from 1.00 (Tth-MCO-C1) to 0.97 (Tth-MCO-C8). Similarly, the T3′Cu atom of the composite structures exhibited high stability to X-rays (Fig. 7 ▸), with a final occupancy of 0.94 when the system reached 100% absorbed dose.
In contrast, the T3Cu and T2Cu atoms of the composite structures again showed a high susceptibility to radiation damage, as we observed in the Tth-MCO structure. In effect, the occupancy of the T3Cu atom was only 0.43 at a dose of 0.2 MGy (Fig. 7 ▸ a). As mentioned above, this observation indicates that several factors converged once more to explain the high susceptibility of the T3Cu atom to radiation damage; for instance, the significant increase in the His137 coordination-bond distance which was observed in Tth-MCO (see inset in Fig. 3 ▸ a and Fig. 9 ▸ a) and all composite structures (Fig. 10 ▸), as well as its location in the entry channel for O2 and at the end of the electron-transfer pathway from T1Cu to TNC (Fig. 11 ▸).
On the other hand, the T2Cu atom showed an occupancy of only 0.16 for Tth-MCO-C1 (Fig. 10 ▸ a) and was totally depleted at a dose of >0.2 MGy (Fig. 10 ▸ b; Tth-MCO-C2). In agreement with this observation, partial occupancy and depletion of the T2Cu atom has previously been reported for the structure of laccase from Coriolopsis gallica (Cg L; De la Mora et al., 2012 ▸). In the latter case, a high susceptibility of T2Cu to radiation damage was shown and a complete depletion of T2Cu at a dose between 0.6 and 4.0 MGy was found. Interestingly, T2Cu of Tth-MCO displays a higher susceptibility to X-rays than T2Cu of Cg L, highlighting the importance of taking radiation effects into account in biochemical interpretations of an MCO structure.
4. Conclusions
We have determined 12 crystallographic structures of Tth-MCO in active and inactive forms and with different absorbed X-ray doses at 1.5–1.8 Å resolution in order to study the X-ray-induced reduction of O2 to 2H2O at the TNC of the enzyme and the proton-relay mechanism involved in this catalytic reaction. To the best of our knowledge, this is the first structural evidence in the MCO family that demonstrates the dynamic behaviour of the proton-donor residue Glu451 in the protonation events at the TNC. In the case of active Tth-MCO structures, the double conformation of Glu451 shows how protons are channelled to the TNC for the reduction of O2 and the importance of the Asp106 residue for the decay of the PI to the NI, but not for protonation events at the TNC. Remarkably, in apo and Hg-bound structures dynamic behaviour of the proton-donor Glu451 residue was not observed, demonstrating that the proton-relay mechanism is not undertaken in these inactive forms.
X-ray-induced reduction experiments of Tth-MCO crystals allowed the systematic study of O2 reduction at the TNC in the crystalline state. Accordingly, at a low absorbed radiation dose of 0.2 MGy the active Tth-MCO-C1 composite structure showed an O2 molecule coordinated at the TNC, demonstrating that in the absence of sufficient electrons in the crystalline state the RS is not the only state that is sufficiently stable to be trapped in Tth-MCO structures and possibly found in solution, but the statistically relevant state has since been observed in six composite structures (Tth-MCO-C2–5, Tth-MCO-C7 and Tth-MCO-C8) as well as in the Tth-MCO structure determined by classical single-crystal data collection. Additionally, when the dose was increased the FR state found in the Tth-MCO-C6 composite structure was also stable enough to be trapped. However, we were unable to isolate composite structures of the PI and the NI coordinated at the TNC owing to their decay rates. Together, these results also demonstrate the high susceptibility of T2Cu and T3Cu to radiation damage, with the former being totally depleted at radiation doses higher than 0.2 MGy.
Finally, a 16-residue methionine-rich β-hairpin motif over the T1Cu site, which appears to have interesting biological implications and is probably involved in extra copper(I)-binding sites, as previously described for CueO (Roberts et al., 2003 ▸; Kataoka et al., 2007 ▸; Singh et al., 2011 ▸), was observed in all structures determined in this work. Both a high concentration of methionines between the top of the β-hairpin motif and T1Cu and a fifth Hg-binding site found in the β-hairpin motif of Hg-bound structures demonstrate that extra metal-coordinating sites may be found in this methionine-rich region; however, their biological role has to be proposed in the future.
