Abstract
Serpins are a structurally conserved family of macromolecular inhibitors found in numerous biological systems. The completion and annotation of the genomes of Schistosoma mansoni and Schistosoma japonicum has enabled the identification by phylogenetic analysis of two major serpin clades. S. mansoni shows a greater multiplicity of serpin genes, perhaps reflecting adaptation to infection of a human host. Putative targets of schistosome serpins can be predicted from the sequence of the reactive center loop (RCL). Schistosome serpins may play important roles in both post-translational regulation of schistosome-derived proteases, as well as parasite defense mechanisms against the action of host proteases.
Keywords: schistosomiasis, inhibitor, protease, serpin, genome
Schistosomiasis
Disease, Distribution, Mortality/Morbidity
The blood flukes Schistosoma mansoni, S. japonicum, and S. haematobium are the three main etiological agents of human schistosomiasis. These parasitic worms infect approximately 207 million individuals, and 700 million are at risk of infection due to exposure to infested waters (Who 2010). The parasites are found throughout the tropical world. S. mansoni is the most widely distributed, affecting people in Africa, the Middle East, South America, and the Caribbean while S. japonicum is confined to China, the Philippines, and Indonesia. S. haematobium is found in Africa and the Middle East.
Schistosomes are digenetic flatworms that spend part of their lifecycle in a freshwater mollusk host and part in a vertebrate host (Box 1). Infected snails release free-swimming cercariae (larvae) into fresh water where humans fish, bathe, wash clothing, and play. Once the cercariae come in contact with the skin of the vertebrate host they begin to invade, stimulated by the lipids on the surface of skin (Shiff et al. 1972). Once in the skin, cercariae transform into schistosomula, degrading the host extracellular matrix by secreting proteolytic enzymes to facilitate entry into the host dermal blood vessels (He et al. 2005). The schistosomula reach the lungs and later migrate to the portal venous system, where they mature into adult worms. Mating pairs migrate either to the mesenteric venules (S. mansoni and S. japonicum) or the venous plexus of the bladder (S. haematobium). Female worms produce eggs that are then released into fresh water and hatch into another larval stage, the miracidium. Miracidia find and penetrate the snail intermediate host and after several rounds of asexual reproduction transform into sporocysts which give rise to the free-swimming cercariae (Box 1).
Box 1.
Schistosome life cycle.
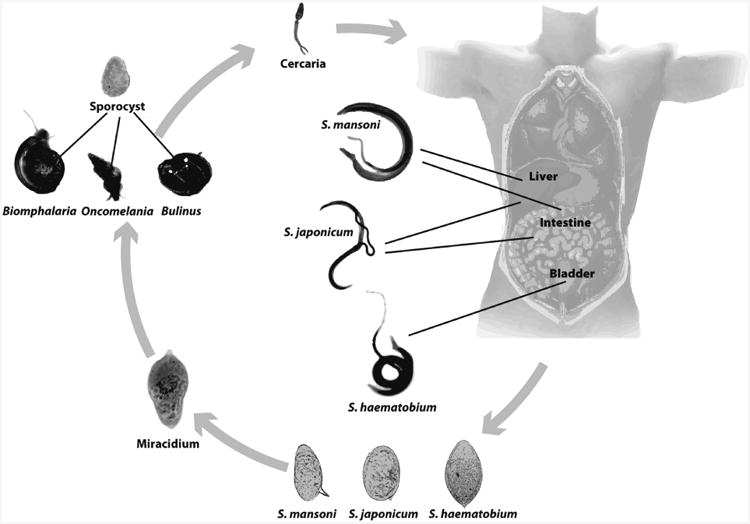
The main developmental stages of schistosomes as they cycle from fresh water mollusks and humans. The intermediate hosts of the parasites are Biomphalaria sp (S. mansoni), Oncomelania sp (S. japonicum), and Bulinus sp (S. haematobium). Shown are the mollusk stage of the parasites (sporocysts), the free swimming cercaria and miracidia, as well as the adults in human circulatory system.
