Abstract
The Cancer Genome Atlas (TCGA) identified 4 groups of endometrial carcinomas based on an integrated genomic characterization: POLE ultramutated (POLE), microsatellite instability-high (MSI-H), copy number-low (CN-L), and copy number-high (CN-H). In that study, CN-H comprised all of the serous carcinoma cases and 25% of all FIGO grade 3 endometrioid carcinoma cases. In this study, two expert gynecologic pathologists undertook a morphologic reassessment of the FIGO grade 3 endometrioid carcinoma subset of the TCGA study cohort, including an analysis for evidence of serous differentiation. Interobserver variability kappa values are reported for the histologic evaluation of all four genomic clusters, and diagnostic discrepancies are discussed. Overall, there were 55 agreements, 6 disagreements, and 14 deferrals. Of the 75 cases analyzed, 6 cases had a consensus morphologic diagnosis of serous carcinoma, but only 2 of these cases had a serous carcinoma genotype while the remaining 4 cases were genotypically endometrioid carcinoma. For the CN-H group, 2 of 15 cases were serous carcinoma by morphology and genotype, whereas at least one pathologist interpreted the remaining 13 cases as endometrioid carcinoma. The interobserver agreement rate was highest in the CN-L group (90%; kappa 0.9), compared to the other genomic groups (POLE: 62%, kappa 0.55; MSI-H: 78%, kappa 0.74; and CN-H: 53%, kappa 0.48). Our review confirms that most high-grade endometrial carcinomas diagnosed by TCGA as FIGO grade 3 endometrioid carcinoma are indeed endometrioid carcinomas by morphology and genotype, and that the reproducibility of histologic diagnosis between pathologists varies between the TCGA integrated genomic clusters.
Keywords: Endometrial cancer, endometrioid, serous, TCGA, interobserver agreement
Introduction
High-grade endometrial carcinomas constitute a heterogeneous group of tumors that show variable histology, molecular abnormalities, and clinical outcomes. Although high-grade carcinomas comprises a minority of cases of endometrial neoplasia, they account for a disproportionate number of endometrial cancer deaths.1,2
Prototypic high-grade endometrial carcinomas can be reproducibly diagnosed on H&E slides alone; however, as many as 30–50% of high-grade endometrial carcinomas are not morphologically prototypic, leading to poor reproducibility in the diagnosis.3 One of the most common diagnostic challenges in the assessment of high-grade endometrial carcinoma is the distinction between FIGO grade 3 endometrioid carcinoma and serous carcinoma. Endometrioid carcinoma is characterized by frequent mutations in PTEN, ARID1A, DNA mismatch repair genes, KRAS, and PIK3CA. Additionally, a subset of high-grade and even rare low-grade endometrioid carcinomas harbors mutations in TP53. In contrast, serous carcinoma is characterized by frequent mutations in TP53 and PPP2R1A, while mutations involving PTEN, ARID1A, DNA mismatch repair genes, and KRAS are very infrequent.4–6
The Cancer Genome Atlas (TCGA), a multi-institutional project sponsored by the National Cancer Institute and the National Human Genome Research Institute, recently reported on the genomic, transcriptomic, and proteomic analysis of 373 endometrial carcinomas (306 endometrioid carcinomas, 66 serous and mixed carcinomas) using massively parallel sequencing and array-based technologies in combination with DNA methylation, reverse phase protein array, and microsatellite instability analyses.7 The TCGA study is the largest comprehensive genomic study of endometrial carcinoma to date. Based on the integration of the somatic gene mutations, microsatellite instability, and somatic copy-number alterations results, endometrial carcinomas were categorized into 4 genomic groups: 1) An “ultramutated” group harboring mutations in the exonuclease domain of the polymerase epsilon (POLE) gene, and characterized by very high mutation rates; 2) A “hypermutated” group characterized by microsatellite instability (MSI) due to MLH1 promoter methylation, with high mutation rates and few copy number alterations; 3) A “copy number-low (CN-low)” group, composed of most of the microsatellite-stable grade 1 and grade 2 endometrioid carcinomas, which demonstrated low mutation rates; and 4) A “copy number-high” (CN-high) group with extensive copy number aberrations, low mutation rates, and recurrent TP53 mutations, composed of all of the serous carcinomas in the study and 25% of all FIGO grade 3 endometrioid carcinomas.7 These results provided a completely novel insight into the genetic and clinical heterogeneity of FIGO grade 3 endometrioid carcinomas. Unlike serous carcinoma, FIGO grade 3 endometrioid carcinomas did not segregate to a single molecular cluster, but were represented in all 4 TCGA subgroups.7
With regards to clinical outcome, while some authors report similar poor clinical outcome among all high-grade endometrial tumors,1,8,9 others report a significantly poorer prognosis for serous carcinoma compared to FIGO grade 3 endometrioid carcinoma.10 It is possible that the variability in the inclusion criteria for FIGO grade 3 endometrioid carcinoma might account for the differences reported. The TCGA study showed that CN-high endometrial carcinomas had the worst progression-free survival of the 4 TCGA genomic groups.7 In contrast, despite their high-grade morphology, POLE-mutated high-grade endometrioid carcinoma had the best progression-free survival of the 4 molecular categories. These results clearly demonstrate the emerging requirement for pathologists to learn to recognize the subset of endometrioid carcinomas that are CN-high, and to reproducibly separate them from ultramutated endometrial carcinomas harboring POLE exonuclease domain somatic mutations (EC-POLE) and MSI-H endometrioid tumors.
