Abstract
Adhesive mussel foot proteins (Mfps) rely in part on DOPA (3,4-dihydroxyphenyl-l-alanine) side chains to mediate attachment to mineral surfaces underwater. Oxidation of DOPA to Dopaquinone (Q) effectively abolishes the adsorption of Mfps to these surfaces. The thiol-rich mussel foot protein-6 (Mfp-6) rescues adhesion compromised by adventitious DOPA oxidation by reducing Q back to DOPA. The redox chemistry and kinetics of foot-extracted Mfp-6 were investigated by using a nonspecific chromogenic probe to equilibrate with the redox pool. Footextracted Mfp-6 has a reducing capacity of ~17 e− per protein; half of this comes from the cysteine residues, whereas the other half comes from other constituents, probably a cohort of four or five nonadhesive, redox-active DOPA residues in Mfp-6 with an anodic peak potential ~500 mV lower than that for oxidation of cysteine to cystine. At higher pH, DOPA redox reversibility is lost possibly due to Q scavenging by Cys thiolates. Analysis by one- and two-dimensional proton nuclear magnetic resonance identified a pronounced β-sheet structure with a hydrophobic core in foot-extracted Mfp-6 protein. The structure endows redox-active side chains in Mfp-6, i.e., cysteine and DOPA, with significant reducing power over a broad pH range, and this power is measurably diminished in recombinant Mfp-6.
Graphical Abstract
A defining adaptation in the evolution of living organisms has been the formation and redox regulation of subcellular compartments. Distinct redox compartments are known to allow aerobic respiration, signaling, and protein disulfide bond formation.1,2 Redox is also regulated outside cells. The once-prevailing view of the extracellular environment as oxidizing and beyond redox control is no longer tenable. Cells and tissues harness their internal redox poise to regulate exterior microenvironments.3,4
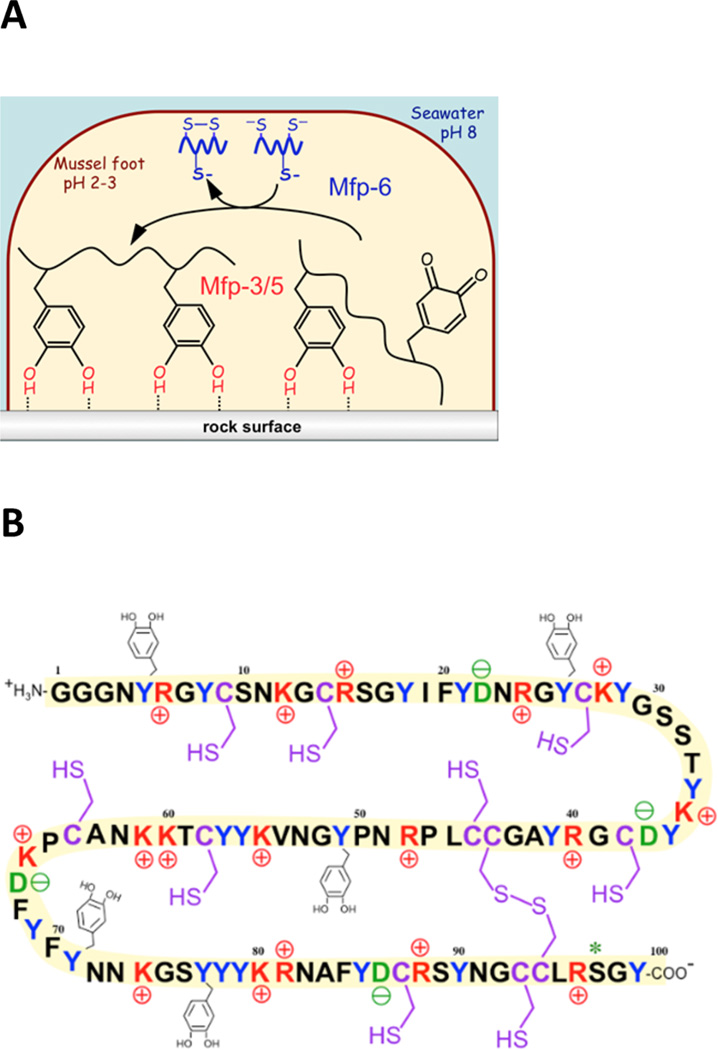
The regulation of redox in the adhesive holdfast or byssus by marine mussels is an exotic and fascinating case in point of extracellular redox. The byssus is essentially a bundle of extra-organismic mini-tendons each tipped with a distal adhesive plaque. Before the formation of each plaque by the foot, a peripherally sealed contact zone between the foot and rock surface is filled with a reducing solution of cysteine-containing and 3,4-dihydroxyphenylalanine (DOPA)-containing proteins (Figure 1A).5,6 The DOPA-rich (20–30 mol % DOPA) mussel foot proteins in Mytilus californianus are known as Mfp-3f and Mfp-5 and provide adhesion that depends critically on the reduced form of DOPA.7,8 Although DOPA-mediated wet adhesion is strong and versatile on different surfaces, it is also vulnerable to DOPA’s tendency to autoxidize at the pH of seawater.8 Mussels significantly reduce the risk of interfacial DOPA oxidation by imposing an acidic pH and strongly reducing environment during secretion.9,10 The reducing environment is provided by Mfp-6, a small (11.6 kDa) basic protein, with high cysteine (11 residues, only two of which are disulfide-linked) and a somewhat lower DOPA (4 or 5 residues) content11 (Figure 1B). Previous studies with the surface forces apparatus determined that adhesion of Mfp-3f to mica depends strongly on pH, decreasing exponentially with an increase in pH. Adhesion loss was attributed to the O2-dependent oxidation of DOPA (QH2) to Dopaquinone (Q), which cannot H-bond to mica.5,8 Adhesion by Mfp-3f was rescued by adding Mfp-6 that restored DOPA,5 apparently driven by the reducing power of Cys in Mfp-6 according to the following two favorable half-reactions under standard conditions (1 M, pH 7, 25 °C, 1 atm):
| (1) |
| (2) |
Figure 1.
Mussel foot adhesive proteins. (A) Scheme of the inverted cup configuration of the mussel foot during the initial deposition of adhesive Mfp-3, Mfp-5, and Mfp-6 (which is not adhesive). Oxidized DOPA or Dopaquinone does not contribute to adhesion. (B) Sequence of a mussel foot protein 6 (Mfp-6) variant. Highlighted are basic residues (red), acidic residues (green), tyrosines (blue), and cysteines (purple). Five of 20 tyrosines are shown as being modified to DOPA. Likewise, 2 of 11 cysteines are coupled as disulfide cystine. Although the implied stoichiometry is correct, the specific position of each modification has yet to be determined.
This study investigates the following specific properties of Mfp-6: (a) reducing capacity, (b) pH dependence of redox, (c) structure dependence of redox, and (d) identification of reducing residues. To measure redox activity, we used a chromogenic redox sensor 1,1-diphenyl-2-picryl-hydrazyl (DPPH),12 a widely used nonspecific oxidant, to quantify the overall pool of reducing residues in Mfp-6.
Our findings suggest that each molecule of Mfp-6 has an average reducing capacity of 17 e−, which come from nine thiolate Cys residues and approximately four or five DOPAs. Reducing capacity is little affected by pH, but the reduction rate is strongly pH-dependent. Both foot-extracted and recombinant Mfp-6 have reducing activity, but the reducing capacity of foot-extracted Mfp-6 is greater.
