ABSTRACT
Aubergine is an RNA-binding protein of the Piwi clade, functioning in germline in the piRNA pathway that silences transposons and repetitive sequences. Several mutations of this gene exist, but they mostly result in truncated proteins or correspond to mutations that also affect neighboring genes. We have generated complete aubergine knock-out mutants that do not disrupt the neighboring genes. These novel mutants are characterized by PCR and sequencing. Their nature is confirmed by female sterility and by the presence of crystals in testes, common to the aubergine loss of function mutations. These mutants provide novel and more appropriate tools for the study of the piRNA pathway that controls genome stability.
KEYWORDS: Aubergine, argonaute, crystal-stellate, knockout mutant, mutagenesis, piRNA, sterility
Introduction
The Aubergine (Aub) protein belongs to the Piwi clade (Piwi, Aubergine, Argonaute 3) of the Argonaute family of proteins. The main role of this protein clade is to prevent transposon movement in germline cells.1 Interacting with the RISC complex too,2 these proteins have been shown to bind Piwi interacting RNAs (piRNAs) and to silence transposable elements,1 in a Dicer independent way,3 as reviewed in Siomi et al.4 piRNAs are 24–30 nucleotide long non-coding RNAs active in germline cells.5,6 They are utilized to ensure genetic stability in animal gonads, as seen in Drosophila,7 rat,8 and zebrafish.9 In this process, the Aub protein also interacts with dFmr1, the Drosophila ortholog of the Fragile X Mental Retardation Protein, which is already known as a translational regulator.10 Besides piRNA maturation, Aub is involved in other RNA-related mechanisms including nanos mRNA localization11 and Poly A tail shortening complex localization,12 epigenetic regulation such as Polycomb group response element clustering,13 and chromosome condensation during mitosis.14 As expected, Aub is highly expressed in ovaries and testes,15 where it is involved in the piRNA pathway occurring in the germline. It is also expressed in the embryo, where it has a role in pole cell formation. Finally, recent studies call for a role of Aub in the nervous system.10 The human ortholog of Piwi clade, HIWI was shown to have a similar role16 and to be expressed in undifferentiated cells.17 These data highlight the importance of the Piwi clade of proteins in the stability of the genome and in development.
Functional analyses tightly rely on the availability of mutations that disrupt gene activity. The BSC213 line serves as an aub mutant;18 however this deficiency comprises several genes including nos, porin, dpr2, SCAR and piwi, in addition to aub. Flies that are homozygous for this deficiency are lethal before the 3rd instar larval stage. Other alleles have also been routinely used: aubQC42, aubHN2,19 aubN11,20 and aubK86,21 all generated by ethyl methanesulfonate (EMS) mutagenesis. aubHN2 and aubK86 carries nonsense mutations and codes for a truncated, supposedly non-functional proteins.20 aubN11 contains a frameshift mutation20 (Fig. 1A). The molecular lesion of aubQC42 is unknown; it is considered a strong hypomorph EMS-induced aub allele,19 confers female sterility, when homozygous.22 At our growing conditions aubQC42 is not completely vital, only few escapers survive; as a consequence we and other groups routinely used the aubHN2/aubQC42 transheterozygotes, in order to analyze aub “loss of function.”3,23-25
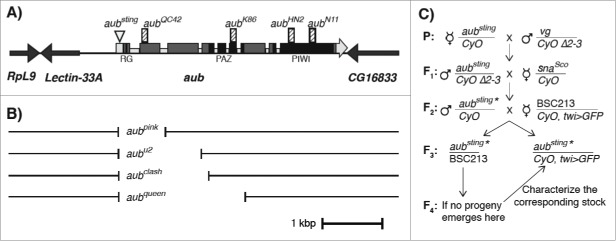
Figure 1.
Genomic organization of the aubergine region. (A) Layout of RpL9, Lectin-33A, aub and CG16833 genes, arrows indicate their orientation. The aubsting insertional mutation and nonsense mutations aubQC42 (Y94, aubK86 (Q377), aubHN2 (Q622) and frameshift mutation aubN11 (G741) are indicated as a triangle and as vertical striped bars on the top of the panel. Light gray boxes represent the 5′, 3′ UTR (big boxes) and the introns (small boxes), dark gray boxes represent the exons; line represents the intergenic regions. Sequences corresponding to RG, PAZ and PIWI domains of aub are indicated by black boxes on the exons. (B) 5′ and 3′ breakpoints of the novel aub mutants. The graph is in-scale. The scale bar is 1 kg base-pair (kbp). aubpink and aubU2 are sequenced using a promoter forward primer (Prom. Fw2 AAATGTGTCCGGAGATTTACAA, see Fig. 2) and the reverse primer in exon 4 (E4 Rv: CCATAATTGCATGCGGAAAT, see Fig. 2). aubclash is sequenced using the forward primer (Prom. Fw GAATTCAACGATGCCTTTTCA see Fig. 2) and the reverse primer in exon 7 (E7 Rv: CAGCTCGATGTTCCAGGACT, see Fig. 2). aubqueen is sequenced using the forward primer Prom. Fw, and the reverse primer in exon 9 (E9 Rv GTGGAGGGGGATCACTACCT, see Fig. 2). (C) Crossing scheme for aub mobilization. Only the 2nd chromosome is represented. Δ2–3 indicates the transposase containing line. vg and snaSco are selectable phenotypic markers. CyO represents the balancer second chromosome and CyO, twi>GFP the fluorescent labeled (twist-Gal4, UAS-GFP) balancer chromosome. BSC213 indicates the deficiency line eliminating aub and many other neighboring genes. aubsting* represents a potential aub mutant upon P element excision.
