ABSTRACT
Understanding regulation of transposon movement in somatic cells is important as mobile elements can cause detrimental genomic rearrangements. Generally, transposons move via one of 2 mechanisms; retrotransposons utilize an RNA intermediate, therefore copying themselves and amplifying throughout the genome, while terminal inverted repeat transposons (TIR Tns) excise DNA sequences from the genome and integrate into a new location. Our recently published work indicates that retrotransposons in Drosophila tissue culture cells are actively transcribed in the antisense direction. Our data support a model in which convergent transcription of retrotransposons from intra element transcription start sites results in complementary RNAs that hybridize to form substrates for Dicer-2, the endogenous small interfering (esi)RNA generating enzyme. Here, we extend our previous analysis to TIR Tns. In contrast to retrotransposons, our data show that antisense TIR Tn RNAs result from transcription of intronic TIR Tns oriented antisense to their host genes. Also, disproportionately less esiRNAs are generated from TIR transcripts than from retrotransposons and transcription of very few individual TIR Tns could be confirmed. Collectively, these data support a model in which TIR Tns are regulated at the level of Transposase production while retrotransposons are regulated with esiRNA post-transcriptional mechanisms in Drosophila somatic cells.
KEYWORDS: Antisense, Dicer-2, inverted repeat, small interfering RNAs, transposon
Introduction
Active transposons (Tns) and transposon derived sequences comprise approximately 22% of the Drosophila melanogaster genome.1-3 Movement of these Tns plays an important role in evolution, but also causes genomic instability;4 therefore, regulation of Tn expansion is important to maintain an appropriate balance. In Drosophila, mutations linked to P element insertions cause hybrid dysgenesis syndrome.5-8 Intensive study of P elements has contributed to a molecular understanding of class II terminal inverted repeat (TIR) transposition mechanisms9,10 and how Tn movement is regulated in vivo.11-14
Transposons are classified based on the identity of their nucleic acid intermediates. Transposons having an RNA intermediate and encoding a reverse transcriptase are retrotransposons (retroTn). Tns utilizing a cut-and-paste mechanism with a DNA intermediate and having inverted repeat end sequences are called terminal inverted repeat (TIR) Tns. Drosophila melanogaster has several classes of both retroTns and TIR Tns,1 although retroTns appear to be more active.3,15
Movement of both retroTns and TIR Tns must be regulated to ensure genomic stability. In Drosophila, 2 non-coding RNA mediated post-transcriptional silencing mechanisms have been elucidated. The piwi-interacting RNA (piRNA) pathway generates small RNAs that suppress Tn mobility by inducing heterochromatin formation in the germline.16-22 These siRNAs are produced from a single stranded RNA precursor. In somatic cells, endogenous small interfering (esi)RNAs silence retroTns using an Argonaute 2 (Ago2)-dependent mechanism.23-27 Most of these esiRNAs are generated from double stranded Tn derived RNA precursors by Dicer-2 (Dcr2).23,28,29 Increased movement of retroTns is observed in somatic tissues from Drosophila mutants lacking RNAi components.27 How these silencing mechanisms function in the fly to suppress Tn movement are poorly understood. One transcriptional regulatory mechanism has been identified for P element transposition. Alternative splicing prevents expression of the P element Transposase (Tnp) in somatic cells.12 Therefore, P elements are only mobile in the Drosophila germline as functional P element Tnp is only produced in this tissue.11,12
To understand the origins of Dcr2 substrates in Drosophila tissue culture cells,30 we performed small RNA-seq and RNA-seq on wild type and Dcr2 depleted samples. Our analyses of these data revealed that many individual retroTns are transcribed in both the sense (S) and antisense (AS) direction from intra element transcription start sites with canonical Drosophila RNA polymerase II promoters. These S and AS RNAs are substrates for Dcr2 as their levels are increased in the Dcr2 depleted sample. Correspondingly, the number esiRNAs generated from these substrates decreases when Dcr2 is knocked down. This work was recently published in Genetics.15
Here we extend this in-depth analysis to include TIR Tns pogo and 1360 (hoppel or ProtoP). The pogo TIR Tn is a member of the Tc1/mariner superfamily of Tns with 21 base pair terminal inverted repeats.31,32 1360 is believed to be derived from an ancient P element-like Tn.33 These elements were chosen for further investigation as S and AS transcription of these Tns was observed previously.15 We conclude that while a few AS RNA-seq reads are observed for these TIR Tns, very few esiRNAs are generated from these transcripts indicating that TIR Tn movement is minimally regulated by esiRNAs in Drosophila melanogaster somatic cells. Additionally, we discovered that unlike retroTns, few individual TIR Tns are transcribed in either the S or AS direction. Finally, analyses of 1360 and pogo transcripts allow insight into the transposition mechanisms of these TIR Tns.
