ABSTRACT
Kidney stone disease is a major health burden with a complex and poorly understood pathophysiology. Drosophila Malpighian tubules have been shown to resemble human renal tubules in their physiological function. Herein, we have used Drosophila as a model to study the proteomic response to crystal formation induced by dietary manipulation in Malpighian tubules. Wild-type male flies were reared in parallel groups on standard medium supplemented with lithogenic agents: control, Sodium Oxalate (NaOx) and Ethylene Glycol (EG). Malpighian tubules were dissected after 2 weeks to visualize crystals with polarized light microscopy. The parallel group was dissected for protein extraction. A new method of Gel Assisted Sample Preparation (GASP) was used for protein extraction. Differentially abundant proteins (p<0.05) were identified by label-free quantitative proteomic analysis in flies fed with NaOx and EG diet compared with control. Their molecular functions were further screened for transmembrane ion transporter, calcium or zinc ion binder. Among these, 11 candidate proteins were shortlisted in NaOx diet and 16 proteins in EG diet. We concluded that GASP is a proteomic sample preparation method that can be applied to individual Drosophila Malpighian tubules. Our results may further increase the understanding of the pathophysiology of human kidney stone disease.
KEYWORDS: Drosophila, gel assisted sample preparation, mass spectrometry, Malpighian tubules, proteome, stone
Introduction
Over the last century, with the increase in prosperity and availability of nutritious food, the incidence of renal stones has progressively increased worldwide.1 The lifetime prevalence of nephrolithiasis is between 5% and 12% in developed countries such as United States and most European states.2 In developing countries, the incidence of stone disease is also increasing in particular as a consequence of hot climate in some geographical regions.3 The incidence of stone disease leads to increased financial burden in the diagnosis and treatment. Despite the advances in minimally invasive surgery in recent decades, little is known as to the fundamental molecular pathophysiology of kidney stone disease. The quest for effective medical treatment and prevention is ongoing.
Previous pilot studies have suggested that Drosophila could be used as a cost-effective model for kidney stone disease.4 The advantages of Drosophila over other animal models include similar anatomical structure and philological function of Malpighian tubule to human renal tubules, brief life cycle and the ease and low cost of rearing.5-8 Drosophila has also been reported to reliably develop calcium oxalate (CaOx) crystals with dietary supplements of stone-forming agents.4 The simplicity and transparent nature of Malpighian tubules allows for direct observation and quantification of crystals, which is not possible in other animal models.
To better understand the underlying biological process of human kidney stone disease, it would be helpful to know how drosophila proteomic profiles are regulated secondary to crystal formation. The objective of this study is to assess the proteomic changes in response to crystal formation in Drosophila Malpighian tubules, using gel-aided sample preparation (GASP)9 combined with nano-liquid chromatography tandem mass spectrometry (nLC-MS/MS) analysis.
Results
Inducing crystallization in Malpighian tubules
Crystal formation in the Malpighian tubule of male Canton S flies was observed as early as one week of ingestion of lithogenic diets, including either sodium oxalate (NaOx) or ethylene glycol (EG). The control group had a basal incidence of crystal formation around 20%. The overall rate of crystal formation was significantly higher in flies fed with NaOx and EG diets, 73% and 84% respectively (Table 1).
Table 1.
Incidence of stone formation in Drosophila Malpighian tubules in 3 experimental conditions.
| Drosophila diet | Overall incidence of crystal formation (%) |
|---|---|
| control | 20 ± 2.2 |
| NaOx (0.05% sodium oxalate) | 73 ± 3.6 |
| EG (0.5% ethylene glycol) | 84 ± 2.2 |
Note. Values were expressed as means ± standard error of mean.
The birefringent crystals were most often observed in the distal segment of the Malpighian tubules. No crystals were observed in the hindgut. Figure 1 shows the different morphology of crystals in 3 experimental conditions. The crystal composition has been shown to be predominantly CaOx monohydrate or CaOx dihydrate by energy-disperse X-ray spectroscopy and scanning electron microscopy in previous studies.4,10
Figure 1.
