Abstract
Alcohols inhibit γ-aminobutyric acid type A ρ1 receptor function. After introducing mutations in several positions of the second transmembrane helix in ρ1, we studied the effects of ethanol and hexanol on GABA responses using two-electrode voltage clamp electrophysiology in Xenopus laevis oocytes. The 6′ mutations produced the following effects on ethanol and hexanol responses: small increase or no change (T6′M), increased inhibition (T6′V) and small potentiation (T6′Y and T6′F). The 5′ mutations produced mainly increases in hexanol inhibition. Other mutations produced small (3′ and 9′) or no changes (2′ and L277 in the first transmembrane domain) in alcohol effects. These results suggest an inhibitory alcohol binding site near the 6′ position. Homology models of ρ1 receptors based on the X-ray structure of GluCl showed that the 2′, 5′, 6’ and 9′ residues were easily accessible from the ion pore, with 5′ and 6′ residues from neighboring subunits facing each other; L3′ and L277 also faced the neighboring subunit. We tested ethanol through octanol on single and double mutated ρ1 receptors [ρ1(I15′S), ρ1(T6′Y) and ρ1(T6′Y,I15′S)] to further characterize the inhibitory alcohol pocket in the wild-type ρ1 receptor. The pocket can only bind relatively short-chain alcohols and is eliminated by introducing Y in the 6’ position. Replacing the bulky 15′ residue with a smaller side chain introduced a potentiating binding site, more sensitive to long-chain than to short-chain alcohols. In conclusion, the net alcohol effect on the ρ1 receptor is determined by the sum of its actions on inhibitory and potentiating sites.
Keywords: ligand-gated ion channel, transmembrane, homology model, 6′ position, pore, oocyte
INTRODUCTION
Pentameric ligand-gated ion channels (pLGICs), a superfamily that includes γ-aminobutyric acid type A (GABAA), nicotinic acetylcholine (nACh), and glycine (Gly) receptors, are crucial for neurotransmission in the central nervous system and are considered likely targets for alcohol action. GABAA are ionotropic receptors composed of five subunits arranged around a central pore. There are multiple GABAA receptor subunits: α1-6, β1-3, γ1-3, δ, ε, π, θ and ρ1-31. GABAA receptors composed of homomeric ρ subunits provide distinct characteristics that differentiate them from receptors with other subunit arrangements, and were previously termed GABAC receptors2.
Previous research3-5 identified amino acids in the transmembrane (TM) domains of inhibitory pLGICs that are critical for alcohol action. For GABAA receptors, these correspond to I307 (15′, TM2) and W328 (TM3) in ρ1 and S270 and A291 in α1 subunits. GABA ρ1 subunits were useful in this identification process because of their distinct alcohol pharmacology5. The ρ subunits were identified in retina6, 7 but detected in much lower levels in specific regions of the central nervous system8, and thus were initially not considered relevant to alcohol action themselves. However, deletion of ρ1 in mice altered the behavioral responses to alcohol9, suggesting that these receptors are important for alcohol action, even though their levels in brain are low. Also, a genetic correlation of the level of ρ1 expression in nucleus accumbens with ethanol consumption and motor activation was found using BxD recombinant inbred mice [r= 0.77, ethanol preference10 and r= −0.48, ethanol-induced motor response11, from genenetwork.org]. Furthermore, single nucleotide polymorphisms (SNPs) in the human genes that encode ρ1 and ρ2 were associated with alcohol dependence12.
Alcohols potentiate other GABA- and glycine-gated inhibitory pLGICs via binding sites located in TM domains13, 14, while alcohols inhibit wild-type ρ1 receptors15. Single mutants, ρ1(I307S) and ρ1(W328A), and the double mutant ρ1(I307S,W328A) were potentiated by long-chain alcohols, but the ability of ethanol to inhibit these mutants was conserved16. This suggested that there was an inhibitory binding site for short-chain alcohols distinct from the binding site at TM2 15’-TM3. An inhibitory binding site for short-chain alcohols was also suggested by a tryptophan scan of α1 TM2 in α1β2γ2 GABAA receptors17 and was described in αβ and αβγ GABAA receptors at the 6’ position of TM218, the α-helix that lines the channel19. Multiple alcohol binding sites that possess opposing effects have been described in pLGICs. For instance, both potentiating and inhibitory sites for alcohols were identified in TM2 of nACh receptors, the effects of which depended on the alcohol chain length20.