In this study, we sought to analyze the clinicopathologic and molecular characteristics of FIGO grade 3 endometrioid carcinomas using the TCGA dataset. No prior morphologic-genomic correlations had ever been performed on this comprehensive dataset. The primary aim of this study was to determine whether the subset of CN-high Gr3-EMC identified in the TCGA study would have histologic features of serous carcinoma on retrospective histologic review by 2 experienced gynecologic pathologists. A secondary aim was to compare the morphologic diagnoses rendered on this tumor cohort by these pathologists in terms of histologic tumor subtyping, molecular profile, and TCGA genomic classes of the cases, particularly in cases with a discordant diagnosis.
Material and Methods
The TCGA database included 116 cases of FIGO grade 3 endometrioid carcinomas, only 75 of which underwent exome sequence analysis. These 75 cases constituted our study cohort. TCGA genomic class was obtained along with the other molecular and clinical data from the cBioPortal website (http://www.cbioportal.org) and the TCGA data portal website (https://tcga-data.nci.nih.gov/tcga).7
Virtual whole slide images of each case were studied by 2 gynecologic subspecialty pathologists (RB & RAS) from 2 academic institutions, and the cases were categorized as endometrioid carcinoma or serous carcinoma according to the World Health Organization classification.11 Tumors with endometrioid differentiation were classified as FIGO grade 3 endometrioid carcinomas if there was >50% solid architecture, or if there was 5–50% solid architecture in the presence of marked nuclear pleomorphism.11 Diagnostic criteria were not reviewed and harmonized between reviewers prior to undertaking the study, in order to simulate “real world” practice. The histologic subtype of a given case was recorded by each pathologist independently without knowing its TCGA genomic class. The cases were categorized as “agreement” when the diagnoses from both reviewers were the same, “disagreement” when they differed, and “indeterminate” when at least one pathologist could not confidently diagnose either serous or endometrioid carcinoma (“deferral”).
Using the criteria from Hoang et al’s study, cases were defined genotypically as endometrioid carcinoma when they had PTEN and/or ARID1A mutations, with or without TP53 mutation, or as serous carcinoma if the tumor harbored a TP53 mutation in the absence of PTEN and ARID1A.12
In order to investigate whether review of a single virtual whole slide image was as diagnostically informative as review of a full set of H&E slides in any one case, all slides pertaining to 12 of the FIGO grade 3 endometrioid carcinoma cases (the virtual slide images of which were already reviewed among the 75 cases of this study) from the TCGA study were retrieved from the archives of Memorial Sloan Kettering Cancer Center for review by one of the study pathologists. The TCGA genomic class and molecular data from these 12 cases were obtained by another study co-author (DAL), while maintaining the patients’ identity. Histologic diagnoses made on the basis of the virtual slide review and review of the complete case slides were compared in the 12 cases.
Pairwise comparison of the agreement between reviewers among the different TCGA genomic groups was done by calculation of Kappa statistics.
Results
In the study cohort of 75 tumors initially diagnosed as FIGO grade 3 endometrioid carcinomas in the TCGA project, median patient age was 64 years (range, 33 to 90 years). The majority of patients presented as FIGO stage I (49 patients; 65%); 5 were stage II, 15 were stage III, and 5 were stage IV.
