Version Changes
Updated. Changes from Version 2
The current version includes changes suggested by two referees, and has two additional tables and three additional figures. Firstly, the DOCLASP predicted binding of phenylthiourea to polyphenol oxidases (PPO) from walnut (JrPPO1) ( http://onlinelibrary.wiley.com/doi/10.1111/tpj.13207/) was corroborated using the recently solved structure of JrPPO1 (PDBid:5CE9). Secondly, a comprehensive analysis of the binding of the antitrypanosomial drug suramin to different non-homologous proteins from the PDB database has been presented, and DOCLASP was used to dock suramin to a phospholipase A2-like protein. This highlighted the non-specific binding of some ligands, and also the complexity of modelling a flexible ligand like suramin.
Abstract
The ability to accurately and effectively predict the interaction between proteins and small drug-like compounds has long intrigued researchers for pedagogic, humanitarian and economic reasons. Protein docking methods (AutoDock, GOLD, DOCK, FlexX and Glide to name a few) rank a large number of possible conformations of protein-ligand complexes using fast algorithms. Previously, it has been shown that structural congruence leading to the same enzymatic function necessitates the congruence of electrostatic properties (CLASP). The current work presents a methodology for docking a ligand into a target protein, provided that there is at least one known holoenzyme with ligand bound - DOCLASP (Docking using CLASP). The contact points of the ligand in the holoenzyme defines a motif, which is used to query the target enzyme using CLASP. If there are significant matches, the holoenzyme and the target protein are superimposed based on congruent atoms. The same linear and rotational transformations are also applied to the ligand, thus creating a unified coordinate framework having the holoenzyme, the ligand and the target enzyme. In the current work, the dipeptidyl peptidase-IV inhibitor vildagliptin was docked to the PI-PLC structure complexed with myo-inositol using DOCLASP. Also, corroboration of the docking of phenylthiourea to the modelled structure of polyphenol oxidase (JrPPO1) from walnut is provided based on the subsequently solved structure of JrPPO1 (PDBid:5CE9). Analysis of the binding of the antitrypanosomial drug suramin to nine non-homologous proteins in the PDB database shows a diverse set of binding motifs, and multiple binding sites in the phospholipase A2-likeproteins from the Bothrops genus of pitvipers. The conformational changes in the suramin molecule on binding highlights the challenges in docking flexible ligands into an already ’plastic’ binding site. Thus, DOCLASP presents a method for ’soft docking’ ligands to proteins with low computational requirements.
Keywords: protein, docking ligand, congruence
Introduction
The ability to computationally predict protein-ligand interactions with accuracy is an invaluable asset, since it allows for large scale screening at minimal costs 1, 2. Consequently, computational methods that predict the favorable conformation of a protein-ligand complex have been the focus of intense research over the last few decades 3. Protein docking methods are a subset of these methods, characterized by their ability to score a large number of possible conformations using fast algorithms. Among these, five programs - AutoDock 4, GOLD 5, DOCK 6, FlexX 7 and Glide 8 - are the most cited, although it should be noted that ‘the number of citations of a given paper is no measure of quality of the corresponding protein-ligand docking software program’ 9. Typically, a protein-ligand docking program has two distinct phases - conformational sampling (or searching) and scoring 10. Despite the significant progress in the field, there are several challenges arising from protein or ligand flexibility, entropic considerations or the presence of water molecules that need to be addressed 11.
Previously, the conservation of spatial and electrostatic properties in cognate pairs of residues in the catalytic site of proteins with the same functionality has been used to develop a computational method (CLASP) for detecting binding and catalytic sites 12– 15. In the current work, this methodology has been extended by proposing a method for docking ligands into target proteins - DOCLASP ( Docking using CLASP). DOCLASP takes as input a set of proteins with known structures which bind a particular ligand, and a target protein into which the ligand is to be docked. Each of these holo structures is used to define a motif consisting of the first four residues making non-hydrophobic interactions. These motifs are used to query the target protein, using an enhanced version of the search engine used by CLASP that uses precompiled databases 16, and significant congruent matches are identified. These significant matches in the target protein are now superimposed to the binding residues (the motif) in the corresponding holoenzyme(s), thus creating a unified coordinate framework formed by the holoenzyme, the ligand and the target enzyme. This gives us the docked ligand to the target protein, which is outputted as a Pymol formatted file. In essence, the holoenzyme is replaced with the target enzyme if the contact points have a good spatial and electrostatic match in the target enzyme by aligning the congruent atoms. Thus, DOCLASP leverages the implicit search and scoring functions in CLASP to rank possible conformations.
The native activity of phosphoinositide-specific phospholipase C (PI-PLC) was previously shown to be inhibited by two dipeptidyl peptidase-IV (DPP4) inhibitors - vildagliptin (LAF-237) at micromolar concentrations, and K-579 at nanomolar concentrations using in vitro experiments based on CLASP analysis 15. Since ‘comparing docking programs can be difficult’ 9, the DOCLASP methodology is validated by docking vildagliptin to the PI-PLC structure in complex with myo-inositol 17. The docked ligand is free from steric clashes and interacts with the exact side chain residues that bind myo-inositol, providing corroboration of the validity of the proposed methodology. Next, an inhibitor of polyphenol oxidase (PPO) 18 was docked to the solved structure of a PPO from walnut 19, corroborating previous docking results from a modelled structure of the same protein 20. Finally, the promiscuous binding of suramin, a well-known antitrypanosomial drug 21, to nine non-homologous proteins in the PDB database revealed diverse binding motifs, and multiple binding sites even within phospholipase A2-like proteins from the Bothrops genus of pitvipers 22. Also, the conformational changes in suramin upon binding underscores the complexity of docking algorithms, which must sample a much larger conformational space created by both the changing binding site residues and ligand 23. Thus, the current work presents a fast methodology for docking ligands into protein structures based on spatial and electrostatic congruence of known binding sites to putative binding targets.
Materials and methods
solvedJrPPO1Docked.pdb: phenylthiourea docked to the solved structure of JrPPO1 (Polyphenol oxidase from walnut, PDBid:5CE9).
modelledJrPPO1Docked.pdb: phenylthiourea docked to the modelled structure of JrPPO1 (Polyphenol oxidase from walnut) using SWISSMODEL based on the PDBid:1BUG, since the solved structure was not available at that time.
PLA2dockedsuramin.p1m: suramin docked to the phospholipase A2-like protein from PDBid:3BJW from Echis carinatus (saw scaled viper), showing that it has multiple sites of suramin binding.
Copyright: © 2016 Chakraborty S
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
DOCLASP uses the basic hypothesis of CLASP - the non-triviality of the spatial and electrostatic congruence in cognate pairs seen across different structures of the same catalytic function, which is extended to the related concept of ligand binding 12. It takes as input a set of Z proteins with known structures ( Equation 1) which bind a particular ligand ( Lig), and a target protein into which Lig is to be docked ( P target). Each of these Z holo structures is used to define a motif consisting of N (=4) residues ( Equation 2), taking the first four closest non-hydrophobic interactions into account ( Algorithm 1).
Algorithm 1. GetMotif(): Choose n closest atoms, excluding hydrophobic interactions.