A series of crystal structures from pLGICs such as GluCl21 and a homomeric GABAA receptor22 have paved the way for molecular models of related pLGICs, allowing the study of binding sites for agonists and modulators located at interfaces in the extracellular domain23. Another technique that has shed light on binding sites was receptor photolabeling using modified allosteric modulators, especially in the TM domains, where intersubunit sites were shown to be of particular interest24.
The objective of this study was to define the inhibitory ethanol binding site in the ρ1 subunit through the introduction of mutations in critical sites. We hypothesized that if a residue forming part of an inhibitory binding site was replaced by a larger residue, n-alcohols would not be able to bind due to the bulkier residue occupying the cavity, and therefore the inhibitory alcohol effect would be decreased or even eliminated. We focused on the 6’ position, but also studied neighboring amino acids. We then combined a mutation that eliminates ethanol inhibition (T6’Y) with a previously described mutation that may provide a potentiating binding site for alcohols (I15’S), and studied the effects of alcohols of different chain length on these mutants. We also built homology models of wild-type and mutated ρ1 receptors, based on the the wild-type GluCl, which has been identified as the best template to model the ρ1 receptor out of five different X-ray crystal structures25.
RESULTS AND DISCUSSION
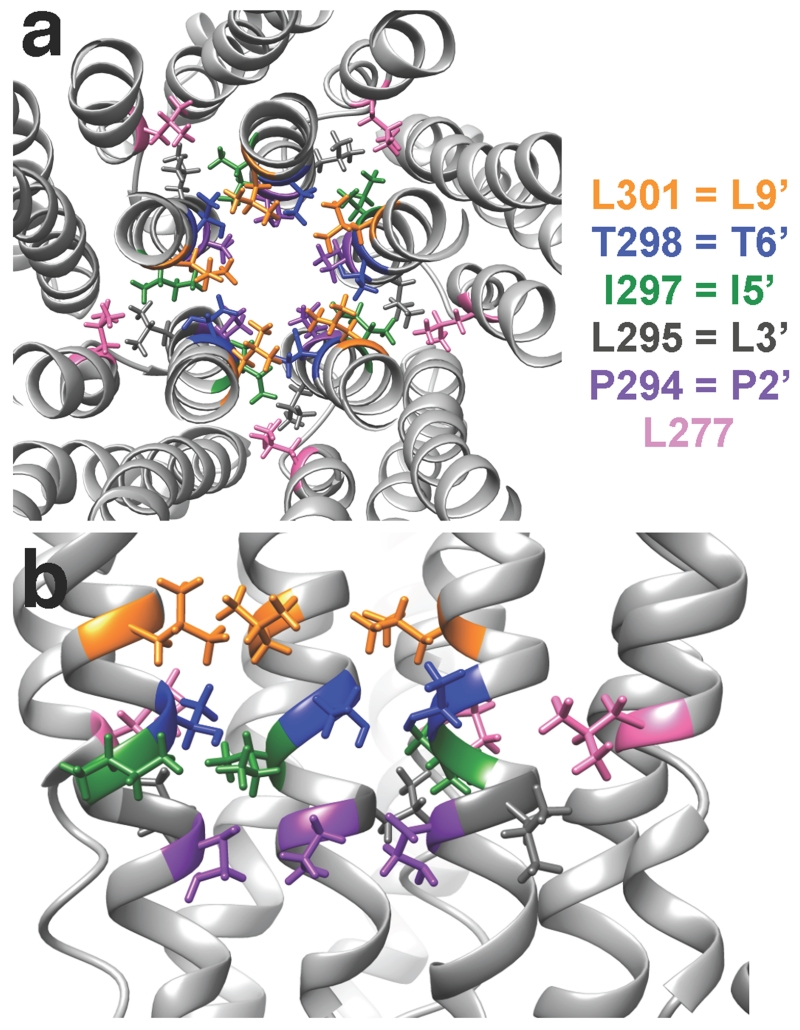
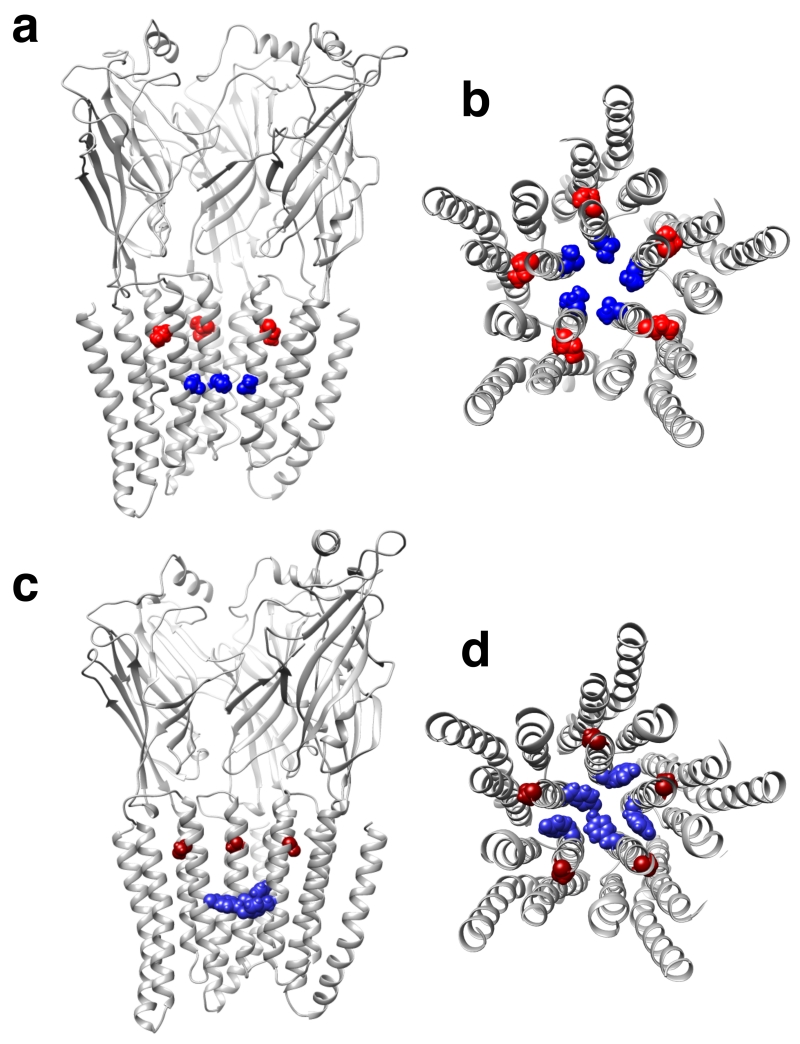
Our approach to identify the inhibitory ethanol binding site in ρ1 was to introduce mutations in the TM2 6’ site, which had been identified as a putative inhibitory binding site for short-chain alcohols in other GABAA receptors17, 18. We hypothesized that a mutation in the region near the inhibitory alcohol binding site would modify the ethanol effect. In addition, we mutated neighboring amino acids in TM2 and TM1. The positions studied are color-coded in the molecular model shown in Fig. 1. The 6′ position is occupied by a threonine in the wild-type receptor. In the model, this residue leans towards the 5′ position in TM2 of the neighboring subunit (7.5 Å Cα-Cα distance), which is essentially facing towards the interface with the neighboring subunit. The leucine in the 9′ position is one turn of the α-helix above T6′; these two residues are quite close (4.4 Å intrasubunit and 7.6 Å intersubunit Cα-Cα distance). The proline in the 2′ position is also lining the pore, and one turn of the α-helix below T6′ (5.7 Å intrasubunit and 9.1 Å intersubunit Cα-Cα distance). The leucine in the 3′ position (TM2) and L277 (TM1) are both facing towards the interface with the neighboring subunit and proximal to I5′ (7.1 Å and 8.7 Å Cα-Cα distance, respectively). Recently, a crystal structure of human α3 glycine receptor bound to strychnine was described26 and the zebrafish α1 glycine receptor was studied using electron cryo-microscopy27. In both cases, the location of the residues homologous to the ones we studied was similar to the one in our model.
Figure 1.
GABAA ρ1 receptor homology model based on GluCl (PDB ID: 3RHW). The second TM helix of each subunit lines the pore of the channel. The amino acids that were mutated are shown as color-coded sticks. All amino acids in the TM2 at the top of panel A and the center of panel B are shown as sticks. A. Extracellular view of the pore from the plane between the extracellular and the TM domains. B. View of three subunits from within the pore.
The GABA concentration-response curves for the ρ1 6′ mutant receptors can be found in Supp. Fig. 1, and the corresponding parameters are in Table 1. Only the T6′F and T6′V mutations significantly changed the GABA sensitivity of the ρ1 receptors.
Table 1.
GABA concentration-response curve parameters. EC50 GABA (effective concentration producing half-maximal response) values are in μM, and 95% confidence intervals are shown in parentheses. Given the variations observed batch to batch, each mutant was compared to WT receptors expressed in the same oocyte batch. nHill is the Hill slope value ± S.E.M.; n is the number of oocytes per receptor. *p< 0.05, ***p< 0.001, ****p< 0.0001 versus WT for logEC50 based on nonlinear regression analysis. The colored text corresponds to the color of the amino acids in the model representations; WT, wild-type.