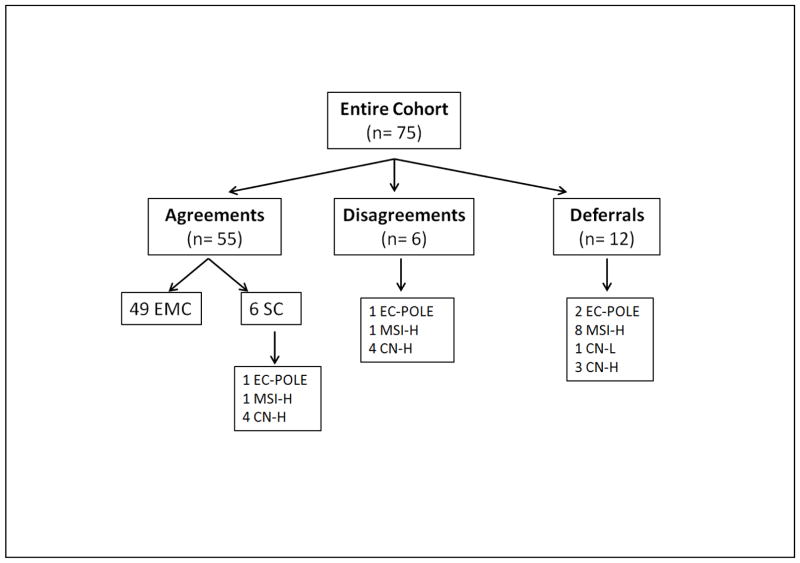
Among the 75 cases, 8 were EC-POLE, 42 MSI-H, 10 CN-low, and 15 CN-high. The diagnostic opinions of the two reviewers and the corresponding TCGA genomic groups are illustrated in Figure 1. Diagnostic agreement was seen in 55 of 75 (73%) cases, while disagreement was noted in 6 of 75 (8%). In 14 (19%) cases, the diagnosis was deferred by at least one pathologist. The TCGA ID list of all the study cases is shown in Supplementary Table 1.
Figure 1.
The different diagnostic opinions between the 2 reviewers (agreement, disagreement, and deferrals) with the corresponding TCGA genomic groups.
Abbreviations: TCGA, The Cancer Genome Atlas; EMC, endometrioid carcinoma; SC, serous carcinoma; EC-POLE, ultramutated endometrioid carcinoma harboring POLE mutation; MSI-H, hypermutated endometrioid carcinoma with microsatellite instability; CN-high, copy number-high serous and serous-like FIGO G3 endometrioid carcinomas
Of the 75 cases analyzed, reviewer 1 diagnosed 53 (71%) cases as endometrioid carcinoma, 8 (11%) as serous carcinoma (3 of which had a serous carcinoma genotype), while the remaining 14 (19%) cases were deferred. Reviewer 2 diagnosed 61 (81%) cases as endometrioid carcinoma and 13 (17%) as serous carcinoma (4 of which had a serous carcinoma genotype); 1 case was deferred.
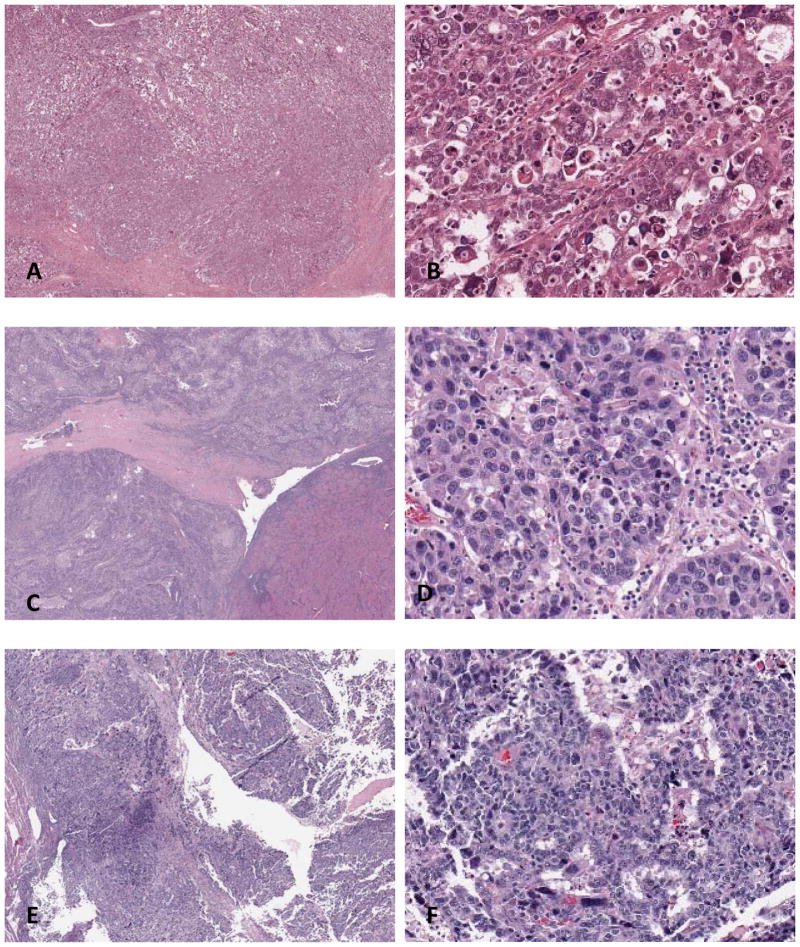
In 6 of the 75 cases, the 2 pathologists had a consensus diagnosis of serous carcinoma (Figure 2); 4 of the 6 cases were CN-high, 1 EC-POLE, and 1 MSI-high. TCGA group and genotype of cases with a consensus diagnosis of serous carcinoma by the 2 reviewers are illustrated in Table 1. Despite the fact that these 6 cases were interpreted as serous carcinoma by both reviewers, only 2 of the 6 cases had a serous carcinoma genotype (i.e., harboring TP53 mutation in the absence of PTEN and/or ARID1A mutation), which were among the CN-high group (TCGA-AP-A053 and TCGA-AP-AOLF). The remaining 4 cases had an endometrioid genotype. Of note, none of the CN-low cases were interpreted as serous carcinoma by any of the reviewers, while 13 of 15 (87%) CN-high cases were interpreted as endometrioid by at least one pathologist.
