Abstract
Background and Aims Arbuscular mycorrhizal (AM) fungi play a key role in the phosphate (P) uptake of many important crop species, but the mechanisms that control their efficiency and their contribution to the P nutrition of the host plant are only poorly understood.
Methods The P uptake and growth potential of two soybean genotypes that differ in their root architectural traits and P acquisition efficiency were studied after colonization with different AM fungi and the transcript levels of plant P transporters involved in the plant or mycorrhizal P uptake pathway were examined.
Key Results The mycorrhizal growth responses of both soybean genotypes ranged from highly beneficial to detrimental, and were dependent on the P supply conditions, and the fungal species involved. Only the colonization with Rhizophagus irregularis increased the growth and P uptake of both soybean genotypes. The expression of GmPT4 was downregulated, while the mycorrhiza-inducible P transporter GmPT10 was upregulated by colonization with R. irregularis. Colonization with both fungi also led to higher transcript levels of the mycorrhiza-inducible P transporter GmPT9, but only in plants colonized with R. irregularis were the higher transcript levels correlated to a better P supply.
Conclusions The results suggest that AM fungi can also significantly contribute to the P uptake and growth potential of genotypes with a higher P acquisition efficiency, but that mycorrhizal P benefits depend strongly on the P supply conditions and the fungal species involved.
Keywords: Arbuscular mycorrhizal symbiosis, Glycine max, Glomus aggregatum, Glomus custos, mycorrhizal growth response, mycorrhizal uptake pathway, gene expression, phosphate acquisition efficiency, phosphate transporter, Rhizophagus irregularis
INTRODUCTION
In addition to nitrogen, phosphate (P) is the nutrient that most often limits crop productivity and plant growth. Although the total soil contents of P may be high, P is often present in forms (e.g. organic P) that are unavailable for plant uptake. Phosphate has a low mobility in the soil (10–12–10–15 m2 s–1; Schachtman et al., 1998), and rapid uptake of inorganic phosphate (Pi) from the rhizosphere leads to depletion zones around the roots that limit further Pi uptake to the slow rate of diffusion. Crop productivity in agricultural systems depends on regular applications of P fertilizers that are produced from processed ground phosphate rock, but the known global reserves of this non-renewable resource are expected to be depleted in 50–100 years (Cordell et al., 2011; Schroder et al., 2011). Therefore, the improvement of the P efficiency of major crops represents an urgent research priority (Wang et al., 2010b; Schroder et al., 2011).
The P efficiency depends on the ability of plants to take up Pi from low-P soils (P acquisition efficiency; PAE) or the ability of plants to produce biomass or yield per unit of acquired P (P utilization efficiency; PUE) (Wang et al., 2010a, b). The PAE of crops can be improved by changes in the root morphology and architecture, an efficient symbiosis with AM fungi or changes in the root exudate composition leading, for example, to an enhanced release of organic acids or phosphatases or to an acidification of the root rhizosphere (Wang et al., 2010b). A better PUE of the plant is characterized, for example, by an increased efficiency with which plants are able to re-translocate P, or to re-use stored P, or to operate with lower tissue P concentrations (Wang et al., 2010a).
The majority of land plants, including soybeans and other crops, form a mutualistic symbiosis with arbuscular mycorrhizal (AM) fungi of the phylum Glomeromycota. The extraradical mycelium (ERM) of the fungus acts as an extension of the root system and is able to explore a large soil matrix in the search for nutrients. The nutrients that are taken up by the fungus are then transferred to the intraradical mycelium (IRM) in the root cortex, where these nutrients are transferred across the mycorrhizal interface to the host. In return for these nutritional benefits, the host plant transfers significant amounts of its photosynthetically fixed carbon to the fungus (Wright et al., 1998).
The AM colonization of the root system leads to a change in the nutrient acquisition strategy of the plant and shifts the surface for nutrient uptake from the root–soil interface to the mycorrhizal interface. In many crops, the mycorrhizal uptake pathway via the ERM and IRM plays a more important role in mycorrhizal roots for P supply than the plant uptake pathway via the epidermis and root hairs (Smith et al., 2003, 2004; Li et al., 2006). The mycorrhizal uptake pathway can, for example, account for 100 % of the total P uptake in AM-responsive flax plants, and even contributes >80 % to the total P uptake in non-responsive tomato or wheat plants (Smith et al., 2003, 2004; Li et al., 2006). High-affinity Pi uptake transporters of the plant that are involved in the Pi uptake from the root–soil interface are often downregulated in mycorrhizal roots (Grunwald et al., 2009), while Pi transporters that are involved in the Pi uptake from the mycorrhizal interface are upregulated. These mycorrhiza-inducible Pi transporters have been identified in many crop species (Rosewarne et al., 2000; Harrison et al., 2002; Nagy et al., 2005; Pumplin et al., 2012; Sisaphaithong et al., 2012), and the expression of MtPT4, the AM-inducible Pi transporter of Medicago truncatula, has been shown to be positively correlated to the P transport activity in colonized roots (Fellbaum et al., 2014). However, there are also contradictory reports in which the transcript levels of these Pi transporters were not related to mycorrhizal P uptake benefits (Grace et al., 2009; Sisaphaithong et al., 2012).
Mycorrhizal growth responses (MGRs) can range from highly beneficial to detrimental, and it has been suggested that they follow a parasitism to mutualism continuum (Johnson and Graham, 2013; Smith and Smith, 2013). The reasons for this high variability are not known, and it has been suggested that negative MGRs are mainly the result of the high carbon costs of the symbiosis for the host plant, particularly under high P supply conditions (Peng et al., 1993). However, it has also been hypothesized that negative MGRs could be the result of the suppression of the plant uptake pathway that is not compensated for by an increase in the P uptake via the mycorrhizal uptake pathway (Smith and Smith, 2011; Smith et al., 2011). The mycorrhizal P responsiveness (improvement of the P nutrition in mycorrhizal plants compared with non-mycorrhizal controls) can also be affected by the P efficiency of the plant, and it has been shown that in barley and wheat, the mycorrhizal P responsiveness was negatively correlated to the PUE of the plant (Baon et al., 1993; Zhu et al., 2001).
We studied here the mycorrhizal growth and P uptake response of two soybean [Glycine max (L.) Merr.] genotypes, HN112 and HN89, after colonization with three different AM fungal species, and examined the effect of these fungi on the transcript levels of Pi transporters involved in the plant or mycorrhizal uptake pathway. The genotypes differ in their root morphology and architecture. The genotype HN89 has a shallower root architecture and shows an increased exudation of malate compared with the more deep-rooted genotype HN112 (Wang et al., 2010b, 2011; Liang et al., 2013). It has been shown that HN89 has a higher PAE than HN112, because this genotype was able to outperform HN112 both under P-deficient conditions on acid soils and under field conditions with P fertilizer applications (Zhao et al., 2004; Wang et al., 2011). The goal of the study is to better understand how P-efficient plant genotypes will respond to the colonization with AM fungi. Hypothetically, it is possible that the AM colonization could nullify the advantage of P-efficient soybean genotypes (e.g. by a downregulation of Pi transporters of the plant uptake pathway, or changes in the root exudate composition), and that P-inefficient soybean genotypes with a successful AM symbiosis could outcompete their P-efficient counterparts on low P soils. Our aim is to better understand how traits of P-efficient genotypes can successfully be combined with beneficial AM fungal communities to increase the P uptake of soybean – one of the most important leguminous crop species worldwide.