Results
Ratios of full-length to truncated Tns differ for TIR and retroTns
1360 or Hoppel is the most abundant TIR Tn in the Drosophila genome with 304 annotated copies,34,35 while 48 pogo TIR Tns have been documented.35 A full-length 1360 element is predicted to be 1107 base pairs (bp) while a full-length pogo element is 2122 bp. As a first step toward insight into the molecular details of TIR Tn mobility in a genomic context, variation in sizes among the annotated 1360 and pogo Tns were first examined. Approximately one-third of 1360 Tns are greater than 1 kb (Table 1), although only 3 are the predicted 1107 bp (data not show). Generally, the 1360 elements vary tremendously in size; often differing by only 1 bp in the 1360 Tns less than 40 bp and by only 10s of bp for the 1360 Tns greater than 40 bp (data not shown). Rarely are 2 1360 elements the same size. In contrast, pogo elements are restricted to 4 sizes: 2120–2123 bp, 1067–1491 bp, 704 bp or 186–187 bp. Examination of length distributions for previously investigated non-LTR retrotransposons (retroTns) Juan and Jockey and LTR retroTns blood, mdg1 and 297 revealed a much higher percentage of full-length elements than observed for pogo and 1360 (data not shown).15 The variability of length distributions may support differences in mechanisms that control retroTn and TIR Tn movement.
Table 1.
Very few TIR Tns greater than 1 kb are transcribed from internal transcription start sites.
| Element | Size | % total | % >1 kb inter | % >1 kb intra | % >1 kb intra (S) | % >1 kb Intra (AS) | %trans. int. tss |
|---|---|---|---|---|---|---|---|
| 1360 | >1 kb | 36.8 | 47.4 | 52.6 | 42.5 | 57.5 | 1.3 |
| 1 kb‐40bp | 52.0 | ||||||
| <40bp | 11.2 | ||||||
| pogo | >2kb | 10.4 | |||||
| 1‐1.5kb | 23.0 | 68.7 | 31.3 | 40.0 | 60.0 | 0.0 | |
| 704bp | 4.2 | ||||||
| 187‐186bp | 58.3 | ||||||
| 186‐40bp | 4.2 |
Note. The percent (% total) of 1360 and pogo TIR Tns in each size class is shown in the left three columns. The remainder of the analysis was only performed on Tns greater than 1 kb. The percentages of intergenic and intragenic 1360 and pogo Tns (%>1 kb inter and %>1 kb intra) are shown in columns three and four. The percent of intragenic 1360 and pogo Tns having mapped sense (S) and antisense (AS) RNA‐seq reads (%>1 kb intra (S) and %>1 kb intra (AS)) are shown in columns five and six. Finally, the percent of 1360 and pogo TIR Tns greater than 1 kb for which S or AS transcription from an internal transcription start site (tss) could be confirmed (%trans. int. tss) is reported in the last column.
AS TIR Tn transcripts are not produced from intraelement tss
To investigate S and AS pogo and 1360 Tn transcription and potential esiRNA biogenesis, small RNA-seq and RNA-seq data sets from control Drosophila tissue culture (Dmel-2) cells15 were mapped to the Drosophila genome followed by visualization of non-unique and unique reads using the UCSC genome browser (http://genome.ucsc.edu, Dm6 assembly, August 2014).36,37 Examples of representative full-length intergenic pogo and 1360 elements show RNA-seq reads mapping to both S and AS strands, although the number of normalized AS reads (reads per million (RPM)) is low (Figs. 1A-B, red).
Figure 1.