Input: Protein :
Input: Ligand :
Input: n: number of closest atoms to choose
Output: ϕ motif = { atom 1 … atom n}
begin
/* Output Motif */
ϕ motif = ∅ ;
/* Accepted atom pairs - exclude hydrophobic interactions*/
ϕ AcceptedAtomPair = [O-N, N-O, O-H, H-O, O-O, N-N, N-H, H-N, S-H, H-S] ;
ATOMS Lig = atoms of all residues of Ligand ;
/* Initial radius in Å */
Radius = 2.5 ;
foreach atom i in ATOMS Lig do
ϕ atoms = ProteinAtomsWithinRadiusOfLigandAtom( Protein,atom i, Radius);
foreach atom j in ϕ atoms do
if atom i-atom j is in ϕ AcceptedAtomPair then
InsertInMotifSet( atom j, ϕ motif);
if (ϕ motif == n) then
last ;
end
end
end
/* increment radius by 0.1 Å */
Radius = Radius + 0.1;
end
return ϕ motif ;
end
Each position of the motif has a set of amino acids specified to allow for stereochemically equivalent matches at that particular position ( Equation 3), such that while matching amino acid type of r i should belong to GROUP i.
Previously, the K sets of N residues were obtained in P target using an exhaustive search procedure similar to the one used in SPASM 24. An enhanced algorithm now precompiles all possible motifs of a set ( N=4 in this case) of predefined amino acid residues from a protein structure that occur within a specified distance 16, and selects the appropriate ones based on each motif ( Equation 4). Any match below a user defined threshold score ( S thresh) is discarded.
In Case is null, the ligand Lig can not be docked to the target protein P target. , the first element, has the minimum CScore and represents the putative binding site in P target based on the holoenzyme P i. The set of putative binding sites Φ bindsite is thus defined ( Equation 5).
In order to compute CScore, the 3D distances and PD are computed and values are then compared with the corresponding feature values obtained from the holoenzyme motif. The distance scores are normalized since that the same pairwise distance deviation should count more when the reference distance is less. While scoring electrostatic potential differences, deviations < 100 are ignored. Similarly for higher potential differences, the deviations are more loosely constrained than for lower potential differences. The differences are absolute values.
Each element of Φ bindsite ( ) is now superimposed to the corresponding holoenzyme, based on the motif binding Lig in P i. In order to superimpose these motifs, linear and rotational transformations are applied on all atoms such that the first three atoms lie on the same plane (Z=0), the first atoms are the origin of the coordinate axis and the second atoms lie on the Y axis. This creates a unified coordinate framework having the holoenzyme, the ligand and the target enzyme, thus providing the docked ligand in the target enzyme. Essentially, the holoenzyme is replaced with the target enzyme if the contact points have a good spatial and electrostatic match in the target enzyme by aligning the congruent atoms. This docked ligand is now outputted as a Pymol formatted file.
The DOCLASP package is written in Perl on Ubuntu. Hardware requirements are modest - all results here are from a simple workstation (2GB ram) and runtimes were a few minutes at the most. Adaptive Poisson-Boltzmann Solver (APBS) and PDB2PQR packages were used to calculate the potential difference between the reactive atoms of the corresponding proteins 25, 26. The APBS parameters and electrostatic potential units were set as described previously in 12. All protein structures were rendered by PyMol ( http://www.pymol.org/).
Previous CLASP analysis of the spatial and electrostatic properties of active site residues in PI-PLC from B. cereus indicated that it is a prolyl peptidase, which was also validated by in vitro experiments 13. Subsequently, it was shown that PI-PLC is inhibited by two dipeptidyl peptidase-IV (DPP4) inhibitors - vildagliptin (LAF-237) at micromolar concentrations, and K-579 at nanomolar concentrations. Since there are no DPP4 structures solved which ligand K-579, a DPP4 protein structure in complex with vildagliptin (PDBid:3W2TA) 28 provided the five closest atoms in the protein (E205, E206, S630, Y662 and Y547) (see Methods) that make non-hydrophobic interactions with the ligand ( Table 1).
Table 1. Residues in dipeptidyl peptidase-IV (DPP4) holoenzyme (PDBid:3W2TA) that have non-hydrophobic interactions with the bound DPP4 inhibitor vildagliptin.
Interactions are sorted based on the distance. R/A/LA/D: Residue number/Atom of the residue/Atom of ligand/distance between the interacting atoms (in Å). For example, ‘S630/OG/N2/2.4’ means that the atom OG from Ser630 is at 2.4 Å from the N2 atom of vildagliptin in PDBid:3W2TA.
| R/A/LA/D | R/A/LA/D | R/A/LA/D | R/A/LA/D | R/A/LA/D |
|---|---|---|---|---|
| S630/OG/N2/2.4 | E205/OE1/N12/2.8 | Y662/OH/O20/3 | E206/OE2/N12/3 | Y547/OH/N2/3.1 |
A subset of these atoms might be sufficient to ligand vildagliptin in the target protein. Thus, motifs, each with four atoms, were created using the five closest atoms in the protein. The binding of ligands is known to induce electrostatic and spatial perturbations in the binding site. The spatial and electrostatic perturbations induced by the vildagliptin binding is shown by comparing the apo (PDBid:2OQIA) and the holoenzyme (PDBid:3W2TA) in Table 2. Hence, the electrostatic and spatial profile of the motif were obtained from the apo DPP4 enzyme (PDBid:2OQIA), and then used for querying the PI-PLC apo structure (PDBid:1PTDA).
Table 2. Changes in the conformation of the binding site due to vildagliptin (a DPP4 inhibitor) binding.
We compare the pairwise distance and electrostatic potential difference (EPD) changes in the apo (PDBid:2OQIA) and holo (PDBid:3W2TA) enzymes. Note that the pairwise distance between these atoms change in the ligand free PDB (2OQIA) as compared to the protein with bound inhibitor (2BUBA). For example, the distance between E205OE2 and E206OE2 (pair ab) changes from 3.9 Å to 5.6 Å. Also, there is a definite change in the EPD between E205OE2 and E205OE2 (pair ab). D = Pairwise distance in Å. PD = Pairwise potential difference. The electrostatic potential are in dimensionless units of kT/e where k is Boltzmann’s constant, T is the temperature in K and e is the charge of an electron.
| PDB | Active site atoms (a,b,c,d) | ab | ac | ad | bc | bd | cd | |
|---|---|---|---|---|---|---|---|---|
| 2OQIA
apo 3W2TA holo |
GLU205OE2,GLU206OE2,SER630OG,TYR662OH,
GLU205OE2,GLU206OE2,SER630OG,TYR662OH, |
D
PD D PD |
3.9
75.7 5.6 -74.4 |
7.9
-193.4 9.1 -307.9 |
4.0
-94.5 6.1 -159.8 |
10.1
-269.1 9.4 -233.5 |
3.5
-170.2 5.1 -85.5 |
7.3
98.9 5.6 148.1 |
These motifs were used to query the PI-PLC structure using an enhanced algorithm (PREMONITION) that precompiles all motifs in a database 16. Table 3 shows the best matches obtained in the PI-PLC structure for the five partial motifs. All these matches have significant electrostatic congruence. The root mean square deviation (RMSD) have low values - however, this is a deceptive metric since these deviations are averaged out. The maximum pairwise distance is another metric to discriminate the spatial congruence, and should be used in combination with the RMSD value. All of these above mentioned matches have significant maximum pairwise distance deviation. Also, three matches do not comprise of active site residues (motifs 2, 3 and 4). However, it can be seen that the first and fifth matches comprises of active site residues (involved in the binding of myo-inositol in PDBid:1PTGA), and have three residues (Asp67, Asp198 and Trp178) in common.