MATERIALS AND METHODS
Plant and fungal materials
The experiments were conducted with the two soybean (Glycine max) genotypes HN112 and HN89 that are characterized by their deep or shallow root architecture, and their low or high PAE, respectively (Zhao et al., 2004; Liao et al., 2006). The seeds were surface sterilized for 15 min in 3 % sodium hypochlorite and rinsed several times in deionized water. The seeds were then sown into pots filled with an autoclaved (121 °C for 1 h) growth substrate of sand, organic soil and perlite (80:10:10, v/v/v) with a pH of 8·2 and a plant-available P level of 0·63 mg kg–1 soil (Olsen et al., 1954). After 1 week of germination, the seedlings were inoculated by adding 1000 spores of Rhizophagus irregularis (previously classified as Glomus intraradices, Schenck & Smith; DAOM 197198; Biosystematics Research Center), Glomus aggregatum [Schenck & Smith; isolate 0165 collected from the Long Term Mycorrhizal Research Site (LTRMS), University of Guelph, Canada] or Glomus custos (Cano & Dalpé; isolate 010 collected from Southwest Spain by Mycovitro S.L., Granada, Spain) into the soil close to the roots. An additional treatment without AM fungal inoculum (NM) was used as a control. The inoculum was produced by growing the fungi in root organ cultures with Ri T-DNA-transformed carrot (Daucus carota, clone DCI) roots in Petri dishes filled with mineral medium (St-Arnaud et al., 1996). The roots were cultured for approx. 8 weeks and the fungal spores were isolated from the growth medium by blending the medium in 10 mm sodium citrate buffer (Doner and Bécard, 1991).
Experimental design and growth conditions
First, the growth response and P efficiency of the two soybean genotypes after colonization with the different AM fungi was tested in a greenhouse experiment. The diurnal air temperatures ranged from 22 to 25 °C, and the natural light intensity from 400 to 1500 μmol m−2 s−1. The pots were randomly placed on a single bench and regularly rearranged when watered. The plants were grown in 3·0 L pots, and supplied with three different P concentrations: 25, 100 or 500 μm P as KH2PO4 that will be referred to as low (LP), medium (MP) or high phosphate (HP) supply, respectively. The different P treatments were applied by watering the plants every week with a solution containing the following nutrients (in μm): KNO3 (2·5 × 103), Ca(NO3)2·4H2O (2·5 × 103), MgSO4·7H2O (1 × 103), K2SO4 (250), Fe-EDTA(Na) (80), H3BO3 (20), MnCl2·4H2O (4·50), ZnSO4·7H2O (0·30), CuSO4·5H2O (0·16) and (NH4)6Mo7O24·4H2O (0·16). The plants were harvested 50 d after transplanting, and prepared for further analysis.
Secondly, a radiotracer experiment was conducted to follow the Pi uptake and transport of R. irregularis and G. aggregatum to both soybean genotypes, and to study the expression of plant Pi transporters that are involved in the plant or mycorrhizal P uptake pathway. For this experiment, the plants were grown in two-compartment systems made out of plastic boxes (series 500/80, Ultraplast, Espergærde, Denmark) with one root compartment (RC) filled with 500 g of substrate (see above) and one hyphal compartment (HC) filled with 100 g of substrate. The HC was separated from the RC by a plastic divider with a central hole closed by two layers of a 50 nm nylon mesh with a 1 mm air gap in between to prevent ion diffusion from the HC to the RC. The membrane allowed fungal hyphae to cross from the RC into the HC, but confined the roots to the RC. Non-mycorrhizal plants were used to confirm that there was no leakage of P from the HC into the RC, and as reference for the gene expression studies. The plants were grown in a growth chamber with a 14 h photoperiod, a 25 °C/20 °C day/night cycle and a light intensity of 280 μmol m–2 s–1. The plants were regularly watered, and every other week a nutrient solution (see above) containing 25 μm P as KH2PO4 (LP) was added to both compartments. The plants were harvested after 7 weeks and, 5 d before the harvest, 1·2 MBq of [33P]orthophosphate (Perkin Elmer, Boston, MA, USA) was added to the HC in a sufficient amount of H2O to ensure a homogeneous penetration of the label into the substrate of the HC.
Plant harvest
After harvest, the plant roots were cleaned with tap water, and root morphological traits, such as total root length, root surface area, root volume and root average diameter, were determined. For the analysis, the computer image analysis software Win-Rhizo Pro (Régent Instruments, Québec, Canada) was used. After the analysis, the roots were cut into segments of 1–2 cm length, and stained with 5 % ink–vinegar solution as described previously (Vierheilig et al., 1998). The mycorrhizal colonization was determined as the percentage of root length colonized by AM fungi using the grid intersection method (McGonigle et al., 1990). The same method was also used to determine the percentage of mycorrhizal roots that were colonized with arbuscules, vesicles or intraradical hyphae. Roots and shoots were dried at 70 °C to determine the dry weight and prepared for the P analysis. The MGRs were calculated using the following formula (Hetrick et al., 1992): MGR = 100 × (AM – NM)/NM, in which AM and NM refer to the total dry biomass of individual mycorrhizal plants or the average of non-mycorrhizal plants, respectively. Similarly, we determined the effect of the different AM fungi on the P content of the plants (mycorrhizal P response; MPR).
For the radiotracer experiment, the plants were harvested 5 d after 33P labelling. At harvest, the shoots were cut off and dried to determine the biomass, and analysed for their 33P content. The roots were washed, weighed and divided into three aliquots; one aliquot was used to determine the fresh to dry weight ratio, to calculate the root biomass and to determine the 33P content of the root; one was used for the AM colonization assay; and one was frozen in liquid nitrogen, and later used for the expression studies of P transporter genes.
P analysis
To measure the non-labelled P in the plant samples, the dried plant material was ground using a tissue homogenizer (Precellys 24, Cayman Chemical Company, Ann Arbor, MI, USA) and an aliquot of the plant material was weighed and digested in 2 m HCl for 2 h at 95 °C. After digestion, P was determined in the samples spectrophotometrically at 436 nm after addition of ammonium molybdate vanadate solution (Fisher Scientific, Pittsburgh, PA, USA). For the 33P analysis, the dried plant material was ground and an aliquot was weighed and digested in the tissue solubilizer TS-2 (Research Product International, Mount Prospect, IL, USA) for 2 h at 45 °C. Then, 150 μL of glacial acetic acid and 2 mL of liquid scintillation cocktail (Biosafe II; Research Product International) were added to each sample. The 33P was measured with a liquid scintillation counter (LS-6500 Multi-Purpose Scintillation Counter, Beckman Instruments, Pullerton, CA, USA), and an internal standard was used to correct the data for differences in counting efficiency.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed according to the manufacturer’s instructions. Briefly, total RNA was extracted using TRIzol (Invitrogen, Grand Island, NY, USA), and the supernatant was purified with the RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA, USA) and eluted into 1 μL of an RNase inhibitor (Murine, New England Biolabs, Ipswich, MA, USA). The first-strand cDNA was synthesized using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). The qRT-PCRs were performed in the ABI 7900 system (Applied Biosystems®, Life Technologies, St Aubin, France) in a final volume of 20 μL using the QuantiTect SYBR Green PCR Kit (Qiagen), 0·6 μm of gene-specific primers and 2 μL of the cDNA template diluted 5-fold. We analysed the expression of all 14 P transporter genes in soybean roots by using the specific primer pairs as shown in Qin et al. (2012). To normalize the data, the soybean housekeeping gene TefS1 (X56856; Qin et al., 2012) was used as a reference gene. The reaction conditions were as follows: 95 °C for 1 min, 40 cycles of 95 °C for 15 s, 58 °C for 30 s and 72 °C for 30 s. Fluorescence data were collected at 72 °C. Changes in gene expression were compared with non-mycorrhizal control plants, and fold induction was calculated using the ΔΔCT method (Winer et al., 1999).