Figure 2.
Tumors interpreted by both reviewers as serous carcinoma. (A & B) TCGA-BS-A0UV: (A) low power, (B) high power. Endometrial carcinoma harboring POLE mutation, interpreted by both pathologists as serous carcinoma. The tumor harbored TP53, PTEN, and ARID1A mutations, an endometrioid genotype. (C & D) TCGA-AP-A0LF: (C) low power, (D) high power. Endometrial carcinoma with high copy-number alteration, interpreted by both pathologists as serous carcinoma. The tumor harbored TP53 mutation in the absence of PTEN and ARID1A mutation, a serous genotype. (E & F) TCGA-AP-A053: (E) low power, (F) high power. Endometrial carcinoma with high copy-number alteration, interpreted by both pathologists as serous carcinoma. The tumor harbored TP53 mutation in the absence of PTEN and ARID1A mutation, a serous genotype.
Table 1.
TCGA group and genotype of cases with a consensus diagnosis of serous carcinoma by the 2 reviewers
| TCGA ID | TCGA genomic group | Mutations | Genotype | |
|---|---|---|---|---|
| TP53 | PTEN +/− ARID1A | |||
| TCGA-BS-A0UV | EC-POLE | + | PTEN&ARID1A | Endometrioid |
| TCGA-BG-A222 | MSI-H | − | None | Endometrioid |
| TCGA-AP-A053 | CN-high | + | None | Serous |
| TCGA-AP-A0LF | CN-high | + | None | Serous |
| TCGA-B5-A0K3 | CN-high | + | ARID1A | Endometrioid |
| TCGA-D1-A1O0 | CN-high | + | PTEN | Endometrioid |
TCGA, The Cancer Genome Atlas; EC-POLE, ultramutated endometrial carcinoma; CN-high, copy-number high.
The extent of phenotype-genotype discrepancies led us to hypothesize that inter-observer diagnostic agreement varies according to TCGA genomic class assignment. In this study we found that while the diagnostic agreement rate between the 2 reviewers was excellent in the CN-low tumors (90%; kappa: 0.9) and good in the MSI-high group (78%; Kappa: 0.74), agreement was moderate in the EC-POLE tumors (62%; Kappa: 0.55), and poor in the CN-high tumors (53%, Kappa: 0.48) (Table 2). Figures 3 and 4 are examples of diagnostically concordant and discordant cases, respectively.
Table 2.
The Interobserver agreement between the 2 gynecologic pathologists for the 75 endometrial carcinoma cases and the correlation with their TCGA genomic groups
| TCGA genomic group | Agreement (n=55) | Disagreement (n=6) | Kappa | Strength | Deferrals (n=14) |
|---|---|---|---|---|---|
| EC-POLE (n=8) | 5 (62%) | 1 (13%) | 0.55 | Moderate | 2 (25%) |
| MSI-H (n=42) | 33 (8%) | 1 (2%) | 0.7 | Good | 8 (19%) |
| CN-low (n=10) | 9 (90%) | 0 | 0.9 | Excellent | 1 |
| CN-high (n=15) | 8 (53%) | 4 (26%) | 0.48 | poor | 3 (20%) |
TCGA, The Cancer Genome Atlas; DX, diagnosis; EC-POLE, ultramutated endometrial carcinoma; MSI-H, hypermutated endometrial carcinoma with microsatellite instability; CN-low, copy-number low; CN-high, copy-number high.