Supplementary Material
PDB reference: Tth-MCO, 2xu9
PDB reference: apo Tth-MCO, 2xuw
PDB reference: Hg-Tth-MCO, 2xvb
PDB reference: Hg-Tth-MCO-2h, 4ai7
PDB reference: Tth-MCO-C1, 2yae
PDB reference: Tth-MCO-C2, 2yaf
PDB reference: Tth-MCO-C3, 2yah
PDB reference: Tth-MCO-C4, 2yam
PDB reference: Tth-MCO-C5, 2yao
PDB reference: Tth-MCO-C6, 2yap
PDB reference: Tth-MCO-C7, 2yaq
PDB reference: Tth-MCO-C8, 2yar
Acknowledgments
HS-P was supported by a Postdoctoral Fellowship from CONACyT. ER-P acknowledges financial support from CONACyT project No. 102370 and PAPIIT IN209114. We are also grateful to the staff of beamline X6A at the BNL NSLS for data-collection facilities, in particular MSc Edwin Lazo and Dr Jean Jakoncic. The X6A beamline is funded by the National Institute of General Medical Sciences, National Institute of Health under agreement GM-0080.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Augustine, A. J., Kjaergaard, C., Qayyum, M., Ziegler, L., Kosman, D. J., Hodgson, K. O., Hedman, B. & Solomon, E. I. (2010). J. Am. Chem. Soc. 132, 6057–6067. [DOI] [PMC free article] [PubMed]
- Augustine, A. J., Quintanar, L., Stoj, C. S., Kosman, D. J. & Solomon, E. I. (2007). J. Am. Chem. Soc. 129, 13118–13126. [DOI] [PMC free article] [PubMed]
- Bello, M., Valderrama, B., Serrano-Posada, H. & Rudiño-Piñera, E. (2012). PLoS One, 7, e40700. [DOI] [PMC free article] [PubMed]
- Bento, I., Martins, L. O., Lopes, G. G., Carrondo, M. A. & Lindley, P. F. (2005). Dalton Trans., pp. 3507–3513. [DOI] [PubMed]
- Bento, I., Silva, C. S., Chen, Z., Martins, L. O., Lindley, P. F. & Soares, C. M. (2010). BMC Struct. Biol. 10, 28. [DOI] [PMC free article] [PubMed]
- Berglund, G. I., Carlsson, G. H., Smith, A. T., Szöke, H., Henriksen, A. & Hajdu, J. (2002). Nature (London), 417, 463–468. [DOI] [PubMed]
- Castañeda, C. A., Fitch, C. A., Majumdar, A., Khangulov, V., Schlessman, J. L. & García-Moreno, B. E. (2009). Proteins, 77, 570–588. [DOI] [PubMed]
- Cha, J.-S. & Cooksey, D. A. (1991). Proc. Natl Acad. Sci. USA, 88, 8915–8919. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Chen, Z., Durão, P., Silva, C. S., Pereira, M. M., Todorovic, S., Hildebrandt, P., Bento, I., Lindley, P. F. & Martins, L. O. (2010). Dalton Trans. 39, 2875–2882. [DOI] [PubMed]
- De la Mora, E., Lovett, J. E., Blanford, C. F., Garman, E. F., Valderrama, B. & Rudino-Pinera, E. (2012). Acta Cryst. D68, 564–577. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Enguita, F. J., Martins, L. O., Henriques, A. O. & Carrondo, M. A. (2003). J. Biol. Chem. 278, 19416–19425. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Felsenfeld, G. (1960). Arch. Biochem. Biophys. 87, 247–251. [DOI] [PubMed]