Table 3. Querying PI-PLC using partial motifs derived from the atoms in vildagliptin that make contact to the DPP4 enzyme (PDBid:3W2TA).
The comparison is done using apo enzymes (PDBid:1PTDA for PI-PLC and PDBid:2OQIA for DPP4), since the binding of a ligand induces spatial and electrostatic changes in the active site. The fifth motif has the least rmsd deviation, and comprises of active site residues (involved in the binding of myo-inositol in PDBid:1PTGA). Out of the best matches in the other four motifs, three do not comprise of active site residues (motifs 2, 3 and 4). Motif 1 has a reasonably significant match, and has three residues (Asp67, Asp198 and Trp178) in common with the best match for Motif 5. These three residues from PI-PLC (Asp67, Asp198 and Trp178) and corresponding three residues from DPP4 (Glu205, Glu206 and Tyr662) were used to superimpose DPP4 and PI-PLC. N = Motif number. D = Pairwise distance in Å. PD = Pairwise potential difference. Rmsd = Root mean square deviation. Max = maximum pairwise distance deviation. APBS writes out the electrostatic potential in dimensionless units of kT/e where k is Boltzmann’s constant, T is the temperature in K and e is the charge of an electron.
| PDB | N | Active site atoms(a,b,c,d) | ab | ac | ad | bc | bd | cd | Rmsd/Max
(Å) |
|
|---|---|---|---|---|---|---|---|---|---|---|
| 2OQIA
1PTDA |
1
|
S630OG,E205OE1,Y662CZ,E206OE1
S113OG,D67OD1,W178CZ2,D198OD1 |
D
PD D PD |
9.5
237 4.3 195 |
7.8
-104 6.7 -79 |
11.2
228 10.1 277 |
7.2
-341 6.7 -275 |
6.6
-9 7.7 81 |
5.8
332 4.9 357 |
0.9/5.1 |
| 2OQIA
1PTDA |
2
|
S630OG,E205OE1,Y662CZ,Y547CZ
S244OG,D274OD1,Y275CZ,Y246CZ |
D
PD D PD |
9.5
237 11.3 312 |
7.8
-104 7.2 -104 |
5.4
-241 8.1 -171 |
7.2
341 7.0 -417 |
11.3
-478 11.6 -484 |
9.3
-137 5.9 -67 |
0.8/3.3 |
| 2OQIA
1PTDA |
3
|
S630OG,E205OE1,E206OE1,Y547CZ
S129OG,D126OD1,E125OE1,Y164CZ |
D
PD D PD |
9.5
237 7.3 172 |
11.2
228 12.5 262 |
5.4
-241 6.7 -232 |
6.6
-9 5.5 90 |
11.3
-478 12.9 -405 |
10.4
-469 17.4 -495 |
1.3/7.0 |
| 2OQIA
1PTDA |
4
|
S630OG,Y662CZ,E206OE1,Y547CZ
S244OG,Y275CZ,D274OD1,Y246CZ |
D
PD D PD |
7.8
-104 7.2 -104 |
11.2
228 11.3 312 |
5.4
-241 8.1 -171 |
5.8
332 7.0 417 |
9.3
-137 5.9 -67 |
10.4
-469 11.6 -484 |
0.8/3.3 |
| 2OQIA
1PTDA |
5
|
E205OE1,Y662CZ,E206OE1,Y547CZ
D67OD1,W178CZ2,D198OD1,Y200CZ |
D
PD D PD |
7.2
-341 6.7 -275 |
6.6
-9 7.7 81 |
11.3
-478 11.6 -318 |
5.8
332 4.9 357 |
9.3
-137 9.6 -42 |
10.4
-469 6.3 -400 |
0.7/4.0 |
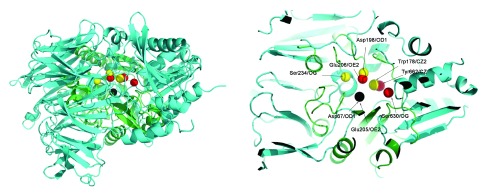
Thus, these three residues from PI-PLC (Asp67, Asp198 and Trp178) and corresponding three residues from DPP4 (Glu205, Glu206 and Tyr662) were used to superimpose DPP4 and PI-PLC. Table 4 shows the congruence of these residues. The corresponding holo structures - PDBid:3W2TA for DPP4, and PDBid:1PTGA for PIPLC - were used for the superimposition. This superimposition applies geometric transformations such that Asp67OD1 and Glu205OE2 were at the center of the coordinate axis (coordinates = [0,0,0]), Asp198OD1 and Glu206OE2 lies on the X-Y axis (i.e. Y coordinate is 0) and Tyr662CZ and Trp178CZ2 were on the X-Y plane (i.e. Z coordinate is 0). Figure 1 shows the superimposed proteins. It is observed that (Asp67, Asp198 and Trp178) overlaps well with (Glu205, Glu206 and Tyr662).
Figure 1. Superimposing PI-PLC (PDBid:3W2TA in green) and DPP4 (PDBid:2OQOA in cyan).
Three residues from PI-PLC (Asp67, Asp198 and Trp178 in yellow) were superimposed to the corresponding three residues from DPP4 (Glu205, Glu206 and Tyr662 in red). Asp67OD1 and Glu205OE2 is at the center of the coordinate axis (coordinates = [0,0,0]) (in black), Asp198OD1 and Glu206OE2 lies on the X-Y axis (i.e. Y coordinate is 0) and Tyr662CZ and Trp178CZ2 are on the X-Y plane (i.e. Z coordinate is 0). Asp67, Asp198 and Trp178 in the PI-PLC protein overlaps well with Glu205, Glu206 and Tyr662 from DPP4, but the Ser234-Ser630 pair is not spatially congruent.
Table 4. Spatial and electrostatic congruence of a three residue partial motif.