Statistical analyses
Means and standard errors of five biological replicates were calculated using Microsoft Excel 2003 (Microsoft Company, Seattle, WA, USA). The statistical software SAS (SAS Institute Inc., Cary, NC, USA) was used for the two- or three-way analyses of variance (ANOVA; the results are given in Supplementary Data Tables S1–S5). Significant differences among treatments were assessed by Duncan’s multiple comparison test (P ≤ 0·05) using SAS. Significant differences are shown in the figures as different letters on the bars.
RESULTS
Effect of AM fungi on biomass of two soybean genotypes under different P supply conditions
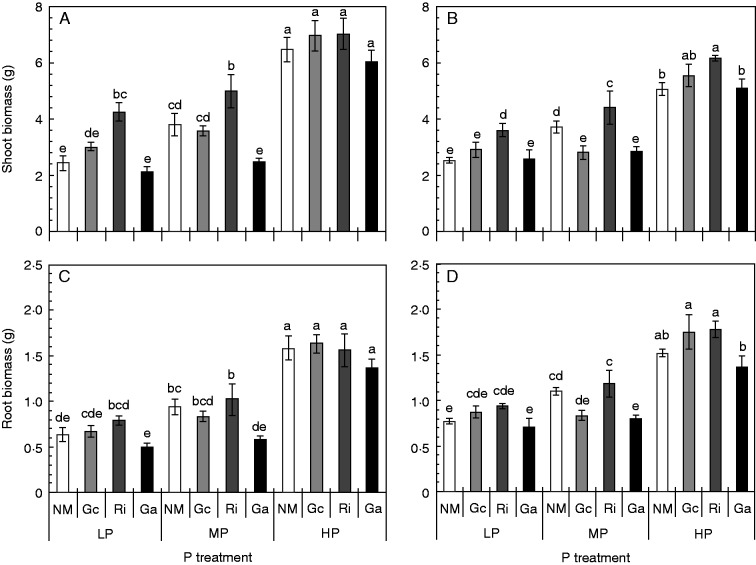
The growth of both soybean genotypes was affected by the P availability and the mycorrhizal fungal species that colonized the root system (Fig. 1). Both shoot and root dry weights significantly increased with higher P availabilities (Fig. 1; Table S1). The genotype HN112 showed a stronger biomass response after an increase in the P supply conditions. Compared with LP conditions, the HP treatment increased the shoot biomass of non-mycorrhizal HN112 plants by 166·5 %, but that of non-mycorrhizal HN89 plants only by 100·7 %. The genotype HN89 had a significantly higher root biomass than HN112 under LP conditions, but the shoot biomass did not differ between the genotypes.
Fig. 1.
Shoot (A, B) and root (C, D) biomass of the two soybean genotypes HN112 (A, C) and HN89 (B, D) after inoculation with three different AM fungal species. The plants were grown for 7 weeks under low (LP), medium (MP) or high (HP) P supply conditions. Shown in the figure are the means of five biological replicates ± s.e.m. NM, non-mycorrhizal controls; Gc, Ri and Ga, plants colonized with Glomus custos, Rhizophagus irregularis or Glomus aggregatum, respectively. ANOVA results are shown in Table S1. Different letters on the bars indicate significant differences among treatments of one soybean genotype according to Duncan’s multiple comparison test (P ≤ 0·05).
Both soybean genotypes showed consistent growth responses after inoculation with the different AM fungi (Fig. 1). Compared with the NM control, R. irregularis led to a significant increase in the shoot biomass of both genotypes under LP and MP availabilities, and of HN89 also under HP supply conditions. In contrast, the inoculation with G. aggregatum had no effect on the shoot biomass under LP conditions, but reduced the shoot biomass of HN112 and HN89 plants under MP conditions. The shoot biomass was generally not affected by an inoculation with G. custos; only at MP did we observe a decrease in the shoot biomass of HN89 plants. The root biomass was only slightly affected by an AM colonization; only plants that were colonized with G. aggregatum had a lower root biomass under MP than non-mycorrhizal control plants (Fig. 1C, D).
The two soybean genotypes differed in their root architectural traits particularly under HP supply conditions, and HN112 was characterized by a longer total root length and a larger root surface area than HN89 (Supplementary Data Fig. S1A, B; Table S2). In contrast, HN89 plants had a slightly larger root average diameter than HN112 plants (Supplementary Fig. S1C). Root architectural traits were affected by the P supply conditions, and HP availabilities led to an increase in the total root length, the root surface area, the root volume and the root average diameter (Fig. S1). In contrast, AM fungal inoculations generally did not have an effect on root architectural traits, and only the colonization of the roots with G. aggregatum led to a reduction in the total root length, the root surface area and the root volume in HN112 under MP conditions (Fig. S1A–D).
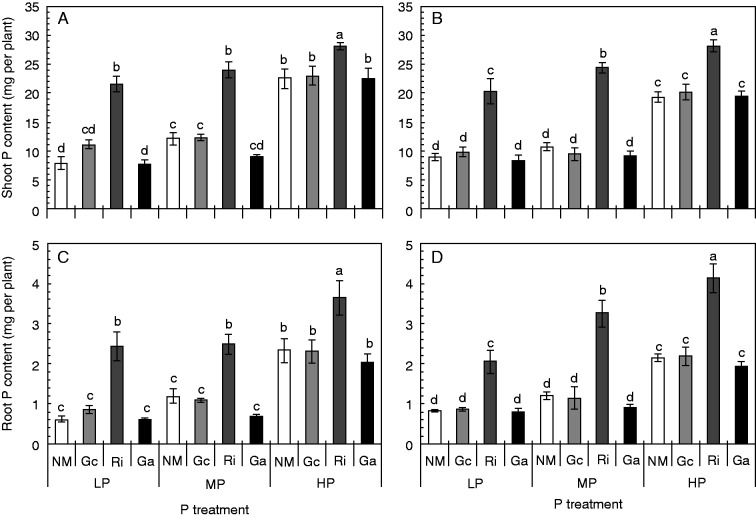
The addition of P significantly increased the total P content in roots and shoots of non-mycorrhizal and mycorrhizal plants of both soybean genotypes (Fig. 2; Table S1). Of the AM fungal species that were tested, only the inoculation with R. irregularis led, under all P supply conditions, to a significant increase in the shoot and root P contents of both soybean genotypes relative to non-mycorrhizal controls. Plants that were colonized with G. aggregatum or G. custos did not differ in their root or shoot P contents from non-mycorrhizal controls (Fig. 2). The mycorrhizal colonization with R. irregularis also led to an increase in the P tissue concentration (P per unit plant biomass) in roots and shoots under LP and MP conditions. However, there was no difference in the shoot P concentrations between non-mycorrhizal or mycorrhizal plants when the plants were supplied with HP (Fig. S2; Table S2).
Fig. 2.