Largely a chronic disease of the poor with inadequate access to a safe water supply, schistosomiasis can affect children's physical, sexual, and mental development as well as impair people's ability to work due to chronic fatigue. Severe schistosomiasis can lead to liver fibrosis, hepatosplenomegaly, and portal hypertension. Urinary schistosomiasis can produce hematuria, cystitis, and is a leading risk factor for squamous cell carcinoma of the bladder. No anti-schistosome vaccine exists. While research to expand treatment options is ongoing (Abdulla et al. 2009), Praziquantel is at present the sole chemotherapeutic treatment recommended for schistosomiasis.
Serpins
Biochemical Function, Involvement in Physiology and Disease Pathogenesis
Serpins are a structurally conserved family of macro-molecular inhibitors found in virtually all biological systems (reviewed in (Gettins 2002), (Law et al. 2006), (Silverman et al. 2001)). The mechanism employed by serpins to inhibit proteases is also highly conserved. Serpins are suicide hemi-substrate inhibitors that are approximately 40-50 kDa in size. The conserved metastable tertiary structure of a serpin features 7 to 9 α-helices and 3 β-sheets (Box 2a). Key to the inhibitory activity of the serpin is the RCL (Crowther et al. 1992). In the native, active form, it lies outside the main body of the serpin, between β-sheets A and C and serves as the protease ‘bait’. The RCL is composed of approximately 20 amino acid residues (P17-P4) with the P1 side chain fitting into the S1 specificity pocket of the target protease, with cleavage of the RCL occurring at the P1-P1′ position. In the initial interaction of the serpin with the target enzyme the active site serine forms a transient covalent ester linkage with the serpin. Following proteolytic cleavage, the RCL inserts into the body of the serpin, completing the antiparallel organization of β-sheet A (Box 2b). The crystal structure of the trypsin-α1-antitrypsin complex reveals that once the complex forms, translocation distorts the structure of the protease by 37%, while the serpin itself remains largely unchanged (Huntington et al. 2000). The crystal structure confirms that translocation of the protease dislocates the active site serine, taking it out of productive alignment with the rest of the catalytic triad, resulting in the destruction of the oxyanion hole.
Box 2.
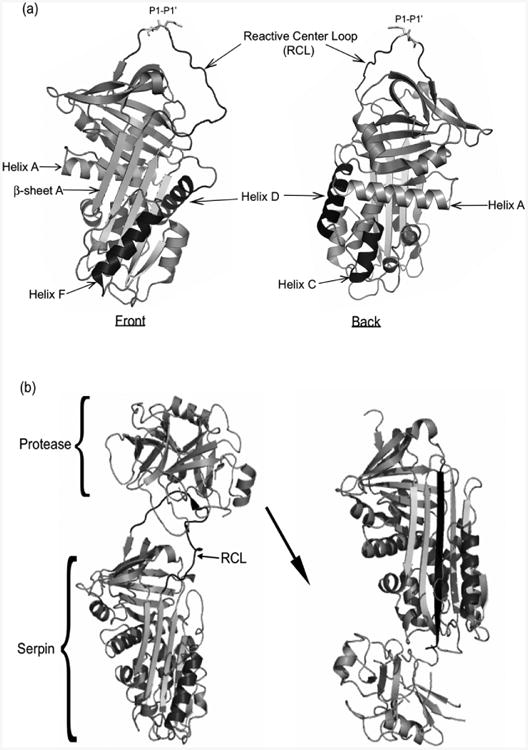
(a) Structure of α1-antitrypsin (adapted from PDB 2QUG) viewed from two angles. Noted structural components are the reactive center loop, helix A, helix F, helix D, helix C, β-sheet A. The P1-P1′ site of the RCL notes the location of the peptide bond cleaved by the cognate protease before inhibition. (b) Mechanism of serpin inhibition. The initial trypsin (protease) and serpin Michaelis complex (adapted from PDB 1K9O). Upon cleavage of the RCL (black), the loop inserts into the β-sheet A of the serpin (PDB 1E2X).
Serpins are found in animals, viruses, plants, archea, and bacteria. Based on their genomes, C. elegans, D. melanogaster, and A. thaliana contain 9, 32, and 13 serpins respectively (Silverman et al. 2001). In mammals serpins are involved in the regulation of proteases in blood coagulation, fibrinolysis, apoptosis, development, and inflammation. Serpins also play an important role in the immune response of insects. In D. melanogaster Spn43Ac regulates the immune response to fungal infection by inhibiting the cleavage of the Toll ligand spaeztle (Levashina et al. 1999). There is evidence that SRPN6 of the malaria mosquito vector Anopheles gambiae is important for parasite clearance and/or lysis (Abraham et al. 2005).