| Receptor | EC50 GABA (μM) | nHill | n |
|---|---|---|---|
| ρ1 WT | 1.02 (0.83 to 1.26) | 1.8 ± 0.2 | 4 |
| ρ1(L9’F) | 0.48 (0.38 to 0.60)*** | 1.3 ± 0.1 | 5 |
| ρ1 WT | 0.69 (0.65 to 0.75) | 2.3 ± 0.1 | 7 |
| ρ1(T6’Y) | 0.69 (0.60 to 0.79) | 2.1 ± 0.2 | 7 |
| ρ1 WT | 0.78 (0.68 to 0.90) | 1.6 ± 0.1 | 8 |
| ρ1(T6’F) | 0.36 (0.27 to 0.47)*** | 1.9 ± 0.3 | 7 |
| ρ1 WT | 1.30 (1.05 to 1.62) | 1.5 ± 0.1 | 7 |
| ρ1(T6’V) | 6.77 (5.98 to 7.68)**** | 1.4 ± 0.1 | 7 |
| ρ1 WT | 1.11 (0.92 to 1.33) | 1.8 ± 0.2 | 4 |
| ρ1(T6’M) | 0.86 (0.60 to 1.24) | 1.6 ± 0.3 | 4 |
| ρ1 WT | 0.78 (0.67 to 0.92) | 1.7 ± 0.2 | 5 |
| ρ1(I5’A) | 2.20 (1.88 to 2.57)**** | 1.7 ± 0.1 | 4 |
| ρ1 WT | 0.87 (0.74 to 1.03) | 1.7 ± 0.2 | 6 |
| ρ1(I5’V) | 0.18 (0.13 to 0.24)**** | 2.5 ± 0.3 | 6 |
| ρ1 WT | 0.69 (0.60 to 0.79) | 2.2 ± 0.2 | 4 |
| ρ1(I5’F) | 7.47 (6.84 to 8.15)**** | 1.5 ± 0.1 | 4 |
| ρ1 WT | 1.02 (0.81 to 1.29) | 1.8 ± 0.2 | 3 |
| ρ1(I5’W) | 0.40 (0.36 to 0.45)**** | 2.4 ± 0.2 | 4 |
| ρ1 WT | 1.04 (0.75 to 1.45) | 1.5 ± 0.2 | 5 |
| ρ1(L3’F) | 0.31 (0.24 to 0.39)**** | 1.5 ± 0.2 | 5 |
| ρ1 WT | 0.78 (0.64 to 0.96) | 2.1 ± 0.3 | 3 |
| ρ1(P2’G) | 1.62 (1.34 to 1.97)*** | 1.5 ± 0.1 | 4 |
| ρ1 WT | 1.37 (0.82 to 2.30) | 1.2 ± 0.2 | 4 |
| ρ1(L277F) | 0.63 (0.42 to 0.94)* | 1.1 ± 0.2 | 6 |
| ρ1 WT | 0.88 (0.69 to 1.11) | 1.7 ± 0.2 | 6 |
| ρ1(I15’S) | 0.27 (0.23 to 0.32)**** | 2.8 ± 0.4 | 6 |
| ρ1(T6’Y,I25’S) | 0.33 (0.29 to 0.37)**** | 2.2 ± 0.2 | 6 |
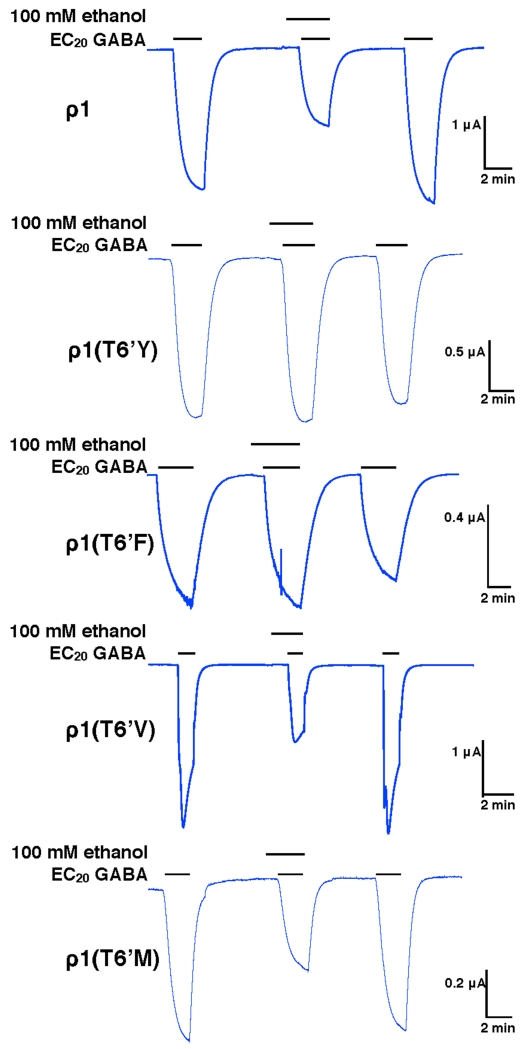
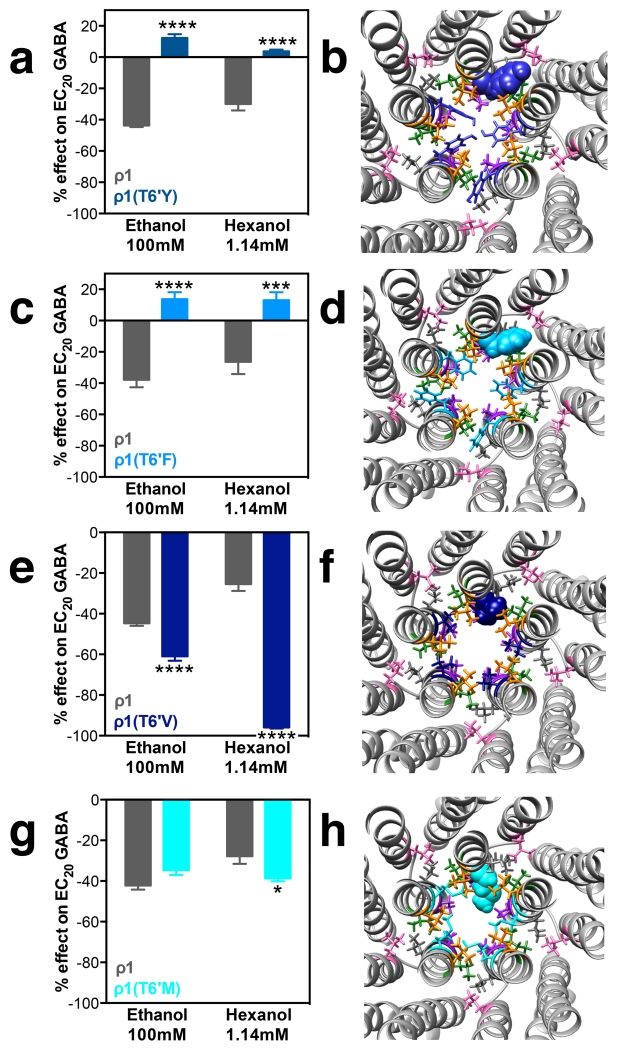
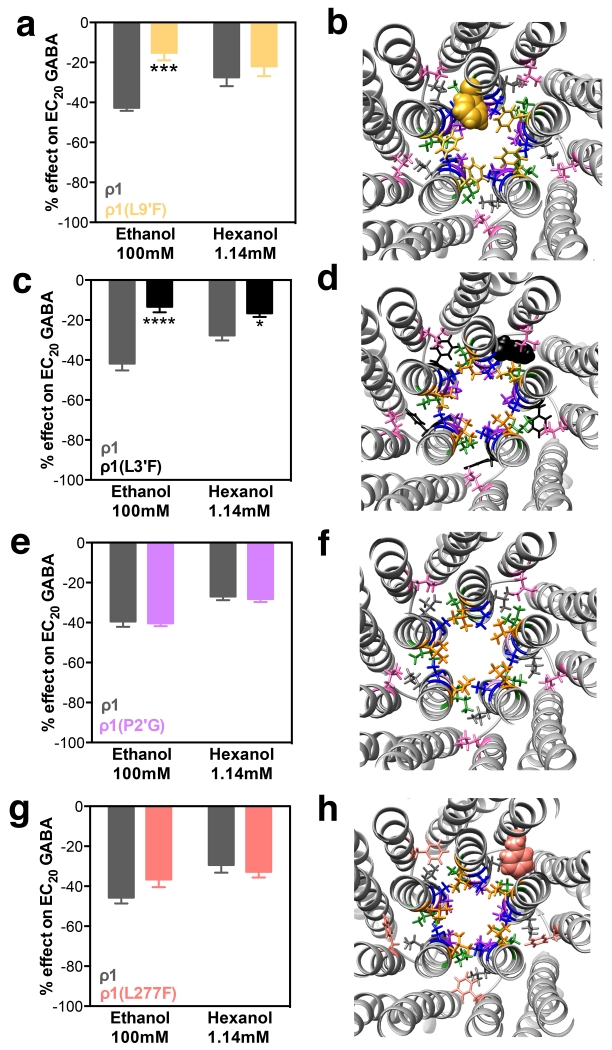
Increasing the residue volume in the 6’ position drastically changed the alcohol effect on GABA-mediated currents in ρ1 receptors (representative tracings can be found in Fig. 2). Instead of inhibition, significant potentiation by both ethanol and hexanol was observed in mutants T6’Y and T6’F (Fig. 3A and C). In the T6’Y mutant model, after optimization of all the side chains in the mutations and relaxation with molecular dynamics, three of the Y residues are protruding into the pore (Fig. 3B) while in the T6’F mutant model, only two F residues are protruding into the pore; the balance of the mutated 6’ residues are tucked between TM2 helices (Fig. 3D). Replacing the T6’ with valine residues had the opposite effect on the alcohol action on GABA currents, increasing the ethanol and hexanol inhibitory effects (Fig. 3E). In the molecular model of this mutant, the small 6’ valine residues are lining the pore (Fig. 3F), similar to the 6′ threonine residues (Fig. 1A). Replacing T6’ with a methionine residue produced small or no changes in alcohol effects compared with wild-type receptors (Fig. 3G). The M6’ residues also line the pore in the molecular model (Fig. 3H). A summary of alcohol effects on 6′ mutants can be found in Table 2.
