Abstract
regionReport is an R package for generating detailed interactive reports from regions of the genome. The report includes quality-control checks, an overview of the results, an interactive table of the genomic regions and reproducibility information. regionReport can easily be expanded with report templates for other specialized analyses. In particular, regionReport has an extensive report template for exploring derfinder results from annotation-agnostic RNA-seq differential expression analyses.
Keywords: Report, Interactive, Reproducibility, Genomics, Sequencing, ChIP-seq, RNA-seq, Software
Introduction
Many analyses of genomic data result in regions along the genome that associate with a covariate of interest. These genomic regions can result from identifying differentially bound peaks from ChIP-seq data 1, identifying differentially methylated regions (DMRs) from DNA methylation data 2, or performing base-resolution differential expression analyses using RNA sequencing data 3, 4. The genomic regions themselves are commonly stored in a GRanges object from GenomicRanges 5 when working with R or the BED file format on the UCSC Genome Browser 6, but other information on these regions, for example summary statistics on the magnitude of effects and statistical significance, also provide useful information. The usage of R in genomics is increasingly common due to the usefulness and popularity of the Bioconductor project 7, and in the latest version (3.0), 181 unique packages use GenomicRanges for many workflows, demonstrating the widespread utility of identifying and summarizing characteristics of genomic regions.
Here we introduce regionReport which allows users to explore genomic regions of interest through interactive stand-alone HTML reports that can be shared with collaborators. These reports are flexible enough to display plots and quality control checks within a given experiment, but can easily be expanded to include custom visualizations and conclusions. The resulting HTML report emphasizes reproducibility of analyses 8 by including all the R code without obstructing the resulting plots and tables. We envision regionReport will provide a useful tool for exploring and sharing genomic region-based results from high throughput genomics experiments.
Methods
Implementation
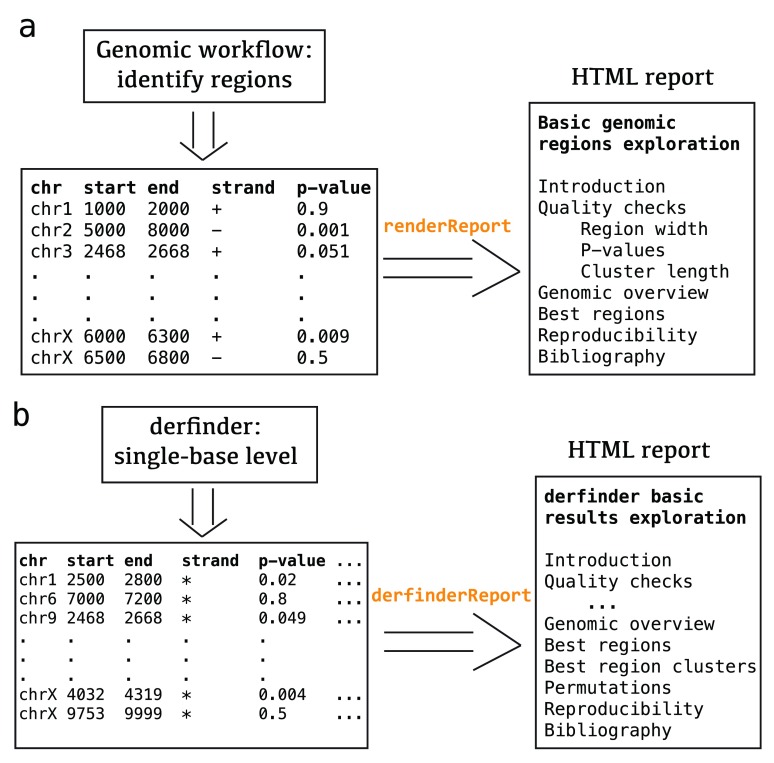
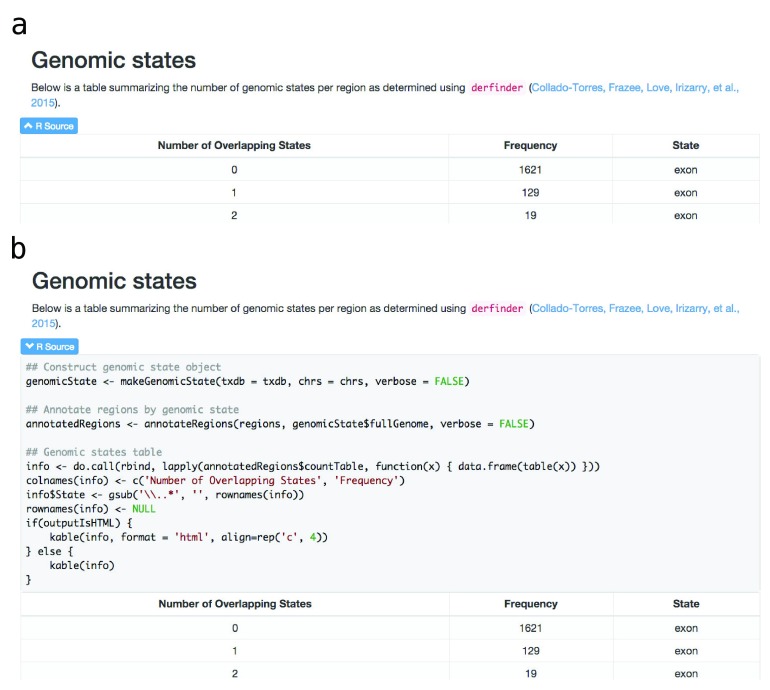
The package includes a R Markdown template which is processed using knitr 9 and rmarkdown 10, then styled using knitrBootstrap 11. This package generates a HTML report that includes a series of plots for checking the quality of the results and browsing the table of regions. Each element of the report has a brief explanation, although actual interpretation of the results is dataset- and workflow-dependent. To facilitate navigation a menu is always included, which is useful for users interested in a particular section of the report. Figure 1, panel a shows the menu of the general report for a set of regions with associated p-values. The code for each plot or table is hidden by default and can be shown by clicking on the appropriate toggle as shown in Figure 2.
Figure 1. regionReport workflow.
Example region input, the appropriate regionReport function to use, and menu of the resulting report for the general use case (panel a) and derfinder results (panel b).