The function of serpins in schistosomes remains speculative and studies of other pathogenic parasites have provided few clues to serpin function. Two serpins have been cloned from the lymphatic filariasis agent Brugia malayi: BmSpn-1 (Yenbutr and Scott 1995) for which there is little functional data, and BmSpn-2 (Zang et al. 1999). BmSpn-2 was originally thought to aid in microfilarial immune evasion by inhibiting neutrophil elastase and cathepsin G (Zang et al. 1999), but a subsequent study showed that it is unable to form a stable complex with either of these proteases (Stanley and Stein 2003). However, BmSpn-2 is released into the blood of the host by microfilariae, suggesting that it plays some role in host-parasite interactions (Stanley and Stein 2003). Investigation of the Onchocerca volvulus serpins Ov-spi-1 and Ov-spi-2 by RNAi showed that knockdown of these serpins led to impaired L3 molting and viability. However, the endogenous targets have not been identified (Ford et al. 2005).
Schistosome Serpins
Phylogeny and Stage Specificity
The completion of the S. mansoni and S. japonicum genomes has made it possible to explore these parasites' proteomes for proteins key to their survival and pathogenesis. By searching parasite databases for proteins homologous to the canonical serpin α1-antitrypsin, eight complete serpin sequences have been identified in the S. mansoni gene database. The S. japonicum database has one partial and 3 complete serpin sequences (Table I). The S. haematobium genome is incomplete as of the writing of this review, but a GenBank search identified one serpin in the organism.
Table I. Serine Protease Inhibitors of Schistosoma mansoni, S. japonicum, and S. haematobium.
| Database gene namea | Alias ref. in literatureb | Size (kDa) | Reactive Center Loop (RCL) P17-P2′ (P1-P1′) | Similar mammalian serpinc | Potential protease targetd | Ref. |
|---|---|---|---|---|---|---|
| S. mansoni | ||||||
| Smp_003300 | Serp_C | 43.7 | EDGVEAAAATVMGIGLR-SA | α1-AT (Bos 53%) | SmCl2, Thrm, CF, KRPs | Curwen et al. 2006 |
| Smp_090080 | AAA29938 SPI | 46.0 | ESGIEATTVTSPIFVPI-SA | Neuroserpin (Mus 48%) | Elastase-2 (Non-inhibitory) | |
| Smp_090090 | None | 46.3 | EVGMEARSVANAMFIPL-SS | Neuroserpin (Homo 52%) | CT, Cat G, CE (Non-inhibitory) | |
| Smp_062080 | AAB86571 | 43.3 | EQGVVAAAASSVEVVQL-SA | SCCA-2 (Mus 53%) | NE, Cat G, CT, CE | Harrop et al. 2000 |
| Smp_155530 | None | 51.0 | EEGVVAAGVTACVFDNC-DS | SCCA-2 (Mus 55%) | Sig. Peptidase complex(SPC) | |
| Smp_155550 | None | 43.5 | EKGAVAAAATATRMIRC-TA | PAI-2 (Rattus 53%) | Thrm, SPC | |
| Smp_155560 | None | 43.6 | EKGAVAAAATATQMVRC-TA | SCCA2/SCCA1 (Bos 52%) | Thrm, CF, SPC | |
| Smp_062120 | None | 43.8 | EKGAVAAAATATQMVRY-SA | PAI-2 (Mus 54%) | DPPIII, Thrm | |
| S. japonicum | ||||||
| Sjp_0113080 | AAW25282 | 43.0 | EKGAEAAAATATKIIPL-SL | PI-6 (Rattus 53%) | CT, Cat G, KRPs, CE | Liu et al. 2006 |
| Sjp_0076600 | None | 45.0 | EEGAVAAAASATVMLKC-SA | SerpinB6c (Mus 57%) | SPC | |
| Sjp_0085750 | AF308366 | 45.6 | ESGIEAASVTSPIIVPI-SA | Neuroserpin (Homo 50%) | Elastase-2 | Yan et al. 2005 |
| Sjp_0113720 | None | 26.4 | EKGVEAAAATAIYSLGR-SL | Ovalbumin (Rattus 52%) | KRPs, Thrm, CFc | |
| S. haematobium | ||||||
| ShSPI | AAA19730 | 45.9 | ESGIEATTVTSPIFVPF-SA | Ovalbumin (Rattus 48%) | CT, chymase, CatG, CE (Non-inhibitory) | Blanton et al. 1994 |
Name given to the gene in the database (www.genedb.org).