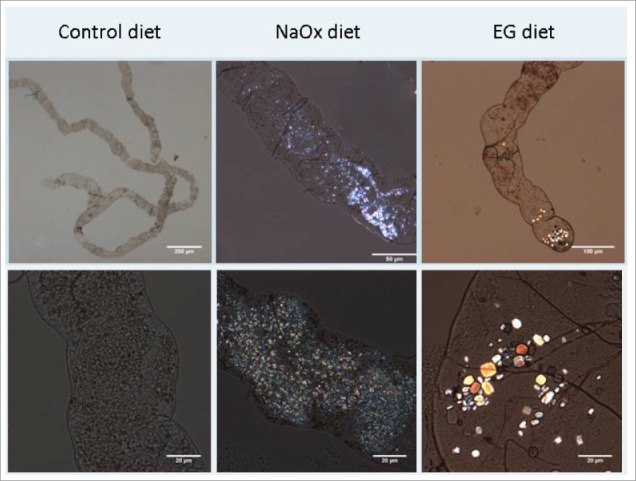
Crystal formation in Malpighian tubules 10-40x magnification. The control diet showed the normal structure of a pair of Malpighian tubules joining into a single ureter, which further secretes into hindgut. The crystal morphology was small and extensive in the NaOx (sodium oxalate) treatment group. In contrast EG (ethylene glycol) enriched diet induced crystals which were bigger, poly-angular with a jewel-like gloss.
Qualitative analysis of detected proteins in 3 biological replicates in Mascot analysis
Initially, the MASCOT (v2.5.1, Matrix Science) search engine was used to interrogate the Malpighian tubule proteome in order to generate a comprehensive list of all detected proteins in 3 experimental conditions. Only proteins common in the 3 biological replicates were further analyzed in sub-categories. We then used LC-Progenesis IQ software as an individual analysis to identify differentially abundant (p<0.05) proteins in control as compared to treated flies and to quantify their fold changes. These proteins identified from MASCOT and Progenesis were screened against Flybase for their molecular functions. We further narrowed down the functionally significant proteins by choosing those functioning as transmembrane ion transporters, calcium or zinc ion binders, which might be relevant in crystal formation.
In the initial experiment using gel-aided sample preparation (GASP), we identified a total of 748 proteins from the Malpighian tubules and hindgut of one fly compared to 859 proteins from 5 flies (Supplementary Fig. 1) from a nLC-MS/MS one hour gradient. In addition, we used a pool of 5 flies and nLC-MS/MS analysis with an optimized two hour gradient, in which more proteins (1217 proteins present in all biological replicates of the control group, 1142 in sodium oxalate group and 983 in ethylene glycol group) were detected in all samples (MASCOT) (Fig. 2A-C). The biological triplicates showed close clustering as shown in the Principal Component Analysis (Supplementary Fig. 2). We then compared the common proteins found in the biological triplicates of the 3 dietary conditions (Fig. 2D).
Figure 2A–D.
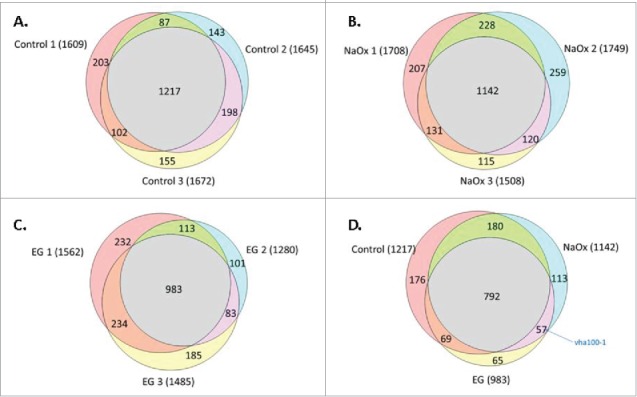
Number of Malpighian tubule proteins identified by mass spectrometry using a two hour LC gradient and MASCOT analysis. We identified 1217 proteins present in all biological replicates in the control group (A), 1142 in NaOx (sodium oxalate) group (B) and 983 in EG (ethylene glycol) group (C), respectively. Venn diagram comparing the proteins identified by MASCOT analysis in 3 experimental conditions (D). Comparing 1142 proteins in NaOx group, 984 in EG group and 1217 in control groups, the majority of proteins identified (792) were common to the 3 dietary conditions. Among the 57 proteins common to both experimental conditions (NaOx and EG), Vha100-1 was one interesting candidate proteins probably related to CaOx crystal formation.
All proteins identified by Mascot analysis were searched in Flybase database for their molecular functions. Candidate proteins which function as transmembrane ion transporters, calcium, zinc or magnesium ion binders that may play a role in crystal formation were shortlisted. Among the 57 proteins which were common to the NaOx and EG groups, only Vha100-1 fulfills this criteria whereas the other 56 proteins do not. Vha100-1(protein accession number: Q6NLA3; Flybase ID: FBgn0028671) functions as proton transporting ATPase which might be of interest in CaOx crystal formation (Fig. 2D).