For the above reasons, we decided to produce novel and null aub alleles by mobilizing the P element transposon present in the P{lacW} aubsting allele, also known as aubsting. P-element mutagenesis is a convenient method to obtain knock-out mutants. Here we report the phenotypic characterization of a novel aub allele that completely eliminates the expression of the Aub protein and leaves the adjacent genes unaffected. This provides a novel and efficient tool to study the role of the Aub protein in genome stability, sterility and neuronal plasticity.
Results
Molecular characterization of the aubQC42 allele
The Aub protein contains typical domains as reviewed by Höck.26 The arginine-glycine rich domain (a.k.a. RG domain, aminoacids 11–17) is necessary to interact with Tudor proteins;27,28 the PAZ domain (Piwi-Argonaut-Zwille) located on the N terminal half (aminoacids 280–413) serves as a docking site for 3′ end of small RNA;29 and the PIWI domain, located on the C-terminal half (amino acids 554–852) of the protein having a function similar to RNase H30 (Fig. 1A).
First, we characterized the molecular lesion contained in the aubQC42 flies. This mutation was isolated in a screen for EMS-induced female sterility.19 FlyBase describes this allele as a point mutation, but no description of the aubQC42 lesion has been so far reported. We amplified the aub cDNA, obtained from RNA extracted from heterozygous animals, with different primer pairs (see Materials and Methods) and sequenced the amplicons. We found a putative point mutation at the 393rd base of cDNA in some fragments, leading to a premature stop at codon 94 (Fig. 1A). In order to confirm the aubQC42 molecular lesion, we cloned and sequenced genomic fragments and the analysis of two independent cDNA clones confirmed the lesion.
P element mediated mutagenesis
The fact that aubQC42, the allele coding for the smallest truncated Aub protein available in the community still codes for the first 93 amino acids that contain the arginine-glycine rich domain prompted us to perform a mutagenesis in order to produce a null allele. aubsting carries a P{lacW} element inserted 58 base upstream to the translational start site of the aub-RA transcript15 (Fig. 1A). This allele behaves as an aub gain of function in somatic cells, due to its ectopic expression in these cells, and as a loss of function in the germline.10 P{lacW} is an engineered31 P element that carries two inverted repeats at the termini, a white+plus; (w+) wild type gene leading to red eye as a phenotypic marker of the insertion32 and no longer contains the gene coding for the transposase. Strains that had lost the w+ marker were selected as potential precise/imprecise excisions (Fig. 1C).
aubsting, the P{lacW} element inserted in the first exon of aub was mobilized to create knockout mutants by imprecise excision in this experiment.
The putative aub mutants were then selected based upon female sterility and subsequently characterized by analytical PCR using primers matching to different regions (Fig. 2). The exact breakpoints were determined by sequencing (Fig. 1B). In total, 103 crosses were set up at F2 with single w males. 44 lines carry internal deletions (where just part of the P element is deleted); 49 lines represent precise excisions (the entire P element is excised out, without disrupting the surrounding sequence), 10 carry imprecise excisions (where the insertion flanking area is disrupted). The interesting lines were isogenized upon crossing with a multiple balancer (Bloomington Stock Center #3703) (Fig. 1C).
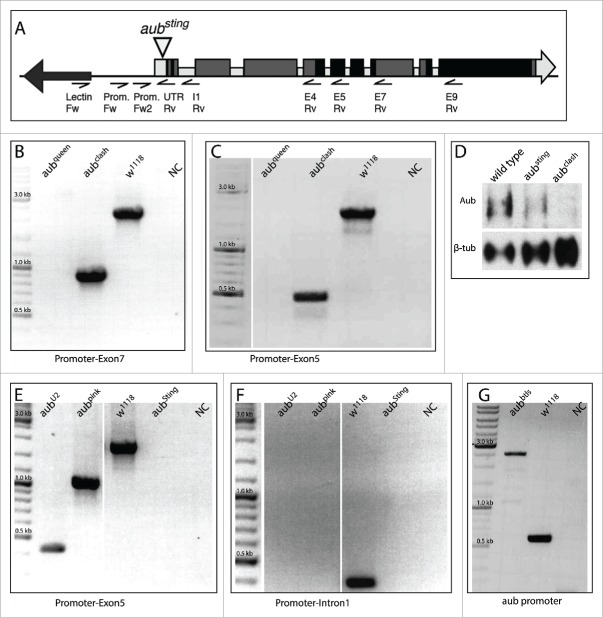
Figure 2.