Other name/annotation given by authors of this or other publications prior to genome annotation.
Obtained using BLASTp non-redundant database (% similarity).
Matches obtained by using the MEROPS database and alignment results.
Phylogenetic analysis of the Schistosoma serpins assigns two major branches to the group (Fig. 1a). Branch 1 consists of Smp_090080, ShSPI, and Sjp_0085750 clustered closely together, with Smp_090090, Sjp_0113720, and Smp_003300 more distantly related but still falling within this grouping. The remaining serpins form the second major branch (Fig. 1a). Smp_15-5560, Smp_155560, Smp_062120 are virtually identical, with most of the variation occurring at the RCL (Table I). Considering the chromosomal arrangement of S. mansoni serpins (Fig. 1b) and the clustering pattern of the phylogenetic tree, the multiplicity of S. mansoni serpin genes compared to those of S. japonicum has likely arisen through several gene duplication events. This is not surprising, given recent findings that S. japonicum appears to be a more ancestral species among mammalian parasites (Snyder and Loker 2000). S. mansoni and S. haematobium are more human-directed parasites than S. japonicum, leading to speculation that serpin gene duplications in S. mansoni reflect adaptations to the human host. It will be interesting to see if S. haematobium also exhibits similar gene duplication events when its genome is sequenced.
Fig. 1.
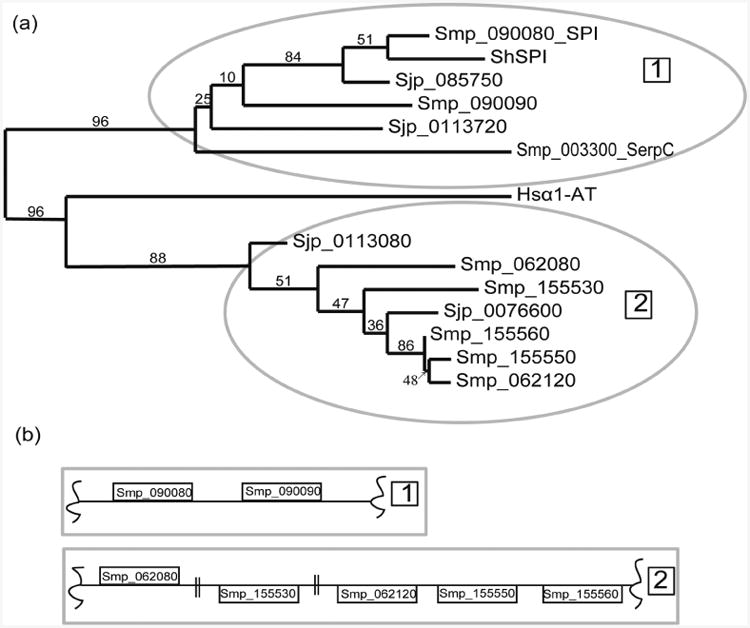
(a) Phylogenetic analysis of Schistosome serpins. The thirteen serpins were aligned using MUSCLE and a bootstrapped maximum likelihood tree was generated using PhyML 3.0. The percentage branch bootstrap support values are shown on branch splits. Method of analysis is fully described in (Dereeper et al. 2008). (b) Serpin gene arrangement in S. mansoni. The location of each gene in there respective supercontig. Double lines denote the location of an unrelated gene.
