Abstract
The rapid rate of influenza virus mutation drives the emergence of new strains that inflict serious seasonal epidemics and less frequent, but more deadly, pandemics. While vaccination provides the best protection against influenza, its utility is often diminished by the unpredictability of new pathogenic strains. Consequently, efforts are underway to identify new antiviral drugs and monoclonal antibodies that can be used to treat recently infected individuals and prevent disease in vulnerable populations. Next Generation Sequencing (NGS) and the analysis of antibody gene repertoires is a valuable tool for Ab discovery. Here, we describe a technology platform for isolating therapeutic monoclonal antibodies (MAbs) by analyzing the IgVH repertoires of mice immunized with recombinant H5N1 hemagglutinin (rH5). As an initial proof of concept, 35 IgVH genes selected using a CDRH3 search algorithm, co-expressed in a murine IgG2a expression vector with a panel of germline murine kappa genes, and culture supernatants screened for antigen binding. Seventeen of the 35 IgVH MAbs (49%) bound rH5VN1203 in preliminary screens and 8 of 9 purified MAbs inhibited 3 heterosubtypic strains of H5N1 virus when assayed by HI, and 2 MAbs demonstrated prophylactic and therapeutic activity in virus-challenged mice. This is the first example in which an NGS discovery platform has been used to isolate anti-influenza MAbs with relevant therapeutic activity.
Keywords: IgVH, next-generation sequencing, repertoire analysis, monoclonal antibodies
1. Introduction
The rapid rate of influenza virus mutation drives the emergence of new strains that inflict serious seasonal epidemics and less frequent, but more deadly, pandemics (Brandenburg et al., 2013). Influenza A viruses fall into discrete lineages based on their expression of 17 divergent hemagglutinin (HA) proteins, each of which combines with one of 10 different subtypes of neuraminidase protein. Vaccination provides the best protection against influenza, although its utility is diminished by the unpredictability of new pathogenic strains, limitations in manufacturing, and their ineffectiveness for the young, the elderly, and the immunecompromised. Another important first-line defense against influenza includes antiviral drugs that treat the recently infected and prevent disease in exposed family members, health caregivers, and vulnerable populations. Unfortunately, these small molecules suffer from limited virus strain specificity, rate-limiting toxicities, and induction of antiviral resistance (de Jong and Hien, 2006). Consequently, there is a need for alternative therapeutics such as monoclonal antibodies (MAbs) that can broadly neutralize most, if not all, strains of influenza (Council, 2006).
The primary target for antibody (Ab) immunotherapy is HA, and an important class of neutralizing MAbs have been identified that binds the HA2 stem region comprising the membrane anchoring domain and fusion peptide, which are conserved across multiple viral subtypes (Corti et al., 2011; Ekiert et al., 2009; Ekiert et al., 2011; Friesen et al., 2010; Sui et al., 2009). Examples of these HA2 binding MAbs include CR6261, which neutralizes 10 strains of virus that belong to the Group 1 HA family [9]; CR8020, which neutralizes the six viruses comprising the Group 2 HA family (Ekiert et al., 2011); and FI6v3, which binds 16 Group 1 and Group 2 HAs (Corti et al., 2011). However, even with FI6v3, significant variability in potency was observed in neutralization assays and only a limited panel of viruses was tested. While encouraging, it seems unlikely that a single MAb will protect against every new isolate at equivalent doses or with the same kinetics and durability. Consequently, it may be necessary to develop a panel of broadly neutralizing MAbs that can provide better coverage to an emerging new virus and protect against subsequent viral escape (Geeraedts and Huckriede, 2011).
Various methodologies have been used to identify broadly neutralizing anti-HA influenza MAbs including phage display (Ekiert et al., 2011; Sui et al., 2009), enriched memory B cells (Throsby et al., 2008), and B cell screening (Beerli et al., 2009; Corti et al., 2011; Whittle et al., 2011). Another promising approach that bypasses traditional high-throughput screening technologies relies on next generation sequencing (NGS) and the analysis of antibody gene repertoires (Friesen et al., 2010; Galson et al., 2015; Georgiou et al., 2014). Here, we describe a technology platform for isolating therapeutic MAbs by analyzing the IgVH repertoire of immunized mice using a CDRH3 search algorithm, whose diversity in length, peptide sequence and IgVH and IgHJ gene usage helps define IgVH clonotype (Galson et al., 2014; Galson et al., 2015). As an initial proof of concept, we produced 35 MAb candidates, 17 (49%) of which specifically bound rH5N1 HA. Further analysis indicated that eight of nine purified MAbs could inhibit H5N1 substrains in vitro using a hemagglutinin inhibition assay (HI). Moreover, two of these MAbs showed strong prophylactic and therapeutic activity in mice challenged with lethal doses of H5N1 virus.
2. Materials and methods
2.1 Animals and viruses
Experiments using female C57BL/6 and CD1 mice (Jackson Laboratories, Bar Harbor, ME) were conducted following approval by the Infectious Disease Research Institute and Colorado State University Animal Care and Use Committees. Immunizations were performed as previously described (Clegg et al., 2012) using 5 μg of recombinant hemagglutinin derived from A/Vietnam/1203/2004 (H5N1) [VN1203] or A/California/07/09 (H1N1) [CA07] (Protein Sciences Corp., Meriden CT), which was adjuvanted with 5 μg of GLA-SE (Immune Design Corp., Seattle, WA). Mice were boosted 4 additional times every two weeks and spleens, femurs, and sera were collected. Virus challenge experiments using the highly pathogenic H5N1 virus, VN1203, were performed under the guidance of the U.S. National Select Agent Program in negative pressure HEPA-filtered BSL-3 laboratories at Colorado State University (Clegg et al, 2012). For the antibody protection assays, an irrelevant mouse anti-FITC antibody control, MAb 2A3, was used (Santa Cruz Biotechnology, Dallas, Tx). Following challenge, animals were monitored for weight loss and general health using a 5 point clinical score and euthanized if body weight loss reached 20%. Comparisons of survival were performed using a nonparametric oneway ANOVA. P values of <0.05 were considered significant. Analyses were performed using GraphPad Prism version 5.00 for Windows.