Figure 2.
Representative tracings of GABA responses in the presence of ethanol in ρ1T6’ mutants expressed in Xenopus laevis oocytes.
Figure 3.
ρ1T6’ mutants: alcohol effects and molecular models. Left panels show ethanol (100 mM) and hexanol (1.14 mM) effects on EC20 GABA responses. Right panels are molecular models of the pore and TM domains of the corresponding ρ1 mutant, viewed from the plane between the extracellular and the TM domains. The graph bars and the corresponding amino acid in the model are the same blue color. A and B, T6’Y; C and D, T6’F; E and F, T6’V; G and H, T6’M. Bars represent means ± SEM, n= 4-9. Two-way ANOVA, Sidak’s multiple comparisons test: *p< 0.05, ***p≤ 0.001, ****p≤ 0.0001.
Table 2.
Summary of alcohol effects on EC20 GABA responses of mutant ρ1 receptors. Symbols: ↑, potentiation; ↓, inhibition; —, no effect. Numbers indicate the percent change of the EC20 GABA responses in the presence of alcohol (+, potentiation; −, inhibition). The colored text corresponds to the color of the amino acids in the model representations; WT, wild-type. The values for ethanol and hexanol effects correspond to the average of all WT receptors in each group. *p< 0.05, **p< 0.01, ***p< 0.001, ****p< 0.000 1 versus WT expressed in oocytes from the same batch as each mutant, as shown in the figures. Analyzed using Two-Way ANOVA with Sidak’s multiple comparisons post-hoc test.
| Receptor | Ethanol | n | Hexanol | n |
|---|---|---|---|---|
| ρ1 WT |
 −42 ± 1 % |
26 |
 −27 ± 3 % |
21 |
| ρ1(T6’Y) |
 12 ± 2 % **** |
7 |
 4 ± 1 % **** |
4 |
| ρ1(T6’F) |
 14 ± 4 % **** |
6 |
 13 ± 5 % *** |
6 |
| ρ1(T6’V) |
 −61 ± 2 % **** |
6 |
 −96 ± 1 % **** |
6 |
| ρ1(T6’M) |
 −35 ± 2 % |
4 |
 −39 ± 1 % * |
6 |
| ρ1 WT |
 −43 ± 1 % |
19 |
 −22 ± 2 % |
18 |
| ρ1(I5’A) |
 −38 ± 3 % |
7 |
 −33 ± 1 % **** |
7 |
| ρ1(I5’V) |
 −50 ± 4 % |
4 |
 −36 ± 1 % |
4 |
| ρ1(I5’F) |
 −40 ± 3 % |
4 |
 −37 ± 2 % ** |
4 |
| ρ1(I5’W) |
 −57 ± 2 % ** |
5 |
 −52 ± 2 % **** |
5 |
When introducing a mutation, there is the possibility of modifying the binding or gating effects of the agonist. To confirm that GABA is still a full agonist in ρ1(T6′Y), we tested 100 mM ethanol on maximal GABA responses in wild-type and mutant ρ1 receptors. If GABA had become a partial agonist at the mutant receptors, we would have seen a differential effect of ethanol. The ethanol modulation at maximal GABA was small and similar in both wild-type and mutant (20 ± 10 and 10 ± 3 % potentiation, respectively; n=4, data not shown).
When the alcohol effect (expressed as % change of the EC20 GABA responses) for each of these mutants was graphed as a function of the amino acid volume present in the 6’ position, about 50-80% of the variance is explained by the size of the amino acids. A positive linear correlation was established for ethanol (r2= 0.79, p< 0.05, Supp. Fig. 2A), but it did not reach significance for hexanol (r2= 0.50, p=0.18, Supp. Fig. 2B). In contrast, there was no correlation between the alcohol effect and the hydrophobicity (r2= 0.24, Supp. Fig. 2C and r2= 0.36, Supp. Fig. 2D). The ethanol effect had no linear correlation with the GABA EC50 for any of the 6’mutants (r2= 0.52, Supp. Fig. 2D), but the hexanol effect was correlated with the GABA EC50 (r2= 0.81, p=0.03, Supp. Fig. 2F). However, this correlation is not meaningful, as it heavily depends on the GABA EC50 value for T6′V, which is markedly different from all the other mutants.
These results define an inhibitory binding site for alcohols at T6’ in the ρ1 receptor; mutation of ρ1T6’ changed both the ethanol and hexanol inhibitory effects (larger amino acids decreased alcohol inhibition, and a smaller amino acid increased alcohol inhibition). This is consistent with larger residues occupying the pocket and preventing alcohol binding and subsequently inhibiting receptor function. There was a correlation between the amino acid residue size and ethanol effect, but not between the GABA affinity (apparent EC50) and ethanol effect. These results suggest a direct effect of the size of amino acids lining an alcohol binding pocket and not an indirect effect of ethanol on GABA binding and channel gating.
The GABA concentration-response curves for the ρ1 5′ mutant receptors are shown in Supp. Fig. 3, and the corresponding parameters appear in Table 1. All the mutations changed the ρ1 receptor GABA sensitivity.
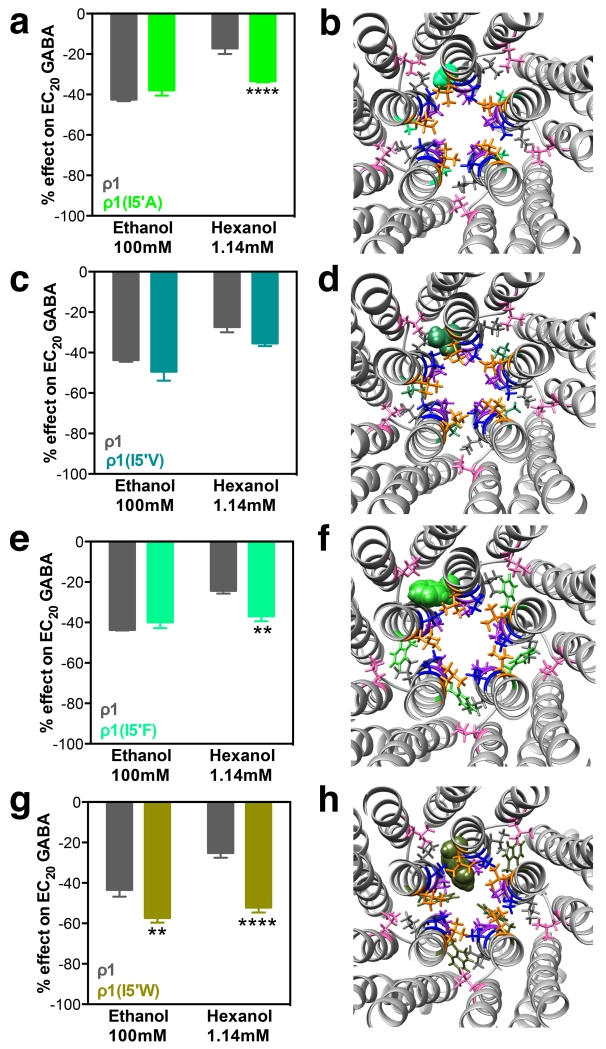
Only the replacement of I5′ with the large residue tryptophan increased the ethanol inhibition; the other mutations at the 5′ position did not alter ethanol action (Fig. 4A, C, E and G). In contrast, almost all substitutions significantly increased the hexanol inhibition (Fig. 4A, C, E and G). In the I5′A, I5′V and I5′F mutant models, after optimization of the side chains in the mutations and relaxation with molecular dynamics, all the residues are tucked into the interface between TM2 helices (Fig. 4B, D and F). In the I5′W mutant model, the residues are still located at the interface, but the larger bulk of the tryptophan causes part of the residue chains to protrude out into the channel. A summary of alcohol effects on 5′ mutants can be found in Table 2.
Figure 4.