EXPERIMENTAL PROCEDURES
Chemicals
2,2-Diphenyl-1-picryl hydrazyl radical (DPPH•), spectrophotometric grade methanol (≥99.9%), and the nonionic detergent Triton X-100 were purchased from Fisher Scientific (Pittsburgh, PA). Citric acid monohydrate (>99% purity) and sodium phosphate monobasic (>98% purity) were purchased from EMD Millipore (San Diego, CA). Acetic acid-d4 (99.5 atom % D), deuterium oxide (99.9 atom % D), and formic acid (~98%) were purchased from Sigma-Aldrich (St. Louis, MO). Doubly distilled water was used throughout the experiments.
Purification of Mfp-6 from Mussel Foot Tissue
Protein isolation was performed as described previously,11 but important steps and quality controls are detailed in the Supporting Information (Figure S1A–E). Briefly, feet from approximately 80–100 mussels were cut at the proximal end and directly transferred to ice. Dissected phenol glands were cut into small pieces, weighed (typically approximately 6–8 g), and then suspended in 10 mL of ice-cold 5% acetic acid buffer per gram of phenol glands, supplemented with protease inhibitors (10 µM leupeptin and pepstatin and 1 mM EDTA). The glands were thoroughly homogenized on ice using a tissue grinder and then centrifuged for 40 min at 20000g and 4 °C. The resulting supernatant (S1) was then acidified using perchloric acid to a final concentration of 1.5% (v/v). After centrifugation for 40 min at 20000g and 4 °C, ammonium sulfate was slowly added to the supernatant (S2) to a final concentration of 20% (w/v). The mixture was stirred on ice for an additional 20 min and then centrifuged for 40 min at 20000g and 4 °C. The resulting supernatant (S3) was dialyzed [molecular weight cutoff (MWCO) of 1000, Spectrum Industries, Los Angeles, CA] overnight at 4 °C in 1% (v/v) acetic acid at a volume ratio of 1:2000. The recovered dialysate was freeze-dried and reconstituted in 2 mL of 5% acetic acid prior to being subjected to Shodex 802.5 gel filtration (Showa Denko America). A final purification was performed using reverse-phase C8 RP-300 chromatography (Applied Biosystems, San Jose, CA) and a linear gradient of aqueous acetonitrile acidified with 0.1% (v/v) trifluoroacetic acid (Figure S1B). All solvents were degassed before use. Amino acid analysis of Mfp-6 was performed on a ninhydrin-based autoanalyzer (Hitachi L-8900) on which disulfide cystine has an elution time of 27 min.
Modified DPPH Assay
We used a modified DPPH assay as described elsewhere.13 The proposed one-electron oxidation of Cys by DPPH to form cystine is shown in reactions 3 and 4:10,14
| (3) |
| (4) |
Consequently, the reducing power (i.e., antioxidant activity) of the cysteine thiols and thiolates in Mfp-6 can be monitored by the reduction of the DPPH• to DPPH-H. Briefly, 1.5 µg of purified Mfp-6 in a 5% acetic acid solution was freeze-dried and dissolved in a final volume of 50 µL of assay buffer, which, when added to the 1 mL final assay volume, corresponds to 2.5 µM Mfp-6. The reaction was started by the addition of 50 µL of a freshly prepared 2 mM DPPH solution (in 100% methanol) to a final volume of 1 mL of citrate/phosphate buffer to reach a final concentration of 100 µM DPPH in the reaction tube. Upon addition of DPPH, the solution was gently mixed by pipetting 3–5 times and adventitious Mfp-6 precipitation at the end of the reaction was removed by centrifugation for 1 min at 17000g prior to measurement. For time-course experiments at different buffer pHs, the absorbance at 515 nm (ε = 15000 M−1 cm−1 in methanol) was monitored at 2 min (right after mixing) and at 5, 10, 15, 30, 45, and 60 min. For the half-maximal effective concentration (EC50) studies, we used Mfp-6 protein concentrations of 2.5, 5, 10, 15, and 20 µM. The absorbance of DPPH without additions in the optimized buffer system was stable for at least 60 min. If not otherwise specified, each plotted datum represents the mean of three independent measurements. Where necessary, the buffer pH was readjusted using glacial acetic acid (17.4 N) or a sodium hydroxide solution (10 N) purchased from Fisher Scientific and followed with a digital pH meter. The antioxidant efficiency at a given buffer pH and time was determined by plotting the DPPH absorbance reduction as a percent of the appropriate control. This was necessary, as DPPH extinction coefficients are pH-dependent, and thus, the control absorbance of 100 µM DPPH had different maxima.13 Bleaching rate Vb and percentage of remaining DPPH radical at infinite time Fi were calculated according to the method described in ref 15. Experimentally determined reduction of DPPH radical as a function of the number of moles of Mfp-6 per mole of DPPH radical was also determined at defined time points. EC50 values were calculated from the fitted curves following a first-order {y = y0 + A1 × exp[−(x − x0)/t1]} or second-order exponential decay {y = y0 + A1 × exp[−(x − x0)/t1] + A2 × exp[−(x − x0)/t2]}, where A1 and A2 are the amplitudes and t1 and t2 the decay constants for the slow and fast decay, respectively. The offset y0 accounts for the unreacted DPPH at the end of the reaction (i.e., at “infinite” time). Antiradical power ARP (=1/EC50), stoichiometric value n (=2 × EC50), and the number of reduced DPPH molecules per target antioxidant molecule (=1/EC100) were calculated according to the method described in ref 16. At pH 7, there is a linear relationship between ARP with respect to DPPH and the oxidation potential of various redox-active compounds up to a limiting oxidation potential of 0.56 V relative to Ag/AgCl.17
Alkylation of Cysteine Residues in Mfp-6
Modification of foot-extracted Mfp-6 thiol groups was performed at low pH in the absence of denaturing agents (i.e., urea and guanidine hydrochloride). Briefly, ~30 µg of purified Mfp-6 protein was reconstituted in 0.1% formic acid (pH ~3) supplemented with 1 mM iodoacetamide (IAM) following incubation for 30 min at room temperature (RT). Unreacted IAM was removed by dialysis (MWCO of 1000, Spectrum Industries) for 2 h at RT in 0.1% (v/v) formic acid at a volume ratio of 1:4000. The recovered dialysate was freeze-dried and reconstituted in 0.1% formic acid prior to ESI Q-TOF2 mass spectrometry.
ESI Q-TOF2 Mass Spectrometry
ESI Q-TOF MS measurements were performed with a Micromass Q-TOF2 quadrupole/time-of-flight tandem mass spectrometer coupled to an Agilent 1100 Nano LC system, or to a homemade syringe pump with a 250 µL gastight Hamilton syringe for sample injection. The ESI Q-TOF2 mass spectrometer was operated at a capillary voltage of 3.5 kV and a cone voltage of 45 V with a desolvation temperature of 100 °C and a source temperature of 80 °C. The mass spectrometer was operated in the positive ion mode. The standard electrospray ion (ESI) source was used to generate the ions. The ESI Q-TOF2 MS instrument was calibrated in the range of m/z 100–2000 using an internal calibration standard (sodium iodide). Protein samples were prepared at a concentration of 10 µg/mL and injected at a constant flow rate (5 µL/min). The solvent was 0.1% (v/v) formic acid (pH 3), and there was no salt addition prior to analysis. Data were processed via MassLynx Data Analysis version 4.1.
Cyclic Voltammetry
Cyclic voltammetry (CV) was performed using a CIH 660D electrochemical workstation (CH Instruments Inc., Austin, TX). These analyses were conducted using a three-electrode cell with a platinum wire as the counter electrode, a glassy carbon working electrode, and a Ag/AgCl reference electrode calibrated with a 5 mM solution of the ferrocene/ferrocenium (Fc/Fc+) redox couple as an external standard in 0.1 M TBAP/acetonitrile (ACN) as an electrolyte solution. The initial potential was 0 V, and a cyclic voltage sweep from −0.2 V to a final potential of 1 V was performed with a scan rate of 50 mV/s. Data were analyzed using CHI software version 12.02.