Figure 2. Interactively display the code for each table/figure in the report.
View by default (panel a) and after clicking on the “R source” toggle (panel b) for a section of the general report. The full report is available at the supplementary website and includes a toggle to hide/show all the R code.
Quality checks
This section of the report includes a variety of quality control steps which help the user determine whether the results are sensible. The quality control steps explore:
P-values, Q-values, and FWER adjusted p-values
Region width
Region area: sum of single-base level statistics (if available)
Mean coverage or other score variables (if available)
A combination of density plots and numerical summaries are used in these quality checks. If there are statistically significant regions, the distributions are compared between all regions and the significant ones. For example, the distribution region widths might have a high density of small values for the global results, but shifted towards higher values for the subset of significant regions.
Genomic overview
The report includes plots to visualize the location of all the regions as well as the significant ones. Differences between them can reveal location biases. The nearest known annotation feature for each region is summarized and visually inspected in the report.
Best regions
An interactive table with the top 500 (default) regions is included in this section. This allows the user to sort the region information according to their preferred ranking option. For example, lowest p-value, longest width, chromosome, nearest annotation feature, etc. The table also allows the user to search and subset it interactively. A common use case is when the user wants to check if any of the regions are near a known gene of their interest.
Reproducibility
At the end of the report, detailed information is provided on how the analysis was performed. This includes the actual function call to generate the report, the path where the report was generated, time spent, and the detailed R session information including package versions of all the dependencies.
The R code for generating the plots and tables in the report is included in the report itself, thus allowing users to manually reproduce any section of the report, customize them, or simply change the graphical parameters to their liking.
derfinder report
When exploring derfinder results, for each of the best 100 (default) DERs a plot showing the coverage per sample is included in the report. These plots allow the user to visualize the differences identified by derfinder along known exons, introns and isoforms. The plots are created using derfinderPlot, also available via Bioconductor.
Due to the intrinsic variability in RNA-seq coverage data or mapping artifacts, in situations where there are two candidate DERs that are relatively close there might be reasons to consider them a single candidate DER and its important to visualize them. This tailored report groups candidate DERs into clusters based on a distance cutoff. After ranking them by their area, for the top 20 (default) clusters it plots tracks with the coverage by sample, the mean coverage by group, the identified candidate DERs colored by whether they are statistically significant, and known alternative transcripts. Figure 1, panel b shows the main categories of the report generated from a richer region data set than in the general case.