Analytical PCR for aub mutants. (A) Map of the primers used to characterize the aub mutants. (B, C) Characterization of aubqueen and aubclash. (B) PCR using primers from the aub promoter (Prom. Fw) and from exon 7 (E7 Rv). w1118 is expected to show a band at 2367 bp. aubqueen is not expected to show a PCR product, since exon 7 is mostly deleted; and aubclash is expected to show a band at 915 bp (due to a deletion of 1452 bp). (C) PCR using primers from the aub promoter (Prom. Fw) and from exon 5 (E5 Rv, AATGGCGTCGATTGAAAGTC). w1118 is expected to show a band at 1903 bp. aubqueen is not expected to give a band, since exon 5 is entirely deleted; and aubclash is expected to show a band at 451 bp. (D, E) Characterization of aubU2 and aubpink. (D) Western Blot on protein extracts from adult testes of the indicated genotypes. Tubulin was used as a loading control. (E) PCR using primers from the aub promoter (Prom. Fw2) and from exon 5 (E5 Rv). w1118 is expected to show a band at 1757 bp. aubU2 is expected to show a band at 412 bp (due to a deletion of 1345 bp), aubpink is expected to show a band at 998 bp (due to a deletion of 759 bp). aubSting is not expected to produce a product using these PCR conditions, since the P element is too large (>10 kb). (F) PCR using primers from aub promoter (Prom. Fw2) and intron 1 (I1 Rv, CAAGGCCAAGCTAATTTTGGA). w1118 is expected to show a band at 333 bp. aubU2 and aubpink are not expected to display a PCR product since intron 1 is entirely deleted in both. aubSting is not expected to display a product in these PCR conditions, due to the large size of the P element. (G) Characterization of aubbtls. PCR using primers from the Lectin-33A promoter (Lectin Fw, TAAACGCTCGGCAGAGAACT) and from aub 5′ UTR (UTR Rv, GTTAGACGCCCAGGGATGT). w1118 is expected to display a band at 575 bp. aubbtls displays a 2.5 kb band due to the presence of P element sequences. For all panels, w1118 is used as wild type control; NC stands for negative control, that is, a PCR reaction with no DNA. The DNA ladder used is Thermo-Fisher GeneRuler™ High Range SM0331.
The 10 imprecise excisions represent independent events and carry different mutations. The largest deletion, named aubqueen, removes 2144 bp (2047 bp after the AUG codon). The border sequences are 5′–ATAACTCACATCCCTGGGCG–3′ on the 5′ UTR and 5′–GGACTCCGCCTTGGTGGAGA–3′ in exon 7. The second largest deletion, aubclash removes 1452 bp (1355 bp after the AUG). The border sequences are 5′–ATAACTCACATCCCTGGGCG–3′ on the 5′ UTR and 5′–CACAAGGTTATGCGAACTGA–3′ in exon 4. The aubU2 allele lacks 1345 bp total (1248 bp after the AUG), from 5′ UTR until mid-intron 3 whereas aubpink lacks 759 bp total (662 bp after the AUG), from 5′ UTR until mid-intron 2. Unlike the 3 other female sterile alleles, aubpink is only partially sterile. aubbtls, an internal deletion, contains 2 kb of the P element present in the promoter region of aub. Although this mutant could not be sequenced, it might serve as a proper aub mutant as female sterility suggests.
We decided to further characterize the aubclash allele. The deletion spans from 11001465 to position 11000010 of the AE014134.6 genomic clone, which carries the aubergine gene. The 1452 bp deletion eliminates the AUG of the Aub protein and no possible protein can be produced from the rearranged region, which we also verified by Western Blot on protein extract from mutant testes (Fig. 2D). Thus, aubclash can be considered a null aubergine allele.
Phenotypic characterization of the aubclash null allele
In order to analyze the new aub allele we assessed several phenotypes that are typical of the aub mutation: 1) sterility,19 2) the presence of crystals made of the Stellate protein in the mutant testes, 3) the presence of morphological abnormalities and variation in the adult, 4) the potential to rescue the crystal phenotype observed in testes that lack the dFmr1 protein.10,15,19
We compared the sterility of aubclash males and females in comparison with that of aubHN2/aubQC42 transheterozygous, aubsting homozygous and control animals. The graph in Figure 3A,B shows that aubclash mutants females are completely sterile as are the aubHN2/aubQC42 transheterozygous females. aubclash mutant males are partially fertile (16% compared to wild type) and this phenotype is more severe than that of aubHN2/aubQC42 transheterozygous animals, whose fertility is 84% of that of wild type animals.
Figure 3.