- Fernandes, A. S., Gaspar, J., Cabral, M. F., Caneiras, C., Guedes, R., Rueff, J., Castro, M., Costa, J. & Oliveira, N. G. (2007). J. Inorg. Biochem. 101, 849–858. [DOI] [PubMed]
- Ferraroni, M., Matera, I., Chernykh, A., Kolomytseva, M., Golovleva, L. A., Scozzafava, A. & Briganti, F. (2012). J. Inorg. Biochem. 111, 203–209. [DOI] [PubMed]
- Ferraroni, M., Myasoedova, N. M., Schmatchenko, V., Leontievsky, A. A., Golovleva, L. A., Scozzafava, A. & Briganti, F. (2007). BMC Struct. Biol. 7, 60. [DOI] [PMC free article] [PubMed]
- Garman, E. F. (2010). Acta Cryst. D66, 339–351. [DOI] [PMC free article] [PubMed]
- Hakulinen, N., Kruus, K., Koivula, A. & Rouvinen, J. (2006). Biochem. Biophys. Res. Commun. 350, 929–934. [DOI] [PubMed]
- Harris, T. K. & Turner, G. J. (2002). IUBMB Life, 53, 85–98. [DOI] [PubMed]
- Hoegger, P. J., Kilaru, S., James, T. Y., Thacker, J. R. & Kües, U. (2006). FEBS J. 273, 2308–2326. [DOI] [PubMed]
- Ji, X., Blaszczyk, J. & Chen, X. (2001). Acta Cryst. D57, 1003–1007. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kataoka, K., Komori, H., Ueki, Y., Konno, Y., Kamitaka, Y., Kurose, S., Tsujimura, S., Higuchi, Y., Kano, K., Seo, D. & Sakurai, T. (2007). J. Mol. Biol. 373, 141–152. [DOI] [PubMed]
- Kataoka, K., Sugiyama, R., Hirota, S., Inoue, M., Urata, K., Minagawa, Y., Seo, D. & Sakurai, T. (2009). J. Biol. Chem. 284, 14405–14413. [DOI] [PMC free article] [PubMed]
- Kjaergaard, C. H., Durand, F., Tasca, F., Qayyum, M. F., Kauffmann, B., Gounel, S., Suraniti, E., Hodgson, K. O., Hedman, B., Mano, N. & Solomon, E. I. (2012). J. Am. Chem. Soc. 134, 5548–5551. [DOI] [PMC free article] [PubMed]
- Komori, H., Miyazaki, K. & Higuchi, Y. (2009). FEBS Lett. 583, 1189–1195. [DOI] [PubMed]
- Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. (2008). Nature Protoc. 3, 1171–1179. [DOI] [PMC free article] [PubMed]
- Lee, S.-K., George, S. D, Antholine, W. E., Hedman, B., Hodgson, K. O. & Solomon, E. I. (2002). J. Am. Chem. Soc. 124, 6180–6193. [DOI] [PubMed]
- Lee, S. M., Grass, G., Rensing, C., Barrett, S. R., Yates, C. J. D., Stoyanov, J. V. & Brown, N. L. (2002). Biochem. Biophys. Res. Commun. 295, 616–620. [DOI] [PubMed]
- Macedo, S., Pechlaner, M., Schmid, W., Weik, M., Sato, K., Dennison, C. & Djinović-Carugo, K. (2009). J. Synchrotron Rad. 16, 191–204. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McNicholas, S., Potterton, E., Wilson, K. S. & Noble, M. E. M. (2011). Acta Cryst. D67, 386–394. [DOI] [PMC free article] [PubMed]
- Miyazaki, K. (2005). Extremophiles, 9, 415–425. [DOI] [PubMed]
- Murphy, M. E. P., Lindley, P. F. & Adman, E. T. (1997). Protein Sci. 6, 761–770. [DOI] [PMC free article] [PubMed]
- Murray, J. W., Garman, E. F. & Ravelli, R. B. G. (2004). J. Appl. Cryst. 37, 513–522.