Operation
Installation. regionReport and required dependencies can be easily installed from Bioconductor with the following commands:
source(“ http://bioconductor.org/biocLite.R”)
biocLite(“regionReport”)
Input. To generate the report, the user first has to identify the regions of interest according to their analysis workflow. For example, by performing bumphunting to identify DMRs with bumphunter. The report is then created using renderReport() which is the main function in this package as shown in Figure 1, panel a. The argument customCode can be used to customize the report if necessary.
For the derfinder use case, the derfinderReport() function creates the recommended report that includes visualizations of the coverage information for the best regions and clusters of regions.
Output. A small example can be generated using:
example(“renderReport”, “regionReport”, ask=FALSE)
The resulting HTML file will open in the users default browser. Note that alternative output formats such as PDF files can also be generated, although they are not as dynamic and interactive as the HTML format.
Use cases
The supplementary website contains reports using DiffBind, bumphunter and derfinder results. The derfinder use case is illustrated with data sets previously described in 3 which span simulation results, a moderately sized data set (25 samples), and a large data set with 487 samples; thus covering a wide range of scenarios.
Summary
regionReport creates interactive reports from a set of regions and can be used in a wide range of genomic analyses. Reports generated with regionReport can easily be extended to include further quality checks and interpretation of the results specific to the data set under study. These shareable documents are very powerful when exploring different parameter values of an analysis workflow or applying the same method to a wide variety of data sets. The reports allow users to visually check the quality of the results, explore the properties of the genomic regions under study, and inspect the best regions and interactively explore them.
Furthermore, regionReport promotes reproducibility of data exploration and analysis. Each report provides R code that can be used as the starting point for other analyses within a dataset. regionReport provides a flexible output for exploring and sharing results from high throughput genomics experiments.
Software availability
Software access
regionReport is freely available via Bioconductor at bioconductor.org.
http://leekgroup.github.io/regionReportSupp/ hosts the code for generating three types of reports as well as the resulting HTML reports generated by regionReport. Versions of all software used are included in the reports.
Latest source code
The latest source code is available at Bioconductor and github.com/leekgroup/regionReport via the git-svn bridge although we recommend users to install regionReport directly from Bioconductor.
Archived source code as at the time of publication
Archived source code available at http://dx.doi.org/10.5281/zenodo.17083
License
Artistic-2.0.
Funding Statement
J.T.L. was partially supported by NIH Grant 1R01GM105705, L.C-T. was supported by Consejo Nacional de Ciencia y Tecnolog’ia México 351535.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 1 approved
References
- 1. Stark R, Brown G: DiffBind: differential binding analysis of ChIP-Seq peak data.2011. Reference Source [Google Scholar]
- 2. Jaffe AE, Murakami P, Lee H, et al. : Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41(1):200–209. 10.1093/ije/dyr238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Torres LC, Frazee AC, Love MI, et al. : derfinder: Software for annotation-agnostic rna-seq differential expression analysis. bioRxiv. 2015;015370 10.1101/015370 [DOI] [Google Scholar]
- 4. Frazee AC, Sabunciyan S, Hansen KD, et al. : Differential expression analysis of RNA-seq data at single-base resolution. Biostatistics. 2014;15(3):413–26. 10.1093/biostatistics/kxt053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lawrence M, Huber W, Pages H, et al. : Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118. 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rosenbloom KR, Armstrong J, Barber GP, et al. : The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43(Database issue):D670–D681. 10.1093/nar/gku1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huber W, Carey VJ, Gentleman R, et al. : Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods. 2015;12(2):115–121. 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sandve GK, Nekrutenko A, Taylor J, et al. : Ten simple rules for reproducible computational research. PLoS Comput Biol. 2013;9(10):e1003285. 10.1371/journal.pcbi.1003285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xie Y: Dynamic Documents with R and knitr. CRC Press.2013;216 Reference Source [Google Scholar]
- 10. Rstudio. RMarkdown: Dynamic Documents for R.2014. Reference Source [Google Scholar]
- 11. Hester J: Knitr Bootstrap framework, R package.2015. Reference Source [Google Scholar]