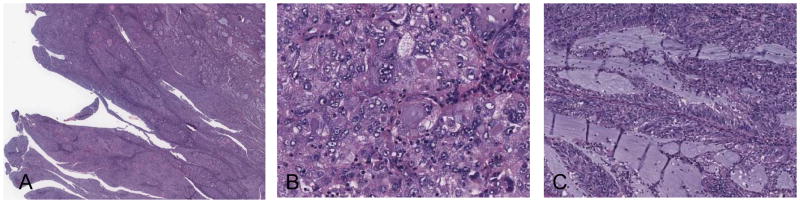
Figure 3.
A tumor (TCGA-BG-A0M8) interpreted as endometrioid carcinoma by reviewer 1 and serous carcinoma by reviewer 2. (A) low power, (B) high power, (C) focal area of mucinous metaplasia. This tumor was an endometrial carcinoma with high copy-number alteration that harbored TP53 mutation in the absence of PTEN and ARID1A mutation, a serous genotype according to Hoang et al (12). Interestingly, this tumor also had KRAS mutation and it was negative for PPP2R1A mutation.
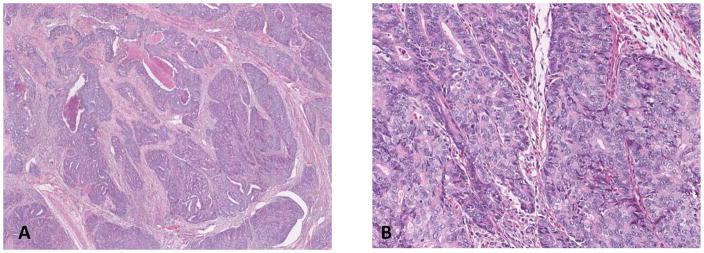
Figure 4.
Tumor interpreted by both pathologists as FIGO grade 3 endometrioid carcinoma. TCGA-B5-A0JR: (A) low power, (B) high power. This tumor was in the low copy-number alteration TCGA genomic group. The molecular profile was consistent with endometrioid genotype.
One of the reviewers (RAS) then examined full H&E slide sets from 12 cases that were part of the TCGA study without knowing the genomic subgroup assignment. Among the 12 cases, 4 were EC-POLE, 7 MSI-H, and 1 CN-high. None of these cases were deferred by the reviewer, unlike the experience of reviewing only 1 representative slide. The single CN-high case was classified histologically as FIGO grade 3 endometrioid carcinoma, despite having a serous carcinoma genomic signature.
Discussion
The TCGA identified 4 groups of endometrial carcinoma based on an integrated genomic characterization, including EC-POLE, MSI-H, CN-low, and CN-high (serous-like).7 Our review of FIGO grade 3 endometrioid carcinomas studied by the TCGA demonstrates that only 13% of CN-high cases would be recognized as serous carcinoma by morphologic consensus and genotype, confirming that a subset of truly endometrioid high-grade carcinomas segregate into the CN-high group with endometrial serous carcinoma. Our findings support previous reports that there is considerable interobserver variability in distinguishing FIGO grade 3 endometrioid carcinoma from serous carcinoma.3,13,14 Only the CN-low tumors (type I endometrial cancer according to Bokman’s model15) were reproducibly and confidently diagnosed as FIGO grade 3 endometrioid carcinoma without any misclassification as serous carcinoma. There were also diagnostic discrepancies in classifying EC-POLE, MSI-H, and CN-high tumors as endometrioid or serous on morphologic grounds. Therefore, assumptions about morphologic, immunohistochemical, and genetic differences between FIGO grade 3 endometrioid carcinoma and serous carcinoma may need to be adjusted, given the fact that a proportion of FIGO grade 3 endometrioid carcinomas cluster with serous carcinomas in terms of both copy number aberrations and p53 mutation status.
Recent studies have identified what are currently thought to be distinct molecular profiles for endometrial endometrioid carcinoma and serous carcinoma. Endometrioid carcinoma is characterized by frequent mutations in PTEN and/or ARID1A, with a subset harboring mutations in TP53 (especially FIGO grade 3 tumors).4–6,12,16 Mutation of PPP2R1A is infrequent in endometrioid carcinomas. In contrast, serous carcinoma is characterized by frequent mutations in TP53 and PPP2R1A, whereas mutations involving PTEN and/or ARID1A are very infrequent.4,5,17,18 PIK3CA mutation is found in a subset of both histologic subtypes. This mutation profile, however, does not address the substantial genetic heterogeneity in FIGO grade 3 endometrioid carcinomas that was subsequently reported by the TCGA, as described above.7 A recent study examined the correlation between tumor histotype and genotype in 36 high-grade previously genotyped endometrial carcinoma cases among 8 gynecologic pathologists.12 When there was high interobserver agreement between the 8 pathologists, cases were genotype-concordant; meanwhile, when there was a lack of interobserver agreement, one-third of cases were genotype-discordant. The recommendation from the investigators of that study was to refine the diagnostic criteria used for distinguishing high-grade endometrioid carcinoma and serous carcinoma as informed by genotype. High nuclear grade distributed diffusely in the setting of glandular or papillary tumor was equated with a serous carcinoma genotype. For tumors with solid architecture and high nuclear grade, a diagnosis of endometrioid carcinoma should be favored when a low-grade endometrioid component, squamous or mucinous differentiation, or endometrial hyperplasia is present. This approach could not be fully applied in the current study, as only a single virtual slide was available for review in most cases.