The match is significant and comprises active site residues (involved in the binding of myo-inositol in PDBid:1PTGA). Pair ‘ab’ is considered to be electrostatically congruent since the PD values are close to zero, and can be considered almost equipotential. This is expected for atoms of the same type from the same residue (GLU205OE1/GLU206OE1 and ASP67OD1/ASP198OD1). These three residues from PI-PLC (Asp67, Asp198 and Trp178) and corresponding three residues from DPP4 (Glu205, Glu206 and Tyr662) were used to superimpose DPP4 and PI-PLC. D = Pairwise distance in Å. PD = Pairwise potential difference. APBS writes out the electrostatic potential in dimensionless units of kT/e where k is Boltzmann’s constant, T is the temperature in K and e is the charge of an electron.
| PDB | Atoms(a,b,c) | ab | ac | bc | |
|---|---|---|---|---|---|
| 2OQIA
1PTDA |
GLU205OE1,GLU206OE1,TYR662CZ,
ASP67OD1,ASP198OD1,TRP178CZ2, |
D
PD D PD |
6.6
-9.3 7.7 81.8 |
7.2
-341.5 6.7 -275.7 |
5.8
-332.2 4.9 -357.6 |
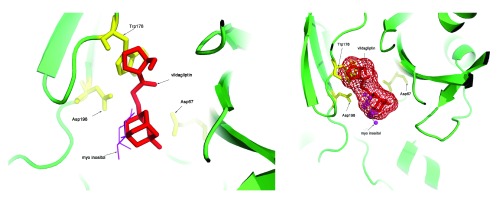
These transformations were also applied to the vildagliptin molecule, and this resulted in a docked structure for this molecule into the PI-PLC protein. Figure 2 shows the vildagliptin docked into the PI-PLC structure which is complexed with myo-inositol (PDBid:1PTGA). The distances of the atoms in vildagliptin and myo-inositol that interact (excluding hydrophobic interactions) to the first ten residues in the PI-PLC structure are shown in Table 5. It is interesting to note that the residues shown in Table 5 are all part of side chain residues in close contact with the myo-inositol ring (shown in Figure 7 13). Further validation was obtained by observing that both Arg69/NH1 and His32/NE2 interact with atom O4 in vildagliptin, and Arg69/NH2 and His32/NE2 interact with O2 in myo-inositol in the PI-PLC structure. The Pymol script for visualizing the docking (SupplementaryPymol.p1m) and a movie (SupplementaryMovie.avi) are also provided as Supplementary information.
Figure 2. Docking vildagliptin to the PI-PLC structure in complex with myo-inositol (PDBid:1PTGA).
It can be seen that vildagliptin fits into the binding site of PI-PLC. It also makes non-hydrophobic contacts to the residues in the protein similar to those made by myo-inositol ( Table 5).
Table 5. Atoms of myo-inositol and vildagliptin that make contact to the residues of the PI-PLC structure (PDBid:1PTGA).
These interactions exclude hydrophobic interactions, and the first closest ten atoms are chosen. Out of ten, seven residues obtained by docking vildagliptin using DOCLASP are seen to be equivalent to those that are known to bind myo-inositol to the PI-PLC structure 13, while two more have the same amino acid type (marked by asterisks). Only one pair has a different amino acid type (Tyr200 for myo-inositol and Glu117 for vildagliptin). Further validation is obtained by observing that both Arg69/NH1 and His32/NE2 both interact with atom O4 in vildagliptin, and Arg69/NH2 and His32/NE2 interact with O2 in myo-inositol in the PI-PLC structure.
| myo-inositol | vildagliptin | Same? |
|---|---|---|
| ASP198/OD1/O3/2.6
ARG163/NH2/O5/2.7 HIS32/NE2/O2/2.9 ARG69/NH2/O2/3.3 LYS115/NZ/O5/3.4 TRP178/NE1/O4/4.0 ASP33/OD1/O2/4.1 SER234/OG/O3/4.8 ASP180/OD1/O6/4.5 TYR200/OH/O6/4.4 |
ASP198/OD2/N12/3.1
ARG163/NE/O20/1.1 HIS32/NE2/O4/4.5 ARG69/NH1/O4/1.0 LYS115/NZ/O4/4.9 TRP178/NE1/N15/2.2 ASP33/OD1/O4/2.8 SER113/OG/O20/5.3 ASP67/OD2/O4/3.8 GLU117/OE1/O4/4.1 |
Y
Y Y Y Y Y Y * * N |
It is important to comment on the previous hypothesis of a nucleophilic serine being responsible for the inhibition of PI-PLC using vildagliptin. The electrostatic and spatial profile of the motif 4 from DPP4 is compared to the electrostatic and spatial profile of matching active site residues in PI-PLC in Table 6, including serine in the comparison. It can be seen that Ser630 in DPP4 has a significant spatial difference as compared to Ser234 in PI-PLC (pair ‘bc’ has a difference of 5 Å), and also a reasonable electrostatic difference (pair ‘ac’ has a difference of 144 PD units). The relatively large distance over which Ser234 in PI-PLC interacts with myo-inositol (4.8 Å) indicates that Ser234 is not directly involved in the binding of the ligand. However, it is responsible for creating the electrostatic milieu that is required for other interacting residues to attain their appropriate potential. Even for DPP4, many inhibitors do not interact with the nucleophilic Ser630 (manuscript in preparation) - although the vildagliptin molecule does 28. Thus, the previous conjecture of a nucleophilic serine being directly responsible for the binding of DPP4 inhibitors to PI-PLC, as implied by the catalytic triad congruence, is incorrect 13. However, this serine is indirectly responsible for driving the neighboring residues to an appropriate state. Spatial constraints are an additional discriminator.
Table 6. Potential and spatial congruence of the residues binding vildagliptin in DPP4 structure (PDBid:2OQIA) to the putative binding site in PI-PLC structure (PDBid:1PTDA).
Both the structures are apo enzymes, since the binding of a ligand induces spatial and electrostatic changes in the active site. Ser630 in DPP4 has a significant spatial difference as compared to Ser234 in PI-PLC (pair ‘bc’ has a difference of 5 Å), and also a reasonable electrostatic difference (pair ‘ac’ has a difference of 144 PD units). D = Pairwise distance in Å. PD = Pairwise potential difference. APBS writes out the electrostatic potential in dimensionless units of kT/e where k is Boltzmann’s constant, T is the temperature in K and e is the charge of an electron.
| PDB | Active site atoms(a,b,c,d) | ab | ac | ad | bc | bd | cd | |
|---|---|---|---|---|---|---|---|---|
| 2OQIA
|
GLU205OE1,GLU206OE2,SER630OG,TYR662CZ,
|
D
PD |
5.2
31.7 |
9.5
-237.4 |
7.2
-341.5 |
10.1
-269.1 |
4.2
-373.2 |
7.8
-104.1 |
| 1PTDA
|
ASP67OD1,ASP198OD1,SER234OG,TRP178CZ2,
|
D
PD |
7.7
81.8 |
8.2
-93.7 |
6.7
-275.7 |
5.7
-175.6 |
4.9
-357.6 |
9.2
-182.0 |
DOCLASP was also used recently 29 to dock human karyopherin to the VP24 30 protein of the Reston Ebola strain using the VP24 from Zaire Ebola 31 as a template, and demonstrate that a single mutation might be one of the critical factors responsible for the non-pathogenic nature of Reston Ebola in humans 32, 33.