ρ1I5’ mutants: alcohol effects and molecular models. Left panels show ethanol (100 mM) and hexanol (1.14 mM) effects on EC20 GABA responses. Right panels are molecular models of the pore and TM domains of the corresponding ρ1 mutant, viewed from the plane between the extracellular and the TM domains. The graph bars and the corresponding amino acid in the model are the same green color. A and B, I5′A; C and D, I5′V; E and F, I5′F; G and H, I5′W. Bars represent means ± SEM, n= 4-9. Two-way ANOVA, Sidak’s multiple comparisons test: **p≤ 0.01, ****p≤ 0.0001.
When the alcohol effect (expressed as % change of EC20 GABA responses) for each of these 5′ mutants was graphed as a function of the amino acid volume present in the 5’ position, there was no correlation for either ethanol (r2= 0.45, Supp. Fig. 4A) or hexanol (r2= 0.35, Supp. Fig. 4B); however, about 35-45% of the variance was explained by the size of the amino acids. For hydrophobicity, there was no correlation for ethanol (r2= 0.41, Supp. Fig. 4C) but a positive linear correlation was established for hexanol (r2= 0.85, p<0.05, Supp. Fig. 4D). In addition, there was no correlation between the alcohol effects and the GABA EC50 for each of the 5’ mutants (r2= 0.34, Supp. Fig. 4E and r2= 0.00, Supp. Fig. 4F).
The GABA concentration-response curves for the neighboring mutations in ρ1 receptors are shown in Supp. Fig. 5, and the corresponding parameters can be found in Table 1. All the mutations modified the ρ1 receptor GABA sensitivity.
Some of the amino acids near the 5′ and 6’ positions also affected alcohol action on ρ1 receptors, but to a lesser degree. A decrease in ethanol inhibition was observed in the L9’F mutant (this particular mutation was chosen because the resulting receptor is not spontaneously open28), but the effect of hexanol was not changed (Fig. 5A). In the molecular model, all 9’F residues were lining the pore (Fig. 5B). The L3’F mutant showed a decrease both in ethanol and hexanol inhibition (Fig. 5C). All 3’F residues were located in the center of the four α-helix bundle within a single subunit (Fig. 5D). Lastly, there were two mutants that did not change either ethanol or hexanol inhibition: ρ1(L277F) and P2’G (Fig. 5E and G). The residue at position 277 is located in TM1, in the interface between α-helices from neighboring subunits for both wild-type (leucine, Fig. 1A) and the mutant (phenylalanine, Fig. 5F). The P2′G mutant lacks a side chain, but it remains lining the pore (Fig. 5H).
Figure 5.
Neighboring ρ1 mutants in TM2 and TM1: alcohol effects and molecular models. Left panels show ethanol (100 mM) and hexanol (1.14 mM) effects on EC20 GABA responses. Right panels are molecular models of the pore and TM domains of the corresponding ρ1 mutant, viewed from the plane between the extracellular and the TM domains. The graph bars and the corresponding amino acid in the model are the same color. A and B, L9’F; C and D, L3’F; E and F, P2′G; G and H, L277F. Bars represent means ± SEM, n= 3-5. Two-way ANOVA, Sidak’s multiple comparisons test: *p< 0.05, ***p≤ 0.001, ****p≤ 0.0001.
In summary, neighboring amino acids also participate in alcohol action sites, and mutation of ρ1L9’ decreased ethanol but not hexanol inhibition. Mutation of ρ1I5’ mainly modified the hexanol effect. Mutation of L3’decreased ethanol inhibition, and to a lesser degree, hexanol inhibition.
The mutations affected alcohol inhibition in a specific manner, indicating that the changes in alcohol effects are not due to general changes in receptor function produced by the mutation itself. In fact, two nearby mutations did not alter alcohol effects: one in TM2 (P2’G) and one located near I5’ [ρ1(L277F) in TM1]. Several of the residues studied (2′, 6′ and 9′) line the pore of the channel in the ρ1 receptor, and had been shown to have an important role in receptor function and pharmacology. For instance, residues at the 6′ and 9′ positions are important for channel gating29. Also, residues at the 2′ and 6′ positions are critical for inhibition by picrotoxinin30, 31 and some neurosteroids32, 33. However, even though P2′ has a profound effect on the ion selectivity, conductance, and pharmacology of ρ1 receptors34-37, its mutation to glycine had no effect on alcohol inhibition, suggesting that the changes observed in alcohol effects due to the studied mutations are not due simply to non-specific changes in the receptor function, but to modifications in sites specific to alcohol action.
The residue in the 15’ position has been shown to be critical for alcohol action, both in ρ1 as well as other inhibitory receptors5, 16, but it appears to fulfill a different role in nACh receptors38. Based on previous research17, 18, we hypothesized that an inhibitory alcohol site in ρ1 involved the 6′ position and a stimulatory alcohol binding site could be introduced in 15′. We tested this hypothesis by introducing mutations at 6’ and/or 15’ (Fig. 6A and B show T6′ and I15′ in the wild-type ρ1, and Fig. 6C and D show Y6′ and S15′ in the ρ1 double mutant). Four different ρ1 receptors were examined: wild-type, single mutants T6’Y and I15’S, and the double mutant T6’Y, I15’S. The GABA concentration-response curves for these mutants can be found in Supp. Fig. 6, and the corresponding parameters in Table 1.
Figure 6.
6’ (blue) and 15’ (red) positions in molecular models of the ρ1 receptor. View of three subunits from the pore for wild-type ρ1 (A) and ρ1(T6′Y,I15′S) (C). Extracellular view of the pore from the plane between the extracellular and the TM domains for wild-type ρ1 (B) and ρ1(T6′Y,I15′S) (D).
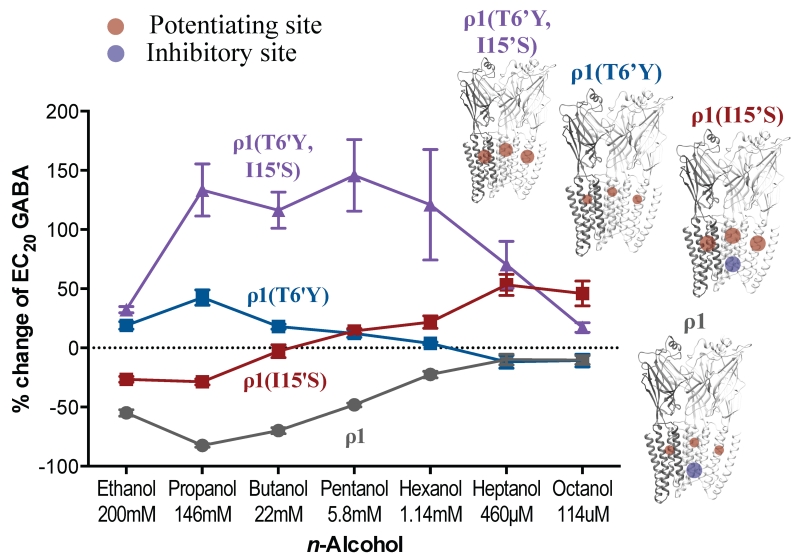
The alcohol effect on EC20 GABA responses was measured in these four receptors (Fig. 7). The wild-type GABAA ρ1 receptor was inhibited by alcohols containing up to six carbons. The ρ1(I15’S) mutant was potentiated by alcohols with chains longer than five carbons, but still inhibited by ethanol and propanol; butanol produced no net effect. The ρ1(T6’Y) mutant showed moderate potentiation by ethanol through pentanol; hexanol had no effect, and heptanol and octanol produced small inhibition. The double mutant ρ1(T6’Y, I15’S) displayed potentiation across all of the n-alcohols tested. A summary of the alcohol effects on these receptors can be found in Table 3.
Figure 7.
Alcohol effect on single mutants, ρ1(T6’Y) and ρ1(I15’S), and double mutant ρ1(T6’Y,I15’S). The circles in the molecular models serve only as a visual representation of the approximate position and relative size of the potentiating (red circle) and inhibitory (blue circle) alcohol sites.