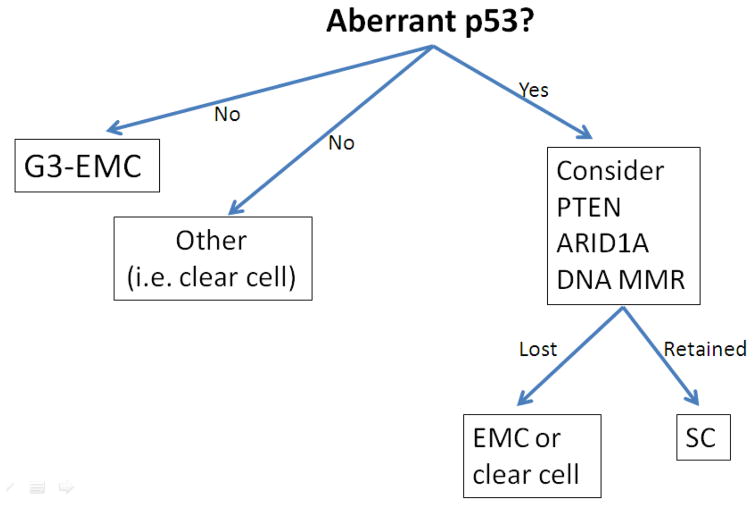
Immunohistochemistry has been recommended as an adjunctive tool to distinguish FIGO grade 3 endometrioid carcinoma from serous carcinoma in diagnostically challenging cases. If defining endometrioid features are not present, a panel of immunohistochemical stains including p53, PTEN, ARID1A, and DNA mismatch repair proteins can be applied. Aberrant p53 immunohistochemical expression without abnormalities in ARID1A or DNA mismatch repair proteins, as well as PTEN, should be sufficient to confirm a serous carcinoma diagnosis if suspected on routine morphological examination. Conversely, aberrant p53 expression along with alteration of one or more of the previously mentioned markers supports endometrioid differentiation, whereas tumors lacking aberrant p53 expression are not likely to be serous carcinoma (Figure 5).19–21
Figure 5.
Immunohistochemistry may aid in the distinction between challenging cases of serous carcinoma and FIGO grade 3 endometrioid carcinoma.
Abbreviations: EMC, Endometrioid carcinoma; DNA MMR, DNA mismatch repair proteins; SC, serous carcinoma.
For the differential diagnosis of EC-POLE and serous carcinoma, our group has recently described the histopathologic features of EC-POLE and demonstrated that EC-POLEs almost always show defining endometrioid features, at least focally. In addition, focal or patchy marked nuclear atypia, but not diffuse nuclear atypia, as seen in serous carcinoma, can be found. Furthermore, in contrast to serous carcinomas, the majority of EC-POLEs show increased tumor-infiltrating lymphocytes and/or peri-tumoral lymphocytes; in addition, these patients are significantly younger.22 A panel of immunohistochemical stains such as p53, PTEN, ARID1A, and p16 may be used to discriminate serous carcinomas from FIGO grade 3 endometrioid carcinoma; however, currently the only definite way to detect EC-POLE tumors is by sequencing the POLE gene. Clinically, both the TCGA and Meng et al found that patients with EC-POLE have excellent disease progression survival.7,23, 24
Regarding the distinction between MSI-H FIGO grade 3 endometrioid carcinomas and serous carcinoma, MSI-H tumors tend to show considerable intra-tumoral heterogeneity and may display dense intra-tumoral and peri-tumoral lymphocytes.25–27 Immunohistochemistry is helpful in this differential diagnosis, as tumors in this category should show abnormal DNA mismatch repair protein expression in more than 90% of cases,28 in contrast to serous carcinoma. According to the TCGA study, patients with MSI-H tumors had progression-free survival that falls between that of EC-POLE and CN-high tumors.7
Future studies are still needed to determine whether CN-high FIGO grade 3 endometrioid carcinoma is clinically separable from serous carcinoma. Further research is also required to find out where endometrial clear cell carcinoma, malignant mixed mullerian tumor, and undifferentiated carcinoma would fit into this new paradigm.