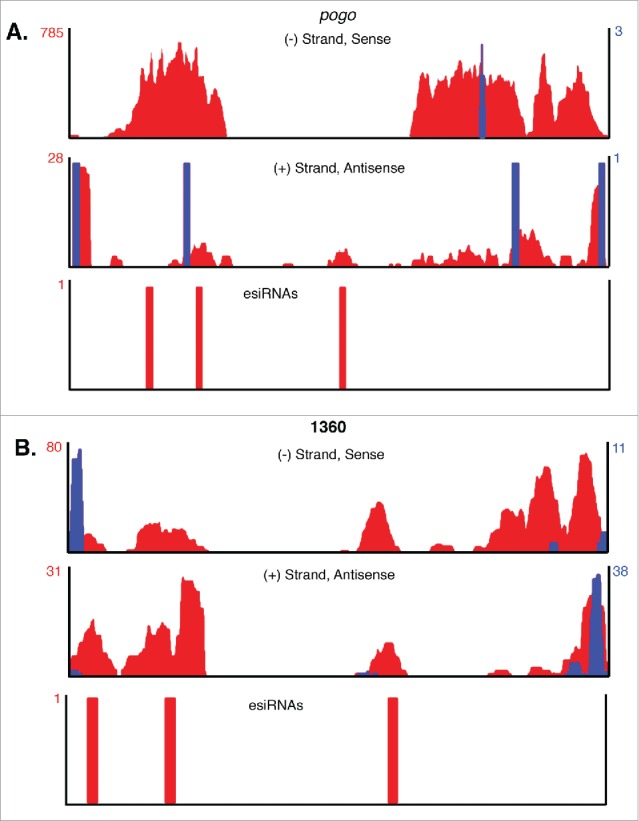
Sense and antisense 1360 and pogo TIR Tn transcripts are not produced by intra element transcription start sites. (A-B) Bedgraphs representing sense (top) and antisense (middle) non-unique RNA-seq reads mapping to a representative full-length pogo TIR Tn (A) or 1360 TIR Tn (B) are shown in red. Peak reads per million (RPM) are listed to the left (red numbers). Non-unique small-capped RNA-seq reads representing transcription start sites are overlaid in blue and RPM values are listed to the right (blue numbers). Non-unique endogenous small-RNA seq reads mapping to pogo and 1360 TIR Tns are shown below the RNA-seq reads (A-B).
We also re-mapped publically available short-capped RNA sequencing data to identify potential S and AS pogo and 1360 transcription start sites (tss).38,39 No intra element tss accounting for S or AS transcripts could be identified for canonical pogo elements (Fig. 1A, blue) indicating that the non-unique reads mapping to this intergenic element originated from a different pogo TIR Tn. While intra element tss are present in canonical 1360 elements, these tss initiate transcription into flanking sequences rather than producing S and AS 1360 TIR Tn RNAs (Fig. 1B, blue). Like pogo, these data show that the observed non-unique reads mapping to this representative intergenic 1360 TIR Tn were produced by a different individual Tn.
If the S and AS TIR Tn RNA-seq reads are not produced from intra element tss, what could be the source of these transcripts? To further investigate the origins of low level AS TIR Tn transcription, we first examined Tn flanking sequences for all elements. No AS tss were observed for pogo and 1360 TIR Tns in regions surrounding the Tns (data not shown). Next we investigated the genomic locations of pogo and 1360 TIR Tns larger than 1 kb. About half of the 112 1360 elements larger than 1 kb are between genes (intergenic) and half are within introns of protein coding genes (intragenic) (Table 1). ∼70% of pogo TIR Tns greater than 1 kb are intergenic while the other ∼30% are intragenic (Table 1). Further examination of intragenic TIR Tns greater than 1 kb revealed that the orientation of the Tn to the mRNA was AS approximately 60% of the time for both pogo and 1360 meaning that transcription of the protein coding gene would produce AS TIR Tn RNAs for 60% of the TIR Tns (Table 1) greater than 1 kb. As the AS RPMs for both pogo and 1360 are low (Figs. 1A-B), we hypothesize that these RNAs are generated indirectly from transcription of protein coding genes with AS oriented intragenic TIR Tns.
Previous experiments show that endogenous small interfering (esi)RNAs are produced from hybridized (double stranded (ds)) retroTn S and AS transcripts by Dicer-2 (Dcr2).15 To investigate the potential for S and AS 1360 and pogo transcripts to generate esiRNAs, we visualized smRNA-seq reads from control Dmel-2 cells corresponding to representative 1360 and pogo TIR Tns (Figs. 1A-B, bottom). Very few esiRNAs were observed for either pogo or 1360.