P contents in shoots (A, B) and roots (C, D) of the two soybean genotypes HN112 (A, C) and HN89 (B, D) after inoculation with three different AM fungal species under different P supply conditions. The plants were grown for 7 weeks under low (LP), medium (MP) and high (HP) P supply conditions. Shown in the figures are the means of five biological replicates ± s.e.m. NM, non-mycorrhizal controls, Gc, Ri and Ga, plants colonized with Glomus custos, Rhizophagus irregularis or Glomus aggregatum, respectively. ANOVA results are shown in Table S1. Different letters on the bars indicate significant differences among treatments of one soybean genotype according to Duncan’s multiple comparison test (P ≤ 0·05).
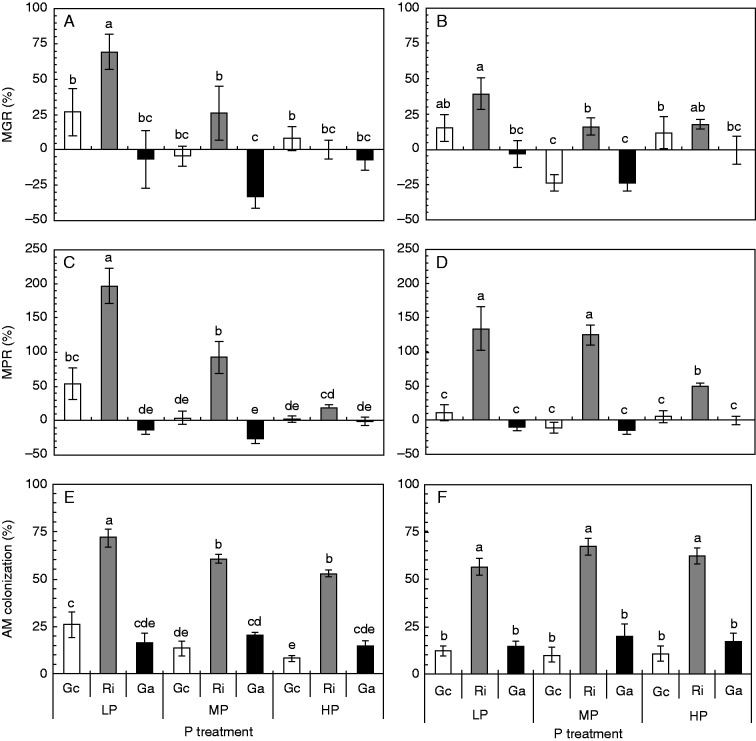
In the first experiment, the non-mycorrhizal plants of both soybean genotypes did not differ in their biomass or their P contents in the shoot under LP conditions (Figs 1 and 2), and both soybean genotypes showed a similar mycorrhizal responsiveness in terms of biomass (Fig. 3A, B) or P contents (Fig. 3C, D). Only at LP, the P-inefficient genotype HN112 showed a slightly higher MGR and MPR than the P-efficient genotype HN89. Rhizophagus irregularis clearly increased the biomass and P uptake of both soybean genotypes under all P supply conditions, while G. aggregatum and G. custos generally led to neutral MGR or MPR (Fig. 3A–D). The colonization with G. aggregatum caused a negative MGR only at MP supply. The positive effects of R. irregularis on soybean growth and P uptake decreased with increasing P availabilities in the soil (Fig. 3A–D).
Fig. 3.
Mycorrhizal growth response (A, B), mycorrhizal phosphate response (C, D) and AM colonization (E, F) of the two soybean genotypes HN112 (A, C, E) and HN89 (B, D, F) after inoculation with three different AM fungal species. The plants were grown for 7 weeks under low (LP), medium (MP) and high (HP) P supply conditions. Shown in the figures are the means of five biological replicates ± s.e.m. Gc, Ri and Ga, plants colonized with Glomus custos, Rhizophagus irregularis or Glomus aggregatum, respectively. ANOVA results are shown in Table S1. Different letters on the bars indicate significant differences among treatments of one soybean genotype according to Duncan’s multiple comparison test (P ≤ 0·05).
We also analysed the percentage root length that was colonized by AM fungi, and found that all AM fungal species formed typical mycorrhizal structures in both soybean genotypes, including ERM, IRM, vesicles and arbuscules. However, while R. irregularis reached colonization rates of up to 70 %, the mycorrhizal colonization rates of G. aggregatum or G. custos were significantly lower in both soybean genotypes (Fig. 3E, F). Increasing P availabilities in the soil led to a reduction in the colonization rates of HN112 with G. custos and R. irregularis (Fig. 3E).
Transport of P to both soybean genotypes via the mycorrhizal uptake pathway
To explore whether the differences in the MGR after the colonization with R. irregularis or G. aggregatum were due to differences in the plant or mycorrhizal P uptake pathway, the transport of P from the ERM to the host plant was followed in two compartment systems. The plants of this experiment were grown in growth chambers under LP conditions, and in this experiment the biomass and the P contents of roots and shoots of HN89 plants were significantly larger than those of HN112 plants (Supplementary Data Fig. S3; Table S1). In contrast to the growth response experiment that was conducted in the greenhouse, the plants in this experiment were exposed to shorter daylengths, and HN112 plants had already started to flower and to develop seed pods. In this experiment, the effect of R. irregularis on biomass and P content was not as large as in the growth response experiment. However, an inoculation with R. irregularis increased the shoot biomass and P content of HN112 plants (Fig. S3A, C; Table S3), and led to a higher root biomass of HN89 plants and higher root P contents in both soybean genotypes compared with NM control plants (Fig. S3B, D; Table S3). Consistent with the previous experiment, an inoculation with G. aggregatum did not have an effect on the biomass and the P contents of both soybean genotypes.
The root colonization rate with R. irregularis was very high in HN112 and HN89 plants (82·5 and 79·5 %, respectively), but the colonization of the root systems with G. aggregatum was significantly lower (12·5 % for HN89 and 67·9 % for HN112) (Supplementary Data Fig. S4A; Table S3). Consistent with the higher root colonization, we found a higher number of arbuscules and vesicles in the mycorrhizal roots of both soybean genotypes that were colonized with R. irregularis (Fig. S4B, C; Table S3), but the root colonization with intraradical hyphae did not differ between both fungi in HN112 (Fig. S4D).
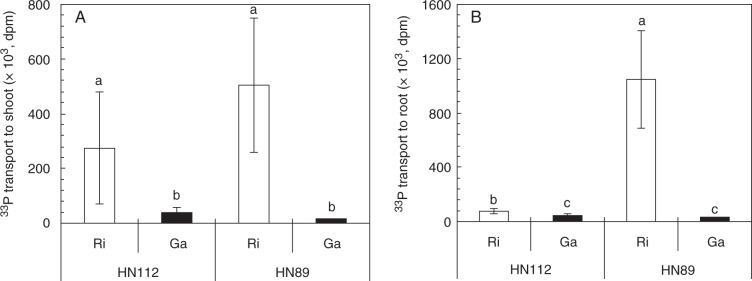
Both fungi translocated P from the ERM to the host plants, but R. irregularis transferred more P than G. aggregatum (Fig. 4) (Table S4). HN112 and HN89 plants that were colonized with R. irregularis had higher count rates in their roots and shoots than plants that were colonized with G. aggregatum. While in HN89 plants a significant proportion of the P that was transferred remained in the root system, the majority of the transferred P was detectable in the shoot of HN112 plants.
Fig. 4.