There are several limitations to this study. First, only a single virtual slide image from each case was available for morphologic evaluation. Review of full H&E slide sets is probably necessary to distinguish high-grade endometrioid carcinoma (particularly in the CN-high group) from mimics, consistent with the fact that these tumors are more heterogenous and difficult to diagnosis, compared to CN-low endometrioid tumors. It is noteworthy that, even when all H&E slides on one CN-high case were reviewed, it was histologically interpreted as FIGO grade 3 endometrioid carcinoma despite having a serous carcinoma genomic signature. A further study limitation is the fact that no immunohistochemical studies were available to the reviewers of this study to aid in the histologic classification, as the TCGA did not perform immunohistochemistry in their study. Thirdly, molecular studies were performed by the TCGA on a section of tumor different from the one studied histologically, suggesting there might be differences in the mutation profile of different parts of FIGO grade 3 endometrioid carcinomas, as these tumors may be morphologically and genetically heterogenous. Finally, the clinical value of genomic-based subtyping of endometrial carcinoma still needs independent validation. Further studies with longer follow-up are necessary to confirm the clinical significance of the TCGA subtyping of endometrioid and serous carcinoma as the TCGA study was retrospective in nature and included heterogeneously treated patients.
In summary, our review confirms that most high-grade endometrial carcinomas diagnosed by TCGA as FIGO grade 3 endometrioid carcinoma are endometrioid by morphology and genotype. Nevertheless, approximately 25% of FIGO grade 3 endometrioid carcinomas cluster with serous carcinoma by virtue of TP53 mutation and high copy number alterations as shown by the TCGA. Prototypic type I endometrial carcinoma (copy number-low by TCGA) are reproducibly recognized as endometrioid, whereas significant interobserver variation in distinguishing FIGO grade 3 endometrioid carcinoma and serous carcinoma remains a problem in the EC-POLE and CN-high groups.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Niamh Conlon for her critical review of the manuscript.
Footnotes
Disclosure/Conflict of Interest
The authors have no conflicts of interest to declare.
This work was presented as a platform presentation at the 103rd United States and Canadian Academy of Pathology (USCAP) Annual Meeting, San Diego, CA.
References
- 1.Creasman WT, Kohler MF, Odicino F, et al. Prognosis of papillary serous, clear cell, and grade 3 stage I carcinoma of the endometrium. Gynecol Oncol. 2004;95:593–596. doi: 10.1016/j.ygyno.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 2.Halperin R, Zehavi S, Langer R, et al. Uterine papillary serous carcinoma (pure and mixed type) compared with moderately and poorly differentiated endometrioid carcinoma. A clinicopathologic study. Eur J Gynaecol Oncol. 2002;23:300–304. [PubMed] [Google Scholar]
- 3.Gilks CB, Oliva E, Soslow RA. Poor interobserver reproducibility in the diagnosis of high-grade endometrial carcinoma. Am J Surg Pathol. 2013;37:874–881. doi: 10.1097/PAS.0b013e31827f576a. [DOI] [PubMed] [Google Scholar]
- 4.McConechy MK, Ding J, Cheang MC, et al. Use of mutation profiles to refine the classification of endometrial carcinomas. J Pathol. 2012;228:20–30. doi: 10.1002/path.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matias-Guiu X, Prat J. Molecular pathology of endometrial carcinoma. Histopathology. 2013;62:111–123. doi: 10.1111/his.12053. [DOI] [PubMed] [Google Scholar]
- 6.Samarnthai N, Hall K, Yeh IT. Molecular profiling of endometrial malignancies. Obstet Gynecol Int. 2010;2010:162363. doi: 10.1155/2010/162363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voss MA, Ganesan R, Ludeman L, McCarthy K, et al. Should grade 3 endometrioid endometrial carcinoma be considered a type 2 cancer-a clinical and pathological evaluation. Gynecol Oncol. 2012;124:15–20. doi: 10.1016/j.ygyno.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Soslow RA, Bissonnette JP, Wilton A, et al. Clinicopathologic analysis of 187 high-grade endometrial carcinomas of different histologic subtypes: similar outcomes belie distinctive biologic differences. Am J Surg Pathol. 2007;31:979–987. doi: 10.1097/PAS.0b013e31802ee494. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton CA, Cheung MK, Osann K, et al. Uterine papillary serous and clear cell carcinomas predict for poorer survival compared to grade 3 endometrioid corpus cancers. Br J Cancer. 2006;94:642–6. doi: 10.1038/sj.bjc.6603012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurman RJ, Carcangiu ML, Herrington CS, Young RH. WHO Clasification of Tumours of Female Reproductive Organs. 4. Lyon: IARC; 2014. p. 9. [Google Scholar]