Pogo{}4759 is the only actively transcribed pogo Tn in the Drosophila genome
Drosophila pogo elements fall into 4 size classes: 2.1 kb, 1.1–1.4 kb, 704 bp and 186–187 bp. Previous analyses indicate that single internal deletions are responsible for the 2.1 kb to 1.1–1.4 kb size reduction resulting in Tns with similar ends, but differing internal structure.31 This observation is confirmed by aligning all annotated Drosophila pogo elements (data not shown). Comparing pogo TIR Tns representative of the 2.1 kb (pogo{}297), 1.1–1.4 kb (pogo{}4759), 704 bp (pogo{}1454), and 186–187 bp (pogo{}718) size classes reveals deletion of pogo{}297 nucleotides (nts) 788 to 1440 in pogo{}4759, deletion of an additional approximately 300 nts flanking the pogo{}4759 internal deletion in pogo{}1454, and loss of all but 93 nts at each end of the Tn in pogo{}718 (Fig. 2A, left).
Figure 2.
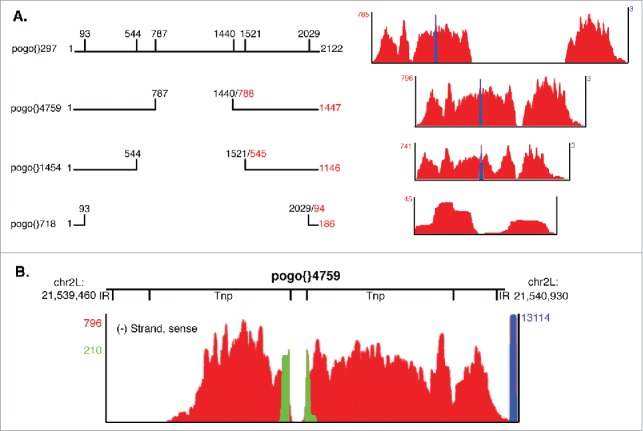
Pogo{}4759 is the only transcribed pogo element in the Drosophila genome. (A) Pogo TIR Tns representing the 4 different size classes of pogo elements are shown. Nucleotide deletion positions are labeled above each schematic. To the right of each Tn, non-unique RNA-seq (red) and small-capped RNA-seq (blue) reads mapping to each pogo TIR Tn are displayed. (B) Non-unique RNA-seq reads (red), unique RNA-seq reads (green) and small-capped RNA-seq (blue) reads mapping to pogo{}4759 are shown with maximum normalized RPM displayed in corresponding colors. Relative locations of specific ORFs are shown above the bedgraphs with the chromosomal location of pogo{}4759.
Visualization of RNA-seq reads corresponding to pogo{}297, pogo{}4759, and pogo{}1454 show no non-unique or unique reads mapping to pogo{}297 nts 788 to 1440 (Fig. 2A, right), nor is there evidence that these sequences have been removed by splicing (data not shown). In contrast, non-unique RNA-seq reads map the entire length of pogo{}4759 and pogo{}1454 (Fig. 2A, right). Additionally, unique reads corresponding to a splice junction are clearly evident for pogo{}4759 and a strong tss (13,114 RPM) is present just upstream of the intergenic pogo{}4759 element (Fig. 2B). These data, together with a lack of observed intra element S pogo tss (Fig. 1A), support a model in which none of the 5 2.1 kb Drosophila pogo elements are transcribed, but that active transcription of pogo{}4759 accounts for all non-unique reads mapping to pogo transposons.
EsiRNAs are generated from 1360 TIR Tns
Canonical 1360 TIR Tns produce very few esiRNAs although AS transcripts are evident that could potentially hybridize with S RNAs (Fig. 1B). Upon further investigation we identified a few 1360 elements with low abundance intra element S and AS tss near the 3′ end of the Tn; the presence of these tss correlates with increased transcription of this 1360 region. An example (1360{}1539) is shown in Figure 3A. Visualization of smRNA-seq reads corresponding to these sequences shows that esiRNAs are generated from these 1360 RNAs, albeit at low frequency (Fig. 3A, bottom).
Figure 3.