Docking phenylthiourea to polyphenol oxidase from walnut
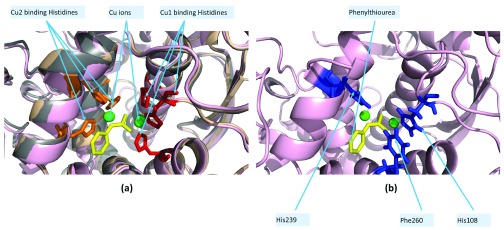
Polyphenol oxidases (PPO/tyrosinases/catechol oxidases) are copper enzymes implicated in the biosynthesis of quinones 34. The recently sequenced walnut genome sequence revealed the presence of two PPO genes (JrPPO1/2) 20, only one of which was previously known (JrPPO1) 18. Since there were no known structures of JrPPO1/2 at the time of writing 20, SWISSMODEL was used to model JrPPO1 (modelJrPPO1) based on the structure of the homologous PPO from Ipomoea batatas (PDBid:1BUG, sweet potato). The structure of JrPPO1 was recently solved (solvedJrPPO1, PDBid:5CE9) 19. DOCLASP docked phenylthiourea (URS) to modelledJrPPO1 and solvedJrPPO1 by aligning conserved copper binding histidines (His87, His108 and His117 in JrPPO1, Figure 3) (see modelledJrPPO1Docked.pdb and solvedJrPPO1Docked.pdb in Dataset1). The binding pose of URS showed a similar configuration in both modelledJrPPO1 and solvedJrPPO1 ( Table 7).
Figure 3. Docking phenylthiourea (URS) to polyphenol oxidase (PPO) from walnut based on the PPO from sweet potato (PDBid:1BUG).
The copper binding histidines (His88/109/118 for Cu1) are in red and (His240/244/274 for Cu2) are in orange. URS in yellow, copper ions in green. ( a) Superimposition of PDBid:1BUG (wheat), modelJrPPO1 (grey) and solvedJrPPO1 (pink). ( b) Three closest residues in solvedJrPPO1 that ligand URS (His108/His239/Phe260) are in blue.
Table 7. Docking phenylthiourea (URS) to modelled (modelJrPPO1) and solved (solved- JrPPO1,PDBid:5CE9A) polyphenol oxidase (PPO) from walnut: JrPPO1 has been modelled using the PPO from Ipomoea batatas (sweet potato), where URS is liganded by His109/His240/Phe261/Asn260.
After DOCLASP docking, modelledJrPPO1 has the analogous His108/His239/Asn259 as binding residues, while solvedJrPPO1 has His108/His239/Phe260 as binding residues.
| PDB | PDBatom | URSatom | Distance°A |
|---|---|---|---|
| 1BUGA
1BUGA 1BUGA 1BUGA 1BUGA |
HIS/109/NE2
HIS/109/CD2 PHE/261/N HIS/240/CE1 ASN/260/C |
N2
N2 C5 N1 C5 |
2.7
2.9 3.1 3.1 3.2 |
| modelJrPPO1
modelJrPPO1 modelJrPPO1 modelJrPPO1 modelJrPPO1 |
HIS/108/NE2
HIS/108/CD2 HIS/239/CE1 HIS/108/NE2 ASN/259/C |
N2
N2 N1 S1 C5 |
2.7
2.9 3.0 3.1 3.2 |
| solvedJrPPO1
solvedJrPPO1 solvedJrPPO1 solvedJrPPO1 solvedJrPPO1 |
PHE/260/CE1
HIS/108/NE2 PHE/260/CD1 HIS/239/CE1 HIS/108/CD2 |
N2
N2 N2 N1 N2 |
2.4
2.4 2.6 2.8 2.9 |
Analysis of suramin binding to a set of nine non-homologous proteins
Human African Trypanosomiasis (HAT), endemic to sub-Saharan Africa, is caused by the parasite Trypanosoma brucei and is transmitted via the tsetse fly 35. Suramin, a hexasulfonated naphthylurea, is a antitrypanosomial drug which has been in clinical use for decades, and more recently for the treatment of malignant tumors 36– 39. It has been known for a while now that suramin binds to several human proteins including cullin-RING E3 ubiquitin ligases 40, serum albumin 41, P2X receptors 42, neutrophil elastases 43, topoisomerase I and II 44, thrombin 45, receptor/G protein 46 and human secreted group IIA phospholipase A2 47. Such interactions may be the rationale for its toxicity in several cases 39. Of late, there have been a plethora of instances highlighting the potential of suramin based therapeutics: treating severe fever with thrombocytopenia syndrome caused by bunyavirus (by inhibiting viral nucleocapsid protein) 48, malaria (by inhibiting falcipain-2, a cysteine protease) 49, gastroenteritis (by inhibiting RNA dependent RNA polymerase in Noroviruses) 50, EV71 infection 51, tuberculosis (by inhibiting RecA) 52 and Dengue virus infection (by inhibiting envelope protein binding to target cell heparan sulfate).
The current PDB database has nine non-homologous protein structures which ligand suramin ( Supplementary Table 1). Positively charged residues (Lys and Arg) in some proteins (PDBids:1Y4LB, 1Y8EA, etc) and negatively charged residues (Asp and Glu) in other proteins (PDBids:2H9TH, 3GANA, etc) interact with suramin ( Table 8). However, all possible hydrogen bonds for negatively charged residues are with the backbone atoms (O or N), while the positively charged residues hydrogen bond to suramin using the sidechain atoms. Furthermore, the non-polar residues (Gly, Ala and Pro) also have (possible) hydrogen bonds to the backbone atoms only. This corroborates the fact that the suramin binds to positively charged parts of the protein 56. Each of these binding sites provides a motif to DOCLASP when suramin is to be docked to a target protein.
Table 8. Suramin binding residues in nine non-homologous proteins from the PDB database: Interactions sorted based on the distance.
R/A/LA/D: Residue number/Atom of the residue/Atom of ligand/distance between the interacting atoms (in Å). For example, ‘LYS53/NZ/O80/2.6’ means that the atom NZ from Lys53 is at 2.6 Å from the O80 of the suramin drug in PDBid:1Y4LB. The non-specific binding of suramin to phospholipase A2-like proteins (PLA2) is demonstrated by different binding sites for the homologous PLA2-X and PLA2-Z. Also, suramin binds to different sites even within the homologous subunits of PLA2-Y (marked with asterisks).