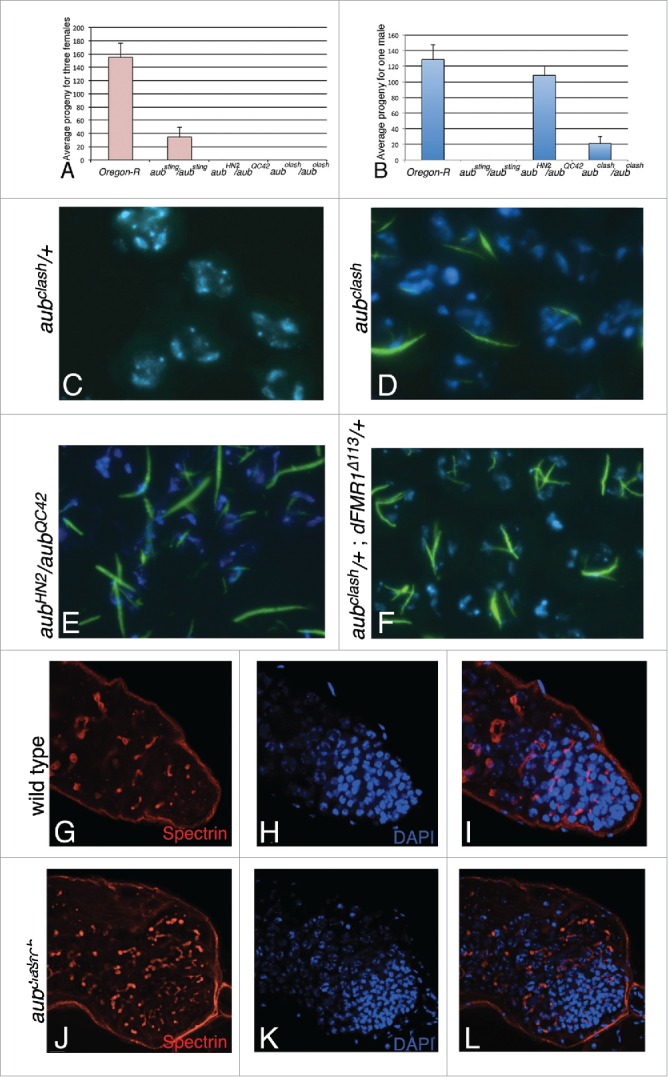
Phenotypic characterization of the aubclash allele (A) Female fertility in the mentioned genotypes. (B) Male fertility in the mentioned genotypes. Results represent mean ± s.d., n = 3. (C-F) Crystal phenotype of adult testes analyzed by immunolabeling: anti-Stellate labeling is in green, DAPI in blue in (C) aubclash /+. (D) aubclash/aubclash. (E) aubHN2/aubQC42. (F) aubclash/+; dFmr1δ113 /+. (G-I) Confocal projections (3 sections) from a wild type testis labeled with anti-α-Spectrin antibody (in red, G) DAPI (in blue, H) and merge (I). (J-L) Confocal projections (3 sections) from an aubclash testis labeled with anti-α-Spectrin antibody, which recognizes the fusome, a germline-specific organelle (in red, J), DAPI (in blue, K) and merge (L).
As the other aub mutants, aubclash testes display Stellate-made crystalline aggregates in their spermatocytes10,15 (Fig. 3C–E) and are enlarged10 (Fig. 3J–L).
It has been previously shown that the reduced levels of the Hsp83 and SpindleE proteins, 2 other components of the piRNA pathway, generate phenotypic variation by transposon-mediated mutagenesis (at 0.9% and 12% frequency, respectively, see33). This includes the lack of bristles on the notum (Sco-like phenotype), a dark notum, notched or abnormally everted wings. We hence tested the possibility that the reduction of Aub levels has a similar effect and analyzed aubclash homozygous as well as aubHN2/aubQC42 transheterozygous adults. Morphological phenotypes are indeed present in a significant fraction of the mutant animals: aubclash homozygotes exhibit a 7.6% frequency (91/1197) of phenotypic variants, the aubHN2/aubQC42 transheterozygotes show a 1.2% frequency (10/800); both frequencies are higher than the one exhibited by the aubclash heterozygotes 0.08% (3/3510).
Finally, the presence of one aubsting allele rescues the “crystal” phenotype of animals mutant for dFmr1, which has been recently defined as a component of the piRNA pathway. This is due to overexpression of the Aub protein in the somatic compartment of aubsting testes. aubHN2 or aubQC42 alleles, which are considered as loss of function mutations, do not rescue the dFmr1-mediated phenotype.10 We set up the appropriate crosses and found that aubclash/+; dFmr1Δ113/+ individuals behave like the aubHN2 or aubQC42 alleles, that is there, is no rescue of the dFmr1-mediated “crystal” phenotype, as expected from a loss of function aub allele10 (Fig. 3F).