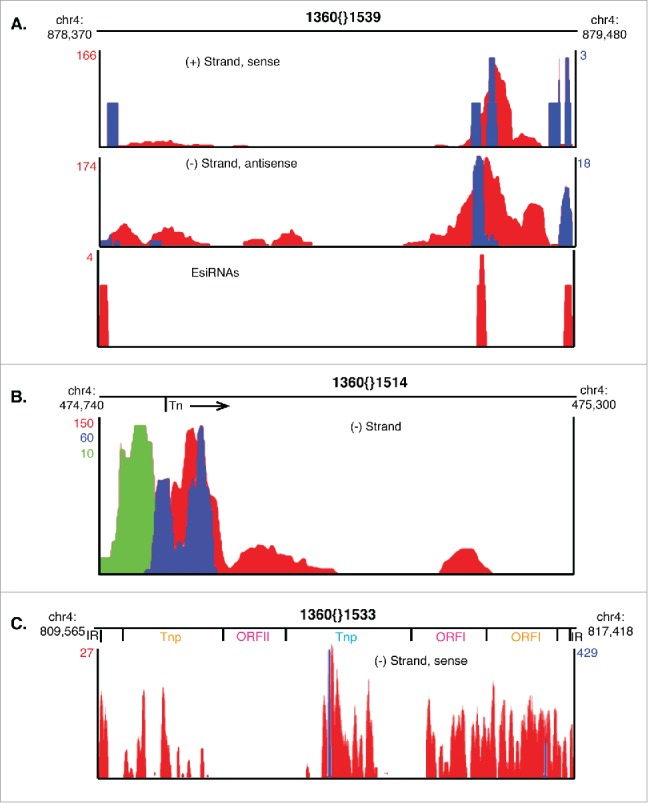
Diverse 1360 TIR Tns produce potential regulatory RNAs. (A-C) Non-unique RNA-seq reads (red), unique RNA-seq reads (green) and small-capped RNA-seq (blue) reads mapping to 1360{}1539 (A), 1360{}1514 (B) or 1360{}1533 (C) are shown with maximum normalized RPM displayed in corresponding colors. Chromosomal locations of each TIR Tn are shown above the bedgraphs. Relative locations of specific ORFs are shown for 1360{}1533.
From these data, we conclude that the number of esiRNAs produced from TIR Tn dsRNA precursors is dramatically less than the expression level of S and AS TIR Tn transcripts. Normalized RNA-seq read counts for S and AS 1360{}1539 are ∼170 while esiRNAs RPMs are 4 (Fig. 3A). Because expression of RNAs from canonical 1360 elements is reduced compared to 1360{}1539 (Fig. 1B), any esiRNAs produced from hybridized S and AS transcripts would be below the limit of detection of our assay.
Transcription from 1360 intra element tss creates fusion RNAs with neighboring sequences
As discussed previously, intra element tss were identified in canonical 1360 elements, but these tss initiate transcription into flanking sequences instead of toward the TIR Tn (Fig. 1B). Further investigation revealed a few individual 1360 elements for which unique RNA-seq reads corresponding to sequences immediately surrounding the Tn could be identified. An example is shown in Fig. 3B (1360{}1514). 1360{}1514 is an intergenic TIR Tn and therefore is not indirectly transcribed as a consequence of being in an intron. 1360{}1514 has 2 tss on the (−) strand that clearly overlap non-unique RNA-seq reads at the 5′ end of the element (Fig. 3B). Further examination reveals unique RNA-seq reads (10 RPM) mapping to the 1360{}1514/flanking sequence indicating that transcription from the observed tss continues beyond the Tn into neighboring sequences (Fig. 3B). To our knowledge, TIR Tn fusion transcripts have not been observed previously.
1360{}1533 may encode a P-element-like Transposase
Previous investigations defined an ancestral Drosophila Tn termed ProtoP from which 1360 elements derive.33 The consensus sequence of this 4480 bp TIR Tn encodes an 864 amino acid Transposase (Tnp) with homology to the modern P-element Tnp.33 Our examinations of the 304 annotated 1360 elements in the Drosophila genome yielded one element that might produce a functional Tnp. 1360{}1533, a 7854 bp 1360 element, is transcribed from the (−) strand and has multiple ORFs in all 3 reading frames (Fig. 3C). The first reading frame encodes an ORF near the 3′ end with homology to the P-element Tnp (582 amino acids) while the third reading frame encodes an ORF with an retroTn RNase H domain and separate homology to retroviral integrases (641 amino acids) (Fig. 3C). Additionally, 2 intra element tss were identified in 1360{}1533 that would allow transcription of the proposed Tnp (Fig. 3C). Unfortunately, all short-capped RNA-seq (defining tss) and RNA-seq reads mapping to 1360{}1533 were non-unique. Therefore, transcription of this specific 1360 element could not be confirmed bioinformatically.