2.2. IgVH database preparation and analysis
The strategy for barcoding, quantifying, and IgVH amplification is based on previous studies (Wiley et al., 2011) with modifications. The primers IgG1 and IgG2 were used to generate the cDNA template from purified lymphocyte mRNA and are shown in Table 1a. The first 21 nucleotides (nt) of each primer include a reverse primer binding site (shown in italics) followed by 12 nt which include a 6 nt fixed barcode (underlined interspersed with a randomly generating 6 nt unique molecular identifier (UMI). The final 18-21 nt include an 18 base gene-specific region. The barcode allows for tagging of individual immunization groups while the UMI allows for quantitating the number of total cDNA amplifications and for calculating IgVH gene frequencies. A total of 32 barcodes (Table 2) were designed in order to correlate specific IgVH sequences with various immunization groups. IgVH amplification was performed using 7 pools of gene specific forward primers, with each pool containing 1 to 5 primers, and a single reverse primer (introduced at the cDNA synthesis step) as shown in Table 1. The reverse primer binding site was within the constant heavy I (CH1) region of IgG1-4 proximal to the junction of the J segment. PCR products were pooled, gel purified, and sequenced using the Illumina MiSeq 2 × 250bp paired-end read technology (SeqWright, Inc.; Houston, TX). Raw MPSS sequences were assembled into full length IgVH sequences (Wiley et al., 2011) and then incorporated into a searchable database within the VSeek bioinformatics software (Imdaptive, Seattle, WA). Total IgVH sequences were sorted into groups or clusters that shared identical CDR3 sequences, each of which contained between 20 and >500 full-length IgVH genes, and the CDR3 cluster frequencies were determined for each immunization group.
Table 1.
cDNA and Murine IgVH Primers.
| a. cDNA primers | ||
| 1 | IgG1 | ACAAGTACATTTCCTCGTCCT ANCNTNCNANCN CMCAGGGGCCAGTGGATA |
| 2 | IgG2 | ACAAGTACATTTCCTCGTCCT ANCNTNCNANCN BCCAGGGACCAAGGGATA |
| b. IgVH Forward PCR primers | ||
| Pool | Primer # | Sequence* |
| 1 | 1.1 | GATAGAGCTGCAGCTTGTTGAGTCTG |
| 1.2 | GATAGAGCTTCAGCTCCAGCAGTCTG | |
| 1.3 | GATAGAGCTGAAGCTTCTCCAGTCTG | |
| 1.4 | GATAGAGCTYCAGCTGCAGCAGTCTG | |
| 2 | 2.1 | GATAGAGCGCTGAGCTKGTGAAGCCT |
| 2.2 | GATAGAGCSCTGAGCTGGTGAAGCCT | |
| 3 | 3.1 | GATAGAGCCTGGCCCTGGKATATTGC |
| 4 | 4.1 | GATAGAGCGTGCACATTGACACAGAG |
| 4.2 | GATAGAGCAGTGCAGCCCTCACAGAG | |
| 5 | 5.1 | GATAGAGCGAACTGTGGTGGTGAAAC |
| 5.2 | GATAGAGCCTGGAGGGTCCCTGAAAC | |
| 5.3 | GATAGAGCGAGCTGAACTGGTGAAAC | |
| 5.4 | GATAGAGCACGCTGAGTTGGTGAAAC | |
| 5.5 | GATAGAGCCTGGACAGTCCCTGAAAC | |
| 6 | 6.1 | GATAGAGCGTGAGGCCTGGAAATTCT |
| 7 | 7.1 | GATAGAGCCTTCAGGAGTCAGGACCT |
| 7.2 | GATAGAGCCTGCARCAGTCTGGACCT | |
| 7.3 | GATAGAGCMTGAAGGAGTCAGGACCT | |
| c. IgVH Reverse PCR Primer | ||
| LP4 | ACAAGTACATTTCCTCGTCCT | |
C, B, K, Y, R, S, or M encode ambiguous nts based on the universal IUPAC Nucleotide Coding System
Table 2.
Barcodes for tagging discrete groups.
| Barcode | Sequence | Barcode | Sequence |
|---|---|---|---|
| 1 | ACTCAC | 17 | CAGTCT |
| 2 | GTCGTA | 18 | ATAGCG |
| 3 | AGTAGC | 19 | ACTACG |
| 4 | ATGCAG | 20 | GCTCTA |
| 5 | GAGTAG | 21 | TAGCTG |
| 6 | AGTGAG | 2 | CGAGAT |
| 7 | TCAGAC | 23 | CACTAC |
| 8 | CTCATC | 24 | TGAGCA |
| 9 | AGCATG | 25 | TCGCAT |
| 10 | ATACGC | 26 | GCTAGT |
| 11 | TCATCG | 27 | TGCAGT |
| 12 | CATCAG | 28 | GTCTGT |
| 13 | TCACGA | 29 | ACGATC |
| 14 | AGAGTC | 30 | TATGCG |
| 15 | AGCTGA | 31 | CTGAGT |
| 16 | ACACTG | 32 | GATGAC |
2.3. MAb cloning and expression
Candidate IgVH sequences were synthesized as GeneBlocks (Integrated DNA Technologies, Inc., Coralville, IA) and cloned into the linearized vector pTR2a (Invitrogen) using two inverse PCR primers; SG101b-UB (5’-AGA GTG CAC CCT GCC GGC TGC GGC) and VH5'FP (5’-GCC AAG ACC ACC GCT CCC TCC GTT TAC CCT). Cloning was performed using the Gibson Assembly Kit (New England BioLabs, Ipswich, MA). Separately, a panel of 20 germline IgVk genes highly expressed in non-immunized mice (Table 4) (Aoki-Ota et al., 2012) were synthesized as GeneBlocks and cloned into pKFM, which is derived from the plasmid pCR2.0 topo (Invitrogen). Each candidate IgVH plasmid was transiently transfected (293-fectin; Invitrogen) in 96-well plates with each IgVk gene to produce mature IgG2a antibodies. Supernatants harvested 3 days later were tested for Ab and antigen binding was measured by ELISA using goat anti-mouse H+L IgG (GaM) (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). For large scale production, MAbs were expressed in 1 L culture volume and purified using MabSelect™ Protein A resin (GE Healthcare). Endotoxin was removed by standard methods.