One-Dimensional (1D) and Two-Dimensional (2D) NOESY NMR
Nuclear magnetic resonance (NMR) spectroscopy was performed on a Varian Inova 600 MHz spectrometer or a Bruker Avance III Ultrashield 800 MHz spectrometer each equipped with a 1H/13C/15N/2H four-channel cryoprobe for optimal 1H detection. All experiments were conducted at 25 °C unless otherwise stated. The NMR sample contained various concentrations of proteins in 85% H2O, 10% D2O, and 5% d4-acetic acid at pH 3. Water suppression was achieved with water flip-back18 and watergate19 techniques. NMR data were processed and analyzed with instrument software and the nmrPipe package.20 Nuclear Overhauser effect spectroscopy (NOESY) was performed with a mixing time of 150 or 180 ms. Quality controls applied to purified Mfp-6 prior to NMR analysis were (1) homogeneity determiined by SDS–PAGE, (2) mass homogeneity determined by MALDI-TOF mass spectrometry after multiple charge deconvolution (Figure S1D), and (3) no more than one disulfide bond determined by amino acid analysis.
RESULTS
pH-Dependent Antioxidant Activity of Mfp-6
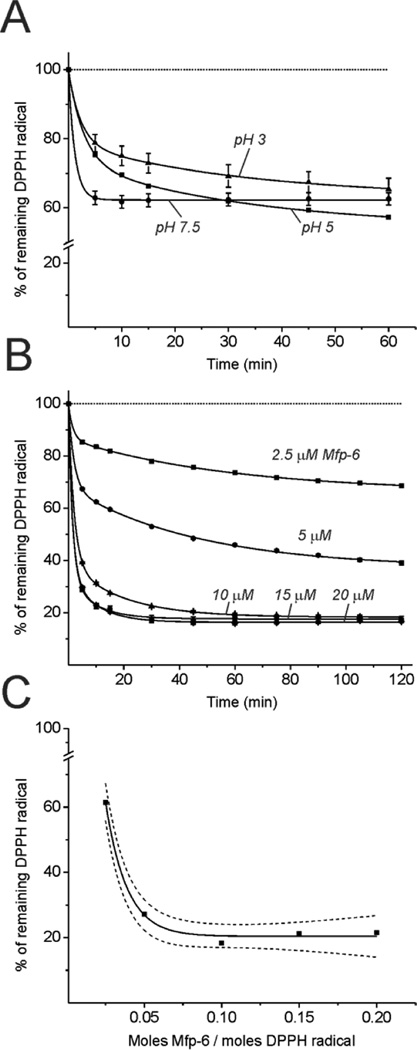
The five variants of Mfp-6 annotated to date are >95% identical and represent small (11.6 kDa) basic proteins that are ~100 amino acids long. Two amino acids, cysteine and DOPA, are present in all variants and prone to oxidation and, hence, are good candidates as reducing residues (Figure 1B). Here, we used the free radical scavenger DPPH to equilibrate with the pool of reducing amino acids in a purified variant of foot-extracted and recombinant Mfp-6, and the kinetics of antioxidant equilibration were measured at three different pHs (3, 5, and 7.5) by following the decrease in the absorbance of the DPPH radical over a 60 min reaction time (Figure 2A); i.e., the purple color of DPPH is bleached upon reduction. These time-lapse experiments were performed with excess DPPH radical at a 1:40 (Mfp-6:DPPH) molar ratio.
Figure 2.
Radical scavenging of Mfp-6 toward DPPH. (A) Buffer pH-dependent reaction kinetics of 100 µM DPPH radical reduction by 2.5 µM Mfp-6 at pH 3 (▲), 5 (■), and 7.5 (●). Absorbance displayed as a percentage of control absorbance using 100 µM DPPH (---). (B) Specific antioxidant activity of Mfp-6 toward DPPH radical quenching at pH 3. (B) Fraction of remaining DPPH radical (%) at infinite time, extrapolated for the range of concentration increments of Mfp-6 from 2.5 to 20 µM.
Expected results were (a) that DPPH reduction would plateau at some level that was proportional to the Mfp-6 concentration, (b) that the asymptotic Mfp-6 concentration was correlated with the content of redox-active amino acids, and (c) that the rate of DPPH reduction might reveal different populations or chemistries of redox-active amino acids in Mfp-6. The kinetics for 100 µM DPPH reduction after adding 2.5 µM Mfp-6 at different pH values showed that at pH 3 and 5 the DPPH radical concentration followed a slow and continuous decay for up to 60 min. At pH 7.5, a fast initial decay occurred within the first 5 min, followed by an asymptotic DPPH radical concentration up to 60 min. The rate of DPPH reduction and, to some extent, the reductive capacity of the foot-extracted Mfp-6 protein were clearly pH-dependent (Figure 2A). Although Mfp-6 remained strongly reducing under all tested conditions, the fastest reaction kinetics of foot-extracted Mfp-6 with the DPPH radical occurred at pH 7.5 (Figure 2A). This pattern of increased reaction rates with increasing pH was also observed for the recombinant Mfp-6 protein (Figure S3A,B). However, at pH 7.5, the fast initial decay and the overall reduction capacity of recombinant protein lacking DOPA were much less pronounced or approximately 40–50% lower than those of foot-extracted Mfp-6 (Figure S3B).