Label-free quantitation of proteins identified in Malpighian tubules of Drosophila
In total 1982 proteins were identified by nLC-MS/MS and Mascot/Progenesis analysis. Using a cutoff of p<0.05, 211 proteins were differentially synthesized between NaOx and control groups (Fig. 3A, Supplementary Table 1), and 314 proteins between EG and control groups (Fig. 3B, Supplementary Table 2). Molecular functions were analyzed and categorized by PANTHER Classification System (Protein ANalysis THrough Evolutionary Relationships, version 10.0) (Fig. 4A and B). Among those differentially abundant proteins, their molecular functions were screened for transmembrane ion transporter, calcium, zinc or magnesium binders that could be relevant in crystal pathogenesis. 11 candidate proteins were selected in NaOx group and 16 in EG group (Table 2A and B). Positive fold changes indicated up regulation compared with control and negative fold changes indicated down regulation.
Figure 3A–B.
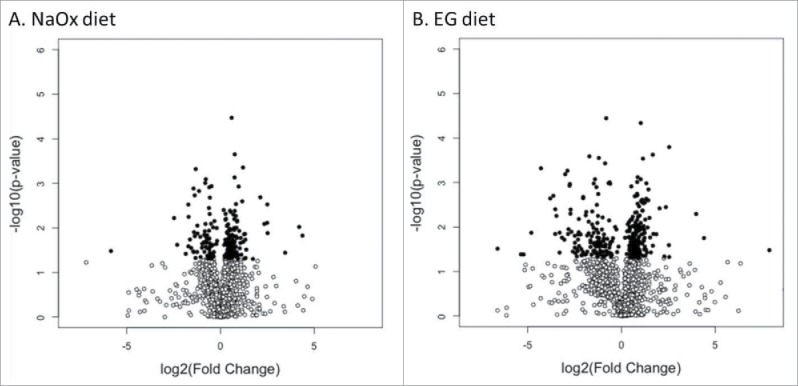
Volcano plot of differentially abundant proteins in Malpighian tubules after treatment. Volcano plots of protein fold changes for control vs. NaOx (A) and vs. EG group (B). The magnitude of the change is plotted on the x-axis (log2 of diet vs. control) against the significance of the change (-log10 of Progenesis P-values, which are equivalent to a 2-tailed Student's t-test of arcsinh-transformed normalized protein abundances) on the y-axis. Proteins were p< = 0.05 are highlighted in dark circles.
Figure 4A–B.
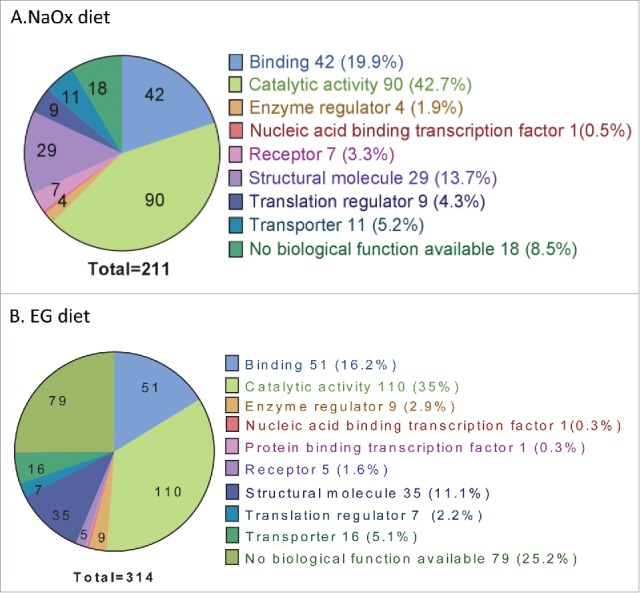
211 proteins were differentially synthesized between NaOx and control groups (A); 314 proteins between EG and control groups (B). Molecular function were analyzed and categorized by PANTHER Classification System (Protein ANalysis THrough Evolutionary Relationships, version 10.0). The functional categories were similar between NaOx and EG group, with proteins with catalytic activity being predominant.
Table 2.