Discussion
RetroTns and TIR Tns are differentially regulated
Recently, we published data supporting a model in which retroTns in Drosophila somatic cells are regulated by esiRNAs.15 RetroTns are convergently transcribed in the sense and antisense direction primarily from intra element transcription start sites (Fig. 4A).15 Many full-length retroTns are present in the Drosophila genome and transcription of individual elements was confirmed for a large percentage of the elements investigated.15 The sense and antisense transcripts produced from retroTns have the potential to hybridize, creating double stranded RNAs that are substrates for esiRNA biogenesis by Dcr2 (Fig. 4A).15 EsiRNAs restrict retroTn movement in Drosophila somatic cells by an unknown mechanism requiring RNAi factors.27 The amount of Dcr2 precursor is determined by the expression level of the least transcribed retroTn strand. As the amount of antisense transcript is usually less, we proposed that the excess sense strand would be translated, providing proteins required for retroTn mobility (Fig. 4A). Therefore, our model indicates that the potential for retroTn amplification is defined by the balance between inhibition by esiRNAs and translation of proteins required for retroTn mobility.
Figure 4.
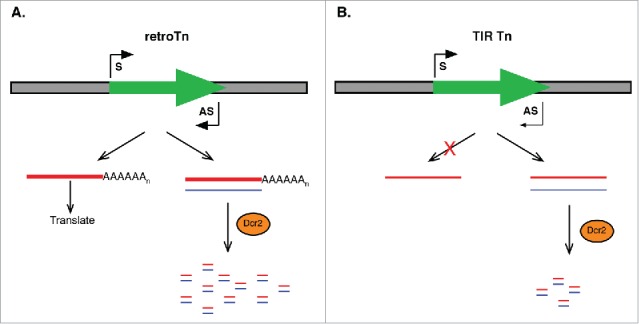
Models depicting Tn regulation in Drosophila somatic cells. (A) RetroTns (green arrow) produce both sense (S, red) and antisense (AS, blue) transcripts by convergent transcription. Hybridization of these RNAs creates a double stranded RNA substrate for biogenesis of endogenous small interfering (esi)RNAs by Dcr2. These esiRNAs repress Tn movement via an unknown mechanism. The retroTn transcript is also translated providing proteins required for Tn movement and balancing Tn repression by esiRNAs. (B) TIR Tns also produce both S and AS transcripts, but the amount of AS transcript is ˜4-fold lower than the lowest expressed retroTn transcript investigated (thin blue line). Additionally, the number of esiRNAs produced from potential TIR Tn dsRNA substrates is dramatically less than for retroTns. While these mechanisms lead to limitations in repressing TIR Tn via the esiRNAs pathway, inhibition is less necessary as S transcription of TIR Tns Tnps is severely restricted.
The analyses described here support a very different mechanism to limit TIR Tn movement in Drosophila somatic cells. Antisense TIR Tn RNAs are produced indirectly from intronic elements oriented antisense to sense mRNAs (Table 1, Fig. 1). Because expression of these transcripts is considerably lower than for retroTns (Figs. 1A-B), the potential for formation of double stranded RNA Dcr2 precursor is greatly reduced (Fig. 4B). Additionally, the number of esiRNAs produced from potential TIR Tn double stranded RNAs is proportionately less than was observed for retroTns (Figs. 3A and 4B).15 Therefore, many less total esiRNAs are generated from TIR Tns than from retroTns. The potential for production of protein(s) required for Tn movement is also dramatically different for TIR Tns. The number of full-length, actively transcribed TIR Tns in the Drosophila genome, is much lower than the number of full-length, actively transcribed retroTns (Table 1).15 Potential Tnp ORFs could only be identified for one pogo TIR Tn (Fig. 2B) and one 1360 element (Fig. 3C). We hypothesize that lower functional TIR Tn Tnp copy number reduces active Tnp concentration.
Collectively, these data support post-transcriptional retroTn regulation, while TIR Tns are inhibited at the transcriptional level. In these models, the potential for retroTn movement is higher because retroTn sense transcript is more highly expressed. To balance this, large numbers of esiRNAs produced from hybridized sense and antisense retroTn RNAs inhibit retroTn mobility. In contrast, because less TIR Tn Tnp transcript is produced, no esiRNAs are required to inhibit TIR Tn movement. Future experiments will be required to test these hypotheses.
TIR Tn 1360 produces fusion transcripts
We observed sequencing reads mapping to junctions between 1360 TIR Tns and flanking sequences indicating that transcription initiates within the 5 prime end of the Tn and continues into neighboring sequences producing hybrid TIR Tn/flanking RNAs. To our knowledge, these fusion Tn/flanking sequence RNAs have not been reported previously in Drosophila. Interestingly, the mammalian LINE-1 retroTn produces a similar fusion transcript by initiating AS transcription near the 5′ end of the element.40-42 LINE-1 elements are often intergenic and oriented AS to their host genes,43 therefore, AS transcription results in LINE-1 RNA/mRNA fusion transcripts known to regulate expression of many genes.40,41,44,45 Further investigation is required to determine if intragenic 1360 TIR Tns could regulate gene expression using a similar mechanism in Drosophila.