Specific Reducing Activity of Mfp-6 at Low pH
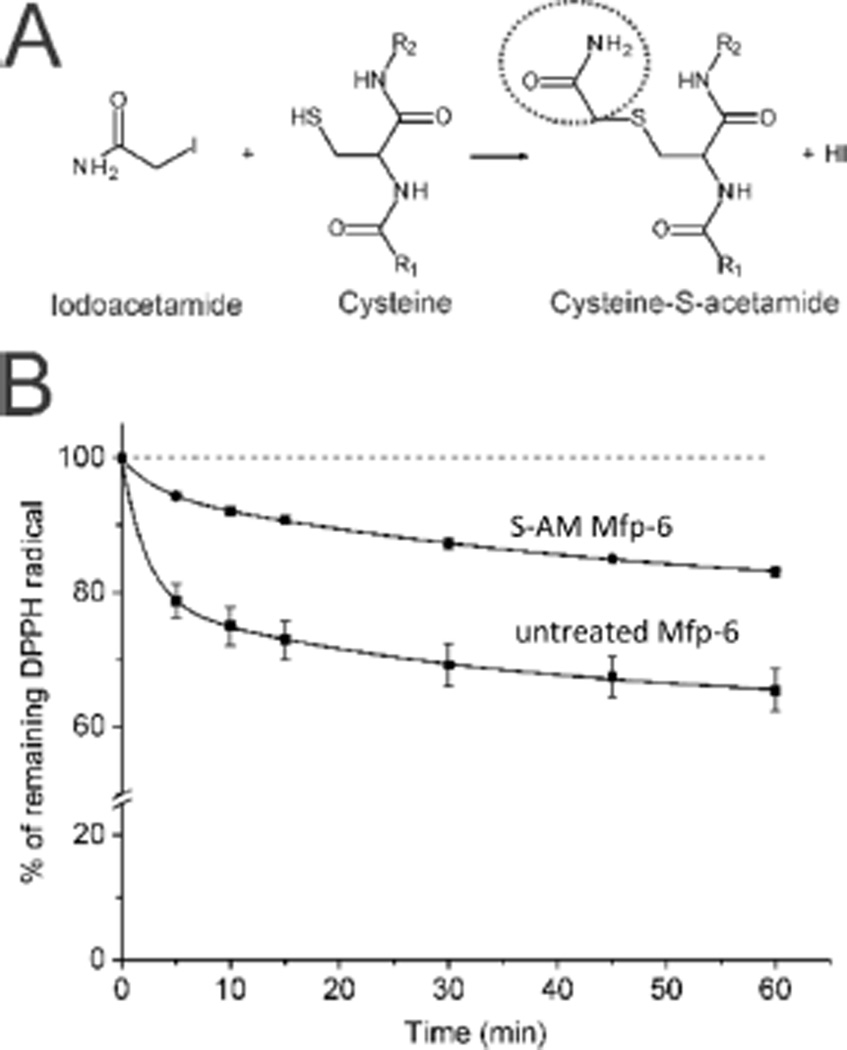
Because of Mfp-6’s tendency to precipitate between pH 5 and 7.5, we determined the specific reducing capacity at pH 3. This is biologically relevant as the pH at which the mussel deposits adhesive proteins is between 2.5 and 3.5.9 For the purposes of the assay, DPPH concentrations were held constant (100 µM), and the decrease in absorption at 515 nm was followed over time at increasing Mfp-6 concentrations. As Mfp-6 concentrations increase, the reaction curve shows a continuous decay with a steady increase in the bleaching rate (Figure 2B, Figure S4A, and Table S1). Notably, the fastest reaction rate with the DPPH radical was observed at the lowest concentration of Mfp-6 (Table S1). Extrapolating this reaction process to “infinite time” as defined previously15 and plotting the percentage of remaining DPPH radical at infinite time (Fi) reveal an end point phase at a molar ratio of 17 DPPHs reduced/mol of Mfp-6 (Figure 2C and Table S1). Recalling that Mfp-6 contains nine Cys thiols with the remaining two Cys residues coupled as a disulfide,11 we speculated that Mfp-6-mediated DPPH reduction at pH 3 might be abolished by blocking all Cys residues in foot-extracted Mfp-6. Both the reducing capacity and alkylation of cysteine are known to depend on the presence of thiolate anions, which with a typical cysteinyl thiol pKa of 8–9 are not expected to be abundant at pH 3. By combining amidomethylation [iodoacetamide (IAM)] of foot-extracted Mfp-6 with ESI Q-TOF2 mass spectrometry at pH 3, we found a +57 Da m/z shift for nine distinct peaks was evident in the mass spectra, indicating that all nine Cys thiols in Mfp-6 are reactive (ionized) at pH 3, though to varying degrees (Figure 3A and Figure S4). The time course of DPPH radical reduction with amidomethylated Mfp-6 is diminished by ~50% compared with that of the untreated Mfp-6 sample (Figure 3B). Because the alkylation of Mfp-6 was incomplete at pH 3, precise apportioning of the remaining redox activity to unblocked Cys, other redox-active residues, or both is precluded.
Figure 3.
Residual antioxidant capacity in amidomethylated Mfp-6. (A) Free sulfhydryl groups in Mfp-6 were labeled at pH 3 using 2-iodoacetamide (IAM). (B) Time course of 100 µM DPPH radical quenching using 2.5 µM pure (■) or thiol group-blocked (●) Mfp-6.
An alternative hypothesis that the redox contribution of non-thiolate side chains to DPPH reduction might be assessed by using recombinant Mfp-6, which does not contain DOPA, was also tested. Notably, the analogous reaction kinetics of reduction of DPPH with rMfp-6 at pH 3 could not be saturated down to a molar ratio of 3.8, despite having the same amount of cysteine (Figure S3B–D and Table S1). This decreased reactivity of rMfp-6 toward the DPPH radical is also reflected in the slower reaction rates for all five tested concentrations as compared with that of the foot-extracted protein (Table S1). Possible reasons for the lower reactivity of thiol groups in rMfp-6 are (i) that foot-extracted Mfp-6 depends on a higher-order protein structure to decrease the thiol pKa (Table S1 and below) and/or (ii) that Mfp-6 depends on a dynamic redox interplay between both thiols and DOPA.
In summary, the DPPH results lead to the following interpretations: (a) The rate of DPPH reduction by Mfp-6 is pH-dependent and faster when using foot-extracted, DOPA-containing Mfp-6 than with the recombinant form. (b) The reducing capacity of Mfp-6 is approximately 17 e−/Mfp-6. (c) Nonreductive alkylation of cysteine suggests that factors other than the total thiol content contribute to the pH-dependent antioxidant capacity of Mfp-6.
Other Redox Functionalities in Foot-Extracted Mfp-6
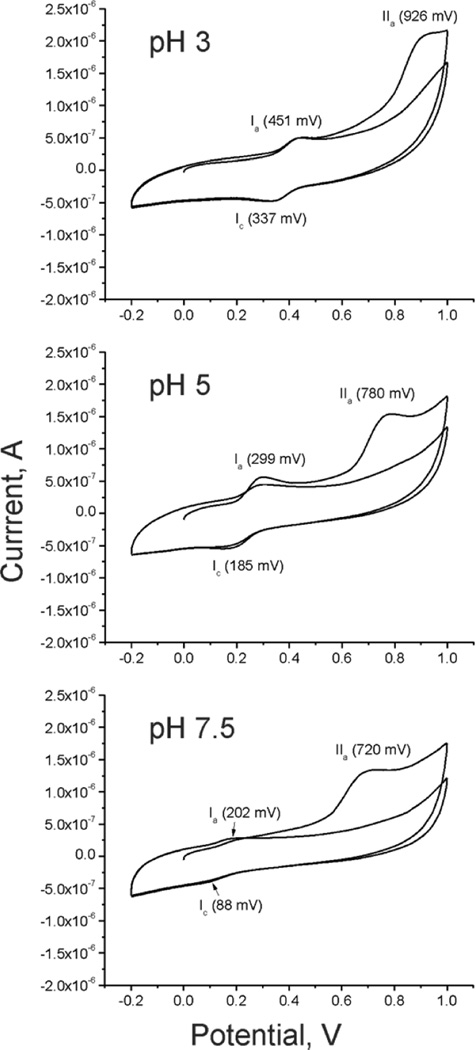
To investigate reducing modalities other than Cys in foot-extracted Mfp-6, we used cyclic voltammetry at three different pHs (Figure 4). The electrochemical behavior of thiols on glassy carbon electrodes is typically poorly defined due to slow heterogeneous electron transfer.21,22 At pH 3, a reversible redox couple was observed with a formal potential E1/2 of 394 mV and ΔEp = Epa − Epc = 451 mV − 337 mV = 114 mV (or 57 mV × 2 for a two-electron redox half-reaction). There is also a second less defined oxidation with an Epc of 926 mV. At pH 5, the estimated E1/2 for Mfp-6 is 242 mV and shows a similar two-electron redox reaction (ΔEp = Epa − Epc = 299 mV − 185 mV = 114 mV = 57 mV × 2), followed by a second oxidation peak at an Epc of 780 mV. At pH 7.5, the estimated E1/2 for Mfp-6 is 141 mV, again involving a two-electron redox reaction (ΔEp = Epa − Epc = 202 mV − 88 mV = 114 mV = 57 mV × 2). In addition, the second, irreversible oxidation peak is visible with an Epc of 720 mV. The reversible redox couple (denoted as I in Figure 4) is consistent with the DOPA → Dopaquinone conversion as has been determined for Mfp-3 in ref 8 and for free L-DOPA in refs 23 and 24. The indistinct oxidation peak at a high positive potential (denoted as II) resembles the reported oxidation of free cysteine at a comparable pH.21 As expected, both redox peak potentials shifted negatively with the increase in solution pH, indicating that protons are involved in the reaction. In addition, with an increase in pH, the peak current of redox couple I decreased and effectively disappeared at pH 7.5. This would be expected as the pH approaches and exceeds the thiol pKa, as cysteine reacts readily with quinones to form cysteinyl-DOPA adducts with oxidation potentials that are lower than that of DOPA but retain strong reducing activity.25,26
Figure 4.