Table 4.
Variable kappa screening panel.
| # | Vk | # | Vk |
|---|---|---|---|
| 1 | XCMZ | 11 | SZ8 |
| 2 | XZ1N | 12 | VZQ |
| 3 | T676 | 13 | E63 |
| 4 | TYEM | 14 | k6-23 |
| 5 | HKPQ | 15 | 4FZ |
| 6 | GL3 v2 | 16 | k14-111 |
| 7 | VVF v2 | 17 | GH4H |
| 8 | ADAN | 18 | OVP4 |
| 9 | J7A2 | 19 | KWW3 |
| 10 | GETA | 20 | k1-135 |
2.4. Hemagglutinin binding and inhibition assays
The specificity of MAb binding was assayed by ELISA using plates coated with 100 ng/well of recombinant HA (rHA) from VN1203, CA07, and Indo05 (A/Indonesia/05/2005 (H5N1); Protein Sciences Corp.), as well as two group 2 rHA proteins, Aichi68 (A/Aichi/2/1968 (H3N2); Sino Biological, Inc. Beijing, China) and Shanghai13 (A/Shanghai/1/2013 (H7N9); Sino Biological, Inc.). Hemagglutination inhibition (HI) activity specific to A/Vietnam/1194/2004 (H5N1), A/Cambodia/RO405050/2007 (H5N1), A/turkey/Turkey/1/2005 (H5N1), and A/H5N1/Anhui/1/05 (H5N1) virus (NIBSC, Hertfordshire England) were determined (Kendal et al., 1982). HI activity specific to A/California/07/09 (H1N1) virus was measured using 1% turkey RBCs. The relative inhibitory activity for candidate IgG2a MAbs was calculated by dividing the MAb concentration by the highest dilution which completely inhibited the agglutination of RBCs.
3. Results
3.1. Induction of broadly-cross reactive Abs in immunized mice
We set out to discover therapeutic MAbs to influenza by analyzing vaccine-induced IgVH repertoires. Normally, the frequency of broadly neutralizing Abs that recognize highly conserved epitopes within the HA2 domain of the protein is very low and has been estimated to occur in only one in 100,000 HA-specific plasma cells (Corti et al., 2011). However, several studies have shown that these types of antibodies can be induced following heterologous primeboost vaccination and by using vaccine adjuvants that broaden heterosubtypic Ab responses (Buricchi et al., 2013; Chiu et al., 2013; Li et al., 2013). Thus, to enrich for cross-reactive Abs, mice were immunized with rHA isolated from 2 Group 1 HA viruses, VN1203 and CA07 virus, in the presence of the TLR4 adjuvant GLA-SE, which increases antibody repertoire diversity and enhances protection against heterosubtypic strains of H5N1 virus in mice and ferrets (Clegg et al., 2014; Clegg et al., 2012; Wiley et al., 2011). Antisera were then tested for their ability to bind to their cognate antigen as well as for cross-binding to heterologous Group 1 and Group 2 rHA proteins. As indicated in Figure 1, mice immunized with either antigen produced cross-reactive antibodies to CA07, VN1203, and Indo05 (group 1), and to a lesser extent Aichi68 and Shanghai13 (Group 2) proteins. To characterize these antisera further, we measured inhibitory antibodies as measured by HI and determined that VN1203-immunized mice produced cross-inhibitory Abs against 3 of 4 clades of H5N1 virus, but not against H1N1 virus (Figure 2). Similarly, the Abs in CA07-immunized mice were strain-specific and failed to inhibit hemagglutination by H5N1 viruses (Figure 2). Collectively, these results suggest that Abs induced in VN1203- and CA07-immunized mice recognized conserved epitopes shared between Group 1 and Group 2, which lie outside of the highly variable receptor binding site (RBS). Additionally, VN1203 mice also produced heterosubtypic inhibitory Abs that bound more restrictive epitopes within the RBS that are shared between different subclades of H5N1 viruses.
Figure 1. Induction of cross-reactive antibodies to group 1 and group 2 HA proteins.

Mice (5/grp) were immunized with either PBS, or 2 different strains of rHA derived from H1CA A/California/07/09 (H1N1) [CA07] or A/VietNam/1203/2004 (H5N1) [VN1203] and adjuvanted with GLA-SE. Sera were assayed by ELISA for binding to rHA proteins derived from (a) CA07, (b) VN1203, (c) A/Indonesia/05/2005 (H5N1) [Indo05]), (d) A/Aichi/2/1968 (H3N2) [Aichi68], and (e) A/Shanghai/1/2013 (H7N9) [Shanghai13]. As indicated, sera from the CA07-immunized mice bound both rH5 proteins and to a lesser extent rHA from Aichi68 or Shanghai13. Similarly, sera from VN1203-immunized mice effectively bound rHA from Indo05 and CA07, and reduced binding to Aichi68 or Shanghai13.
Figure 2. Induction of heterosubtypic inhibitory antibodies.