Lists of candidate proteins differentially expressed in NaOx and EG groups as compared to control. The candidate proteins were shortlisted by screening their molecular function. The log2 fold change values of treated groups to control were shown in the last column respectively.
| Protein | ||||||
|---|---|---|---|---|---|---|
| Protein accession number |
Peptide counts |
Gene name |
FlyBase ID |
Protein function |
p-Value |
log2 fold change |
| A. Sodium oxalate diet | ||||||
| A1Z876 | 3(3) | Ndg | FBgn0026403 | Zinc ion binding | 0.032 | 0.453 |
| Q9V3J1-2 | 6(6) | VhaSFD | FBgn0027779 | Vacuolar H[+]-ATPase SFD subunit | 0.017 | 0.449 |
| Q07171-2 | 5(5) | Gel | FBgn0010225 | Calcium ion binding | 0.041 | 0.427 |
| A4V0N4 | 42(19) | Vha68-2 | FBgn0263598 | Vacuolar H[+] ATPase 68 kDa subunit 2 | 0.042 | 0.328 |
| Q9VLI1 | 7(6) | CG9465 | FBgn0032067 | Zinc ion binding | 0.035 | −1.425 |
| Q0E9G4 | 1(1) | Drat | FBgn0033188 | Zinc ion binding | 0.012 | −1.408 |
| Q8SXQ8 | 1(1) | CG32335 | FBgn0063667 | Zinc ion binding | 0.045 | −1.057 |
| A8QI34 | 2(1) | Jyalpha | FBgn0267363 | sodium: potassium-exchanging ATPase | 0.036 | −0.563 |
| M9PJN5 | 9(8) | HDAC6 | FBgn0026428 | Zinc ion binding | 0.008 | −0.500 |
| Q8IPZ3 | 1(1) | CG17221 | FBgn0031500 | Zinc ion binding | 0.026 | −0.496 |
| Q95T12 | 1(1) | flower | FBgn0261722 | Calcium channel activity | 0.049 | −0.474 |
| B. Ethylene glycol diet | ||||||
| Q9VUB5 | 1(1) | Upset | FBgn0036398 | Zinc ion binding | 8.82E-08 | infinity |
| B5RIR1 | 1(1) | nrv1 | FBgn0015776 | Na: potassium transporting ATPase | 0.003 | 1.254 |
| Q9W141-2 | 3(2) | CG4692 | FBgn0035032 | Hydrogen-exporting ATPase activity | 0.013 | 1.238 |
| Q9VNF8 | 5(5) | sec23 | FBgn0262125 | Zinc ion binding | 0.044 | 1.167 |
| Q8SXQ8 | 1(1) | CG32335 | FBgn0063667 | Zinc ion binding | 0.011 | 0.935 |
| Q9VKM3 | 3(3) | ATPsynG | FBgn0010612 | Hydrogen-exporting ATPase activity | 0.005 | 0.737 |
| Q7JYV3 | 1(1) | CG12374 | FBgn0033774 | Zinc ion binding | 0.027 | −2.262 |
| Q9VLI0 | 7(6) | CG9466 | FBgn0032068 | Zinc ion binding | 0.015 | −2.176 |
| Q9VS66 | 3(3) | CG8562 | FBgn0035779 | Zinc ion binding | 0.002 | −2.042 |
| Q9VBP9-2 | 2(2) | NpL4 | FBgn0039348 | Zinc ion binding | 0.004 | −1.555 |
| Q9VLI1 | 7(6) | CG9465 | FBgn0032067 | Zinc ion binding | 0.025 | −1.434 |
| Q9VLI2 | 7(7) | CG9463 | FBgn0032066 | Zinc ion binding | 0.027 | −1.368 |
| Q961J8 | 2(2) | CG8560 | FBgn0035781 | Zinc ion binding | 0.028 | −1.166 |
| Q9VNF9 | 3(3) | elm | FBgn0037358 | Calcium ion binding | 0.001 | −0.867 |
| Q9W197 | 2(2) | CG3362 | FBgn0034988 | Magnesium ion binding | 0.04 | −0.721 |
| Q8IPI3 | 2(2) | Ndae1 | FBgn0259111 | Na: bicarbonate symporter activity | 0.043 | −0.510 |
Discussion
Kidney stone disease is a major health problem worldwide but the underlying pathophysiology remain largely unknown and effective medical treatment is scarce. Using the Drosophila as a model for kidney stone disease, we have revealed a comprehensive profile of altered proteome in response to induced crystal formation in Malpighian tubules. To our knowledge, this is the first study to identify the possible candidate proteins that may play an important role in tubular crystal formation.
The recently described Gel-aided sample preparation (GASP) method9 has been applied for the first time on Drosophila Malpighian tubules. The GASP method is very reliable especially with low protein amounts compared to the known in-gel, in-solution digestion methods in mass spectrometry. In our hands the GASP protocol, shows more consistency with biological replicates due to lesser samples loss during preparation. Using the GASP method, we were able to identify similar number of proteins from 5 drosophila Malpighian tubules and 10 tubules (859 compared to 867, respectively, on a one hour gradient). The sensitivity of the GASP method for 1, 5 and 10 tubules are shown in Supplementary Figure 1.