Table 3.
Summary of alcohol effects on EC20 GABA responses of mutant ρ1 receptors. Symbols: ↑, potentiation; ↓, inhibition; —, no effect. Numbers indicate the percent change of the EC20 GABA responses in the presence of alcohol (+, potentiation; −, inhibition). The colored text corresponds to the color of the amino acids in the model representations; WT, wild-type.
| Receptor | Ethanol | Propanol | Butanol | Pentanol | Hexanol | Heptanol | Octanol |
|---|---|---|---|---|---|---|---|
| ρ1 WT |
 −55% |
 −83% |
 −70% |
 −48% |
 −23% |
— −10% |
— −10% |
| ρ1(T6’Y) |
 +19% |
 +43% |
 +18% |
— +12% |
— +4% |
— −12% |
— −11% |
| ρ1(I15’S) |
 −27% |
 −29% |
— −3% |
— +14% |
 +22% |
 +53% |
 +46% |
| ρ1(T6’Y,I15’S) |
 +32% |
 +133% |
 +116% |
 +146% |
 +121% |
 +70% |
 +17% |
Previous studies16 have shown that if the residue in the 15’ position is large (e.g., isoleucine in the wild-type), all alcohols inhibit the ρ1 receptor; in contrast, if the residue is small (e.g., serine, as in the α1 GABAA and Gly receptor subunits), ethanol inhibits but longer chain alcohols potentiate the GABA responses in the ρ1 mutant. Considering those results together with the present ones, the existence of two different binding sites for alcohols is a strong possibility: an inhibitory binding site for alcohols lined by the 6’ residue and the possibility of introducing an excitatory binding site (where mainly long-chain alcohols act) when the 15’ residue is replaced by a smaller one. Replicating earlier results15, alcohols up to six carbons long inhibited the wild-type GABAA ρ1 receptor in our study. This could be the result of alcohols up to hexanol binding in the inhibitory binding site at 6’. Alcohols showed a dual effect on the ρ1(I15’S) mutant, with short-chain alcohols inhibiting and long-chain alcohols potentiating the responses; the crossover point was for butanol, which produced no net effect. In our model, reducing the size of the 15’ residue creates a potentiating binding site where alcohols can also bind, in addition to the inhibitory site at 6’; therefore, the alcohol effect will be the combined result of its effect on both sites. Alcohols like ethanol and propanol act mainly at the 6’ inhibitory site, while butanol has an equal effect on both sites, and pentanol and longer alcohols act mainly at the 15’ potentiating site. Short-chain alcohols produced moderate potentiation in the ρ1(T6’Y) mutant, with long-chain alcohols producing no effect or small inhibition.
In terms of our model, a large residue at 6’ would eliminate the inhibitory site, leaving only a small and/or not very efficient potentiating site at 15’. Therefore, the combined effects are very small and pertain only to short-chain alcohols. In support of our hypothesis, all the n-alcohols tested produced potentiation of the double mutant ρ1(T6’Y, I15’S). According to our model, the 6’ inhibitory site has been eliminated in this mutant and a 15’ potentiating site has been introduced; therefore, all alcohols act through the 15’ potentiating site, including the long-chain alcohols like hexanol, that have little to no effect on the wild-type and other mutant receptors. Evidence for a similar situation, with dual and opposing sites of action for alcohols, was found in GABAA18, GLIC39-41 and nACh receptors20.
The molecular simulations often yielded a pentamer without symmetry in the pore region, where the homologous side chains are located in different rotamer positions even though the backbone of TM2 is conserved. In the case of the residues of interest, some of the side chain residues are in the channel and others are between the subunits. Different approaches were tested, including using an algorithm to find the “best” rotamer for each side chain versus not adjusting rotamers, and using both short and long optimizations, etc.; however, the asymmetry persisted. Because of this, any pore profile or evaluation of changes in intrasubunit dimension spaces would be arbitrary.
This molecular modeling provides a static model of the protein, but it is worth keeping in mind that conformational movements result in very dynamic pLGICs, even in regions such as TM239, 42. The mobility of TM2 in response to different agonists and modulators has been explored using labeling with fluorophores43 and crosslinking of mutated cysteines44, including asymmetric rotations in the area of the 6′ position45. These studies also revealed a surprising degree of mobility in this domain.
A recent study in ELIC (a bacterial pLGIC) showed that isoflurane, a volatile anesthetic that presents functional similarities to alcohol, inhibited this receptor46. Co-crystallization of ELIC with isoflurane revealed that the anesthetic occupied sites inside the pore near the 6′ and 13′ positions. Electrophysiological recordings showed that mutations at these positions modify isoflurane inhibition, providing further support to the idea of an inhibitory binding site located in the pore.
In summary, we have identified an inhibitory alcohol binding site on the GABAA receptor near the pore of the channel. Not only it is possible to eliminate this inhibitory binding site through mutagenesis, it is also possible to introduce a potentiating alcohol binding site analogous to those present in other GABAA and Gly receptors, such that a net effect of alcohol on the ρ1 receptor is determined by the sum of its actions on inhibitory and potentiating sites. This suggestion is consistent with dual sites found in other pLGICs, such as GABAA18, GLIC39-41 and nACh receptors20. The present results warrant construction of mice with mutated ρ1 subunits that either lack ethanol inhibition or display ethanol potentiation, enabling behavioral studies of the role of ethanol on ρ1 function in vivo.
METHODS
Clones
Mutations in the ρ1 cDNAs were made through site-directed mutagenesis using QuikChange (Agilent Technologies, Santa Clara, CA, USA).
Transcription and oocyte injection
The in vitro transcription of wild-type and mutant ρ1 subunits was performed using mMessage mMachine (Life Technologies, Grand Island, NY, USA). Surgery of Xenopus laevis frogs was performed according to an approved institutional protocol. After manual isolation of Xenopus laevis oocytes, they were injected with complementary RNAs (cRNAs, 5 ng/oocyte) encoding wild-type or mutant ρ1 subunits. The injected oocytes were incubated at 15°C in sterilized Barth’s solution for 3-7 days before recording.
Electrophysiological recordings
The responses of GABAA ρ1 receptors expressed in oocytes were studied through two-electrode voltage clamp. The oocyte was placed in a chamber perfused with ND96 buffer (96 mM NaCl, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, pH 7.5), and voltage-clamped at −70 mV. GABA applications lasted for 30-150 s and the interval between applications was 5-15 min.
Concentration-response curves
Increasing concentrations of GABA were applied (0.1-100 μM) and responses were expressed as percentages of the maximal current.
Alcohol application protocol
Alcohols were first pre-applied for 1 min and then co-applied with GABA. The application sequence for the wild-type and mutants ρ1 receptors was as follows: maximal GABA (to obtain maximal response, EC100), EC20 GABA, EC20 GABA, pre-application of ethanol (100 mM) immediately followed by a co-application with EC20 GABA, EC20 GABA, pre-application of hexanol (1.14 mM) immediately followed by a co-application with EC20 GABA, EC20 GABA. The response to GABA in the presence of the modulator was expressed as a % change compared to the mean of the previous and subsequent GABA responses. In the past, we have used EC5 to assess alcohol effects, as the alcohol effect is larger in the presence of lower EC values. However, we used EC20 in these studies because currents in some of the mutant receptors were greatly diminished.
Statistical analysis
Statistical analysis was performed using Prism 6 (GraphPad Software, La Jolla, CA, USA). Pooled data are represented as mean ± S.E.M. Statistical significance was determined using two-way analysis of variance (ANOVA). Nonlinear regression analysis for the concentration-response curves was performed after normalizing the agonist responses in each cell to the maximal current that could be elicited by GABA. The concentration-response curves were fitted to the following equation:
where I/IMAX is the fraction of the maximally-obtained GABA response, EC50 (effective concentration 50) is the concentration of GABA producing a half-maximal response, [GABA] is GABA concentration, and nH is the Hill coefficient. Linear correlation analysis was also carried out using Prism 7, using the following sources for the independent variables: amino acid volume (calculated by J.R. Trudell), hydrophobicity47, and EC50 GABA (determined in this study, Table 1).