33P transport by the ERM of R. irregularis (Ri) or G. aggregatum (Ga) to the shoot (A) or the roots (B) of the two soybean genotypes HN112 or HN89. The plants were inoculated with Rhizophagus irregularis (Ri) or Glomus aggregatum (Ga). Shown in the figures are the means of three biological replicates ± s.e.m. ANOVA results are shown in Table S4. Different letters on the bars indicate significant differences among different AM treatments and soybean genotypes according to Duncan’s multiple comparison test (P ≤ 0·05).
Transcript levels of Pi transporter (PHT1) genes
We measured the expression of 14 Pi transporter genes in mycorrhizal and non-mycorrhizal roots of both soybean genotypes under LP conditions and found that the transcript levels of 13 of these transporter genes were affected by the AM colonization. Several transporter genes were downregulated (GmPT1, GmPT2 and GmPT11) by both AM fungi, while others were upregulated by R. irregularis alone (GmPT10) or by both AM fungi (GmPT9) (Fig. 5; Supplementary Data Fig. S5). Other transporter genes such as GmPT4 were induced by a colonization with G. aggregatum, but suppressed by a colonization with R. irregularis in both soybean genotypes (Fig. 5A). The mycorrhiza-inducible Pi transporter gene GmPT9 on the other hand was upregulated in the mycorrhizal roots of both soybean genotypes, but the transcript levels were much higher in HN89 than in HN112 (Fig. 5B). While we found an up to 1100-fold induction of GmPT9 in HN89, the transcript levels were only induced by up to 42-fold in HN112. In contrast, GmPT10, another mycorrhiza-inducible P transporter, was only upregulated (up to 17-fold) in roots that were colonized with R. irregularis, but not in roots that were colonized with G. aggregatum (Fig. 5C). Several other Pi transporter genes, including GmPT3, GmPT5, GmPT6, GmPT8, GmPT12, GmPT13 and GmPT14, were induced by a colonization with G. aggregatum in the P-efficient soybean genotype HN89, but were downregulated or unaffected by R. irregularis (Supplementary Data Fig. S5; Table S5).
Fig. 5.
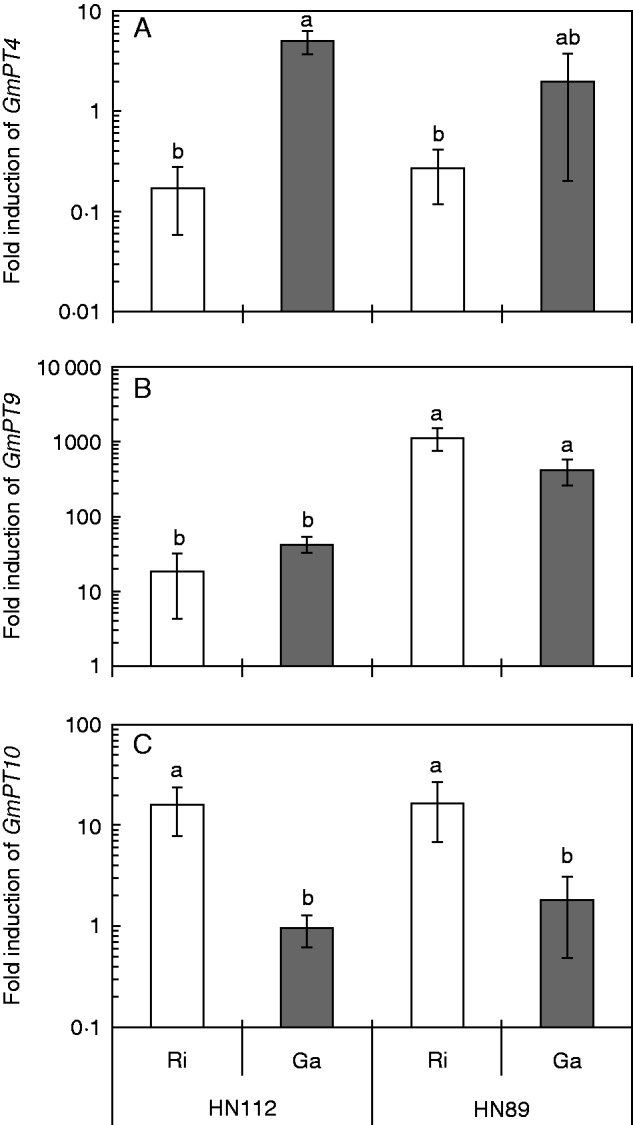
Expression of the high affinity Pi transporter genes GmPT4 (A), and the two mycorrhiza-inducible plant Pi transporter genes GmPT9 (B) and GmPT10 (C) in mycorrhizal roots of the two soybean genotypes HN112 and HN89 after inoculation with Rhizophagus irregularis (Ri) or Glomus aggregatum (Ga). Shown in the figures are the means of three biological replicates ± s.e.m. Different letters on the bars indicate significant differences among different AM treatments and soybean genotypes according to Duncan’s multiple comparison test (P ≤ 0·05). ANOVA results are shown in Table S5. Values <1 represent a decrease and values >1 an increase in transcript levels relative to the NM control.
DISCUSSION
Mycorrhizal growth responses can range from highly beneficial to detrimental, and have been suggested to follow a mutualism to parasitism continuum (Johnson et al., 1997; Johnson and Graham, 2013; Smith and Smith, 2013). Mycorrhizal growth responses are context dependent and are affected by the nutrient supply conditions (Peng et al., 1993), and/or the fungal or plant species involved (Smith et al., 2004; Koch et al., 2006), but the mechanisms that are responsible for the high variability in mycorrhizal benefits are only poorly understood. We studied the effect of different AM fungi on the mycorrhizal growth response and P uptake of two soybean genotypes that differ in their root architecture and PAE to determine whether and how the mycorrhizal dependency of the host affects mycorrhizal growth and P uptake benefits. The MGRs under our experimental conditions ranged from highly positive to negative, but the mycorrhizal growth or P uptake benefits demonstrated a stronger dependency on the fungal partner involved or the P supply conditions than on the PAE of the host (Figs 1–3).
It has been suggested that HN89 with its shallow root system has a higher PAE, because HN89 outperformed HN112 under P-deficient conditions on acid soils (Zhao et al., 2004; Wang et al., 2011) and in soils with toxic aluminium concentrations (Liao et al., 2006). In our experiments, the growth or P uptake responses of non-mycorrhizal HN89 plants were higher than those of HN112 plants under LP conditions (Supplementary Data Fig. S3). Accordingly, the P-inefficient genotype HN112 showed, after an inoculation with R. irregularis, a slightly higher growth and P uptake response under LP supply conditions than the P-efficient genotype HN89 (Fig. 3; Fig. S3A). This is consistent with the findings of Wang et al. (2011), who reported that HN112 benefited more strongly from the co-inoculation with N-fixing bacteria and AM fungi than HN89. Mycorrhizal growth benefits, however, depended more strongly on the fungal species involved, and we found that only R. irregularis increased the plant biomass and P uptake of both soybean genotypes under all P supply conditions. In contrast, G. custos and G. aggregatum did not lead to positive growth or P uptake benefits of either soybean genotype, and G. aggregatum even led to growth depression under MP supply conditions. Fungal species or isolates of one species differ in the efficiency with which they provide nutritional benefits to their host (Koch et al., 2006; Avio et al., 2009; Ehinger et al., 2012; Mensah et al., 2015), and it has been suggested that the compatibility between the fungus and the host plant plays a significant role in these performance differences among various fungal species (Smith et al., 2004). Glomus custos and G. aggregatum, however, also led to smaller growth responses than R. irregularis in Allium porrum and M. truncatula. This supports the results of Kiers et al. (2011), who characterized these G. custos and G. aggregatum isolates as less beneficial compared with R. irregularis. Root organ culture experiments demonstrated that G. aggregatum transfers less P per unit carbon to mycorrhizal roots than R. irregularis (Kiers et al., 2011).