GASP shows high reproducibility of biological replicates and gave more protein identifications in our hands than the in-solution method (data not shown). As GASP has been previously optimized for scalability, ease of use and sensitivity, it appears to be well suited for low abundant samples that require a flexible application to other body parts and/or fluids of flies and will have a significant impact of future applications in fly research.
The chosen label-free method for Mass Spectrometry relies on a good alignment of the chromatograms for quantitation but is more time and cost effective compared to a SILAC approach.11
Among the 57 proteins present in both NaOx and EG group but not in control group in MASCOT analysis, Vha100-1 should be pointed out as an interesting candidate protein. Vha100-1 which is known to be involved in the pathogenesis of renal tubular acidosis and calcium homeostasis, both being recognized pathways of calcium crystal formation. None of the other 56 candidate proteins identified have previously been shown to have a role in recognized pathways of calcium crystal formation.
Vha100-1 stands for vacuolar H+ ATPase 100 kD subunit 1, which is one of the many isoforms of V-type proton transporting ATPase, mainly involved in reduction of intracellular pH and ATP hydrolysis coupled proton transport.12 The human ortholog of Vha100-1 is Hsap\ATP6V0A4. Located on chromosome 7q33-34, Hsap\ATP6V0A4 is known to play a key role in the pathogenesis of renal tubular acidosis. The homozygous mutation of ATP6V0A4 results in distal renal tubular acidosis, which is a hereditary disorder of renal stone disease.13 The kidney fails to produce an appropriately acid urine in the presence of systemic metabolic acidosis, due to failure of hydrogen ion secretion or bicarbonate reabsorption in the distal nephron. This results in hyperchloremic metabolic acidosis and is usually accompanied by nephrocalcinosis or nephrolithiasis.14 Heterozygous mutation of V-ATPase B1 subunit in human has been shown to cause incomplete distal renal tubular acidosis.15 Vha100-1 is also involved in the process of calmodulin binding, a calcium-binding proteins with many roles, indicating Vha100-1 may have a modulating effect in calcium metabolism and calcium related metabolic pathways. It has been reported that insect calcitonin-like diuretic peptides stimulated V-ATPase activity in Drosophila Malpighian tubules resulting urine acidification hence crystallization.16
V-ATPase plays a central role in many aspects of cellular function in the kidney as well in other organs.17,18 These range from proton secretion in order to acidify the extracellular milieu, to intracellular acidification of vesicles and regulation of a variety of processes that range from lysosomal degradation, ligand receptor dissociation and intracellular trafficking.19 The holoenzyme is comprised of many different subunits, each having different functions that are involved not only in the activity of the enzyme but also to its intracellular regulation and targeting. It might be an important agent in the common pathways in kidney crystal pathogenesis as several other V-ATPase subunits has been identified in the Progenesis analysis as well. We have observed both Vha68-2 (Vacuolar H+ ATPase 68 kDa subunit 2) and VhaSFD (Vacuolar H+ ATPase SFD subunit) were up-regulated in NaOx group.
Among the differentially abundant proteins, we have identified 11 proteins in NaOx groups and 16 proteins in EG group that function as transmembrane ion transporters, calcium, zinc or magnesium ion binders. We made the hypothesis these groups of proteins are more important in the process of crystal formation based on previous published work.20-23 We have previously looked into a different cohort of flies for the corresponding transcriptive changes (data not shown), but the candidate proteins cannot be correlated at the mRNA level.
A recent study using the Drosophila model suggested that zinc and possibly magnesium had an essential role in driving heterogeneous nucleation that eventually results in crystal formation.24 Chi et al. proposed that zinc facilitates calcification given the presence of non-trace levels of zinc in fly crystals, human xanthine stones and CaOx plaques. By inhibiting zinc transporter genes a suppression of Drosophila crystals formation were observed. In this study, numerous zinc binding proteins were differentially abundant in both NaOx and EG diets. Although the particle structure and mechanism of crystallization reported by Chi et al. are different from the CaOx crystals induced in the current experiment, our results also support the hypothesis that zinc pathways may be important in crystal formation.
JYalpha, as well as nervana 1, are proteins that function as sodium: potassium exchanging ATPase. We found this group of protein of interest because sodium and potassium transport has been implicated in calcium ion transport across renal tubules and hemostasis.25 It was shown in rat and rabbit model that the basal-lateral plasma membrane contain sodium/calcium exchange system which mediates the counter-transport of calcium and sodium across the basal cell border.26 Decreased Na+/K+ ATPase activity were described in idiopathic hypercalciuria.27 This transporter might be differentially synthesized as a result of CaOx crystal formation.