Reversible and irreversible oxidation of Mfp-6 at pH 3, 5, and 7.5 using cyclic voltammetry. The voltammograms show reversible oxidation from DOPA to Dopaquinone (Ia and Ic). A pH-dependent, second irreversible oxidation peak (IIa) is visible, possibly attributed to the interaction of cysteines with Dopaquinone. Note that the lower the pH, the higher the peak potential and the anodic peak current of IIa.
Mfp-6 Exhibits Defined Secondary and Tertiary Structure Elements
The overall structure of Mfp-6 was investigated by solution NMR spectroscopy for possible insights about structure-dependent redox activity. Foot-extracted Mfp-6 variants isolated from mussels are not ideal for detailed structure determination because they include subtle sequence differences and variable post-translational conversion of Tyr and Ser to DOPA and pSer, respectively.11 Notwithstanding these limitations, foot-extracted and recombinant Mfp-6 were purified and shown to be essentially homogeneous and free of contaminants by SDS–PAGE and MALDI mass spectrometry (Figure S1). The search for secondary structure in Mfp-6 was first conducted by circular dichroism spectrophotometry in the far-ultraviolet region (Figure S5). The spectra revealed similar maximal and minimal ellipticities in both the foot-extracted and recombinant proteins, with a particularly strong positive ellipticity at 230 nm, typically attributed to aromatic excitation energy transfers27 and consistent with 20 Tyr residues per Mfp-6. One suggestion of enhanced β-structure in foot-extracted Mfp-6 is the much less negative ellipticity at 200 nm compared with that of recombinant Mfp-6 (Figure S6A).
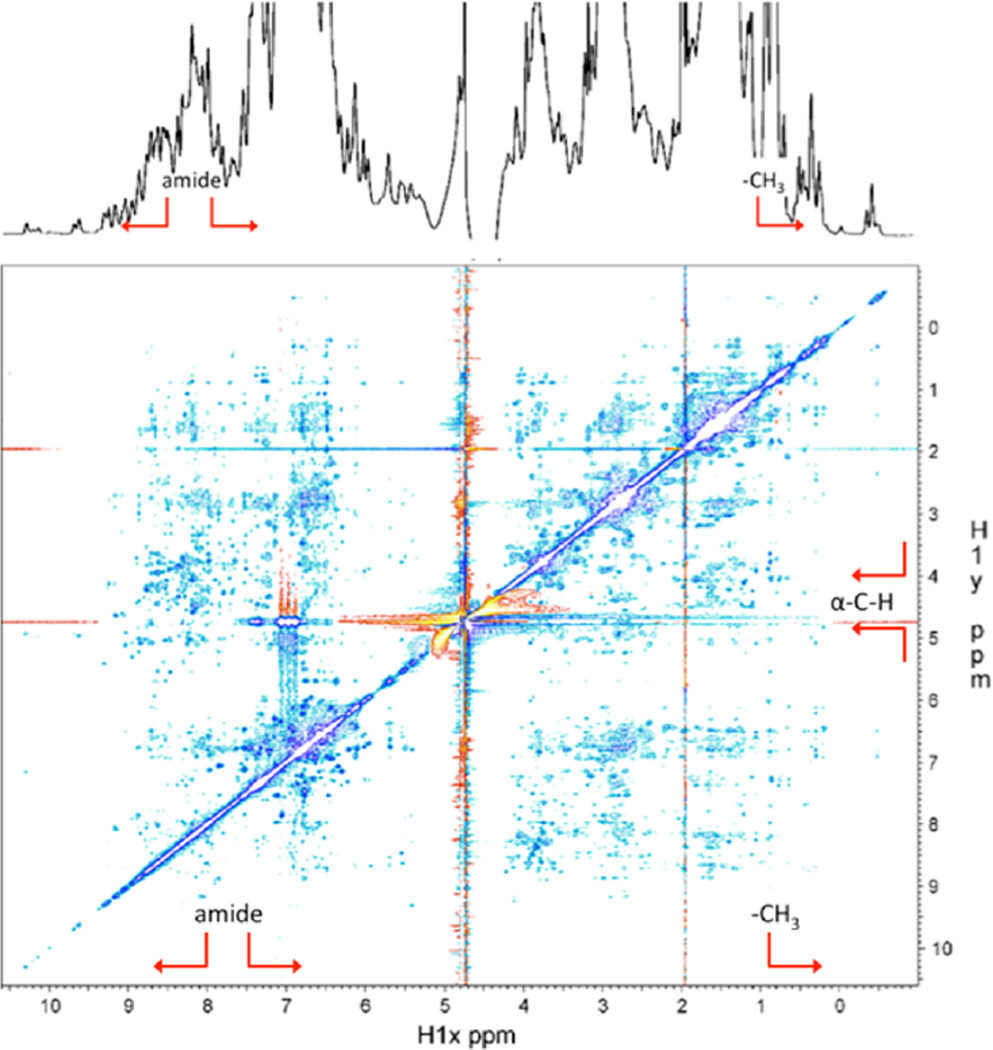
It is well-known that helical and β-sheet secondary structures are manifested by deviations in 1H chemical shifts from values for random coil peptides: an upper-field (toward lower parts per million values) shift in α-helical regions and a downfield (toward higher parts per million values) shift of the side chain α-protons (1Hα) in β-sheets.28 A tertiary fold in the protein is indicated from the overall spread and deviations of amide proton chemical shifts and significant upper field shifts of methyl protons (e.g., A42, A63, A83, I19, L46, and L95). Furthermore, a stable tertiary fold is strongly indicated by the presence of extensive 1H–1H cross peaks in 2D NOESY experiments.29 Such extensive NOE cross peaks, which only occur when protons are within 5 Å of one another, indicate packing of the protein chain into a compact defined foot-extracted fold with a hydrophobic core. An upper field shift of methyl protons often indicates packing of the methyl groups near aromatic groups, leading to a localized change in chemical shielding from the aromatic ring current effect.29,30
Both 1D and 2D NMR spectra of foot-extracted Mfp-6 show clear signatures of a defined foot-extracted fold, even though overall peak crowding prevents detailed deconvolution (Figure 5). The spectra show an expansion of the range of chemical shifts typically associated with protein folding, intense and extensive NOEs across the spectra among the backbone amide and aromatic ring protons (~6–10 ppm) and side chain aliphatic protons (~0–6 ppm) that are strongly indicative of a folded globular structure with a hydrophobic core (Figure 5). Specifically, the upfield shift of methyl peaks, typically between 0.9 and 2.2 ppm in random coil peptides, toward and beyond 0 ppm indicates tight hydrophobic packing and aromatic ring contacts that likely involve the 18–20 tyrosine residues in the protein.29 Furthermore, the spread of amide protons outside the 8.0–8.5 ppm region is typical of a protein with a foot-extracted fold formed with a significant presence of amide-related H-bonds.28,29 The downfield shifts of some Cα protons to the 4.8–6.0 ppm region from a typical 4.0–4.8 ppm range in random coil peptides indicate the presence of β-sheet structure in the foot-extracted Mfp-6 protein. The additional presence of α-helical structure is shown by strong amide NH–NH proton NOEs,29 but the relatively smaller number of strong NH–NH NOEs as compared to the NOEs in the β-strand regions suggests that on average the β-sheet content is more prominent than the α-helical content. Few, if any, of these features exist in recombinant Mfp-6, indicating much less secondary structure (Figure S6B). Overall, the data suggest that the foot-extracted Mfp-6 protein exhibits a well-defined structure primarily with β-strands and a low helical content and that this structure persists in the presence of DPPH (Figure S7A,B).