- Olsson, M. H. M., Søndergaard, C. R., Rostkowski, M. & Jensen, J. H. (2011). J. Chem. Theory Comput. 7, 525–537. [DOI] [PubMed]
- Owen, R. L., Holton, J. M., Schulze-Briese, C. & Garman, E. F. (2009). J. Synchrotron Rad. 16, 143–151. [DOI] [PMC free article] [PubMed]
- Paithankar, K. S., Owen, R. L. & Garman, E. F. (2009). J. Synchrotron Rad. 16, 152–162. [DOI] [PubMed]
- Palmer, A. E., Lee, S.-K. & Solomon, E. I. (2001). J. Am. Chem. Soc. 123, 6591–6599. [DOI] [PubMed]
- Palmer, A. E., Quintanar, L., Severance, S., Wang, T.-P., Kosman, D. J. & Solomon, E. I. (2002). Biochemistry, 41, 6438–6448. [DOI] [PubMed]
- Quintanar, L., Stoj, C., Taylor, A. B., Hart, P. J., Kosman, D. J. & Solomon, E. I. (2007). Acc. Chem. Res. 40, 445–452. [DOI] [PubMed]
- Quintanar, L., Stoj, C., Wang, T.-P., Kosman, D. J. & Solomon, E. I. (2005). Biochemistry, 44, 6081–6091. [DOI] [PubMed]
- Ravel, B. & Newville, M. (2005). J. Synchrotron Rad. 12, 537–541. [DOI] [PubMed]
- Roberts, S. A., Weichsel, A., Grass, G., Thakali, K., Hazzard, J. T., Tollin, G., Rensing, C. & Montfort, W. R. (2002). Proc. Natl Acad. Sci. USA, 99, 2766–2771. [DOI] [PMC free article] [PubMed]
- Roberts, S. A., Wildner, G. F., Grass, G., Weichsel, A., Ambrus, A., Rensing, C. & Montfort, W. R. (2003). J. Biol. Chem. 278, 31958–31963. [DOI] [PubMed]
- Sakuraba, H., Koga, K., Yoneda, K., Kashima, Y. & Ohshima, T. (2011). Acta Cryst. F67, 753–757. [DOI] [PMC free article] [PubMed]
- Serrano-Posada, H., Valderrama, B., Stojanoff, V. & Rudiño-Piñera, E. (2011). Acta Cryst. F67, 1595–1598. [DOI] [PMC free article] [PubMed]
- Sharma, P., Goel, R. & Capalash, N. (2007). World J. Microbiol. Biotechnol. 23, 823–832.
- Silva, C. S., Damas, J. M., Chen, Z., Brissos, V., Martins, L. O., Soares, C. M., Lindley, P. F. & Bento, I. (2012). Acta Cryst. D68, 186–193. [DOI] [PubMed]
- Silva, C. S., Durão, P., Fillat, A., Lindley, P. F., Martins, L. O. & Bento, I. (2012). Metallomics, 4, 37–47. [DOI] [PubMed]
- Singh, S. K., Roberts, S. A., McDevitt, S. F., Weichsel, A., Wildner, G. F., Grass, G. B., Rensing, C. & Montfort, W. R. (2011). J. Biol. Chem. 286, 37849–37857. [DOI] [PMC free article] [PubMed]
- Solomon, E. I., Augustine, A. J. & Yoon, J. (2008). Dalton Trans., pp. 3921–3932. [DOI] [PMC free article] [PubMed]
- Solomon, E. I., Chen, P., Metz, M., Lee, S.-K. & Palmer, A. E. (2001). Angew. Chem. Int. Ed. 40, 4570–4590. [DOI] [PubMed]
- Solomon, E. I., Sundaram, U. M. & Machonkin, T. E. (1996). Chem. Rev. 96, 2563–2606. [DOI] [PubMed]
- Taylor, A. B., Stoj, C. S., Ziegler, L., Kosman, D. J. & Hart, P. J. (2005). Proc. Natl Acad. Sci. USA, 102, 15459–15464. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yoon, J., Liboiron, B. D., Sarangi, R., Hodgson, K. O., Hedman, B. & Solomon, E. I. (2007). Proc. Natl Acad. Sci. USA, 104, 13609–13614. [DOI] [PMC free article] [PubMed]
- Yoon, J. & Solomon, E. I. (2007). J. Am. Chem. Soc. 129, 13127–13136. [DOI] [PMC free article] [PubMed]
- Zhukhlistova, N. E., Zhukova, Y. N., Lyashenko, A. V., Zăitsev, V. N. & Mikhăilov, A. M. (2008). Crystallogr. Rep. 53, 92–109.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Tth-MCO, 2xu9
PDB reference: apo Tth-MCO, 2xuw
PDB reference: Hg-Tth-MCO, 2xvb
PDB reference: Hg-Tth-MCO-2h, 4ai7
PDB reference: Tth-MCO-C1, 2yae
PDB reference: Tth-MCO-C2, 2yaf
PDB reference: Tth-MCO-C3, 2yah
PDB reference: Tth-MCO-C4, 2yam
PDB reference: Tth-MCO-C5, 2yao
PDB reference: Tth-MCO-C6, 2yap
PDB reference: Tth-MCO-C7, 2yaq
PDB reference: Tth-MCO-C8, 2yar