The altered proteome probably represents changes at a cellular level rather than in the extracellular environment. These differentially abundant proteins could be directly involved in CaOx formation or could be a regulated response to the obstructive uropathy, metabolic acidosis and failure to maintain electrolyte or acid-base balance in Malpighian tubules. A significant proportion of detected proteomic changes such as those with binding, catalytic and enzyme regulator activity could as well be the result of inflammatory responses or cellular adaptation to the lithogenic agents or crystal formation.
During the screening of candidate proteins, we made the hypothesis that those functioning as transmembrane ion transporter, calcium or zinc binding proteins might be more critical in crystal formation but other proteins might also be relevant. More studies will be desirable to elucidate the biological process of how these differentially abundant proteins function as a whole in the process of CaOx crystal formation. Though previous researchers 4,10 have shown the crystals inside the Malpighian tubules to be CaOx, there was the possibility given that the relative elemental composition was altered during prolonged feeding. One of the other limitation of the current study is the lack of functional validation of the candidate proteins. Further work will be needed to evaluate if changing the expression of candidate proteins will have any impact on the crystal formation process. Assisted by powerful transgenic resources, these proteins could be targeted to allow manipulation of stone formation and develop therapeutic agents in human kidney stone disease in the future.
To conclude, our study has applied a new proteomic sample preparation method GASP on Drosophila model to narrow down the potential protein targets involved in human stone pathogenesis. These results may further increase the understanding of the pathophysiology of human kidney stone disease.
Material and methods
Drosophila stocks
Wild-type (Canton S) D. melanogaster were used. Male flies were selected and maintained in plastic vials containing standard medium (maize 18.6%, yeast 3.8%, soya 2.2%, agar 3.1% and water 72.3%) in a 25°C, 50-60% humidity incubator.
Lithogenic agents
Different concentrations of crystal forming agents at a physiological dose were added into standard fly medium. Three conditions were prepared: control (standard fly medium), 0.05% sodium oxalate and 0.5% Ethylene Glycol. Biological triplicates were raised in each condition.
Fly maintenance
New emergent flies were collected from eclosion under light anesthesia. Flies were housed at a density of 25 flies per vial. At day 3 male flies were randomly divided into 3 diet groups (control, NaOx, EG) and kept on these diets for 2 weeks. Every third day the male flies were transferred into fresh medium. The proteomic study contains 5 male flies for each diet group in biological triplicates of Malpighian tubules and hindgut (Fig. 5).
Figure 5.
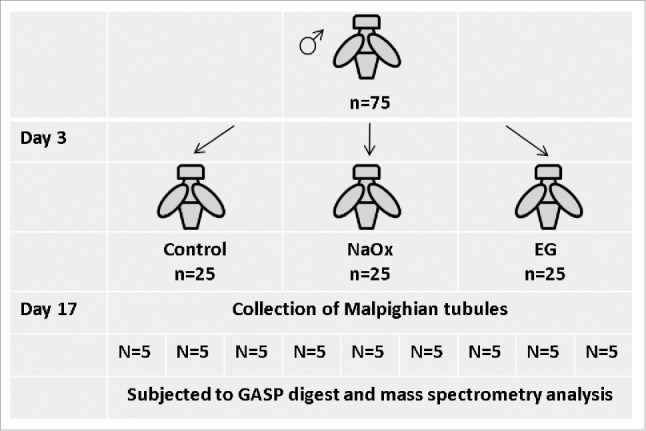
Workflow for fly maintenance.
Malpighian tubule preparation
After CO2 anesthesia the excretory organs (Malpighian tubules and hindgut) were dissected in Schneider's Drosophila medium. Freshly dissected tubules and hindgut from 15 flies were prepared for polarized light microscopy examination. Groups were dissected in parallel and processed for protein extraction with 5 flies pooled from each biological triplicate and analyzed separately.
Birefringence microscopy
Malpighian tubules and hindgut were prepared fresh on SuperFrost slides and observed immediately under normal and polarized white light with an Olympus BX60 microscope. The tubules with crystal formation were photographed and scaled.