The mycorrhizal colonization of both soybean genotypes with R. irregularis was higher than that with G. custos or G. aggregatum. It seems unlikely, however, that the lack of positive MGR and MPR by an inoculation with G. custos or G. aggregatum was the result of the lower mycorrhizal colonization rates with these fungi. Though R. irregularis reached higher colonization rates than G. custos or G. aggregatum in both experiments (Fig. 3E, F; Supplementary Data Fig. S4A), the colonization rate of HN112 with G. aggregatum was higher and reached 67·9 % in the radiotracer experiment. The higher colonization with G. aggregatum, however, also had no effect on biomass or P content in this experiment (Fig. S3). The results of a recent meta-analysis suggest that while the extent of mycorrhizal colonization can lead to higher MGR or MPR, fungal taxa differ considerably in their degree of host plant benefit per root length colonized (Treseder, 2013). This is also supported by the fact that despite the relatively low colonization rate in the growth response experiment, G. aggregatum caused a significant growth depression under MP conditions (Fig. 1). Other authors have also observed a large negative MGR in plants with relatively low colonization rates (Hetrick et al., 1992; Li et al., 2006; Grace et al., 2009). The negative MGR could be a result of the changes in root architectural traits (Fig. S1) that we observed after an inoculation with G. aggregatum under MP conditions. While there were no significant effects of the AM colonization on the root architectural traits of both genotypes under low or high P supply conditions, an inoculation with G. aggregatum significantly reduced the total root length, the root surface area and the root volume of HN112 roots under MP conditions (HN89 roots showed the same trends, but the differences from the NM controls were not significant).
We observed a decrease in the MGR by R. irregularis with increasing P availabilities in the soil, and a mycorrhizal growth depression in both soybean genotypes after an inoculation with G. aggregatum when more P became available (Figs 1 and 3). Decreasing or negative MGRs have often been described as a result of high P availabilities in the soil (Peng et al., 1993; Kahiluoto et al., 2012). Negative MGRs at high P availabilities have often been attributed to the high carbon costs of the symbiosis (Wright et al., 1998; Graham, 2000) that are not counterbalanced by a net gain in P (Valentine et al., 2001; Jifon et al., 2002). However, to what extent the carbon costs of the symbiosis can explain negative or decreasing MGR under high P availabilities is still under debate (Li et al., 2008; Johnson and Graham, 2013; Smith and Smith, 2013), because the AM fungal colonization also increases the photosynthetic rate (Wright et al., 1998; Zhu et al., 2012), and it has been suggested that the host is able to regulate the carbon costs of the symbiosis, e.g. by a premature degradation of arbuscules under high P supply conditions (Javot et al., 2007; Breuillin et al., 2010). A shorter arbuscular life span could, for example, explain the decrease in the mycorrhizal colonization of HN112 plants with R. irregularis under HP supply conditions (Fig. 3E).
However, it has also been hypothesized that a negative MGR may be the result of a reduced P uptake via the plant uptake pathway (via the epidermis and root hairs) that is not compensated for by an increase in P uptake via the mycorrhizal uptake pathway (via the ERM and IRM), leading to a reduction in total P uptake and P deficiency of the plant (Li et al., 2008; Smith and Smith 2011; Smith et al., 2011). We studied this hypothesis by following the P uptake and transport in multicompartment systems and examined the expression of plant Pi transporters that are involved in the plant uptake pathway or the mycorrhizal uptake pathway. Consistent with the results of the growth response experiment, R. irregularis took up more P from the fungal compartment and transferred more P to both HN89 and HN112 plants than G. aggregatum (Fig. 4). The lower P contents in the shoots and roots of the plants that were colonized with G. aggregatum suggest that the lower efficiency with which this fungus takes up P from the soil is at least in part responsible for the lack of an effect of this fungus on P nutrition.
In soybean, 14 genes of the Pht1 family have been identified (Qin et al., 2012), and the results demonstrate that the expression of most of these transporter genes was affected by the AM symbiosis. Several of these Pi transporter genes, including GmPT1, GmPT2, GmPT4 and GmPT11, are downregulated in mycorrhizal roots, particularly by the AM fungus R. irregularis (Fig. 5A; Supplementary Data Fig. S5). The Pi transporter gene GmPT4, for example, was upregulated in roots that were colonized with G. aggregatum, but downregulated in roots that were colonized with R. irregularis (Fig. 5A). This suggests that the plants that were colonized with G. aggregatum were P deficient compared with the plants that were colonized with R. irregularis. The Pi transporter genes GmPT1, GmPT2, GmPT3, GmPT4, GmPT6, GmPT7, GmPT11, GmPT12 and GmPT13 are predominantly expressed in soybean roots under P starvation (Qin et al., 2012; Fan et al., 2013). The downregulation of several of these Pi transporter genes particularly by R. irregularis could be associated with the P benefits of this fungus and confirms the findings of other authors who demonstrated that Pi transporters that are involved in the plant uptake pathway are often downregulated in mycorrhizal roots (Chiou et al., 2001; Grunwald et al., 2009; Tamura et al., 2012). Consistent with the low P status of these plants, six GmPT genes were particularly induced by G. aggregatum in the P-efficient soybean genotype HN89, but downregulated in the P-inefficient soybean genotype HN112. However, the upregulation of these transporters by G. aggregatum did not lead to an increase in P uptake via the direct uptake pathway, and the P contents in these plants did not differ from that of the NM controls (Figs S3 and S5).
We also studied the expression of the mycorrhiza-inducible Pi transporter genes GmPT8, GmPT9 and GmPT10. These transporter genes are particularly expressed in mycorrhizal roots under P limitation, and cluster with the mycorrhiza-inducible Pi transporter genes OsPT11 from Oryza sativa (rice) and MtPT4 from M. truncatula (Qin et al., 2012; Tamura et al., 2012). GmPT8 and GmPT9 were induced by colonization with R. irregularis and G. aggregatum (Fig. 5), but GmPT10 was only upregulated by R. irregularis. The upregulation of GmPT8, GmPT9 and GmPT10 by R. irregularis is consistent with the positive impact of this AM fungus on the P nutrition of soybeans and suggests that these transporters play a role in the P uptake by the plant from the mycorrhizal interface. Interestingly, GmPT8 was only induced in the P-efficient genotype HN89, but was downregulated in the P-inefficient genotype HN112 (Fig. S5), and the transcript levels of GmPT9 were more strongly induced in HN89 than in HN112 (Fig. 5). This indicates that the higher transcript levels of these transporters could contribute to the efficient AM symbiosis and P uptake of this genotype. However, GmPT9 was also upregulated by G. aggregatum, a fungus that did not have a positive impact on P nutrition, and the genotype HN112 showed a slightly higher mycorrhizal growth and P uptake benefit than HN89 under our experimental conditions, but also in the experiments of other authors (Wang et al., 2011). Tamara and co-authors (2012) examined the expression of GmPT8, GmPT9 and GmPT10 in a time course experiment, and found that GmPT9 was induced much earlier than GmPT8 and GmPT10; they concluded that these three transporter genes play different roles in AM roots. Our results also suggest that GmPT8, GmPT9 and GmPT10 may play different roles in the P transport across the mycorrhizal interface. Of all Pi transporter genes, the expression levels of GmPT10 are most tightly aligned with the mycorrhizal P benefits, suggesting that GmPT10 could serve as a molecular marker for the mycorrhizal growth or P uptake benefit of soybeans. However, more experimental work is needed to support this hypothesis.