Molecular models
Homology models of ρ1 GABAA receptors were built by threading the primary sequence of five subunits onto the X-ray structure of GluCl (Protein Data Bank ID code 3RHW21). The Modeler module of Discovery Studio 4.1 (DS 4.1Biovia, San Diego, CA, USA) was used to build 50 models of the wild-type and each mutation. We chose the ‘best’ models based on the force field potential energies measured by the Discovery Studio version of CHARMm. The ‘side-chain refinement’ module of DS 4.1 was used to optimize the side chain rotamers in the model while the backbone atoms were fixed. Then all backbone atoms were tethered with a quadratic restraint of 10 kcal/Å2 and optimized the resulting models to a gradient of 0.001 kcal/ (mol × Å). We performed an optimization of each model and then a molecular dynamics ‘relaxation’ at 300 K using the default parameters of DS 4.1. All of the molecular graphics were created using the UCSF Chimera package version 1.748.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by the Waggoner Center for Alcohol and Addiction Research and by grants from the National Institutes of Health: AA006399 (RAH) and R01AA020980 (JRT). We thank Dr. Jody Mayfield for excellent editorial assistance.
ABBREVIATIONS
- EC
effective concentration
- cRNA
complementary RNA
- GABA
γ-aminobutyric acid
- GLIC
Gloeobacter violaceus ligand-gated ion channel
- GluCl
Caenorhabditis elegans glutamate-gated chloride channel
- Gly
glycine
- nACh
nicotinic acetylcholine
- pLGIC
pentameric ligand-gated ion channel
- TM
transmembrane
Footnotes
SUPPORTING INFORMATION: Figure 1: GABA concentration-response curves for ρ1T6′ mutants. Figure 2: Linear correlation between alcohol effects on ρ1T6′ mutants and several variables. Figure 3: GABA concentration-response curves for ρ1I5′ mutants. Figure 4: Linear correlation between alcohol effects on ρ1I5′ mutants and several variables. Figure 5: GABA concentration-response curves for neighboring ρ1 mutants in TM2 and TM1. Figure 6: GABA concentration-response curves for ρ1T6′Y and/or I15′S mutants.
REFERENCES
- [1].Sigel E, Steinmann ME. Structure, function, and modulation of GABA(A) receptors. J Biol Chem. 2012;287:40224–40231. doi: 10.1074/jbc.R112.386664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Martínez-Delgado G, Estrada-Mondragón A, Miledi R, Martínez-Torres A. An Update on GABAρ Receptors. Curr Neuropharmacol. 2010;8:422–433. doi: 10.2174/157015910793358141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lobo IA, Trudell JR, Harris RA. Cross-linking of glycine receptor transmembrane segments two and three alters coupling of ligand binding with channel opening. J Neurochem. 2004;90:962–969. doi: 10.1111/j.1471-4159.2004.02561.x. [DOI] [PubMed] [Google Scholar]
- [4].Mascia MP, Trudell JR, Harris RA. Specific binding sites for alcohols and anesthetics on ligand-gated ion channels. Proc Natl Acad Sci U S A. 2000;97:9305–9310. doi: 10.1073/pnas.160128797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mihic SJ, Ye Q, Wick MJ, Koltchine VV, Krasowski MD, Finn SE, Mascia MP, Valenzuela CF, Hanson KK, Greenblatt EP, Harris RA, Harrison NL. Sites of alcohol and volatile anaesthetic action on GABA(A) and glycine receptors. Nature. 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- [6].Cutting GR, Curristin S, Zoghbi H, O’Hara B, Seldin MF, Uhl GR. Identification of a putative γ-aminobutyric acid (GABA) receptor subunit rho2 cDNA and colocalization of the genes encoding rho2 (GABRR2) and rho1 (GABRR1) to human chromosome 6q14-q21 and mouse chromosome 4. Genomics. 1992;12:801–806. doi: 10.1016/0888-7543(92)90312-g. [DOI] [PubMed] [Google Scholar]
- [7].Cutting GR, Lu L, O’Hara BF, Kasch LM, Montrose-Rafizadeh C, Donovan DM, Shimada S, Antonarakis SE, Guggino WB, Uhl GR, et al. Cloning of the γ-aminobutyric acid (GABA) ρ1 cDNA: a GABA receptor subunit highly expressed in the retina. Proc Natl Acad Sci U S A. 1991;88:2673–2677. doi: 10.1073/pnas.88.7.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Boue-Grabot E, Roudbaraki M, Bascles L, Tramu G, Bloch B, Garret M. Expression of GABA receptor ρ subunits in rat brain. J Neurochem. 1998;70:899–907. doi: 10.1046/j.1471-4159.1998.70030899.x. [DOI] [PubMed] [Google Scholar]
- [9].Blednov YA, Benavidez JM, Black M, Leiter CR, Osterndorff-Kahanek E, Johnson D, Borghese CM, Hanrahan JR, Johnston GA, Chebib M, Harris RA. GABA(A) receptors containing ρ1 subunits contribute to in vivo effects of ethanol in mice. PLoS One. 2014;9:e85525. doi: 10.1371/journal.pone.0085525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gill K, Liu Y, Deitrich RA. Voluntary alcohol consumption in BXD recombinant inbred mice: relationship to alcohol metabolism. Alcohol Clin Exp Res. 1996;20:185–190. doi: 10.1111/j.1530-0277.1996.tb01063.x. [DOI] [PubMed] [Google Scholar]
- [11].Demarest K, Koyner J, McCaughran J, Jr., Cipp L, Hitzemann R. Further characterization and high-resolution mapping of quantitative trait loci for ethanol-induced locomotor activity. Behav Genet. 2001;31:79–91. doi: 10.1023/a:1010261909853. [DOI] [PubMed] [Google Scholar]
- [12].Xuei X, Flury-Wetherill L, Dick D, Goate A, Tischfield J, Nurnberger J, Jr., Schuckit M, Kramer J, Kuperman S, Hesselbrock V, Porjesz B, Foroud T, Edenberg HJ. GABRR1 and GABRR2, encoding the GABA-A receptor subunits ρ1 and ρ2, are associated with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:418–427. doi: 10.1002/ajmg.b.30995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Horani S, Stater EP, Corringer PJ, Trudell JR, Harris RA, Howard RJ. Ethanol modulation is quantitatively determined by the transmembrane domain of human α1 glycine receptors. Alcohol Clin Exp Res. 2015;39:962–968. doi: 10.1111/acer.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Howard RJ, Trudell JR, Harris RA. Seeking structural specificity: direct modulation of pentameric ligand-gated ion channels by alcohols and general anesthetics. Pharmacol Rev. 2014;66:396–412. doi: 10.1124/pr.113.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mihic SJ, Harris RA. Inhibition of ρ1 receptor GABAergic currents by alcohols and volatile anesthetics. J Pharmacol Exp Ther. 1996;277:411–416. [PubMed] [Google Scholar]
- [16].Wick MJ, Mihic SJ, Ueno S, Mascia MP, Trudell JR, Brozowski SJ, Ye Q, Harrison NL, Harris RA. Mutations of gamma-aminobutyric acid and glycine receptors change alcohol cutoff: evidence for an alcohol receptor? Proc Natl Acad Sci U S A. 1998;95:6504–6509. doi: 10.1073/pnas.95.11.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ueno S, Lin A, Nikolaeva N, Trudell JR, Mihic SJ, Harris RA, Harrison NL. Tryptophan scanning mutagenesis in TM2 of the GABA(A) receptor α subunit: effects on channel gating and regulation by ethanol. Br J Pharmacol. 2000;131:296–302. doi: 10.1038/sj.bjp.0703504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Johnson WD, 2nd, Howard RJ, Trudell JR, Harris RA. The TM2 6′ position of GABA(A) receptors mediates alcohol inhibition. J Pharmacol Exp Ther. 2012;340:445–456. doi: 10.1124/jpet.111.188037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xu M, Akabas MH. Identification of channel-lining residues in the M2 membrane-spanning segment of the GABA(A) receptor α1 subunit. J Gen Physiol. 1996;107:195–205. doi: 10.1085/jgp.107.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Borghese CM, Henderson LA, Bleck V, Trudell JR, Harris RA. Sites of excitatory and inhibitory actions of alcohols on neuronal α2β4 nicotinic acetylcholine receptors. J Pharmacol Exp Ther. 2003;307:42–52. doi: 10.1124/jpet.102.053710. [DOI] [PubMed] [Google Scholar]