Methods
rRNA depletion, library preparation, and next generation sequencing
Performed as described.15
Next generation sequencing analysis
Performed as described.15
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Lon Chubiz and Dr. Ambrose R. Kidd III for critical review of the manuscript.
Funding
This work was supported by NIH grant R15GM107931 to M. S. and UMSL start up funds to M. S.
References
- [1].Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al.. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol 2002; 3:1-20; http://dx.doi.org/ 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lerat E, Rizzon C, Biémont C. Sequence divergence within transposable element families in the Drosophila melanogaster genome. Genome Res 2003; 13:1889-96; PMID:12869581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kofler R, Nolte V, Schlötterer C. Tempo and mode of transposable element activity in drosophila. PLoS Genet 2015; 11:e1005406; PMID:26186437; http://dx.doi.org/ 10.1371/journal.pgen.1005406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kazazian HH. Mobile elements: drivers of genome evolution. Science 2004; 303:1626-32; PMID:15016989; http://dx.doi.org/ 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- [5].Kidwell MG, Kidwell JF, Sved JA. Hybrid dysgenesis in Drosophila melanogaster: A syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 1977; 86:813-833; PMID:17248751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Picard G, Bregliano JC, Bucheton A, Lavige JM, Pélisson A, Kidwell MG. Non-mendelian female sterility and hybrid dysgenesis in Drosophila melanogaster. Genet Res 1978; 32:275-87; PMID:109354; http://dx.doi.org/ 10.1017/S0016672300018772. [DOI] [PubMed] [Google Scholar]
- [7].Rubin GM, Kidwell MG, Bingham PM. The molecular basis of P-M hybrid dysgenesis: The nature of induced mutations. Cell 1982; 29:987-94; PMID:6295640; http://dx.doi.org/ 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- [8].Bingham PM, Kidwell MG, Rubin GM. The molecular basis of P-M hybrid dysgenesis: The role of the P element, a P-strain-specific transposon family. Cell 1982; 29:995-1004; PMID:6295641; http://dx.doi.org/ 10.1016/0092-8674(82)90463-9. [DOI] [PubMed] [Google Scholar]
- [9].Kaufman PD, Rio DC. P element transposition in vitro proceeds by a cut-and-paste mechanism and uses GTP as a cofactor. Cell 1992; 69:27-39; PMID:1313335; http://dx.doi.org/ 10.1016/0092-8674(92)90116-T. [DOI] [PubMed] [Google Scholar]
- [10].Tang M, Cecconi C, Bustamante C, Rio DC. Analysis of P element transposase protein-DNA interactions during the early stages of transposition. J Biol Chem 2007; 282:29002-12; PMID:17644523; http://dx.doi.org/ 10.1074/jbc.M704106200. [DOI] [PubMed] [Google Scholar]
- [11].Rio DC, Laski FA, Rubin GM. Identification and immunochemical analysis of biologically active Drosophila P element transposase. Cell 1986; 44:21-32; PMID:2416475; http://dx.doi.org/ 10.1016/0092-8674(86)90481-2. [DOI] [PubMed] [Google Scholar]
- [12].Laski FA, Rio DC, Rubin GM. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell 1986; 44:7-19; PMID:3000622; http://dx.doi.org/ 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- [13].Spradling AC, Bellen HJ, Hoskins RA. Drosophila P elements preferentially transpose to replication origins. Proc Natl Acad Sci U S A 2011; 108:15948-53; http://dx.doi.org/ 10.1073/pnas.1112960108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 2008; 322:1387-92; PMID:19039138; http://dx.doi.org/ 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Russo J, Harrington AW, Steiniger M. Antisense transcription of retrotransposons in drosophila: An origin of endogenous small interfering RNA precursors. Genetics 2016; 202:107-21; PMID:26534950; http://dx.doi.org/ 10.1534/genetics.115.177196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006; 313:320-4; PMID:16809489; http://dx.doi.org/ 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- [17].Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007; 128:1089-103; PMID:17346786; http://dx.doi.org/ 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- [18].Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 2007; 318:761-4; PMID:17975059; http://dx.doi.org/ 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- [19].Sentmanat MF, Elgin SCR. Ectopic assembly of heterochromatin in Drosophila melanogaster triggered by transposable elements. Proc Natl Acad Sci U S A 2012; 109:14104-9; http://dx.doi.org/ 10.1073/pnas.1207036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Tóth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 2013; 27:390-9; PMID:23392610; http://dx.doi.org/ 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gu T, Elgin SCR. Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in Drosophila. PLoS Genet 2013; 9:e1003780; PMID:24068954; http://dx.doi.org/ 10.1371/journal.pgen.1003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Haynes KA, Caudy AA, Collins L, Elgin SCR. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr Biol 2006; 16:2222-7; PMID:17113386; http://dx.doi.org/ 10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler ELW, Zapp ML, Weng Z, et al.. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 2008; 320:1077-81; PMID:18403677; http://dx.doi.org/ 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chung W-J, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol 2008; 18:795-802; PMID:18501606; http://dx.doi.org/ 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al.. An endogenous small interfering RNA pathway in Drosophila. Nature 2008; 453:798-802; PMID:18463631; http://dx.doi.org/ 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Saito K, Siomi MC. Small RNA-mediated quiescence of transposable elements in animals. Dev Cell 2010; 19:687-97; PMID:21074719; http://dx.doi.org/ 10.1016/j.devcel.2010.10.011. [DOI] [PubMed] [Google Scholar]
- [27].Xie W, Donohue RC, Birchler JA. Quantitatively increased somatic transposition of transposable elements in Drosophila strains compromised for RNAi. PLoS ONE 2013; 8:e72163; PMID:23940807; http://dx.doi.org/ 10.1371/journal.pone.0072163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell 2007; 130:299-308; PMID:17662944; http://dx.doi.org/ 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Marques JT, Kim K, Wu P-H, Alleyne TM, Jafari N, Carthew RW. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat Struct Mol Biol 2010; 17:24-30; PMID:20037596; http://dx.doi.org/ 10.1038/nsmb.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol 1972; 27:353-65; PMID:4625067. [PubMed] [Google Scholar]
- [31].Tudor M, Lobocka M, Goodell M, Pettitt J, O'Hare K. The pogo transposable element family of Drosophila melanogaster. Mol Gen Genet 1992; 232:126-34; PMID:1313144; http://dx.doi.org/ 10.1007/BF00299145. [DOI] [PubMed] [Google Scholar]
- [32].Plasterk RH, Izsvák Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet 1999; 15:326-32; PMID:10431195; http://dx.doi.org/ 10.1016/S0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- [33].Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci USA 2003; 100:6569-74; PMID:12743378; http://dx.doi.org/ 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bartolomé C, Maside X, Charlesworth B. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol Biol Evol 2002; 19:926-37; http://dx.doi.org/ 10.1093/oxfordjournals.molbev.a004150. [DOI] [PubMed] [Google Scholar]
- [35].Celniker SE, Dillon LAL, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al.. Unlocking the secrets of the genome. Nature 2009; 459:927-30; PMID:19536255; http://dx.doi.org/ 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 2002; 12:996-1006; PMID:12045153; http://dx.doi.org/ 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Santos dos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM, FlyBase Consortium. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res 2015; 43:D690-7; PMID:25398896; http://dx.doi.org/ 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nechaev S, Fargo DC, Santos dos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 2010; 327:335-8; PMID:20007866; http://dx.doi.org/ 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell 2013; 52:517-28; PMID:24184211; http://dx.doi.org/ 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 2001; 21:1973-85; PMID:11238933; http://dx.doi.org/ 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nigumann P, Redik K, Mätlik K, Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 2002; 79:628-34; PMID:11991712; http://dx.doi.org/ 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- [42].Li J, Kannan M, Trivett AL, Liao H, Wu X, Akagi K, Symer DE. An antisense promoter in mouse L1 retrotransposon open reading frame-1 initiates expression of diverse fusion transcripts and limits retrotransposition. Nucleic Acids Res 2014; 42:4546-62; PMID:24493738; http://dx.doi.org/ 10.1093/nar/gku091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Molecular archeology of L1 insertions in the human genome. Genome Biol 2002; 3:research0052; PMID:12372140; http://dx.doi.org/ 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mätlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol 2006; 2006:71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cruickshanks HA, Tufarelli C. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics 2009; 94:397-406; PMID:19720139; http://dx.doi.org/ 10.1016/j.ygeno.2009.08.013. [DOI] [PubMed] [Google Scholar]