| PDBid | R/A/LA/D | R/A/LA/D | R/A/LA/D | R/A/LA/D |
|---|---|---|---|---|
| 1Y4LB,PLA2-X
3BJWA,PLA2-Y* 3BJWB,PLA2-Y* 4YV5A,PLA2-Z |
LYS53/NZ/O80/2.6
PHE124/CD2/O4/2.6 SER21/OG/O25/2.7 ASN114/ND2/O35/2.7 |
LYS69/NZ/O54/2.8
TRP125/O/O29/3.0 SER17/OG/N19/2.8 LYS116/NZ/O35/2.7 |
TYR52/CD2/C60/2.8
LYS127/N/O30/3.4 LYS115/NZ/O36/2.8 TYR121/CD2/N44/3.1 |
ARG34/CD/C70/2.9
PRO36/CD/O28/3.5 LYS16/NZ/O34/2.8 LYS115/C/O23/3.3 |
| 1Y8EA
2H9TH 2NYRB 3GANA 3PP7B 3UR0B 4J4VA 4X3UB |
ARG244/NH1/O23/2.7
GLU247/N/O77/2.5 TYR255/OH/O45/2.6 PRO152/O/N44/2.8 LYS335/NZ/O84/3.1 ARG413/NE/O84/2.7 THR63/O/N53/2.7 SER40/O/N63/2.6 |
LYS241/NZ/O24/2.7
ARG93/NE/O35/3.0 TYR102/OH/O30/2.8 ASP151/O/N41/3.0 ASN51/ND2/O81/3.1 PRO39/O/O79/2.8 ASN66/N/O78/2.9 HIS47/NE2/O23/2.8 |
LYS214/NZ/O84/2.7
ARG101/NE/O36/3.0 ARG105/NH2/O29/2.9 TYR48/OH/N53/3.0 THR26/OG1/O80/3.1 TRP42/NE1/O84/3.3 LYS67/NZ/O77/3.1 ARG20/NH1/O86/2.8 |
LYS220/NZ/O78/2.7
ASP243/O/N44/3.3 ARG71/NE/O23/3.0 GLY79/O/O54/3.8 GLY331/N/O82/3.3 ALA41/N/O79/3.3 GLU94/O/O85/3.4 TRP42/NE1/O81/3.0 |
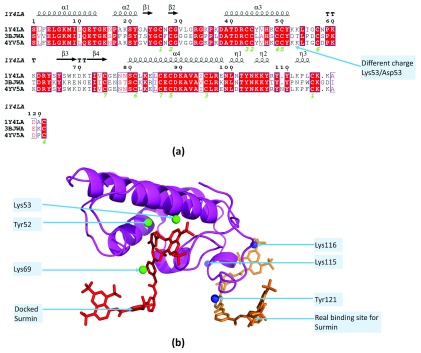
The extensive data available on suramin binding to different proteins highlights the complex nature of ligand docking. Three phospholipase A2-like proteins (PLA2) from poisonous vipers exist in the current PDB database which have suramin as a ligand: (i) PLA2-X, PDBid:1Y4L from Bothrops asper (pit viper), (ii) PLA2-Y, PDBid:3BJW from Echis carinatus (saw scaled viper) and (iii) PLA2-Z, PDBid:4YV5 from B. moojeni (Brazilian lancehead viper). Suramin has different binding sites in the two homologous subunits ( Table 8) of PLA2-Y, which is significantly different from PLA2-X and PLA2-Z ( Figure 4a). Interestingly, inspite of conserved residues in PLA2-X and PLA2-Z, suramin binds to different sites in these proteins ( Table 8). DOCLASP docking of suramin to PLA2-Z based on the binding residues from PLA2-X (Lys53, Lys69 and Tyr52) shows that this is reasonable binding site (see PLA2dockedsuramin.p1m in Dataset 1, Figure 4b).
Figure 4. Non-specific binding of suramin to phospholipase A2-like proteins (PLA2).
( a) MSA of three PLA2 from poisonous vipers: (i) PLA2-X, PDBid:1Y4L from Bothrops asper, (ii) PLA2-Y, PDBid:3BJW from Echis carinatus and (iii) PLA2-Z, PDBid:4YV5 from Bothrops moojeni shows that PLA2-Y is significantly different from the other two proteins in the charge composition of the residues. For example, the positively charged Lys53 which makes contact with suramin in PLA2-X, and is conserved in PLA2-Z, is replaced by the negatively charged Asp53 in PLA2-Y.( b) The DOCLASP docked suramin to PLA2-Z based on the binding residues obtained from PLA2-X (Lys53, Lys69 and Tyr52) shows that there are two possible binding sites of suramin within PLA2-X/Z.
The suramin molecule itself undergoes a significant conformational change on binding, and different proteins induce different conformational changes. Two molecules of suramin bound to different non homologous proteins (PDBid:1Y4LB and PDBid:1Y8EA - Table 1) is superimposed using DECAAF 57 by aligning three atoms - S17, S21 and S31 ( Figure 5). Different atoms of suramin are involved in the ligand binding for different proteins, and induce significant conformational changes in the drug. For example, the maximum distance between any two atoms in SVR:PDBid:1Y8EA is 30.211 Å (between O80 and O29), while the maximum distance in SVR:PDBid:1Y4LB is 26.4 Å (between O82 and O30). This underscores the complexity of docking algorithms, which must sample a much larger conformational space created by both the flexible binding residues and ligand 58.
Figure 5. Different conformational changes in the suramin molecule on binding to different proteins.
Suramin molecules bound to heparin-binding site of a complement control protein from Vaccinia virus (poxvirus, PDBid:1Y8EA, in green) and the myotoxin II from Bothrops asper (venomous pit viper, PDBid:1Y4LB, in blue) shows the different conformational changes induced in the drug molecule upon binding. This significantly increases the conformational space to be sampled by computational docking methods. Different atoms of SVR are involved in the ligand binding for different proteins, although these are predominantly positively charged residues (SVR:PDBid:1Y4LB in red, SVR:PDBid:1Y8EA in mageneta).
The number of docking methods available is such that even a detailed review could only provide a partial list of currently available docking methods 9. The current work presents a template based, static method that leverages the spatial and electrostatic properties of the binding site. The definitive advantage of a static method can only be highlighted by emphasizing the known limitations of conformational sampling of the protein structure 59, 60. The problem is indeed exacerbated by the plasticity of the drug itself 61, 62. While DOCLASP is completely ineffectual in the absence of such a database, unlike de novo methods, it benefits from the burgeoning database of protein-ligand structures 63. The conservation of electrostatic properties, extracted using APBS/PDB2PQR, is the strongest argument in favor of DOCLASP.
There are several limitations in the method. Firstly, it can be applied to those compounds which are bound to proteins whose structures have been solved. Additionally, it is requires the structure of the apoenzyme, as this is used to extract the query motif considering the structural and electrostatic changes induced by ligand binding. However, with an ever increasing number of protein structures being solved, this is not a severe limitation since most proteins with ligands also have their apo structures solved. Furthermore, the lack of congruent matches leads DOCLASP to return a null result. This can be overcome by relaxing constraints, for example by checking for spatial congruence only. Finally, it is required to develop an energy function which will be able to discriminate poorly docked structures that have either significant steric clashes or are docked on the surface of the protein.
To summarize, this work presents an implicit method for docking ligands to proteins, in which the search and scoring are implicit in the CLASP algorithm. One significant limitation of this method is the requirement of template protein structures in complex with the given compound. As future work, I intend to incorporate the flexibility of the ligand and protein to add further discrimination.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2016 Chakraborty S
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
F1000Research: Dataset 1. Data for “DOCLASP - Docking ligands to target proteins using spatial and electrostatic congruence extracted from a known holoenzyme and applying simple geometrical transformations”, 10.5256/f1000research.5145.d125646 64
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 3; referees: 2 approved]
Supplementary materials
Pymol script for visualizing the docking and movie.
The file “SupplementaryPymol.p1m” contains the Pymol file for viewing vildagliptin docked to PIPLC and the file “SupplementaryMOvie.avi” contains a movie showing a 360 rotation of the ligand docked to PIPLC.
Click here to access the data.