Our results confirm that soybeans with a higher P efficiency can also benefit greatly from an AM symbiosis. The P-efficient genotype HN89 showed a slightly lower mycorrhizal MGR or MPR than the P-inefficient genotype HN112 but only under low P availabilities. This is in contrast to results in wheat and barley, in which a decrease in the MPR was found in genotypes with a higher PUE (Baon et al., 1993; Zhu et al., 2001). Our results should have important implications for the development of nutrient-efficient soybean cultivars. The Pi transporter expression clearly demonstrates that the colonization with R. irregularis shifts the P-absorbing surface of the plant from the soil–root interface with epidermis and root hairs to the plant–fungal interface inside the root in both soybean genotypes. There is increasing evidence suggesting that mycorrhizal- or other rhizophere-related traits should be integrated into modern breeding programmes to identify plant and fungal traits that can be used synergistically to increase the P and nutrient uptake efficiency of important crop species, such as soybean.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: effect of AM fungi and P supply conditions on root traits. Figure S2: effect of AM fungi and P supply conditions on the P tissue concentration. Figure S3: biomass and P content of roots and shoots of the two soybean genotypes HN112 and HN89 in the 33P transport experiment. Figure S4: AM colonization characteristics of the two soybean genotypes HN112 and HN89 in the 33P transport experiment. Figure S5: expression of 11 high affinity P transporters in mycorrhizal roots of the two soybean genotypes HN112 and HN89. Table S1: results of the statistical analysis of the effect of P levels or arbuscular mycorrhizal treatments or interactions of the P level and the arbuscular mycorrhizal colonization on growth parameters of the two soybean genotypes HN112 and HN89 in the greenhouse experiment. Table S2: results of the statistical analysis of the effects of P supply conditions, genotypes and arbuscular mycorrhizal treatments and interactions among these treatments on root architectural traits, and shoot or root P concentration in the greenhouse experiment. Table S3: results of the statistical analysis of the effects of soybean genotypes or arbuscular mycorrhizal treatments and interactions between soybean genotypes and mycorrhizal treatments on plant growth, mycorrhizal colonization and 33P transport in the 33P transport experiment. Table S4: results of the statistical analysis of the effects of the soybean genotype or the arbuscular mycorrhizal treatments or the interaction between soybean variety and arbuscular mycorrhizal treatment on 33P transport and expression of plant phosphate transporter genes in the 33P experiment. Table S5: results of the statistical analysis of the effects of the soybean genotypes or the arbuscular mycorrhizal treatments or the interaction between soybean genotype and arbuscular mycorrhizal treatment on expression of plant P transporter genes.
ACKNOWLEDGEMENTS
This work was supported by the China Scholarship Council (CSC, 201208440278) and the National Science Foundation (IOS award 1051397). We thank Carl Fellbaum, Jerry Mensah and Arjun Kafle (South Dakota State University) for technical assistance.
LITERATURE CITED
- Avio L, Cristani C, Strani P, Giovannetti M. 2009. Genetic and phenotypic diversity of geographically different isolates of Glomus mosseae. Canadian Journal of Microbiology 55: 242–253. [DOI] [PubMed] [Google Scholar]
- Baon JB, Smith SE, Alston AM. 1993. Mycorrhizal responses of barley cultivars differing in P efficiency. Plant and Soil 157: 97–105. [Google Scholar]
- Breuillin F, Schramm J, Hajirezaei M, et al. 2010. Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. The Plant Journal 64: 1002–1017. [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Liu H, Harrison MJ. 2001. The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. The Plant Journal 25: 281–293. [DOI] [PubMed] [Google Scholar]
- Cordell D, Rosemarin A, Schroder JJ, Smit AL. 2011. Towards global phosphorus security: a systems framework for phosphorus recovery and reuse options. Chemosphere 84: 747–758. [DOI] [PubMed] [Google Scholar]
- Doner LW, Bécard G. 1991. Solubilization of gellan gels by chelation of cations. Biotechnological Techniques 5: 25–29. [Google Scholar]
- Ehinger MO, Croll D, Koch AM, Sanders IR. 2012. Significant genetic and phenotypic changes arising from clonal growth of a single spore of an arbuscular mycorrhizal fungus over multiple generations. New Phytologist 196: 853–861. [DOI] [PubMed] [Google Scholar]
- Fan CM, Wang X, Hu RB, et al. 2013. The pattern of phosphate transporter 1 genes evolutionary divergence in Glycine max L. BMC Plant Biology 13: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellbaum CR, Mensah JA, Cloos AJ, et al. 2014. Fungal nutrient allocation in common mycelia networks is regulated by the carbon source strength of individual host plants. New Phytologist 203: 645–656. [DOI] [PubMed] [Google Scholar]
- Grace EJ, Cotsaftis O, Tester M, Smith FA, Smith SE. 2009. Arbuscular mycorrhizal inhibition of growth in barley cannot be attributed to extent of colonization, fungal phosphorus uptake or effects on expression of plant phosphate transporter genes. New Phytologist 181: 938–949. [DOI] [PubMed] [Google Scholar]
- Graham JH. 2000. Assessing costs of arbuscular mycorrhizal symbiosis in agroecosystems In: Podila GK, Douds DD, eds. Current advances in mycorrhizae research. University of Michigan: APS Press, 127–140. [Google Scholar]
- Grunwald U, Guo WB, Fischer K, et al. 2009. Overlapping expression patterns and differential transcript levels of phosphate transporter genes in arbuscular mycorrhizal, Pi-fertilised and phytohormone-treated Medicao truncatula roots. Planta 229: 1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J. 2002. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. The Plant Cell 14: 2413–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick BAD, Wilson GWT, Cox TS. 1992. Mycorrhizal dependence of modern wheat varieties, landraces, and ancestors. Canadian Journal of Botany 70: 2032–2040. [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. 2007. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proceedings of the National Academy of Sciences, USA 104 (5): 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jifon JL, Graham JH, Drouillard DL, Syvertsen JP. 2002. Growth depression of mycorrhizal citrus seedlings grown at high phosphorus supply is mitigated by elevated CO2. New Phytologist 153: 133–142. [Google Scholar]
- Johnson NC, Graham JH. 2013. The continuum concept remains a useful framework for studying mycorrhizal functioning. Plant and Soil 363: 411–419. [Google Scholar]
- Johnson NC, Graham JH, Smith FA. 1997. Functioning of mycorrhizal associations along the mutualism–parasitism continuum. New Phytologist 135: 575–585. [Google Scholar]
- Kahiluoto H, Ketoja E, Vestberg M. 2012. Plant-available P supply is not the main factor determining the benefit from arbuscular mycorrhiza to crop P nutrition and growth in contrasting cropping systems. Plant and Soil 350: 85–98. [Google Scholar]
- Kiers ET, Duchamel M, Beesetty Y, et al. 2011. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333: 880–882. [DOI] [PubMed] [Google Scholar]
- Koch AM, Croll D, Sanders IR. 2006. Genetic variability in a population of arbuscular mycorrhizal fungi causes variation in plant growth. Ecology Letters 9: 103–110. [DOI] [PubMed] [Google Scholar]
- Li HY, Smith SE, Holloway RE, Zhu YG, Smith FA. 2006. Arbuscular mycorrhizal fungi contribute to phosphorus uptake by wheat grown in a phosphorus-fixing soil even in the absence of positive growth responses. New Phytologist 172: 536–543. [DOI] [PubMed] [Google Scholar]
- Li H, Smith FA, Dickson S, Holloway RE, Smith SE. 2008. Plant growth depressions in arbuscular mycorrhizal symbioses: not just caused by carbon drain? New Phytologist 178: 852–862. [DOI] [PubMed] [Google Scholar]
- Liang C, Piñeros MA, Tian J, et al. 2013. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiology 161: 1347–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Wan H, Shaff J, Wang X, Yan X, Kochian LV. 2006. Phosphorus and aluminum interactions in soybean in relation to aluminum tolerance. Exudation of specific organic acids from different regions of the intact root system. Plant Physiology 141: 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. 1990. A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytologist 115: 495–501. [DOI] [PubMed] [Google Scholar]
- Mensah JA, Koch AM, Antunes PM, Kiers ET, Hart M, Bücking H. 2015. High functional diversity within species of arbuscular mycorrhizal fungi is associated with differences in phosphate and nitrogen uptake and fungal phosphate metabolism. Mycorrhiza 25: 533–546. [DOI] [PubMed] [Google Scholar]
- Nagy R, Karandashov V, Chague V, et al. 2005. The characterization of novel mycorrhiza specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. The Plant Journal 42: 236–250. [DOI] [PubMed] [Google Scholar]
- Olsen S, Cole C, Watanabe F, Dean L. 1954. Estimation of available phosphorus in soils by extraction with sodium bicarbonate. USDA Circular Nr. 939, US Government Printing Office, Washington, USA.