Discussion
Aub belongs to the Argonaute family of proteins, which are necessary for keeping germline genomic integrity. In addition, recent data also suggest a role for Aub in tumorigenesis,34-37 stem cell identity renewal 17 and in the nervous system.38 Characterizing the role and the genetic interactions of aub in the different tissues is essential. Knock-out mutants and targeted overexpression are indispensible for such characterization. Some alleles generated by EMS mutagenesis (aubHN2, aubQC42 etc.) have been used as loss of function, however, they contain premature stop codons so that truncated proteins or alternative transcripts from a downstream translation start site might be expressed. The fact that these alleles may not be nulls is in line with the finding that homozygous aubclash males are much less fertile than the transheterozygous aubHN2/aubQC42 males. Other alleles (e.g. aubsting-3a) represent large deletions that completely eliminate Aub expression, but adjacent genes are disrupted as well. RNA interference has been used to affect the activity of Aub, however, this condition represents a knockdown and in addition we cannot exclude off target effects, due to the high sequence similarity among the members of the Argonaute family of proteins.
We have generated clean mutants of aub, where the neighboring genes are not affected and we have characterized at least one null mutation, aubclash by molecular and phenotypic means. This allele will provide the scientific community with an efficient and specific tool to study the role of Aub in RNA biology.
Materials and methods
Drosophila strains
The aub gene is on the 2nd chromosome. The aubsting allele has been described in.15 w1118; aubQC42 cn1 bw1/CyO, P{sevRas1.V12}FK1 is ordered from Bloomington Stock Center, number 4968. The transposase carrying line is a gift from P. Heitzler (Strasbourg), with the genotype w1118 ; cn, vgu, bw, sp/CyO, H[w+, Δ2–3] Ho 2.1 on the 2nd chromosome. The fluorescent balancer abbreviated as CyO, twi>GFP is CyO, twist-Gal4, UAS-GFP on the 2nd chromosome. The multiple balancer line, with Bloomington Stock Center number 3703, is w1118/Dp(1;Y)y+ ; CyO/nub1 b1 snaSco lt1 stw3; MKRS/TM6B, Tb1, bearing mutations on the 1st, 2nd and 3rd chromosomes. dFmr1Δ113 is a null allele.10
Mutagenesis
aubsting flies can be recognized by orange eyes, while all the balancers and deficiency chromosomes are in a white eye background. The mobilization started by crossing aubsting/CyO virgins (orange eyes) with vg/CyO Δ2–3 males that carry the transposase. Bringing the P element and transposase enzyme together allows P element mobilization in aubsting/CyO Δ2-3 flies of the F1. aubsting/CyO Δ2–3 males were mated with virgin females carrying the balancer. In the F2, which likely contains mutants, the snaSco negative and CyO flies with white eyes were collected, since the white eye indicates that the P element had been excised. Individual aubsting* (aub mutant candidate) males were crossed with 6 virgins of the following genotype BSC213/CyO, twi>GFP (aub deficiency over the fluorescent balancer line); 103 such crosses were set. In the F3, an aubsting*/CyO, twi>GFP line was established as a stable stock from each cross, which also allowed the identification of homozygous mutant candidates. From the same vials, CyO negative flies were also kept to establish an aubsting*/BSC213 line to test for sterility at the same time (Fig. 1C). The sterile strains were selected as primary candidates for imprecise excision. The matching stocks were characterized by CyO negative selection from their corresponding aubsting*/CyO, twi>GFP adults. Deletion size and location were calculated on the bases of aub transcript-RA (FBtr0080165, release r6.09).
DNA analyses
The DNA isolation buffer contains 50 mM Tris-HCl pH 8.0, 100 mM EDTA pH 8.0, 100 mM NaCl, 1% SDS. DNA was extracted from 5 anesthetized adults from each genotype, in presence of 1 mg/ml proteinase K. The solution was centrifuged at 15000 g for 10 min; the supernatant was transferred to clean 1.5 ml tubes. NaCl to final concentration 0.24 M and half volume isopropanol was added. The DNA was pelleted with centrifuge at 13000 g, 4 C°, for 10 minutes. The pellets were washed with 70% ethanol and air-dried. DNA was resuspended in 100 µl ddH2O.
Total RNA extraction, cDNA production and cloning
Total RNA was extracted from 30 mg of male or female gonadal tissues using the RNAqueos-4 PCR Kit (AMBION) reagent, following the manufacturer's protocol as described previously.39 Samples were incubated with DNase I RNase free (AMBION) (2U DNase up to 2 μg RNA) at 37°C for 30 min (100 μl). DNase-treated RNA was precipitated at −80°C overnight and after centrifugation (10,000 g for 15 min) it was dissolved in 50 μl of nuclease-free water. The RNA concentration and purity were determined photometrically. 5 μg of total RNA were used as a template for oligonucleotide dT-primed reverse transcription using SuperScriptIII RNaseH-reverse transcriptase (Invitrogen), according to manufacturer's instructions.
For the cDNA preparation the M-MLV Reverse Transcriptase kit (Invitrogen) was used following the manufacturer's protocol.