- [21].Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Miller PS, Aricescu AR. Crystal structure of a human GABA(A) receptor. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bergmann R, Kongsbak K, Sorensen PL, Sander T, Balle T. A unified model of the GABA(A) receptor comprising agonist and benzodiazepine binding sites. PLoS One. 2013;8:e52323. doi: 10.1371/journal.pone.0052323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chiara DC, Jayakar SS, Zhou X, Zhang X, Savechenkov PY, Bruzik KS, Miller KW, Cohen JB. Specificity of intersubunit general anesthetic-binding sites in the transmembrane domain of the human α1β3γ2 γ-aminobutyric acid type A [GABA(A)] receptor. J Biol Chem. 2013;288:19343–19357. doi: 10.1074/jbc.M113.479725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Naffaa MM, Chebib M, Hibbs DE, Hanrahan JR. Comparison of templates for homology model of rho1 GABA receptors: More insights to the orthosteric binding site’s structure and functionality. J Mol Graph Model. 2015;62:43–55. doi: 10.1016/j.jmgm.2015.09.002. [DOI] [PubMed] [Google Scholar]
- [26].Huang X, Chen H, Michelsen K, Schneider S, Shaffer PL. Crystal structure of human glycine receptor-alpha3 bound to antagonist strychnine. Nature. 2015;526:277–280. doi: 10.1038/nature14972. [DOI] [PubMed] [Google Scholar]
- [27].Du J, Lu W, Wu S, Cheng Y, Gouaux E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature. 2015;526:224–229. doi: 10.1038/nature14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chang Y, Weiss DS. Substitutions of the highly conserved M2 leucine create spontaneously opening ρ1 γ-aminobutyric acid receptors. Mol Pharmacol. 1998;53:511–523. doi: 10.1124/mol.53.3.511. [DOI] [PubMed] [Google Scholar]
- [29].Pan ZH, Zhang D, Zhang X, Lipton SA. Agonist-induced closure of constitutively open γ-aminobutyric acid channels with mutated M2 domains. Proc Natl Acad Sci U S A. 1997;94:6490–6495. doi: 10.1073/pnas.94.12.6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Carland JE, Johnston GA, Chebib M. Relative impact of residues at the intracellular and extracellular ends of the human GABA(C) ρ1 receptor M2 domain on picrotoxinin activity. Eur J Pharmacol. 2008;580:27–35. doi: 10.1016/j.ejphar.2007.10.036. [DOI] [PubMed] [Google Scholar]
- [31].Zhang D, Pan ZH, Zhang X, Brideau AD, Lipton SA. Cloning of a γ-aminobutyric acid type C receptor subunit in rat retina with a methionine residue critical for picrotoxinin channel block. Proc Natl Acad Sci U S A. 1995;92:11756–11760. doi: 10.1073/pnas.92.25.11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li P, Khatri A, Bracamontes J, Weiss DS, Steinbach JH, Akk G. Site-specific fluorescence reveals distinct structural changes induced in the human ρ1 GABA receptor by inhibitory neurosteroids. Mol Pharmacol. 2010;77:539–546. doi: 10.1124/mol.109.062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Eaton MM, Lim YB, Covey DF, Akk G. Modulation of the human ρ1 GABA(A) receptor by inhibitory steroids. Psychopharmacology (Berl) 2014;231:3467–3478. doi: 10.1007/s00213-013-3379-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Carland JE, Moore AM, Hanrahan JR, Mewett KN, Duke RK, Johnston GA, Chebib M. Mutations of the 2′ proline in the M2 domain of the human GABA(C) ρ1 subunit alter agonist responses. Neuropharmacology. 2004;46:770–781. doi: 10.1016/j.neuropharm.2003.11.027. [DOI] [PubMed] [Google Scholar]
- [35].Carland JE, Moorhouse AJ, Barry PH, Johnston GA, Chebib M. Charged residues at the 2′ position of human GABA(C) ρ1 receptors invert ion selectivity and influence open state probability. J Biol Chem. 2004;279:54153–54160. doi: 10.1074/jbc.M410625200. [DOI] [PubMed] [Google Scholar]
- [36].Zhu Y, Ripps H, Qian H. A single amino acid in the second transmembrane domain of GABA ρ receptors regulates channel conductance. Neurosci Lett. 2007;418:205–209. doi: 10.1016/j.neulet.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Qian H, Dowling JE, Ripps H. A single amino acid in the second transmembrane domain of GABA ρ subunits is a determinant of the response kinetics of GABA(C) receptors. J Neurobiol. 1999;40:67–76. doi: 10.1002/(sici)1097-4695(199907)40:1<67::aid-neu6>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- [38].Borghese CM, Ali DN, Bleck V, Harris RA. Acetylcholine and alcohol sensitivity of neuronal nicotinic acetylcholine receptors: mutations in transmembrane domains. Alcohol Clin Exp Res. 2002;26:1764–1772. doi: 10.1097/01.ALC.0000042012.58231.D9. [DOI] [PubMed] [Google Scholar]
- [39].Bromstrup T, Howard RJ, Trudell JR, Harris RA, Lindahl E. Inhibition versus potentiation of ligand-gated ion channels can be altered by a single mutation that moves ligands between intra- and intersubunit sites. Structure. 2013;21:1307–1316. doi: 10.1016/j.str.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Howard RJ, Murail S, Ondricek KE, Corringer PJ, Lindahl E, Trudell JR, Harris RA. Structural basis for alcohol modulation of a pentameric ligand-gated ion channel. Proc Natl Acad Sci U S A. 2011;108:12149–12154. doi: 10.1073/pnas.1104480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Murail S, Howard RJ, Broemstrup T, Bertaccini EJ, Harris RA, Trudell JR, Lindahl E. Molecular mechanism for the dual alcohol modulation of Cys-loop receptors. PLoS Comput Biol. 2012;8:e1002710. doi: 10.1371/journal.pcbi.1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yoluk O, Bromstrup T, Bertaccini EJ, Trudell JR, Lindahl E. Stabilization of the GluCl ligand-gated ion channel in the presence and absence of ivermectin. Biophys J. 2013;105:640–647. doi: 10.1016/j.bpj.2013.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pless SA, Dibas MI, Lester HA, Lynch JW. Conformational variability of the glycine receptor M2 domain in response to activation by different agonists. J Biol Chem. 2007;282:36057–36067. doi: 10.1074/jbc.M706468200. [DOI] [PubMed] [Google Scholar]
- [44].Rosen A, Bali M, Horenstein J, Akabas MH. Channel opening by anesthetics and GABA induces similar changes in the GABA(A) receptor M2 segment. Biophys J. 2007;92:3130–3139. doi: 10.1529/biophysj.106.094490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Horenstein J, Wagner DA, Czajkowski C, Akabas MH. Protein mobility and GABA-induced conformational changes in GABA(A) receptor pore-lining M2 segment. Nat Neurosci. 2001;4:477–485. doi: 10.1038/87425. [DOI] [PubMed] [Google Scholar]
- [46].Chen Q, Kinde MN, Arjunan P, Wells MM, Cohen AE, Xu Y, Tang P. Direct pore binding as a mechanism for isoflurane inhibition of the pentameric ligand-gated ion channel ELIC. Sci Rep. 2015;5:13833. doi: 10.1038/srep13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- [48].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera — a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.