With the exception of Sjp_0113720, alignment of the Schistosoma serpins with α1-antitrypsin, and super-imposition of the structural components of α1-antitrypsin, show that these proteins contain all elements necessary for functionality (Fig. 2). Their predicted structures reveal the interesting phenomenon of elongation of the regions surrounding helix D. Branch 1 sequences (Fig. 1) indicate a 9-16 amino acid extension of the helix D region across all three species. This could indicate either an elongation of helix D or an extension of the region connecting helix C to helix D. In either case, this extension could have implications for the biochemical properties and functionality of these serpins. Previous work on helix D has suggested it is involved in the allosteric control of the binding and release of glucocorticoids by corticoid-binding globulin, a non-inhibitory serpin (Lin et al. 2010), and heparin activation of antithrombin is mediated through its binding site on helix D (Belzar et al. 2002). On the other hand, MENT, a serpin important in heterochromatin formation, contains an extension in the region between helix C and D which forms a loop outside the body of the serpin (Grigoryev et al. 1999). The authors speculate it may be involved in DNA binding.
Fig. 2. Primary amino acid sequence alignment.
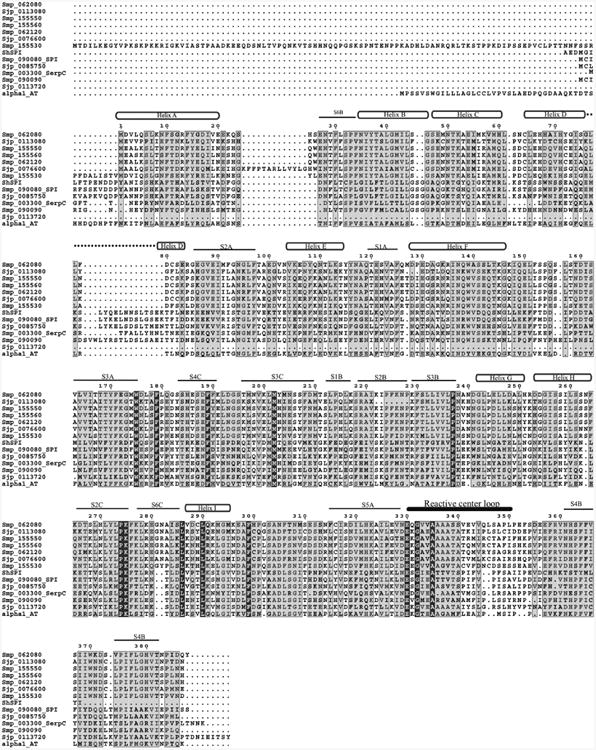
The predicted or known sequences were aligned with α1-antitrypsin using Clustal W v1.8. Highlighted regions show at least a 50% sequence identity. The location of the helices (cylinders), β-sheets (black lines), and RCL (bold black line) were approximated using the known α1-antitrypsin structure.
The RCLs of ShSPI, Smp_090080 and Smp_090090 contain a feature unusual for inhibitory serpins (Table I). The P12-P9 region of the RCLs of most inhibitory serpins is characterized by amino acids with short side-chains (Ala, Gly, Ser) (Hopkins et al. 1993), but this feature is not present in ShSPI, Smp_090080 and Smp_090090. Whether the unusual RCL amino acid sequence signifies a non-inhibitory function, or if elongation of the helix D region is important for cofactor binding, is a subject for future study.
ShSPI was discovered using a λgt11 cDNA library from adult worms (Blanton et al. 1994). Immunolocalization showed ShSPI on the surface of adult worms. Although one study suggested that ShSPI could bind to trypsin but not elastase or chymotrypsin (Huang et al. 1999), there was no corroborating evidence to validate active site binding. Closely related to ShSPI, Sjp_0085-750 was the subject of another study. Transcripts of the Sjp_0085750 gene were found in male and female adult worms, and to a lesser extent in cercariae (Yan et al. 2005). Yan et al. showed that this serpin localized to the surface of the adult worms, much like ShSPI, as well as the gut epithelium of the worm. When the authors evaluated the immune response to Sjp_0085750, they noted a Th2-type humoral response in mice. Protection studies with the serpin antigen showed a decrease in worm burden, suggesting a potential use of this parasite serpin as a vaccine candidate.
The most distantly related serpin in branch 1, Smp_003300, was initially identified as Serp_C by a proteomic analysis of the secreted products of cercariae (Curwen et al. 2006). Release of this serpin into the host was validated by mass spectrometry analysis of proteins released by cercariae invading human skin (Hansell et al. 2008). The gene is not found in the same contig as Smp_090090 and Smp_090080 nor is it flanked by any other serpin.