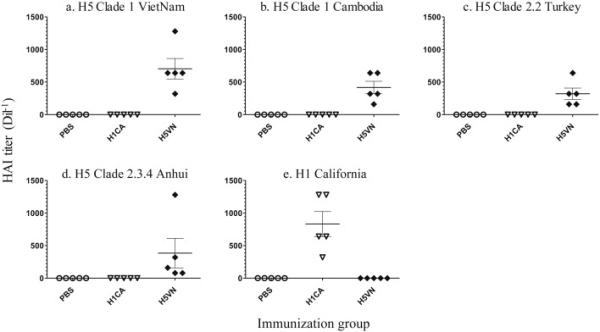
Mice (5/grp) were immunized with either PBS, rH1CA + GLA-SE, or rH5VN + GLA-SE, and serum collected on day 42 was assayed for cross-inhibitory Abs by HAI. The viruses tested included clade 1 viruses A/Vietnam/1194/2004 (H5N1) (panel a) and A/Cambodia/RO405050/2007 (H5N1) (panel b), clade 2.2 virus A/turkey/Turkey/1/2005 (H5N1) (panel c), clade 2.3.4 virus A/H5N1/Anhui/1/05 (H5N1) (panel d), or the H1N1 virus A/California/07/09 (H1N1) (panel e).
3.2. IgVH repertoire analysis
To explore changes in IgVH gene expression between these three groups of immunized mice, splenocyte and bone marrow RNA were prepared and cDNA synthesized using a barcoding strategy (Tables 1a and 2) to allow for correlation of specific sequences to their respective antigen groups. IgVH genes were PCR-amplified using pools of degenerate forward primers (Table 1b) corresponding to >300 murine IgVH gene family members (Wiley et al., 2011) and a single universal reverse primer (LP4) homologous to the 5’ end of the gene specific DNA primer as shown in Table 1c. The final PCR-products were pooled, normalized, and subjected to 2 × 250bp MiSeq paired-end sequencing (Table 3). A total of 5.4M paired end reads were obtained, of which ~60% (~3.2M) passed quality control and were able to be included in the IgVH assemblies. Approximately 40% of these assemblies corresponded to additional immunization groups not discussed herein. A total of 1.07M cDNA transcripts were identified as determined by the UMI counts. The final sequence count reports the number of unique IgVH genes expressed in each immunization group plus a subset of genes that were amplified in spleens as much as 2000-fold as a result of clonal expansion.
Table 3.
MiSeq results.
| Group | PBS | CA07 | VN1203 | Total |
|---|---|---|---|---|
| UMI counts(A) | 331,058 | 228,419 | 510,101 | 1,069,578 |
| Sequence count(B) | 832,447 | 349,534 | 822,950 | 2,011,931 |
The number of cDNA transcripts defined by the 6 base unique molecular identifier.
The absolute number of IgVH sequences amplified.
Standard Chothia numbering was used to assign framework regions, complementarity determining regions (CDR1-3), and IgH isotypes (Abhinandan and Martin, 2008; Alamyar et al., 2012). Gene families were determined by alignment with germline genes, which also identified the likely VDJ recombination events and somatic mutations. We then determined the total number of complete IgVH sequences represented more than once in mice immunized with PBS (126,556), or with VN1203 (143,359) or CA07 (77,167). It has been shown that the IgVH sequences with the highest frequency CDR3 following immunization are likely to encode antigen-specific Abs (Reddy et al., 2010). Consequently, we sorted IgVH genes from each immunization group into clusters on the basis of sharing an identical CDR3. The frequencies of these clusters, each containing roughly 20-500 different IgVH genes, were then plotted (Figure 3). As indicated, the vast majority of CDR3 sequences were specific to each immunization group, although a low frequency of shared CDR3 clusters were detected. Presumably, these clusters contain related IgVH lineages that were selected by highly conserved epitopes in distant HA proteins. Similar convergent IgVH rearrangements have previously been reported (Jackson et al., 2014; Parameswaran et al., 2013).
Figure 3. CDR3 sequences shared between immunization groups.
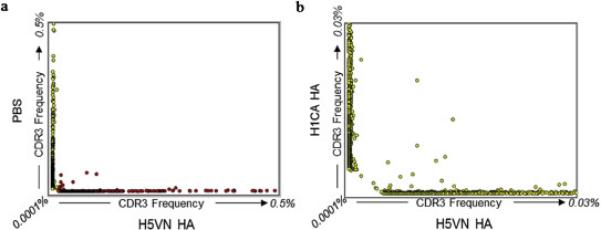
Panel A: The frequencies of specific CDR3 sequences, unique or shared, between PBS (y-axis) and H5VN (x-axis) immunization groups. Each circle contains between 20 and 500 IgVH genes that share an identical CDR3, although sequence identity was variable outside this region. Panel B: Frequency plot comparing unique and shared CDR3s between H1CA HA (y-axis) and H5VN HA (x-axis) immunization groups following subtraction of CDR3s from PBS treated mice.
3.3. MAb screening and in vitro analysis
As an initial proof for isolating HA-specific MAbs, we selected the most highly expressed IgVH gene within the 35 most frequent CDR3 clusters within the VN1203 data set. Thirty-five candidate IgVH genes were synthesized as GeneBlocks (Integrated DNA Technologies, Inc., Coralville, Iowa) and cloned by Gibson assembly (Gibson et al., 2009) into a murine IgG2a expression vector (pTR2a). Each candidate IgVH plasmid was co-transfected into 293F cells with a panel of 20 different germline murine kappa genes (Table 4);, and day 3 culture supernatants were assayed for IgG production by SDS-PAGE and for VN1203 HA binding by ELISA. All 35 IgVH candidates in a preliminary screen produced mature IgG2a, 17 of which bound VN1203 protein. Overall, the IgVH candidates bound antigen when paired with six of the panel of 20 IgVKs. The 17 HA-binding IgVH genes were then co-transfected in large scale with their preferred Vk partner, purified by Protein A chromatography, and tested for binding to an expanded panel of HA antigens. Nine of these 17 MAbs were further characterized (Table 5). Interestingly, the heavy chains for 8 of the 9 MAbs were derived from the VH1-39*01 gene family and bound antigen most effectively using the K1-135 light chain, the outlier being MAb 632G which was derived from VH1-82*01 and paired most effectively with GETA light chain. Three of the 9 MAbs paired effectively with two additional light chains resulting in specific, albeit lower levels, binding to HA. All 9 heavy chains completed class switching and contained multiple CDR and framework mutations.