Proteomic sample preparation and quality control
For protein extraction, Malpighian tubules and hindgut samples were homogenized by tissue grinder (4 minutes) and vortexing (2 minutes) respectively in 180 µl of cocktail buffer consisting of 7M Urea, 2M Thiourea, 4% CHAPS and 0.2% Triton X-100 with freshly added 10 µl of 1M tris (2-carboxyethyl) phosphine (TCEP) and 10 µl of 1M iodoacetamide (IAA) and incubated for 30 min at room temperature. During method development we compared the Gel-aided sample preparation (GASP) (6) to an in-solution digestion protocol on one Malpighian tubules and hindgut. Briefly, the steps for the GASP protocol includes (i) protein extraction, (ii) co-polymerization of proteins with monomeric acrylamide, (iii) shredding of gel, (iv) depletion of small molecules, (v) proteolysis, and (vi) peptide recovery. The other steps for Gel-aided sample preparation (GASP), an in house developed trypsin digestion and peptide extraction procedure, were performed according to the protocol described elsewhere (6). Alternatively, for the in-solution protocol, samples were subjected to a Methanol/Chloroform extraction.28 The precipitated protein pellet was resuspended in 6M Urea and diluted to 2M Urea prior to the addition of trypsin as described (6). Digested samples were analyzed on a Dionex Ultimate 3000 UPLC coupled to a hybrid quadrupole-orbitrap instrument (Q Exactive, Thermo Scientific).29 Samples were desalted online (PepMAP C18, 300 µm × 5 mm, 5 µm particle, Thermo) for 1 minute at a flow rate of 20 µl/min and separated on a nEASY column (PepMAP C18, 75 µm × 500 mm, 2 µm particle, Thermo) over 120 Minutes using a gradient of 2%-35% Acetonitrile in 5% DMSO with 0.1% Formic acid at 250 nl/min. Scans were acquired at a resolution of 70,000 @ 200 m/z and the 15 most abundant precursors were selected for HCD fragmentation.
Mass spectrometry data analysis
Peak list files were generated with MSConvert (Proteowizard v3.0.5211) using the 200 most abundant peaks/spectrum. The SwissProt D. melanogaster reference proteome (retrieved 30/03/2015) (21,361 sequences; 14,358,849 residues) database was used for searches in Mascot (v2.5.1, Matrix Science), PEAKS (v7, Bioinformatics Solutions). Searches were performed with fixed modification for Propionamide (C) and variable modifications for Oxidation (M) and Deamidated (N). A mass deviation of 10 ppm for MS1 and 0.05 Da for MS2 spectra were applied for all analysis. A decoy database search was implemented in order to get a probability score threshold regarding search engines and a general false discovery rate of 1% was set. Raw data were individually searched in Mascot or Peaks. All proteins were searched against Flybase30 to investigate reported molecular function. Ion transporters, water channels and calcium dependent proteins were short listed and compared to the results from Progenesis (v2.0.5556.29015). For label-free quantification raw data were submitted to Progenesis.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We appreciate the help of Dr. Markus Toegel, Dr. Tudor Fulga from Weatherall Institute of Molecular Medicine, University of Oxford for the generous support during the experiments. We also appreciate the help of Mr. Mark Sullivan for reviewing the manuscript.
References
- [1].Ansari MS, Gupta NP. Impact of socioeconomic status in etiology and management of urinary stone disease. Urologia Internationalis 2003; 70:255-61; PMID:12776701; http://dx.doi.org/ 10.1159/000070130 [DOI] [PubMed] [Google Scholar]
- [2].Preminger GM, Tiselius HG, Assimos DG, Alken P, Buck C, Gallucci M, Knoll T, Lingeman JE, Nakada SY, Pearle MS, et al.. 2007 guideline for the management of ureteral calculi. J Urol 2007; 178:2418-34; PMID:17993340; http://dx.doi.org/ 10.1016/j.juro.2007.09.107 [DOI] [PubMed] [Google Scholar]
- [3].Trinchieri A. Epidemiological trends in urolithiasis: impact on our health care systems. Urol Res 2006; 34:151-6; PMID:16440192; http://dx.doi.org/ 10.1007/s00240-005-0029-x [DOI] [PubMed] [Google Scholar]
- [4].Chen YH, Liu HP, Chen HY, Tsai FJ, Chang CH, Lee YJ, Lin WY, Chen WC. Ethylene glycol induces calcium oxalate crystal deposition in Malpighian tubules: a Drosophila model for nephrolithiasis/urolithiasis. Kidney Int 2011; 80:369-77; PMID:21451462; http://dx.doi.org/ 10.1038/ki.2011.80 [DOI] [PubMed] [Google Scholar]