Figure 5.
1D and 2D NOESY NMR spectra of Mfp-6 at pH 3. The chemical shifts are dispersed over a wide range and show an extensive network of NOEs for the amide (above 8–8.5 ppm, red arrow brackets) and methyl protons (below ~1 ppm, red arrow), indicative of a well-folded domain. The downfield shifts of the Cα protons above ~5 ppm (red arrow brackets) further point to the presence of extended β-strand structure. The 1D spectrum was recorded on a Varian Inova 600 MHz spectrometer. The 2D NOESY spectrum was recorded with a mixing time of 150 ms on a Bruker 800 MHz spectrometer. The protein concentration was 0.5 mM in 85% H2O, 10% D2O, and 5% d4-acetic acid.
DISCUSSION
In contrast to other Mfps, which are intrinsically unstructured, Mfp-6 exhibits secondary structure and has significant reducing activity toward the redox sensor DPPH over a broad pH range, with the fastest reaction kinetics occurring close to the pH of seawater (~8.2). The increasing rate of DPPH reduction from low to high pH is accompanied by a change in the solubility of the protein, with facile precipitation of Mfp-6 at pH 7.5 and perhaps cross-link formation (Figure S8). Recombinant Mfp-6, which has little to no secondary structure and no DOPA, also reduces DPPH, but the rates and redox capacity are both lower by half. Combined CV and DPPH reaction kinetics and stoichiometry with Mfp-6 and its recombinant DOPA-less homologue (rMfp-6) were described to provide a better understanding of redox cycling outside of living cells.
On the basis of combined carboxymethylation and thiolate titration of Mfp-6 with Ellman’s reagent, Yu et al.5 earlier suggested that Cys residues are the major contributors to Mfp-6 reducing activity even at acidic pH and must therefore have unusually low pKas. Given the unprecedented Gly-, Lys-, Tyr-, and Cys-rich composition of Mfp-6, it is unclear how such low Cys pKas are achieved. Comparisons to other redox protein sequences are interesting, but their relevance is premature. Strategies for lowering Cys pKa (typical pKa of 8–9) in well-known redox proteins include nesting in Lys- or Arg-rich sequences and the use of electron-withdrawing effects.31 Buried Cys in a folded protein can also be shielded and controlled by aryl–thiol interactions, e.g., between tyrosines and cysteines, as is known for Tyr84 and Cys34 in human albumin.32 The strong remaining reducing activity following Cys amidomethylation in Mfp-6 and the 17 equiv of DPPH reduced per foot-extracted Mfp-6 (Table S1), however, indicate that reducing groups besides Cys exist. At pH 7.5, this reservoir undergoes autoxidation before being able to contribute to DPPH reduction, thereby decreasing the reducing capacity of Mfp-6. The DOPA-deficient recombinant Mfp-6 also has a lowered reducing capacity. A strategic DOPA oxidation potential is readily evident in cyclic voltammograms of Mfp-6 at Eox + 0.329 V (pH 3) (Figure 4) and is typically associated with catechols including DOPA.33 However, DOPA is usually identified with mussel adhesion, so its presence in Mfp-6 is puzzling. Mfp-6 exhibited no adhesion to mica,5 suggesting that DOPA in Mfp-6 may be adapted for other functions such as redox cycling. The fact that DOPA and AM-Mfp-6 both reduce DPPH supports the reservoir hypothesis, but the Mfp-6 DOPAs are corralled in a special nonadhesive environment or conformation.
Why does Mfp-6 need a reducing resource beyond the nine thiolates? There is no compelling answer to this at present. DOPA in Mfp-6 could be involved in electron tunneling or serve as an alternative sacrificial antioxidant that is easily (ΔEo = 0 V) recycled by thiolates:
| (5) |
| (6) |
The reverse, that is, reduction of disulfides by DOPA, however, has a similar ΔEo:
| (7) |
| (8) |
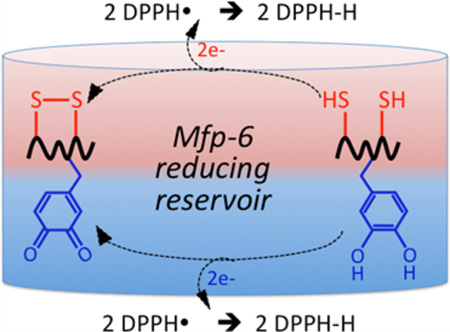
Thus, even though DOPA reduces the redox sensor DPPH,34 it is less strongly reducing than the antioxidant thiolates and roughly equal in redox to the catechols undergoing oxidation in Mfp-3 (Figure 6). DOPA may be even better adapted in Mfp-6 as a cross-linker (Figure S8) than as a reservoir for reducing disulfides; however, the relationship between cross-links and antioxidants is a tangled one, and there are many instances in which a kinetically favored reaction is not the same one predicted by thermodynamics. As suggested previously,10 a known intermediate in the thiolate reduction of Dopaquinone is a thiol–DOPA adduct that tautomerizes to 5-S-cysteinyl-DOPA. These cross-linking adducts get trapped and accumulate as the thiolates are depleted and have been detected in insoluble plaques.11 The Mfp-6 oligomers that form at pH 7.5 (Figure S5) may be cross-linked by 5-S-Cys-DOPA, but this has yet to be proven. In any case, 5-S-Cys-DOPA cross-links remain redox-active, with an oxidation potential of ~50 mV at pH 7,35 making them more reducing than DOPA.
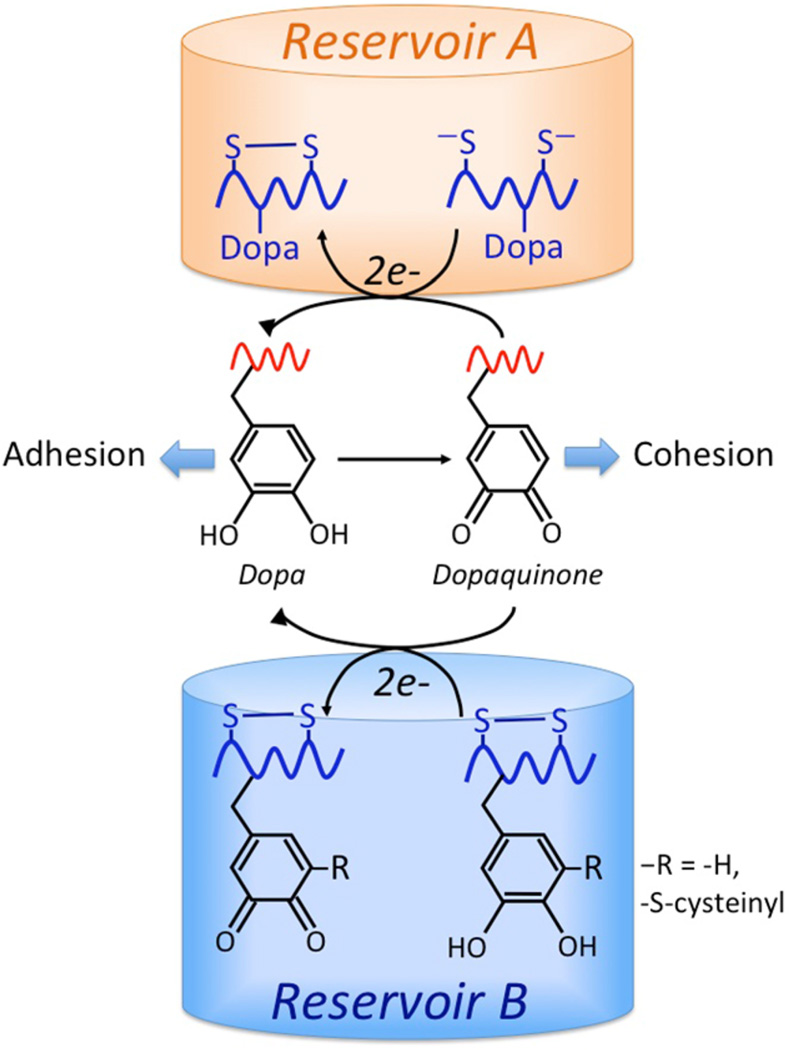
Figure 6.