Fewer studies have been conducted on the serpins clustered in the second phylogenetic branch (Fig. 1a). An antisera screen against cercarial secreted products from a cercarial cDNA library identified a partial gene product, clone 8, with homology to serpin MNEI (Harrop et al. 2000). The authors found the protein product in cercarial and lung stage parasite lysates but not in adult worms. Based on the sequence, clone 8 is Smp_062080. A proteomic study of skin lipid-induced cercarial secretion products also found a unique peptide belonging to clone 8/ Smp_062080 (Knudsen et al. 2005).
Schistosome Serpins
Putative Targets
A putative serpin, Smpi56, was partially purified from extracts of adult S. mansoni (Ghendler et al. 1994). Smpi56 was able to form an SDS-stable complex with neutrophil elastase, pancreatic elastase, and an endogenous cercarial protease. Because the sequence of Smpi56 is not known, the authors were unable to positively identify it as a serpin. It did exhibit the biochemical characteristics of an inhibitory serpin and could therefore be Smp_090090 which has a Leu-Ser P1-P1′ motif (Table I). Cercarial elastases as well as mammalian elastases favor hydrophobic residues in their S1 pockets, making Smp_090090 potentially able to form the complexes Smpi56 was reported to form. However, the unconventional RCL of Smp_090090 makes it difficult predict its ability to form an inhibitory complex with a protease.
The predicted RCL sequence of Sjp_0113080 also has a Leu-Ser at the P1-P1′ specificity determining region. The S. japonicum genome contains one cercarial elastase gene. S. japonicum cercarial elastase antibodies were reported to localize in infected mouse skin, suggesting that the S. japonicum protease was released by the parasite during skin invasion (Consortium 2009). However, neither a protein corresponding to the putative protease nor enzymatic activity were found in the cercarial secretions of S. japonicum (Dvorak et al. 2008). Less is known about the S. haematobium cercarial elastase (ShCE). ShCE transcripts can be found in the sporocyts and it is closely related to SmCE (Salter et al. 2002). ShSPI, Smp_090090, Smp_062080, and Sjp_0113080 could be involved in the regulation of these elastases, although in the absence of further studies host proteases cannot be ruled out as potential targets.
Smp_003300/ Serp_C contains an Arg at the P1 position, making it a good candidate for trypsin-like protease inhibition. Considering its release into the host during invasion, thrombin, coagulation factors, and kallikrein-related peptidases are all potential targets.
Concluding Remarks
The role of serpins in schistosomiasis was last reviewed 14 years ago (Modha et al. 1996). Although there are caveats and limitations to using of the RCL sequence as an accurate predictor of protease specificity, such as the existence of exosites in and outside the RCL region (Whisstock et al. 2010) and the differing binding specificities of protease substrate binding pockets beyond the S1-S1′ sites, RCL-based specificity predictions can still lead to testable hypotheses about the function of serpins in these parasites. With 102 cysteine proteases and 65 serine proteases identified in the S. japonicum genome (Consortium 2009) and 97 cysteine and 78 serine proteases in the S. mansoni genome (Berriman et al. 2009), the 12 predicted schistosome serpins may perform specific, focused functions in these organisms. It is to be hoped that new lines of inquiry building on the findings detailed in this review will advance our understanding of host-parasite biology and lead to the recognition of new drug targets and vaccine candidates against the schistosome parasites.
Acknowledgments
We would like to express our gratitude to Potter Wickware (Sandler Center for Drug Discovery, UCSF) and Mohammed Sajid (Leiden University Medical Center, Netherlands) for their editorial comments. This work was supported by the National Institute of General Medical Science training grant, R25 GM56847, the UCSF Microbial Pathogenesis Host Defense training grant, T32-AI060537, and the Sandler Center for Drug Discovery.