- Peng S, Eissenstat DM, Graham JH, Williams K, Hodge NC. 1993. Growth depression in mycorrhizal citrus at high-phosphorus supply. Plant Physiology 101: 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N, Zhang XC, Noar RD, Harrison MJ. 2012. Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proceedings of the National Academy of Sciences, USA 109: E665–E672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Guo Y, Chen L, Liang R, Gu M, Xu G, Zhao J, Walk T, Liao H. 2012. Functional characterization of 14 Pht1 family genes in yeast and their expressions in response to nutrient starvation in soybean. PLoS One 7: e47726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosewarne GM, Barker SJ, Smith SE, Smith FA, Schachtman DP. 2000. A Lycopersicon esculentum phosphate transporter (LePT1) involved in phosphorus uptake from a vesicular-arbuscular mycorrhizal fungus. New Phytologist 144: 507–516. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Reid RJ, Ayling SM. 1998. Phosphorus uptake by plants: from soil to cell. Plant Physiology 116: 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder JJ, Smit AL, Cordell D, Rosemarin A. 2011. Improved phosphorus use efficiency in agriculture: a key requirement for its sustainable use. Chemosphere 84: 822–831. [DOI] [PubMed] [Google Scholar]
- Sisaphaithong T, Kondo D, Matsunaga H, Kobae Y, Hata S. 2012. Expression of plant genes for arbuscular mycorrhiza-inducible phosphate transporters and fungal vesicle formation in sorghum, barley and wheat roots. Bioscience Biotechnology and Biochemistry 76: 2364–2367. [DOI] [PubMed] [Google Scholar]
- Smith FA, Smith SE. 2013. How useful is the mutualism–parasitism continuum of arbuscular mycorrhizal functioning? Plant and Soil 363: 7–18. [Google Scholar]
- Smith SE, Smith FA. 2011. Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annual Review of Plant Biology 62: 227–250. [DOI] [PubMed] [Google Scholar]
- Smith SE, Smith FA, Jakobsen I. 2003. Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiology 133: 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Smith FA, Jakobsen I. 2004. Functional diversity in arbuscular mycorrhizal (AM) symbioses: the contribution of the mycorrhizal P uptake pathway is not correlated with mycorrhizal responses in growth or total P uptake. New Phytologist 162: 511–524. [Google Scholar]
- Smith SE, Jakobsen I, Grønlund M, Smith FA. 2011. Roles of arbuscular mycorrhizas in plant phosphorus nutrition: interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiology 156: 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Arnaud M, Hamel C, Vimard B, Caron M, Fortin JA. 1996. Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycological Research 100: 328–332. [Google Scholar]
- Tamura Y, Kobae Y, Mizuno T, Hata S. 2012. Identification and expression analysis of arbuscular mycorrhiza-inducible phosphate transporter genes of soybean. Bioscience, Biotechnology, and Biochemistry 76: 309–313. [DOI] [PubMed] [Google Scholar]
- Treseder KK. 2013. The extent of mycorrhizal colonization of roots and its influence on plant growth and phosphorus content. Plant and Soil 371: 1–13. [Google Scholar]
- Valentine AJ, Osborne BA, Mitchell DT. 2001. Interactions between phosphorus supply and total nutrient availability on mycorrhizal colonization, growth and photosynthesis of cucumber. Scientia Horticulturae 88:177–189. [Google Scholar]
- Vierheilig H, Coughlan AP, Wyss U, Piche Y. 1998. Ink and Vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Applied and Environmental Microbiology 64: 5004–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shen JB, Liao H. 2010a. Acquisition or utilization, which is more critical for enhancing phosphorus efficiency in modern crops? Plant Science 179: 302–306. [Google Scholar]
- Wang X, Yan XL, Liao H. 2010b. Genetic improvement for phosphorus efficiency in soybean: a radical approach. Annals of Botany 106: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pan Q, Chen F, Yan X, Hong L. 2011. Effects of co-inoculation with arbuscular mycorrhizal fungi and rhizobia on soybean growth as related to root architecture and availability of N and P. Mycorrhiza 21: 173–181. [DOI] [PubMed] [Google Scholar]
- Winer J, Jung CKS, Shackel I, Williams PM. 1999. Development and validation of real-time quantitative reverse transcriptase–polymerase chain reaction for monitoring gene expression in cardiac myocetes in vitro. Analytical Biochemstry 270: 41–49. [DOI] [PubMed] [Google Scholar]
- Wright DP, Read DJ, Scholes JD. 1998. Mycorrhizal sink strength influences whole plant carbon balance of Trifolium repens L. Plant Cell and Environment 21: 881–891. [Google Scholar]
- Zhao J, Fu J, Liao H, et al. 2004. Characterization of root architecture in an applied core collection for phosphorus efficiency of soybean germplasm. Chinese Science Bulletin 49: 1611–1620. [Google Scholar]
- Zhu XC, Song FB, Liu SQ, Liu TD, Zhou X. 2012. Arbuscular mycorrhizae improves photosynthesis and water status of Zea mays L. under drought stress. Plant, Soil and Environment 58: 186–191 [Google Scholar]
- Zhu YG, Smith SE, Barritt AR, Smith FA. 2001. Phosphorus (P) efficiencies and mycorrhizal responsiveness of old and modern wheat cultivars. Plant and Soil 237: 249–255. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.