Reducing reservoirs in Mfp-6 that maintain DOPA-containing adhesive proteins Mfp-3 and Mfp-5 during surface deposition. The reduced form of DOPA is required for the first, bidentate adsorption step of adhesion, whereas the oxidized form (Dopaquinone) contributes to the covalent pathways of cohesion, e.g., aryl coupling and Michael additions. Reservoir A consists of 9 equiv of thiolate Cys residues/Mfp-6 that provides strongly reducing electrons (9e− + 9H+). Reservoir B, in contrast, consists of 4–5 equiv of DOPA/Mfp-6 that provides weakly reducing electrons (8–10e− + 8–10H+). There may be electron shuttling between the two populations.
The 1D and 2D proton NMR spectra of Mfp-6 at pH 3 provide evidence that this protein has a well-defined domain with a hydrophobic core and extended β-sheets (Figure 5). Even when Mfp-6 was saturated with DPPH (at molar ratios of 1:50), the resulting 2D NOESY spectra indicate that the overall structure of Mfp-6 was conserved (Figure S6). Although the analysis is incomplete, the β-sheet fold may be bifunctional, offering, on one hand, a blueprint for self-assembly already widely associated with the β-sheet-rich amyloid fibrils34 and the cysteine and β-sheet-rich β-keratins36 and, on the other, a structure conducive to electron shuttling as in α-crystallins.37–39
Perhaps Mfp-6 solubility at low and high pHs can be tuned by constructing mutants consisting of only one of the putative domains or by truncation experiments. Investigative aims such as these must await successful expression of recombinant Mfp-6 homologues with higher fidelity to foot-extracted structure.40 Eventual determination of Mfp-6 crystal structures under different pH conditions will help us understand how Mfp-6 undergoes the transition from a chemically reactive globular protein with bulky and reactive side chains, including cysteine, tyrosine, and DOPA, to a load-bearing structural protein that mediates H-bonded or cross-linked tight packing in the mature plaque matrix.
In conclusion, the catecholic moiety of DOPA has been previously shown to endow polypeptides with wet adhesion by chemisorbing to a variety of oxide surfaces, e.g., titania and mica, by bidentate chelation and/or H-bonding.5,8 Once DOPA is oxidized to quinone, however, Mfp adhesion is effectively abolished on all but nucleophilic surfaces, where the Dopaquinone still has a cross-linking option with other proteins, for example. Mussels retard oxidation by depositing Mfp-3 and Mfp-5 under acidic conditions where the catecholic form of DOPA is favored.10 However, a low plaque pH can be only briefly maintained in an adhesive plaque surrounded by seawater at pH 8.2, and surface binding by DOPA needs time to overcome the conformational entropy of the Mfps. An antioxidant DOPA salvage mechanism may function to increase the size of the window for surface binding. Mfp-6 contains a reservoir of reducing electrons capable of recovering DOPA in Mfp-3 from the quinone. These results confirm that at least nine Cys thiols in Mfp-6 participate in the reduction of quinones41 but suggest that up to eight additional electrons may be provided by four hostage DOPA residues in Mfp-6 that are not available for adhesion (Figure 6). This means that although pristine Mfp-6 is not an enzyme, each protein is capable of approximately eight quinone to DOPA turnovers. The estimate is a minimum as cysteinyl–DOPA adducts continue to possess reducing capacity, though their reducing power is lower than that of thiols.35 The reducing and low-pH conditions imposed during plaque deposition are unlikely to last very long after the foot retracts, given equilibration of the plaque with ambient seawater at pH ~8 and Eo = 0.7 V (SHE). The question is how long. Recent live mussel studies suggest that plaques remain reducing even after 21 days in seawater.42 How mussels contrive to maintain such a long lasting reducing environment in plaques with limited reservoirs and no enzymes, ATP, or NADH remains an important question for science and technology.
Supplementary Material
Acknowledgments
We thank E. G. Ionita for providing helpful insights into the DPPH radical chemistry and V. D. Vacquier for critical suggestions.
Funding
The National Institutes Health (R01 DE 018468) and the Materials Research Science and Engineering Centers Program of the National Science Foundation (Grant DMR 1121053) funded this research.
ABBREVIATIONS
- CD
circular dichroism
- CV
cyclic voltammetry
- DLS
dynamic light scattering
- DOPA
3,4-dihydroxyphenylalanine
- Q
Dopaquinone
- DPPH
2,2-diphenyl-1-picrylhydrazyl
- ESI
electrospray ionization
- IAM
iodoacetamide
- Mfp
Mytilus foot protein
- NOESY
nuclear Overhauser effect spectroscopy
- NMR
nuclear magnetic resonance spectroscopy
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SHE
standard hydrogen electrode
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Purification and quality control of Mfp-6, antioxidant properties and NOE NMR of recombinant and modified recombinant Mfp-6, and details regarding circular dichroism and mass spectrometry of iodoacetamide-modified foot-extracted Mfp-6 and its cross-linking properties (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Ritz D, Beckwith J. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 2001;55:21–48. doi: 10.1146/annurev.micro.55.1.21. [DOI] [PubMed] [Google Scholar]
- 2.Ortenberg R, Beckwith J. Functions of thiol-disulfide oxidoreductases in E. coli: Redox myths, realities, and practicalities. Antioxid. Redox Signaling. 2003;5:403–411. doi: 10.1089/152308603768295140. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee R. Redox outside the box: linking extracellular redox remodeling with intracellular redox metabolism. J. Biol. Chem. 2012;287:4397–4402. doi: 10.1074/jbc.R111.287995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ottaviano BS, Handy DE, Loscalzo J. Redox regulation in the extracellular environment. Circ. J. 2008;72:1–16. doi: 10.1253/circj.72.1. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, Wei W, Danner E, Ashley RK, Israelachvili JN, Waite JH. Mussel protein adhesion depends on interprotein thiol-mediated redox modulation. Nat. Chem. Biol. 2011;7:588–590. doi: 10.1038/nchembio.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H, Scherer NF, Messersmith PB. Single-molecule mechanics of mussel adhesion. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12999–13003. doi: 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danner E, Kan Y, Hammer M, Israelachvili JN, Waite JH. Adhesion of mussel foot protein Mefp-5 to mica: an underwater superglue. Biochemistry. 2012;51:6511–6518. doi: 10.1021/bi3002538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J, Wei W, Danner E, Israelachvili JN, Waite JH. Effects of interfacial redox in mussel adhesive protein films on mica. Adv. Mater. 2011;23:2362–2366. doi: 10.1002/adma.201003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez Rodriguez NR, Das S, Kaufman Y, Israelachvili JN, Waite JH. Interfacial pH during mussel adhesive plaque formation. Biofouling. 2015;31:221–227. doi: 10.1080/08927014.2015.1026337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicklisch SCT, Waite JH. Mini-review: The role of redox in Dopa-mediated marine adhesion. Biofouling. 2012;28:865–877. doi: 10.1080/08927014.2012.719023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao H, Waite JH. Linking adhesive and structural proteins in the attachment plaque of Mytilus californianus. J. Biol. Chem. 2006;281:26150–26158. doi: 10.1074/jbc.M604357200. [DOI] [PubMed] [Google Scholar]