Abbreviations
- Serpin
serine protease inhibitor
- RCL
reactive center loop
- MENT
myeloid and erythroid nuclear termination stage-specific protein or heterochromatin associated serpin
- MS
mass spectrometry
- MNEI
monocyte / neutrophil elastase inhibitor
- CT
chymotrypsin
- CE
cercarial elastase
- α1-AT
alpha 1 antitrypsin
- SCCA
squamous cell carcinoma antigen
- PAI
plasminogen activator inhibitor
- PI
protease inhibitor
- Cl
cathepsin L
- Thrm
thrombin
- CF
coagulation factor
- Cat G
cathepsin G
- NE
neutrophil elastase
- SPC
signal peptidase complex
- DPP
dipeptidyl aminopeptidase
- KRPs
kallikrein related peptidases
References
- Abdulla MH, Ruelas DS, Wolff B, Snedecor J, Lim KC, Xu F, Renslo AR, Williams J, McKerrow JH, Caffrey CR. Drug discovery for schistosomiasis: hit and lead compounds identified in a library of known drugs by medium-throughput phenotypic screening. PLoS Negl Trop Dis. 2009;3:e478. doi: 10.1371/journal.pntd.0000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham EG, Pinto SB, Ghosh A, Vanlandingham DL, Budd A, Higgs S, Kafatos FC, Jacobs-Lorena M, Michel K. An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proc Natl Acad Sci USA. 2005;102:16327–16332. doi: 10.1073/pnas.0508335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzar KJ, Zhou A, Carrell RW, Gettins PG, Huntington JA. Helix D elongation and allosteric activation of antithrombin. J Biol Chem. 2002;277:8551–8558. doi: 10.1074/jbc.M110807200. [DOI] [PubMed] [Google Scholar]
- Berriman M, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton RE, Licate LS, Aman RA. Characterization of a native and recombinant Schistosoma haematobium serine protease inhibitor gene product. Mol Biochem Parasitol. 1994;63:1–11. doi: 10.1016/0166-6851(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Consortium SG. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460:345–351. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther DC, Evans DL, Carrell RW. Serpins: implications of a mobile reactive centre. Curr Opin Biotechnol. 1992;3:399–407. doi: 10.1016/0958-1669(92)90169-j. [DOI] [PubMed] [Google Scholar]
- Curwen RS, Ashton PD, Sundaralingam S, Wilson RA. Identification of novel proteases and immunomodulators in the secretions of schistosome cercariae that facilitate host entry. Mol Cell Proteomics. 2006;5:835–844. doi: 10.1074/mcp.M500313-MCP200. [DOI] [PubMed] [Google Scholar]
- Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J, et al. Differential use of protease families for invasion by schistosome cercariae. Biochimie. 2008;90:345–358. doi: 10.1016/j.biochi.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Ford L, Guiliano DB, Oksov Y, Debnath AK, Liu J, Williams SA, Blaxter ML, Lustigman S. Characterization of a novel filarial serine protease inhibitor, Ov-SPI-1, from Onchocerca volvulus, with potential multifunctional roles during development of the parasite. J Biol Chem. 2005;280:40845–40856. doi: 10.1074/jbc.M504434200. [DOI] [PubMed] [Google Scholar]
- Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- Ghendler Y, Arnon R, Fishelson Z. Schistosoma mansoni: isolation and characterization of Smpi56, a novel serine protease inhibitor. Exp Parasitol. 1994;78:121–131. doi: 10.1006/expr.1994.1013. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Bednar J, Woodcock CL. MENT, a heterochromatin protein that mediates higher order chromatin folding, is a new serpin family member. J Biol Chem. 1999;274:5626–5636. doi: 10.1074/jbc.274.9.5626. [DOI] [PubMed] [Google Scholar]
- Hansell E, Braschi S, Medzihradszky KF, Sajid M, Debnath M, Ingram J, Lim KC, McKerrow JH. Proteomic analysis of skin invasion by blood fluke larvae. PLoS Negl Trop Dis. 2008;2:e262. doi: 10.1371/journal.pntd.0000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrop R, Jennings N, Mountford AP, Coulson PS, Wilson RA. Characterization, cloning and immunogenicity of antigens released by transforming cercariae of Schistosoma mansoni. Parasitology. 2000;121(Pt 4):385–394. doi: 10.1017/s003118209900640x. [DOI] [PubMed] [Google Scholar]