Non-homologous set of nine proteins in the current PDB database with bound suramin:
References
- 1. Cosconati S, Forli S, Perryman AL, et al. : Virtual Screening with AutoDock: Theory and Practice. Expert Opin Drug Discov. 2010;5(6):597–607. 10.1517/17460441.2010.484460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tanrikulu Y, Krüger B, Proschak E: The holistic integration of virtual screening in drug discovery. Drug Discov Today. 2013;18(7–8):358–364. 10.1016/j.drudis.2013.01.007 [DOI] [PubMed] [Google Scholar]
- 3. Seddon G, Lounnas V, McGuire R, et al. : Drug design for ever, from hype to hope. J Comput Aided Mol Des. 2012;26(1):137–150. 10.1007/s10822-011-9519-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morris GM, Huey R, Lindstrom W, et al. : AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jones G, Willett P, Glen RC, et al. : Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267(3):727–748. 10.1006/jmbi.1996.0897 [DOI] [PubMed] [Google Scholar]
- 6. Ewing TJ, Makino S, Skillman AG, et al. : DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des. 2001;15(5):411–428. 10.1023/A:1011115820450 [DOI] [PubMed] [Google Scholar]
- 7. Schellhammer I, Rarey M: FlexX-Scan: fast, structure-based virtual screening. Proteins. 2004;57(3):504–517. 10.1002/prot.20217 [DOI] [PubMed] [Google Scholar]
- 8. Friesner RA, Banks JL, Murphy RB, et al. : Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–1749. 10.1021/jm0306430 [DOI] [PubMed] [Google Scholar]
- 9. Sousa SF, Ribeiro AJ, Coimbra JT, et al. : Protein-ligand docking in the new millennium--a retrospective of 10 years in the field. Curr Med Chem. 2013;20(18):2296–2314. 10.2174/0929867311320180002 [DOI] [PubMed] [Google Scholar]
- 10. Grinter SZ, Zou X: Challenges, applications, and recent advances of protein-ligand docking in structure-based drug design. Molecules. 2014;19(7):10150–10176. 10.3390/molecules190710150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yuriev E, Agostino M, Ramsland PA: Challenges and advances in computational docking: 2009 in review. J Mol Recognit. 2011;24(2):149–164. 10.1002/jmr.1077 [DOI] [PubMed] [Google Scholar]
- 12. Chakraborty S, Minda R, Salaye L, et al. : Active site detection by spatial conformity and electrostatic analysis--unravelling a proteolytic function in shrimp alkaline phosphatase. PLoS One. 2011;6(12):e28470. 10.1371/journal.pone.0028470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rendon-Ramirez A, Shukla M, Oda M, et al. : A Computational Module Assembled from Different Protease Family Motifs Identifies PI PLC from Bacillus cereus as a Putative Prolyl Peptidase with a Serine Protease Scaffold. PLoS One. 2013;8(8):e70923. 10.1371/journal.pone.0070923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chakraborty S, Asgeirsson B, Minda R, et al. : Inhibition of a cold-active alkaline phosphatase by imipenem revealed by in silico modeling of metallo-β-lactamase active sites. FEBS Lett. 2012;586(20):3710–3715. 10.1016/j.febslet.2012.08.030 [DOI] [PubMed] [Google Scholar]
- 15. Chakraborty S, Rendon-Ramirez A, Asgeirsson B, et al. : Dipeptidyl peptidase-iv inhibitors used in type-2 diabetes inhibit a phospholipase c: a case of promiscuous scaffolds in proteins [v1; ref status: approved 1, approved with reservations 1, http://f1000r.es/2hw]. F1000Res. 2013;2:286. 10.12688/f1000research.2-286.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chakraborty S, Rao BJ, Asgeirsson B, et al. : Premonition - preprocessing motifs in protein structures for search acceleration [v1; ref status: awaiting peer review, http://f1000r.es/492]. F1000Res. 2014;3:217 10.12688/f1000research.5166.1 [DOI] [Google Scholar]
- 17. Heinz DW, Ryan M, Bullock TL, et al. : Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol. EMBO J. 1995;14(16):3855–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Escobar MA, Shilling A, Higgins P, et al. : Characterization of polyphenol oxidase from walnut. J Am Soc Hortic Sci. 2008;133(6):852– 858 Reference Source [Google Scholar]
- 19. Bijelic A, Pretzler M, Molitor C, et al. : The structure of a plant tyrosinase from walnut leaves Reveals the importance of Substrate-Guiding Residues for Enzymatic Specificity. Angew Chem Int Edit. 2015;54:14677–14680. 10.1002/anie.201506994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martínez-García PJ, Crepeau MW, Puiu D, et al. : The walnut (juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of nonstructural polyphenols. Plant J. 2016. 10.1111/tpj.13207 [DOI] [PubMed] [Google Scholar]
- 21. Steverding D: The development of drugs for treatment of sleeping sickness: a historical review. Parasit Vectors. 2010;3(1):15. 10.1186/1756-3305-3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salvador GH, Cavalcante WL, Dos Santos JI, et al. : Structural and functional studies with mytoxin ii from Bothrops moojeni reveal remarkable similarities and differences compared to other catalytically inactive phospholipases A 2-like. Toxicon. 2013;72:52–63. 10.1016/j.toxicon.2013.06.013 [DOI] [PubMed] [Google Scholar]
- 23. Maximova T, Moffatt R, Ma B, et al. : Principles and Overview of Sampling Methods for Modeling Macromolecular Structure and Dynamics. PLoS Comput Biol. 2016;12(4):e1004619. 10.1371/journal.pcbi.1004619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kleywegt GJ: Recognition of spatial motifs in protein structures. J Mol Biol. 1999;285(4):1887–1897. 10.1006/jmbi.1998.2393 [DOI] [PubMed] [Google Scholar]
- 25. Baker NA, Sept D, Joseph S, et al. : Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dolinsky TJ, Nielsen JE, McCammon JA, et al. : PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32(Web Server issue):W665–667. 10.1093/nar/gkh381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arnold K, Bordoli L, Kopp J, et al. : The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195– 201. 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- 28. Nabeno M, Akahoshi F, Kishida H, et al. : A comparative study of the binding modes of recently launched dipeptidyl peptidase IV inhibitors in the active site. Biochem Biophys Res Commun. 2013;434(2):191–196. 10.1016/j.bbrc.2013.03.010 [DOI] [PubMed] [Google Scholar]
- 29. Chakraborty S, Rao B, Asgeirsson B, et al. : Correlating the ability of VP24 protein from Ebola and Marburg viruses to bind human karyopherin to their immune suppression mechanism and pathogenicity using computational methods [v1; ref status: awaiting peer review, http://f1000r.es/4o3]. F1000Res. 2014;3 10.12688/f1000research.5666.1 [DOI] [Google Scholar]
- 30. Zhang AP, Abelson DM, Bornholdt ZA, et al. : The Ebolavirus VP24 interferon antagonist: Know your enemy. Virulence. 2012;3(5):440– 5. 10.4161/viru.21302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu W, Edwards MR, Borek DM, et al. : Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe. 2014;16(2):187– 200. 10.1016/j.chom.2014.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miranda ME, White ME, Dayrit MM: Seroepidemiological study of filovirus related to Ebola in the Philippines. Lancet. 1991;337(8738):425– 426. 10.1016/0140-6736(91)91199-5 [DOI] [PubMed] [Google Scholar]