PCR amplification was conducted using specific pair of primers and the Platinum Taq Polymerase (Invitrogen) for fragment up to 1000 bp. To amplify fragment longer than 1000 bp, the Expand Long Template PCR System kit (Roche) was used. All the primers used for the amplification are reported below. The product of single PCR was then purified using QIAquick PCR Purification Kit (Qiagen) following the manufacturer's protocol. It was directly used for sequencing or used for the cloning in the TA-vector (Strata Clone PCR Cloning Kit (Stratagene) and the StrataCloneTM SoloPack®CompetentCells, following the manufacturer's protocol. We used this vector for the cloning of the cDNA amplified fragments and for the genomic fragments as well. Primers used in the PCR reactions:
1F5′sting upper 5′ CTGAACGGCATTTGTGACGA 3′
1Fsting upper 5′ CGTGGTCGAGGAAGAAAGCC 3′
2Usting upper 5′ GAGGCAATGGTGGTGGTGGT 3′
3Usting upper 5′ CGTGCTGGCGAAAACATTGA 3′
6Usting upper 5′ CGGGAATGACGGACGCTATG 3′
8Usting upper 5′ CGCAACGGCACTTACTCCCA 3′
3Lsting lower 5′ GGTTCCGTCAAAGATGTAGC 3′
5Lsting lower 5′ ATGCGATAGGTTTTGTTATT 3′
9Lsting lower 5′ TGCTGTCGAGGCGCGATAAC 3′
9L-2sting lower 5′ TGCGATGCCCAGTAAAGTAG 3′
Immunofluorescence of Stellate-made crystals
Testes were dissected in Ringer's modified solution (182 mM KCl, 46 mM NaCl, 3 mM CaCl2, 10 mM Tris-HCl pH 7.5), fixed in methanol, washed in PBST (1x PBS, 1% Triton X-100, 0.5% acetic acid) for 15 min, washed in 1X PBS for 5 min 3 times and incubated with the polyclonal mouse anti-Stellate antibody (1:100).40 Samples were washed in 1X PBS for 5 min 3 times, incubated 2 h with 1:100 FITC-conjugated anti-mouse-IgG antibody (Jackson) and examined by epifluorescence microscopy (Nikon-Optiphot 2); DAPI was used at 100 ng/ml for nuclear labeling.
Antibody production and Western blot analyses
The anti-Aub antibody was made in rabbit against the C-terminal peptide (847–866) also used by Brennecke.1 The antibody was affinity purified according to the sulfolink coupling gel protocol (Pierce #20401) using the peptide employed to immunize the animals. 10 ml of centrifuged sera were washed with 20 ml of PBS and eluted with 5 ml of 0.1M Glycine pH2.8. 0.5 ml fractions were collected and neutralized with 25 μl of Tris pH 9.5 1M. They were then were quantified by Bradford assay.
Adult testes from 10 flies (control and mutant) were dissected in cold PBS buffer and lysed in 30 μl Laemli Sample Buffer 2X (60 mM Tris-Cl pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue), with the addition of the protease inhibitor (Roche), using small pestle, on ice, and then the samples were boiled (8 min, 80°C). Proteins were transferred from 8% SDS-polyacrylamide gels to the membrane using constant amperage (200mA), for 1 h, in transfer buffer and the membrane was subsequently rinsed in TBS1X/0.1% Tween 20 buffer (TBST). To avoid non-specific binding, the membrane was placed in a 5 % milk solution (in TBST) and incubated for 1 h at room temperature (RT) with slow shaking. The filter was then incubated with the affinity purified rabbit anti-Aubergine diluted (1:500) and mouse monoclonal anti−β−tubulin (Millipore) diluted (1:4.000) in TBST at 4°C overnight. Upon incubation, the membrane was rinsed 3 x with TBST and washed 3 x for 10 min at RT with shaking. Secondary antibody conjugated to horseradish peroxidase (HRP) 1:5000 (Jackson Immunoresearch), was added and incubated for 1h at RT. Colorimetric analysis was performed with the detection system for the HRP, as described by the company.
Male and female fertility testing
One young male was mated to 3 control virgin females, and 3 virgin females were mated with 3 control males. 10 individual males and 30 females were tested for each genotype. After 4 days the crosses were transferred to a fresh vial. The parental flies were removed from the last vial after an additional 4 days. The number of the adult progeny from each vial was counted.
Immunofluorescence of the gonads and confocal microscopy
Drosophila gonads were dissected in Ringer's solution and fixed in 4% paraformaldehyde for 20 min, washed in PBT (1X PBS with 0.5% Triton X-100), blocked in 5% NGS for 1 h and incubated with rabbit mouse anti-α-Spectrin (DSHB) antibody at 4°C overnight. Samples were washed in PBST and then incubated for 2 h with the secondary antibodies against mouse IgG, conjugated to Cy3 dyes (1:500, Jackson). All the samples were examined and captured using a laser-scanning confocal microscope (Zeiss LSM 700 on Axio imager M2).
Abbreviations and Acronyms
- aub
aubergine
- AGO
Argonaute
- bp
base-pair
- FMRP
Fragile X Mental Retardation Protein
- GFP
Green Fluorescent Protein
- piRNA
Piwi interacting RNA
- RISC
RNA-induced silencing complex
- UTR
untranslated region
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Bloomington center as well as U Schaefer and P Heitzler for fly strains. We also thank the IGBMC facilities for sequencing and fly food.