Table 5.
IgVH Characteristics.
| MAb | CDR3 | Frequency (%) | IgVH Family | IgH isotype | Mutations in CDR1, 2, 3 | Mutations in FW1, 2, 3, 4 | Optimal Vk pairing | Alternative Vk pairing |
|---|---|---|---|---|---|---|---|---|
| 808K | EGFTTVEGPDY | 0.43 | 1-39*01 | IgG2C | 2, 2, 6 | 0, 0, 0, 1 | k1-135 | none |
| 24415K | GSWSVYEAY | 0.18 | 1-39*01 | IgG2C | 2, 4, 8 | 2, 1, 9, 1 | k1-135 | none |
| 2407K | GSWSDYEAY | 0.41 | 1-39*01 | IgG2C | 1, 2, 5 | 0, 4, 4, 1 | k1-135 | none |
| 1620K | GGFTTGSY | 0.18 | 1-39*01 | IgG2C | 1, 1, 6 | 0, 0, 1, 1 | k1-135 | none |
| 1968K | EGFVSDGGYAMDY | 0.24 | 1-39*01 | IgG2C | 1, 2, 7 | 0, 0, 0, 1 | k1-135 | GH4H>GETA |
| 491K | EERLRHF | 0.30 | 1-39*01 | IgG2C | 2, 3, 5 | 2, 2, 10, 1 | k1-135 | ADAN>k14-111 |
| 1166K | LYGTYYFDY | 0.32 | 1-39*01 | IgG2B | 2, 2, 3 | 1, 0, 3, 1 | k1-135 | none |
| 447K | DGWNNNYNDY | 0.36 | 1-39*01 | IgG1 | 1, 3, 6 | 1, 0, 2, 1 | k1-135 | none |
| 632G | GAVPFYFDY | 0.32 | 1-82*01 | IgG2B | 2, 3, 4 | 1, 0, 8, 1 | GETA | ADAN>k6-23 |
Consistent with the bioinformatic analysis, all 9 MAbs derived from VN1203 CDR3 clusters bound VN1203, but not CA07 (Table 6a). In addition, all bound a truncated VN1203 protein encompassing the HA1 head domain, which is the most antigenic region of the protein (Dormitzer et al., 2011). To determine function, each MAb was assessed for its ability to inhibit hemagglutination mediated by four different strains of H5N1 virus. As indicated in Table 6b, 8 of 9 MAbs inhibited A/Vietnam/1194/2004 (H5N1) (clade 1.0) and A/Cambodia/RO405050/2007 (H5N1) (clade 1.1) antigen; 6 of 9 also inhibited A/Turkey/Turkey/1/2005 (H5N1) (clade 2.2), whereas no MAbs inhibited A/Anhui/1/05 (H5N1) (clade 2.3.4). To define the nature of the bound epitope, we compared MAb binding to native and denatured (boiled and reduced) VN1203 by ELISA (Table 6c). Five MAbs appeared to bind a linear epitope whereas four recognized a conformational epitope.
Table 6.
Functional analysis of head-binding IgG MAbs.
| a. Screening ELISA profile (1450) | b. HI Titer (H5 clades) (ng/uL) | c. Nature of Epitope | ||||||
|---|---|---|---|---|---|---|---|---|
| MAbs | H5VN | H1CA | HA1 | A/VietNam (clade 1) | A/Cambodia (clade 1) | A/Turkey (clade 2.2) | A/Anhui (clade 2.2.4) | Linear or Conformational |
| 808K | 2.918 | nb | 2.999 | 2.07 | 2.07 | 2.07 | NA | Linear |
| 24415K | 3.129 | nb | 3.269 | 1.64 | 1.64 | 1.64 | NA | Linear |
| 2407K | 3.099 | nb | 3.057 | 0.43 | 0.87 | 0.43 | NA | Linear |
| 1620K | 2.599 | nb | 2.931 | 0.94 | 0.94 | 0.00 | NA | Conformational |
| 1968K | 3.003 | nb | 2.951 | 3.13 | 3.13 | 3.13 | NA | Linear |
| 491K | 2.704 | nb | 3.102 | 1.72 | 0.86 | 13.75 | NA | Conformational |
| 1166K | 2.844 | nb | 3.067 | 0.82 | 1.64 | 1.64 | NA | Linear |
| 447K | 2.245 | nb | 2.921 | 6.88 | 0.86 | 0.00 | NA | Conformational |
| 632G | 2.779 | nb | 2.612 | NA | NA | NA | NA | Conformational |
nb = no binding; NA = no HI activity
3.4. Antiviral protection in mice
To assess MAb activity in vivo, CD-1 mice (6 / grp) were injected with MAb 808K or 24415K (5 mg kg−1) and were then challenged 12 hours later with a lethal dose (10 LD50) of A/Vietnam/1203/04 (H5N1) virus (Figure 4a). As indicated, all mice pretreated with either MAb continued to gain weight and survived viral challenge, while all PBS injected mice died (Figure 4b).
Figure 4. MAb-mediated protection against a lethal challenge of H5N1 virus in mice.