- 33. Miranda ME, Miranda NL: Reston ebolavirus in humans and animals in the Philippines: a review. J Infect Dis. 2011;204(Suppl 3):S757– S760. 10.1093/infdis/jir296 [DOI] [PubMed] [Google Scholar]
- 34. Araji S, Grammer TA, Gertzen R, et al. : Novel roles for the polyphenol oxidase enzyme in secondary metabolism and the regulation of cell death in walnut. Plant Physiol. 2014;164(3):1191– 1203. 10.1104/pp.113.228593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bacchi CJ: Chemotherapy of human african trypanosomiasis. Interdiscip Perspect Infect Dis. 2009;2009: 195040. 10.1155/2009/195040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stein CA, LaRocca RV, Thomas R, et al. : Suramin: an anticancer drug with a unique mechanism of action. J Clin Oncol. 1989;7(4):499–508. [DOI] [PubMed] [Google Scholar]
- 37. Bhargava S, Hotz B, Hines OJ, et al. : Suramin inhibits not only tumor growth and metastasis but also angiogenesis in experimental pancreatic cancer. J Gastrointest Surg. 2007;11(2):171–178. 10.1007/s11605-006-0081-z [DOI] [PubMed] [Google Scholar]
- 38. Borges S, Döppler HR, Storz P: A combination treatment with DNA methyltransferase inhibitors and suramin decreases invasiveness of breast cancer cells. Breast Cancer Res Treat. 2014;144(1):79–91. 10.1007/s10549-014-2857-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Phillips MA: Stoking the drug target pipeline for human African trypanosomiasis. Mol Microbiol. 2012;86(1):10–14. 10.1111/mmi.12001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu K, Chong RA, Yu Q, et al. : Suramin inhibits cullin-RING E3 ubiquitin ligases. Proc Natl Acad Sci U S A. 2016;113(14):E2011–8. 10.1073/pnas.1601089113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bos OJ, Vansterkenburg EL, Boon JP, et al. : Location and characterization of the suramin binding sites of human serum albumin. Biochem Pharmacol. 1990;40(7):1595–1599. 10.1016/0006-2952(90)90460-3 [DOI] [PubMed] [Google Scholar]
- 42. Leff P, Wood BE, O’Connor SE: Suramin is a slowly-equilibrating but competitive antagonist at P 2x-receptors in the rabbit isolated ear artery. Br J Pharmacol. 1990;101(3):645–649. 10.1111/j.1476-5381.1990.tb14134.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cadène M, Duranton J, North A, et al. : Inhibition of neutrophil serine proteinases by suramin. J Biol Chem. 1997;272(15):9950–9955. 10.1074/jbc.272.15.9950 [DOI] [PubMed] [Google Scholar]
- 44. Yamazaki H, Dilworth A, Myers CE, et al. : Suramin inhibits DNA damage in human prostate cancer cells treated with topoisomerase inhibitors in vitro. Prostate. 1993;23(1):25–36. 10.1002/pros.2990230104 [DOI] [PubMed] [Google Scholar]
- 45. Vijayalakshmi J, Padmanabhan KP, Mann KG, et al. : The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin. Protein Sci. 1994;3(12):2254–2271. 10.1002/pro.5560031211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beindl W, Mitterauer T, Hohenegger M, et al. : Inhibition of receptor/G protein coupling by suramin analogues. Mol Pharmacol. 1996;50(2):415–423. [PubMed] [Google Scholar]
- 47. Aragão EA, Chioato L, Ferreira TL, et al. : Suramin inhibits macrophage activation by human group IIA phospholipase A 2, but does not affect bactericidal activity of the enzyme. Inflamm Res. 2009;58(4):210–217. 10.1007/s00011-008-8137-z [DOI] [PubMed] [Google Scholar]
- 48. Jiao L, Ouyang S, Liang M, et al. : Structure of severe fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential. J Virol. 2013;87(12):6829–6839. 10.1128/JVI.00672-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marques AF, Esser D, Rosenthal PJ, et al. : Falcipain-2 inhibition by suramin and suramin analogues. Bioorg Med Chem. 2013;21(13):3667–3673. 10.1016/j.bmc.2013.04.047 [DOI] [PubMed] [Google Scholar]
- 50. Croci R, Pezzullo M, Tarantino D, et al. : Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds. PLoS One. 2014;9(3):e91765. 10.1371/journal.pone.0091765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Y, Qing J, Sun Y, et al. : Suramin inhibits EV71 infection. Antiviral Res. 2014;103:1–6. 10.1016/j.antiviral.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 52. Aragao EA, Vieira DS, Chioato L, et al. : Characterization of suramin binding sites on the human group IIA secreted phospholipase A 2 by site-directed mutagenesis and molecular dynamics simulation. Arch Biochem Biophys. 2012;519(1):17–22. 10.1016/j.abb.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 53. Chen Y, Maguire T, Hileman RE, et al. : Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3(8):866–871. 10.1038/nm0897-866 [DOI] [PubMed] [Google Scholar]
- 54. Feixas F, Lindert S, Sinko W, et al. : Exploring the role of receptor flexibility in structure-based drug discovery. Biophys Chem. 2014;186:31–45. 10.1016/j.bpc.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Naviaux JC, Schuchbauer MA, Li K, et al. : Reversal of autism-like behaviors and metabolism in adult mice with single-dose antipurinergic therapy. Transl Psychiatry. 2014;4:e400. 10.1038/tp.2014.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chakraborty S: An automated flow for directed evolution based on detection of promiscuous scaffolds using spatial and electrostatic properties of catalytic residues. PLoS One. 2012;7(7):e40408. 10.1371/journal.pone.0040408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nautiyal A, Patil KN, Muniyappa K: Suramin is a potent and selective inhibitor of Mycobacterium tuberculosis RecA protein and the SOS response: RecA as a potential target for antibacterial drug discovery. J Antimicrob Chemother. 2014;69(7):1834–1843. 10.1093/jac/dku080 [DOI] [PubMed] [Google Scholar]
- 58. Naviaux RK, Zolkipli Z, Wang L, et al. : Antipurinergic therapy corrects the autism-like features in the poly(IC) mouse model. PLoS One. 2013;8(3):e57380. 10.1371/journal.pone.0057380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Durrant JD, McCammon JA: Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9:71. 10.1186/1741-7007-9-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Borhani DW, Shaw DE: The future of molecular dynamics simulations in drug discovery. J Comput Aided Mol Des. 2012;26(1):15–26. 10.1007/s10822-011-9517-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Perola E, Charifson PS: Conformational analysis of drug-like molecules bound to proteins: an extensive study of ligand reorganization upon binding. J Med Chem. 2004;47(10):2499–2510. 10.1021/jm030563w [DOI] [PubMed] [Google Scholar]
- 62. Mobley DL, Dill KA: Binding of small-molecule ligands to proteins: “what you see” is not always “what you get”. Structure. 2009;17(4):489–498. 10.1016/j.str.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu T, Lin Y, Wen X, et al. : BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucl Acids Res. 2007;35(Database issue):D198–D201. 10.1093/nar/gkl999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chakraborty S: Dataset 1 in: DOCLASP - Docking ligands to target proteins using spatial and electrostatic congruence extracted from a known holoenzyme and applying simple geometrical transformations. F1000Research. 2016. Data Source [DOI] [PMC free article] [PubMed]