Funding
The work in the Giangrande laboratory was funded by the Institut National de la Santé et de la Recherche Médicale; Center National de la Recherche Scientifique, Université de Strasbourg; Hôpital de Strasbourg; Association pour la Recherche sur le Cancer; Institut National du Cancer; Agence Nationale de la Recherche (ANR); Région Alsace; and the FRAXA foundation. It was also supported by a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir labeled ANR-10-IDEX-0002-02 [grant number ANR-10-LABX-0030-INRT]. H.B.S. was supported by the AFM, the FRM and the Région Alsace. V.S. acknowledges the grant from the MIUR for the project ‘Futuro in ricerca 2010’ [grant number RBFR10V8K6], as well as the EMBO for the short-term fellowship in 2009 to visit the lab of A.G. SDT. was supported by the MIUR [grant number PONa3_00334]. The financial support of Telethon ‐ Italy (Grant no. GG14181) is gratefully acknowledge.
References
- [1].Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007; 128:1089-103; PMID:17346786; http://dx.doi.org/ 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- [2].Dufourt J, Dennis C, Boivin A, Gueguen N, Théron E, Goriaux C, Pouchin P, Ronsseray S, Brasset E, Vaury C. Spatio-temporal requirements for transposable element piRNA-mediated silencing during Drosophila oogenesis. Nucleic Acids Res 2014; 42:2512-24; PMID:24288375; http://dx.doi.org/ 10.1093/nar/gkt1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006; 313:320-4; PMID:16809489; http://dx.doi.org/ 10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- [4].Siomi MC, Miyoshi T, Siomi H. piRNA-mediated silencing in Drosophila germlines. Semin Cell Dev Biol 2010; 21:754-9; PMID:20080197; http://dx.doi.org/ 10.1016/j.semcdb.2010.01.011 [DOI] [PubMed] [Google Scholar]
- [5].Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev 2010; 24:636-46; PMID:20360382; http://dx.doi.org/ 10.1101/gad.1899210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol: CB 2001; 11:1017-27; PMID:11470406; http://dx.doi.org/ 10.1016/S0960-9822(01)00299-8 [DOI] [PubMed] [Google Scholar]
- [7].Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 2007; 315:1587-90; PMID:17322028; http://dx.doi.org/ 10.1126/science.1140494 [DOI] [PubMed] [Google Scholar]
- [8].Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 2009; 23:1749-62; PMID:19584108; http://dx.doi.org/ 10.1101/gad.1814809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al.. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 2007; 129:69-82; PMID:17418787; http://dx.doi.org/ 10.1016/j.cell.2007.03.026 [DOI] [PubMed] [Google Scholar]
- [10].Bozzetti MP, Specchia V, Cattenoz PB, Laneve P, Geusa A, Sahin HB, Di Tommaso S, Friscini A, Massari S, Diebold C, et al.. The Drosophila fragile X mental retardation protein participates in the piRNA pathway. J Cell Sci 2015; 128:2070-84; PMID:25908854; http://dx.doi.org/ 10.1242/jcs.161810 [DOI] [PubMed] [Google Scholar]
- [11].Becalska AN, Kim YR, Belletier NG, Lerit DA, Sinsimer KS, Gavis ER. Aubergine is a component of a nanos mRNA localization complex. Dev Biol 2011; 349:46-52; PMID:20937269; http://dx.doi.org/ 10.1016/j.ydbio.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010; 467:1128-32; PMID:20953170; http://dx.doi.org/ 10.1038/nature09465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell 2006; 124:957-71; PMID:16530043; http://dx.doi.org/ 10.1016/j.cell.2006.01.036 [DOI] [PubMed] [Google Scholar]
- [14].Pek JW, Kai T. A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr Biol: CB 2011; 21:39-44; PMID:21185189; http://dx.doi.org/ 10.1016/j.cub.2010.11.051 [DOI] [PubMed] [Google Scholar]
- [15].Schmidt A, Palumbo G, Bozzetti MP, Tritto P, Pimpinelli S, Schäfer U. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics 1999; 151:749-60; PMID:9927466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Keam SP, Young PE, McCorkindale AL, Dang TH, Clancy JL, Humphreys DT, Preiss T, Hutvagner G, Martin DI, Cropley JE, et al.. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res 2014; 42:8984-95; PMID:25038252; http://dx.doi.org/ 10.1093/nar/gku620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP, Hoffman R. Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood 2001; 97:426-34; PMID:11154219; http://dx.doi.org/ 10.1182/blood.V97.2.426 [DOI] [PubMed] [Google Scholar]
- [18].Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, Kaufman TC, Cook KR. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol 2012; 13:R21; PMID:22445104; http://dx.doi.org/ 10.1186/gb-2012-13-3-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 1991; 129:1119-36; PMID:1783295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 2001; 128:2823-32; PMID:11526087 [DOI] [PubMed] [Google Scholar]