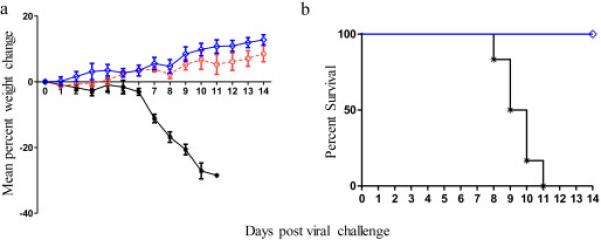
CD1 mice (6/grp) were injected with either PBS (black line) or 5 mg kg-1 of MAb 808k (blue line, open diamonds) or MAb 24415k (red line, open circles). Animals were challenged 12 hours later with 10 LD50 A/Vietnam/1194/2004 (H5N1) virus and then monitored for the next 14 days. Panel A: average weight gain. Panel B: Survival kinetics.
To test for therapeutic activity, mice were inoculated with a lethal dose of virus (10 LD50) and then injected after 4h, 24h, or 48h with either MAb 24415k or 808k, or with an irrelevant control mouse MAb, 2A3 (5 mg kg−1). As shown in Figure 5, all mice injected with PBS or the control MAb lost weight within 3 days, developed severe disease within 9 days, and were euthanized to minimize suffering. By contrast, treatment with MAbs 24415K or 808K 4 hrs after virus inoculation prevented substantial weight loss and protected all the animals from virus. Even when treatment was delayed 24h (Figs. 5b and 5e) or 48h (Figs. 5c and 5f), these two MAbs protected, collectively, 8 of 12 mice (67%) from viral challenge. This is a convincing demonstration that protective anti-H5N1 MAbs can be selected using an IgVH discovery platform.
Figure 5. Therapeutic protection of MAbs in mice challenged with H5N1 virus.
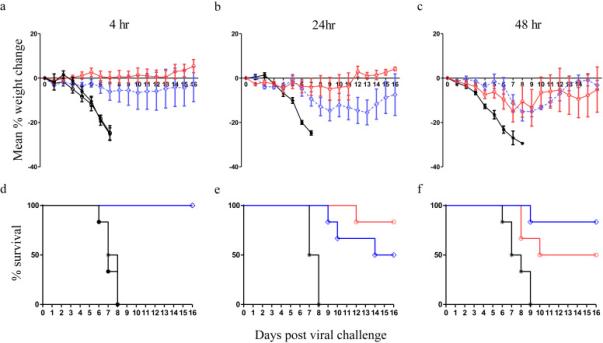
CD1 mice (6/grp) received a lethal dose (10LD50) of A/Vietnam/1194/2004 (H5N1) virus and were then injected after 4h (A & D), 24h (B & E), or 48h (C & F) with either PBS (black line, closed black circles) or 5 mg kg-1 808k MAb (blue line, open diamonds), 24415k MAb (red line, open circles) or the irrelevant control MAb 2A3 (black line, asterisk).
4. Discussion
The high rate of influenza virus mutation is responsible for seasonal epidemics with a worldwide prevalence of 5-15%, and rare but more deadly pandemics that have caused the death of millions of people. Avian influenza viruses are normally species-specific, but five strains (H5N1, H7N3, H7N7, H7N9, and H9N2) have repeatedly infected humans, and two (H5N1 and H7N9) cause fatality rates of roughly 50% (Keitel and Atmar, 2007; Poland et al. 2007; Stephenson, 2006). H5N1 viruses are evolving rapidly and there is great concern that they will eventually acquire the ability to transmit between humans (World Health Organization/World Organisation for Animal and Agriculture Organization, 2014). This worry is compounded by the fact that a relatively small number of mutations permit aerosol H5N1 transmission between ferrets without causing a significant change in virulence (Herfst et al., 2012; Imai et al., 2012). While vaccination is the most effective tool for preventing influenza, health organizations worldwide have advocated for the development of MAbs that could be deployed in the early stages of an outbreak and used to prevent or treat infections in recently exposed individuals.
NGS permits analysis of millions of B cell receptor sequences in a single experiment. This technology has been used to characterize the Ab repertoires in disease states and following vaccination, and is a valuable tool for Ab discovery (Galson et al., 2014; Galson et al., 2015; Georgiou et al., 2014; Herfst et al., 2012; Imai et al., 2012; Reddy et al., 2010; Saggy et al., 2012). Here, we explored the feasibility of building an NGS-based antibody discovery platform with the ultimate goal of isolating MAbs that can broadly cross-neutralize multiple strains of virus. Since the frequency of these types of antibodies are normally very low in humans (Corti and Lanzavecchia, 2013), we reasoned that functionally relevant MAbs could be enriched in immunized mice, which could then be adapted for the clinic using standard antibody technologies.
Evidence supporting the induction of broadly cross-reactive antibodies was presented in Figure 1, where immunization with two different group 1 rHA proteins (CA07 and VN1203) resulted in the production of antisera that could cross-bind each other, recognize rHA from a second strain of H5N1 (Indo05), and even bound two group 2 rHA proteins (Aichi68 and Shanghai13). Most likely these broadly conserved epitopes reside in the HA2 domain of the protein (Corti and Lanzavecchia, 2013). This experiment also highlighted the induction of heterosubtypic inhibitory Abs that bound conserved epitopes within the RBS domain of Clade 1 and Clade 2 H5N1 viruses (Fig 2). IgVH repertoires were sequenced by MiSeq paired-end technology, and approximately 1.07M full length IgVH genes were assembled in silco into the IgVH database. To identify HA-specific IgVH sequences, we adopted search criteria based on previous observations that the IgVH and IgVL genes containing the most frequently expressed CDR3 regions following immunization encoded antigen-specific Abs (Reddy et al., 2010, Fig 3). We demonstrated that 17 of 35 candidate IgVH proteins bound VN1203 protein when expressed as mature MAb, 8 out of 9 MAbs exhibited HI activity with 3 heterosubtypic strains of H5N1 virus, and 2 MAbs demonstrated prophylactic and therapeutic activity in vivo in virus-challenged mice. This is the first example in which an NGS discovery platform has been used to isolate anti-influenza MAbs with a relevant therapeutic activity.