- 12.Antolovich M, Prenzler PD, Patsalides E, McDonald S, Robards K. Methods for testing antioxidant activity. Analyst. 2002;127:183–198. doi: 10.1039/b009171p. [DOI] [PubMed] [Google Scholar]
- 13.Nicklisch SCT, Waite JH. Optimized DPPH assay in a detergent-based buffer system for measuring antioxidant activity of proteins. MethodsX. 2014;1:233–238. doi: 10.1016/j.mex.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villaño D, Fernández-Pachón MS, Moyá ML, Troncoso aM, García-Parrilla MC. Radical scavenging ability of polyphenolic compounds towards DPPH free radical. Talanta. 2007;71:230–235. doi: 10.1016/j.talanta.2006.03.050. [DOI] [PubMed] [Google Scholar]
- 15.Campos A, Duran N, Lopez-Alarcon C, Lissi E. Kinetic and stoichiometric evaluation of free radicals scavengers activities based on diphenyl-picryl hydrazyyl (DPPH) consumption. J. Chil. Chem. Soc. 2012;57:1381–1384. [Google Scholar]
- 16.Brand-Williams W, Cuvelier ME, Berset C. Use of a free radical method to evaluate antioxidant activity. LWT - Food Sci. Technol. 1995;28:25–30. [Google Scholar]
- 17.Arteaga JF, Ruiz-Montoya M, Palma A, Alonso-Garrido G, Pintado S, Rodriguez-Mellado JM. Comparison of the simple voltammetry and DPPH assays for the determination of antioxidant capacity of active principles. Molecules. 2012;17:5126–5138. doi: 10.3390/molecules17055126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grzesiek S, Bax A. The importance of not saturating water in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 1993;115:12593–12594. [Google Scholar]
- 19.Piotto M, Saudek V, Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 20.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 21.Raoof JB, Ojani R, Beitollahi H. L-Cysteine voltammetry at a carbon paste electrode bulk-modified with ferrocenedicarboxylic acid. Electroanalysis. 2007;19:1822–1830. [Google Scholar]
- 22.Maree S, Nyokong T. Electrocatalytic behavior of substituted cobalt phthalocyanines towards the oxidation of cysteine. J. Electroanal. Chem. 2000;492:120–127. [Google Scholar]
- 23.Liu X, Zhang Z, Cheng G, Dong S. Spectroelectrochemical and voltammetric studies of L-DOPA. Electroanalysis. 2003;15:103–107. [Google Scholar]
- 24.Eslami M, Zare HR, Namazian M. Thermodynamic parameters of electrochemical oxidation of L-DOPA: Experimental and theoretical studies. J. Phys. Chem. B. 2012;116:12552–12557. doi: 10.1021/jp3054229. [DOI] [PubMed] [Google Scholar]
- 25.Shen X, Xia B, Wrona MZ, Dryhurst G. Synthesis, redox properties, in vivo formation, and neurobehavioral effects of N-acetylcysteinyl conjugates of dopamine: possible metabolites of relevance to Parkinson’ s disease. Chem. Res. Toxicol. 1996;9:1117–1126. doi: 10.1021/tx960052v. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F, Dryhurst G. Effects of L-cysteine on the oxidation chemistry of dopamine: new reaction pathways of potential relevance to idiopathic Parkinson’s disease. J. Med. Chem. 1994;37:1084–1098. doi: 10.1021/jm00034a006. [DOI] [PubMed] [Google Scholar]
- 27.Pain R. Determining the CD spectrum of a protein. Curr. Protoc. Protein Sci. 2004 doi: 10.1002/0471140864.ps0706s38. 7.6.1. [DOI] [PubMed] [Google Scholar]
- 28.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 29.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: John Wiley & Sons, Inc.; 1986. [Google Scholar]
- 30.Pople JA. Proton magnetic resonance of hydrocarbons. J. Chem. Phys. 1956;24:1111. [Google Scholar]
- 31.Jensen KS, Hansen RE, Winther JR. Kinetic and thermodynamic aspects of cellular thiol-disulfide redox regulation. Antioxid. Redox Signaling. 2009;11:1047. doi: 10.1089/ars.2008.2297. [DOI] [PubMed] [Google Scholar]
- 32.Stewart AJ, Blindauer CA, Berezenko S, Sleep D, Tooth D, Sadler PJ. Role of Tyr84 in controlling the reactivity of Cys34 of human albumin. FEBS J. 2005;272:353–362. doi: 10.1111/j.1742-4658.2004.04474.x. [DOI] [PubMed] [Google Scholar]
- 33.Gülçin I. Comparison if in vitro antioxidant and antiradical activities of L-Tyrosine and L-Dopa. Amino Acids. 2007;32:431–438. doi: 10.1007/s00726-006-0379-x. [DOI] [PubMed] [Google Scholar]
- 34.Proudfoot GM, Ritchie IM. A cyclic voltammetric study of some 4-substituted Benzene-1,2-diols. Aust. J. Chem. 1983;36:885–894. [Google Scholar]
- 35.Shen X, Dryhurst G. Further insights into the influence of L-cysteine on the oxidation chemistry of Dopamine: reaction pathways of potential relevance to Parkinson’s disease. Chem. Res. Toxicol. 1996;9:751–763. doi: 10.1021/tx960008f. [DOI] [PubMed] [Google Scholar]
- 36.Dzwolak W, Loksztejn A, Smirnovas V. New insights into the self-assembly of insulin amyloid fibrils: an HD exchange FT-IR study. Biochemistry. 2006;45:8143–8151. doi: 10.1021/bi060341a. [DOI] [PubMed] [Google Scholar]
- 37.Dalla Valle L, Nardi A, Bonazza G, Zuccal C, Emera D, Alibardi L. Forty keratin-associated beta-proteins (beta-keratins) form the hard layers of scales, claws, and adhesive pads in the green anole lizard, Anolis carolinensis. J. Exp. Zool., Part B. 2010;314:11–32. doi: 10.1002/jez.b.21306. [DOI] [PubMed] [Google Scholar]
- 38.Slingsby C, Clout N. Structure of the crystallins. Eye. 1999;13:395–402. doi: 10.1038/eye.1999.113. [DOI] [PubMed] [Google Scholar]
- 39.Manzanares D, Bauby C, de la Peña R, Garcia JC, Sanchez R, Martinez S, Romay CH, López-Reconde JL, Pino E, Lissi EA. Antioxidant properties of alpha-Crystallin. J. Protein Chem. 2001;20:181–189. doi: 10.1023/a:1010996528884. [DOI] [PubMed] [Google Scholar]
- 40.Chen SJ, Sun TX, Akhtar NJ, Liang JJ. Oxidation of human lens recombinant alpha A-Crystallin and cysteine-deficient mutants. J. Mol. Biol. 2001;305:969–976. doi: 10.1006/jmbi.2000.4348. [DOI] [PubMed] [Google Scholar]
- 41.Nicklisch SCT, Das S, Martinez Rodriguez NR, Waite JH, Israelachvili JN. Antioxidant efficacy and adhesion rescue by a recombinant mussel foot protein-6. Biotechnol. Prog. 2013;29:1587–1593. doi: 10.1002/btpr.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller DR, Spahn JA, Waite JH. The staying power of adhesion associated anti-oxidant activity in Mytilus californianus. J. R. Soc., Interface. 2015;12:20150614. doi: 10.1098/rsif.2015.0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.