- [21].Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development 1996; 122:1631-9; PMID:8625849 [DOI] [PubMed] [Google Scholar]
- [22].Reiss D, Josse T, Anxolabehere D, Ronsseray S. aubergine mutations in Drosophila melanogaster impair P cytotype determination by telomeric P elements inserted in heterochromatin. Mol Genet Genomics: MGG 2004; 272:336-43; PMID:15372228; http://dx.doi.org/ 10.1007/s00438-004-1061-1 [DOI] [PubMed] [Google Scholar]
- [23].Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 2004; 116:817-29; PMID:15035984; http://dx.doi.org/ 10.1016/S0092-8674(04)00250-8 [DOI] [PubMed] [Google Scholar]
- [24].Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009; 137:522-35; PMID:19395010; http://dx.doi.org/ 10.1016/j.cell.2009.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sato K, Iwasaki YW, Shibuya A, Carninci P, Tsuchizawa Y, Ishizu H, Siomi MC, Siomi H. Krimper enforces an antisense bias on piRNA pools by binding AGO3 in the Drosophila germline. Mol Cell 2015; 59:553-63; PMID:26212455; http://dx.doi.org/ 10.1016/j.molcel.2015.06.024 [DOI] [PubMed] [Google Scholar]
- [26].Hock J, Meister G. The Argonaute protein family. Genome Biol 2008; 9:210; PMID:18304383; http://dx.doi.org/ 10.1186/gb-2008-9-2-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, et al.. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 2009; 28:3820-31; PMID:19959991; http://dx.doi.org/ 10.1038/emboj.2009.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 2009; 11:652-8; PMID:19377467; http://dx.doi.org/ 10.1038/ncb1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 2004; 429:318-22; PMID:15152257; http://dx.doi.org/ 10.1038/nature02519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004; 305:1434-7; PMID:15284453; http://dx.doi.org/ 10.1126/science.1102514 [DOI] [PubMed] [Google Scholar]
- [31].Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, et al.. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev 1989; 3:1273-87; PMID:2558049; http://dx.doi.org/ 10.1101/gad.3.9.1273 [DOI] [PubMed] [Google Scholar]
- [32].Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, et al.. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 2004; 167:761-81; PMID:15238527; http://dx.doi.org/ 10.1534/genetics.104.026427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Specchia V, Bozzetti MP. Different aubergine alleles confirm the specificity of different RNAi pathways in Drosophila melanogaster. Fly 2009; 3:170-2; PMID:19242123; http://dx.doi.org/ 10.4161/fly.8054 [DOI] [PubMed] [Google Scholar]
- [34].Li S, Meng L, Zhu C, Wu L, Bai X, Wei J, Lu Y, Zhou J, Ma D. The universal overexpression of a cancer testis antigen hiwi is associated with cancer angiogenesis. Oncol Rep 2010; 23:1063-8; PMID:20204292 [PubMed] [Google Scholar]
- [35].Liang D, Dong M, Hu LJ, Fang ZH, Xu X, Shi EH, Yang YJ. Hiwi knockdown inhibits the growth of lung cancer in nude mice. Asian Pacific J Cancer Prevent: APJCP 2013; 14:1067-72; PMID:23621188; http://dx.doi.org/ 10.7314/APJCP.2013.14.2.1067 [DOI] [PubMed] [Google Scholar]
- [36].Litwin M, Dubis J, Arczyńska K, Piotrowska A, Frydlewicz A, Karczewski M, Dzięgiel P, Witkiewicz W. Correlation of HIWI and HILI expression with cancer stem cell markers in colorectal cancer. Anticancer Res 2015; 35:3317-24; PMID:26026091 [PubMed] [Google Scholar]
- [37].Taubert H, Greither T, Kaushal D, Würl P, Bache M, Bartel F, Kehlen A, Lautenschläger C, Harris L, Kraemer K, et al.. Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene 2007; 26:1098-100; PMID:16953229; http://dx.doi.org/ 10.1038/sj.onc.1209880 [DOI] [PubMed] [Google Scholar]
- [38].Perrat PN, DasGupta S, Wang J, Theurkauf W, Weng Z, Rosbash M, Waddell S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science 2013; 340:91-5; PMID:23559253; http://dx.doi.org/ 10.1126/science.1231965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Specchia V, Piacentini L, Tritto P, Fanti L, D'Alessandro R, Palumbo G, Pimpinelli S, Bozzetti MP. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 2010; 463:662-5; PMID:20062045; http://dx.doi.org/ 10.1038/nature08739 [DOI] [PubMed] [Google Scholar]
- [40].Bozzetti MP, Massari S, Finelli P, Meggio F, Pinna LA, Boldyreff B, Issinger OG, Palumbo G, Ciriaco C, Bonaccorsi S, et al.. The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the beta subunit of casein kinase 2. Proc Natl Acad Sci U S A 1995; 92:6067-71; PMID:7597082; http://dx.doi.org/ 10.1073/pnas.92.13.6067 [DOI] [PMC free article] [PubMed] [Google Scholar]