Not unexpectedly, a search criteria that focuses on high frequency IgVH genes biases the selection of antibodies that recognize the most immunogenic region of the protein, which includes the HA1 head domain and RBS domain (Dormitzer et al., 2011). Consequently, alternative search criteria will be required for identifying broadly cross-reactive Abs, which appear in much lower frequencies. Based on a variety of sequence signatures identified in broadly neutralizing anti-influenza and anti-HIV MAbs (Avnir et al., 2014; Zhu et al., 2013b), these bioinformatics tools would search for strong biases in germline IgVH gene family usage, test for substantial changes in CDR sequences with particular emphasis on increased CDR3 length, and look for genes with unusually high rates of somatic mutation post-immunization. Collectively, these molecular signatures have been used to build IgVH lineages and phylogenetic trees (Wu et al., 2011; Zhu et al., 2013a). Another powerful tool that our approach can exploit is based on the convergent IgVH rearrangements that occur between individuals that target the same epitopes following vaccination or infection (Jackson et al., 2014; Parameswaran et al., 2013). Thus, it should be possible to identify broadly cross-reactive Abs by immunizing mice with distantly related HA proteins and then searching for IgVH sequences that share these common signatures. Another important innovation is the assemblage of paired heavy- and light-chain Ab repertoires by VH-VL amplicon sequencing of single B cells (DeKosky et al., 2015). Light chains have a much lower sequence diversity than heavy chains and, as demonstrated in our analysis, germline light chains can functionally pair with a variety of heavy chains. However, the light chain rearrangements associated with broadly neutralizing Abs also fall into distinct phylogenetic trees that can be used in concert with IgVH analyses (Wu et al., 2011; Zhu et al., 2013a). In conclusion, we have described a technology platform for the discovery of novel MAbs that relies on the manipulation and interrogation of mouse IgVH gene repertoires. With further refinement this approach should prove useful in the identification of important therapeutic MAbs for influenza and other infectious diseases.
Highlights.
The IgVH repertoires of mice immunized with influenza antigen were assembled following next generation sequencing.
Candidate IgVH sequences were selected using a CDRH3 search algorithm and expressed in cells to produce mature IgG.
These MAbs demonstrated anti-H5N1 antigen binding and neutralization activity in vitro.
Selected MAbs protected virus-challenged mice successfully following prophylactic and therapeutic delivery.
Acknowledgements
This work was funded by National Institute of Allergy and Infectious Diseases, National Institutes of Health Grant R43AI098290.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abhinandan KR, Martin AC. Analysis and improvements to Kabat and structurally correct numbering of antibody variable domains. Molecular immunology. 2008;45:3832–3839. doi: 10.1016/j.molimm.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Alamyar E, Duroux P, Lefranc MP, Giudicelli V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods in molecular biology. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- Aoki-Ota M, Torkamani A, Ota T, Schork N, Nemazee D. Skewed primary Igkappa repertoire and V-J joining in C57BL/6 mice: implications for recombination accessibility and receptor editing. Journal of immunology. 2012;188:2305–2315. doi: 10.4049/jimmunol.1103484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avnir Y, Tallarico AS, Zhu Q, Bennett AS, Connelly G, Sheehan J, Sui J, Fahmy A, Huang CY, Cadwell G, Bankston LA, McGuire AT, Stamatatos L, Wagner G, Liddington RC, Marasco WA. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS pathogens. 2014;10:e1004103. doi: 10.1371/journal.ppat.1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Bauer M, Schmitz N, Buser RB, Gwerder M, Muntwiler S, Renner WA, Saudan P, Bachmann MF. Prophylactic and therapeutic activity of fully human monoclonal antibodies directed against influenza A M2 protein. Virology journal. 2009;6:224. doi: 10.1186/1743-422X-6-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg B, Koudstaal W, Goudsmit J, Klaren V, Tang C, Bujny MV, Korse HJ, Kwaks T, Otterstrom JJ, Juraszek J, van Oijen AM, Vogels R, Friesen RH. Mechanisms of hemagglutinin targeted influenza virus neutralization. PloS one. 2013;8:e80034. doi: 10.1371/journal.pone.0080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buricchi F, Bardelli M, Malzone C, Capecchi B, Nicolay U, Fragapane E, Castellino F, Del Giudice G, Galli G, Finco O. Impact of preexisting memory to seasonal A/H1N1 influenza virus on the immune response following vaccination against avian A/H5N1 virus. European journal of immunology. 2013;43:641–648. doi: 10.1002/eji.201242563. [DOI] [PubMed] [Google Scholar]
- Chiu C, Wrammert J, Li GM, McCausland M, Wilson PC, Ahmed R. Cross-reactive humoral responses to influenza and their implications for a universal vaccine. Annals of the New York Academy of Sciences. 2013;1283:13–21. doi: 10.1111/nyas.12012. [DOI] [PubMed] [Google Scholar]
- Clegg CH, Roque R, Van Hoeven N, Perrone L, Baldwin SL, Rininger JA, Bowen RA, Reed SG. Adjuvant solution for pandemic influenza vaccine production. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17585–17590. doi: 10.1073/pnas.1207308109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg CH, Roque R, Perrone LA, Rininger JA, Bowen R, Reed SG. GLA-AF, an emulsion-free vaccine adjuvant for pandemic influenza. PloS one. 2014;9:e88979. doi: 10.1371/journal.pone.0088979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annual review of immunology. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- Council HS. National Strategy for Pandemic Influenza Implementation Plan. 2006 http://www.flu.gov/planning-preparedness/federal/pandemic-influenza-implementation.pdf.