- He YX, Salafsky B, Ramaswamy K. Comparison of skin invasion among three major species of Schistosoma. Trends Parasitol. 2005;21:201–203. doi: 10.1016/j.pt.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Hopkins PC, Carrell RW, Stone SR. Effects of mutations in the hinge region of serpins. Biochemistry. 1993;32:7650–7657. doi: 10.1021/bi00081a008. [DOI] [PubMed] [Google Scholar]
- Huang W, Haas TA, Biesterfeldt J, Mankawsky L, Blanton RE, Lee X. Purification and crystallization of a novel membrane-anchored protein: the Schistosoma haematobium serpin. Acta Crystallogr D Biol Crystallogr. 1999;55:350–352. doi: 10.1107/S0907444998008658. [DOI] [PubMed] [Google Scholar]
- Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- Knudsen GM, Medzihradszky KF, Lim KC, Hansell E, McKerrow JH. Proteomic analysis of Schistosoma mansoni cercarial secretions. Mol Cell Proteomics. 2005;4:1862–1875. doi: 10.1074/mcp.M500097-MCP200. [DOI] [PubMed] [Google Scholar]
- Law RH, et al. An overview of the serpin superfamily. Genome Biol. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffmann JA, Reichhart JM. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- Lin HY, Muller YA, Hammond GL. Molecular and structural basis of steroid hormone binding and release from corticosteroid-binding globulin. Mol Cell Endocrinol. 2010;316:3–12. doi: 10.1016/j.mce.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Liu F, et al. New Perspectives on Host-Parasite Interplay by Comparative Transcriptomic and Proteomic Analyses of Schistosoma japonicum. PLoS Pathog. 2006;2(4):e29. doi: 10.1371/journal.ppat.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modha J, Roberts MC, Kusel JR. Schistosomes and serpins: a complex business. Parasitol Today. 1996;12:119–121. doi: 10.1016/0169-4758(96)80672-7. [DOI] [PubMed] [Google Scholar]
- Salter JP, Choe Y, Albrecht H, Franklin C, Lim KC, Craik CS, McKerrow JH. Cercarial elastase is encoded by a functionally conserved gene family across multiple species of schistosomes. J Biol Chem. 2002;277:24618–24624. doi: 10.1074/jbc.M202364200. [DOI] [PubMed] [Google Scholar]
- Shiff CJ, Cmelik SH, Ley HE, Kriel RL. The influence of human skin lipids on the cercarial penetration responses of Schistosoma haematobium and Schistosoma mansoni. J Parasitol. 1972;58:476–480. [PubMed] [Google Scholar]
- Silverman GA, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- Snyder SD, Loker ES. Evolutionary relationships among the Schistosomatidae (Platyhelminthes : Digenea) and an Asian origin for Schistosoma. J Parasitol. 2000;86:283–288. doi: 10.1645/0022-3395(2000)086[0283:ERATSP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Stanley P, Stein PE. BmSPN2, a serpin secreted by the filarial nematode Brugia malayi, does not inhibit human neutrophil proteinases but plays a noninhibitory role. Biochemistry. 2003;42:6241–6248. doi: 10.1021/bi0271650. [DOI] [PubMed] [Google Scholar]
- Whisstock JC, Silverman GA, Bird PI, Bottomley SP, Kaiserman D, Luke CJ, Pak SC, Reichhart JM, Huntington JA. Serpins flex their muscle: II. Structural insights into target peptidase recognition, polymerization, and transport functions. J Biol Chem. 2010;285:24307–24312. doi: 10.1074/jbc.R110.141408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Schistosomiasis Fact Sheet #115 2010 [Google Scholar]
- Yan Y, Liu S, Song G, Xu Y, Dissous C. Characterization of a novel vaccine candidate and serine proteinase inhibitor from Schistosoma japonicum (Sj serpin) Vet Parasitol. 2005;131:53–60. doi: 10.1016/j.vetpar.2005.04.038. [DOI] [PubMed] [Google Scholar]
- Yenbutr P, Scott AL. Molecular cloning of a serine proteinase inhibitor from Brugia malayi. Infect Immun. 1995;63:1745–1753. doi: 10.1128/iai.63.5.1745-1753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang X, Yazdanbakhsh M, Jiang H, Kanost MR, Maizels RM. A novel serpin expressed by blood-borne microfilariae of the parasitic nematode Brugia malayi inhibits human neutrophil serine proteinases. Blood. 1999;94:1418–1428. [PubMed] [Google Scholar]