- 12.Hoang LN, McConechy MK, Kobel M, et al. Histotype-genotype correlation in 36 high-grade endometrial carcinomas. Am J Surg Pathol. 2013;37:1421–1432. doi: 10.1097/PAS.0b013e31828c63ed. [DOI] [PubMed] [Google Scholar]
- 13.Garg K, Leitao MM, Jr, Wynveen CA, et al. p53 overexpression in morphologically ambiguous endometrial carcinomas correlates with adverse clinical outcomes. Mod Pathol. 2010;23:80–92. doi: 10.1038/modpathol.2009.153. [DOI] [PubMed] [Google Scholar]
- 14.Lomo L, Nucci MR, Lee KR, et al. Histologic and immunohistochemical decision-making in endometrial adenocarcinoma. Mod Pathol. 2008;21:937–942. doi: 10.1038/modpathol.2008.97. [DOI] [PubMed] [Google Scholar]
- 15.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 16.O’Hara AJ, Bell DW. The genomics and genetics of endometrial cancer. Adv Genomics Genet. 2012;2012:33–47. doi: 10.2147/AGG.S28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McConechy MK, Anglesio MS, Kalloger SE, et al. Subtype-specific mutation of PPP2R1A in endometrial and ovarian carcinomas. J Pathol. 2011;223:567–573. doi: 10.1002/path.2848. [DOI] [PubMed] [Google Scholar]
- 18.Shih Ie M, Panuganti PK, Kuo KT, et al. Somatic mutations of PPP2R1A in ovarian and uterine carcinomas. Am J Pathol. 2011;178:1442–1447. doi: 10.1016/j.ajpath.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alkushi A, Clarke BA, Akbari M, et al. Identification of prognostically relevant and reproducible subsets of endometrial adenocarcinoma based on clustering analysis of immunostaining data. Mod Pathol. 2007;20:1156–1165. doi: 10.1038/modpathol.3800950. [DOI] [PubMed] [Google Scholar]
- 20.Alkushi A, Kobel M, Kalloger SE, et al. High-grade endometrial carcinoma: serous and grade 3 endometrioid carcinomas have different immunophenotypes and outcomes. Int J Gynecol Pathol. 2010;29:343–350. doi: 10.1097/PGP.0b013e3181cd6552. [DOI] [PubMed] [Google Scholar]
- 21.Alvarez T, Miller E, Duska L, et al. Molecular profile of grade 3 endometrioid endometrial carcinoma: is it a type I or type II endometrial carcinoma? Am J Surg Pathol. 2012;36:753–761. doi: 10.1097/PAS.0b013e318247b7bb. [DOI] [PubMed] [Google Scholar]
- 22.Hussein YR, Weigelt B, Levine DA, et al. Clinicopathologic Analysis of Endometrial Carcinomas Harboring Somatic POLE Mutations. Mod Pathol. 2014 doi: 10.1038/modpathol.2014.143. [Epub Nov 14, 2014] [DOI] [PubMed] [Google Scholar]
- 23.Meng B, Hoang LN, McIntyre JB, et al. POLE exonuclease domain mutation predicts long progression-free survival in grade 3 endometrioid carcinoma of the endometrium. Gynecol Oncol. 2014;134:15–9. doi: 10.1016/j.ygyno.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Church DN, Stello E, Nout RA, Valtcheva N, et al. Prognostic significance of POLE proofreading mutations in endometrial cancer. J Natl Cancer Inst. 2014;107(1):402. doi: 10.1093/jnci/dju402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shia J, Black D, Hummer AJ, et al. Routinely assessed morphological features correlate with microsatellite instability status in endometrial cancer. Hum Pathol. 2008;39:116–125. doi: 10.1016/j.humpath.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Garg K, Leitao MM, Jr, Kauff ND, et al. Selection of endometrial carcinomas for DNA mismatch repair protein immunohistochemistry using patient age and tumor morphology enhances detection of mismatch repair abnormalities. Am J Surg Pathol. 2009;33:925–933. doi: 10.1097/PAS.0b013e318197a046. [DOI] [PubMed] [Google Scholar]
- 27.Broaddus RR, Lynch HT, Chen LM, et al. Pathologic features of endometrial carcinoma associated with HNPCC: a comparison with sporadic endometrial carcinoma. Cancer. 2006;106:87–94. doi: 10.1002/cncr.21560. [DOI] [PubMed] [Google Scholar]
- 28.Modica I, Soslow RA, Black D, et al. Utility of immunohistochemistry in predicting microsatellite instability in endometrial carcinoma. Am J Surg Pathol. 2007;31:744–751. doi: 10.1097/01.pas.0000213428.61374.06. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.