- de Jong MD, Hien TT. Avian influenza A (H5N1). Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2006;35:2–13. doi: 10.1016/j.jcv.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nature medicine. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- Dormitzer PR, Galli G, Castellino F, Golding H, Khurana S, Del Giudice G, Rappuoli R. Influenza vaccine immunology. Immunological reviews. 2011;239:167–177. doi: 10.1111/j.1600-065X.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen RH, Koudstaal W, Koldijk MH, Weverling GJ, Brakenhoff JP, Lenting PJ, Stittelaar KJ, Osterhaus AD, Kompier R, Goudsmit J. New class of monoclonal antibodies against severe influenza: prophylactic and therapeutic efficacy in ferrets. PloS one. 2010;5:e9106. doi: 10.1371/journal.pone.0009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galson JD, Pollard AJ, Truck J, Kelly DF. Studying the antibody repertoire after vaccination: practical applications. Trends in immunology. 2014;35:319–331. doi: 10.1016/j.it.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Galson JD, Truck J, Fowler A, Munz M, Cerundolo V, Pollard AJ, Lunter G, Kelly DF. In-Depth Assessment of Within-Individual and Inter-Individual Variation in the B Cell Receptor Repertoire. Frontiers in immunology. 2015;6:531. doi: 10.3389/fimmu.2015.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeraedts F, Huckriede A. Influenza vaccines: what do we want and how can we get it? Advances in experimental medicine and biology. 2011;780:161–174. doi: 10.1007/978-1-4419-5632-3_13. [DOI] [PubMed] [Google Scholar]
- Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nature biotechnology. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KJ, Liu Y, Roskin KM, Glanville J, Hoh RA, Seo K, Marshall EL, Gurley TC, Moody MA, Haynes BF, Walter EB, Liao HX, Albrecht RA, Garcia-Sastre A, Chaparro-Riggers J, Rajpal A, Pons J, Simen BB, Hanczaruk B, Dekker CL, Laserson J, Koller D, Davis MM, Fire AZ, Boyd SD. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell host & microbe. 2014;16:105–114. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keitel WA, Atmar RL. Preparing for a possible pandemic: influenza A/H5N1 vaccine development. Current opinion in pharmacology. 2007;7:484–490. doi: 10.1016/j.coph.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Kendal AP, Pereira MS, Skehel JJ. Hemagglutination inhibition. In: Kendal AP, Pereira MS, Skehel JJ, editors. concepts and procedures for laboratory based influenza surveillance. Centers for Disease Control and Pan-American Health Organization; Atlanta, GA: 1982. pp. B17–35. [Google Scholar]
- Li CK, Rappuoli R, Xu XN. Correlates of protection against influenza infection in humans--on the path to a universal vaccine? Current opinion in immunology. 2013;25:470–476. doi: 10.1016/j.coi.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang KK, Yang Y, Chen X, Gao F, Munshaw S, Kepler TB, Denny T, Moody MA, Haynes BF. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. Journal of virological methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran P, Liu Y, Roskin KM, Jackson KK, Dixit VP, Lee JY, Artiles KL, Zompi S, Vargas MJ, Simen BB, Hanczaruk B, McGowan KR, Tariq MA, Pourmand N, Koller D, Balmaseda A, Boyd SD, Harris E, Fire AZ. Convergent antibody signatures in human dengue. Cell host & microbe. 2013;13:691–700. doi: 10.1016/j.chom.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland GA, Jacobson RM, Targonski PV. Avian and pandemic influenza: an overview. Vaccine. 2007;25:3057–3061. doi: 10.1016/j.vaccine.2007.01.050. [DOI] [PubMed] [Google Scholar]
- Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, Hunicke-Smith SP, Iverson BL, Tucker PW, Ellington AD, Georgiou G. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nature biotechnology. 2010;28:965–969. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]
- Saggy I, Wine Y, Shefet-Carasso L, Nahary L, Georgiou G, Benhar I. Antibody isolation from immunized animals: comparison of phage display and antibody discovery via V gene repertoire mining. Protein engineering, design & selection : PEDS. 2012;25:539–549. doi: 10.1093/protein/gzs060. [DOI] [PubMed] [Google Scholar]
- Stephenson I. H5N1 vaccines: how prepared are we for a pandemic? Lancet. 2006;368:965–966. doi: 10.1016/S0140-6736(06)69340-9. [DOI] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature structural & molecular biology. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PloS one. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley SR, Raman VS, Desbien A, Bailor HR, Bhardwaj R, Shakri AR, Reed SG, Chitnis CE, Carter D. Targeting TLRs expands the antibody repertoire in response to a malaria vaccine. Science translational medicine. 2011;3:93ra69. doi: 10.1126/scitranslmed.3002135. [DOI] [PubMed] [Google Scholar]
- World Health Organization/World Organisation for Animal, H.F., Agriculture Organization, H.N.E.W.G. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respir Viruses. 2014;8:384–388. doi: 10.1111/irv.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Program NCS, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Ofek G, Yang Y, Zhang B, Louder MK, Lu G, McKee K, Pancera M, Skinner J, Zhang Z, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Simek M, Burton DR, Koff WC, Program NCS, Mullikin JC, Mascola JR, Shapiro L, Kwong PD. Mining the antibodyome for HIV-1-neutralizing antibodies with next- generation sequencing and phylogenetic pairing of heavy/light chains. Proceedings of the National Academy of Sciences of the United States of America. 2013a;110:6470–6475. doi: 10.1073/pnas.1219320110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wu X, Zhang B, McKee K, O'Dell S, Soto C, Zhou T, Casazza JP, Program NCS, Mullikin JC, Kwong PD, Mascola JR, Shapiro L. De novo identification of VRC01 class HIV-1-neutralizing antibodies by next-generation sequencing of B-cell transcripts. Proceedings of the National Academy of Sciences of the United States of America. 2013b;110:E4088–4097. doi: 10.1073/pnas.1306262110. [DOI] [PMC free article] [PubMed] [Google Scholar]