- [5].Miller J, Chi T, Kapahi P, Kahn AJ, Kim MS, Hirata T, Romero MF, Dow JA, Stoller ML. Drosophila melanogaster as an emerging translational model of human nephrolithiasis. J Urol 2013; 190:1648-56; PMID:23500641; http://dx.doi.org/ 10.1016/j.juro.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Weavers H, Prieto-Sanchez S, Grawe F, Garcia-López A, Artero R, Wilsch-Bräuninger M, Ruiz-Gómez M, Skaer H, Denholm B. The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature 2009; 457:322-6; PMID:18971929; http://dx.doi.org/ 10.1038/nature07526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang J, Kean L, Yang J, Allan AK, Davies SA, Herzyk P, Dow JA. Function-informed transcriptome analysis of Drosophila renal tubule. Genome Biol 2004; 5:R69; PMID: 15345053; http://dx.doi.org/ 10.1186/gb-2004-5-9-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chien S, Reiter LT, Bier E, Gribskov M. Homophila: human disease gene cognates in Drosophila. Nucleic Acids Res 2002; 30:149-51; PMID:11752278; http://dx.doi.org/ 10.1093/nar/30.1.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fischer R, Kessler BM. Gel-aided sample preparation (GASP)–a simplified method for gel-assisted proteomic sample generation from protein extracts and intact cells. Proteomics 2015; 15:1224-9; PMID:25515006; http://dx.doi.org/ 10.1002/pmic.201400436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hirata T, Cabrero P, Berkholz DS, Bondeson DP, Ritman EL, Thompson JR, Dow JA, Romero MF. In vivo Drosophilia genetic model for calcium oxalate nephrolithiasis. Am J Physiol Renal Physiol 2012; 303:F1555-62; PMID:22993075; http://dx.doi.org/ 10.1152/ajprenal.00074.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sury MD, Chen JX, Selbach M. The SILAC fly allows for accurate protein quantification in vivo. Mol Cell Proteomics: MCP 2010; 9:2173-83; PMID:20525996; http://dx.doi.org/ 10.1074/mcp.M110.000323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].FlyBase C. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res 2003; 31:172-5; PMID:12519974; http://dx.doi.org/ 10.1093/nar/gkg094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Karet FE, Finberg KE, Nayir A, Bakkaloglu A, Ozen S, Hulton SA, Sanjad SA, Al-Sabban EA, Medina JF, Lifton RP. Localization of a gene for autosomal recessive distal renal tubular acidosis with normal hearing (rdRTA2) to 7q33-34. Am J Hum Genet 1999; 65:1656-65; PMID:10577919; http://dx.doi.org/ 10.1086/302679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Smith AN, Skaug J, Choate KA, Nayir A, Bakkaloglu A, Ozen S, Hulton SA, Sanjad SA, Al-Sabban EA, Lifton RP, et al.. Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat Gen 2000; 26:71-5; PMID:10973252; http://dx.doi.org/ 10.1038/82492 [DOI] [PubMed] [Google Scholar]
- [15].Zhang J, Fuster DG, Cameron MA, Quiñones H, Griffith C, Xie XS, Moe OW. Incomplete distal renal tubular acidosis from a heterozygous mutation of the V-ATPase B1 subunit. Am J Physiol Renal Physiol 2014; 307:F1063-71; PMID:25164082; http://dx.doi.org/ 10.1152/ajprenal.00408.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Coast GM, Webster SG, Schegg KM, Tobe SS, Schooley DA. The Drosophila melanogaster homologue of an insect calcitonin-like diuretic peptide stimulates V-ATPase activity in fruit fly Malpighian tubules. J Exp Biol 2001; 204:1795-804; PMID:11316500 [DOI] [PubMed] [Google Scholar]
- [17].Allan AK, Du J, Davies SA, Dow JA. Genome-wide survey of V-ATPase genes in Drosophila reveals a conserved renal phenotype for lethal alleles. Physiol Genomics 2005; 22:128-38; PMID:15855386; http://dx.doi.org/ 10.1152/physiolgenomics.00233.2004 [DOI] [PubMed] [Google Scholar]
- [18].Davies SA, Goodwin SF, Kelly DC, Wang Z, Sözen MA, Kaiser K, Dow JA. Analysis and inactivation of vha55, the gene encoding the vacuolar ATPase B-subunit in Drosophila melanogaster reveals a larval lethal phenotype. J Biol Chem 1996; 271:30677-84; PMID:8940044; http://dx.doi.org/ 10.1074/jbc.271.48.30677 [DOI] [PubMed] [Google Scholar]
- [19].Brown D, Paunescu TG, Breton S, Marshansky V. Regulation of the V-ATPase in kidney epithelial cells: dual role in acid-base homeostasis and vesicle trafficking. J Exp Biol 2009; 212:1762-72; PMID:19448085; http://dx.doi.org/ 10.1242/jeb.028803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Soleimani M. SLC26 Cl-/HCO3- exchangers in the kidney: roles in health and disease. Kidney Int 2013; 84:657-66; PMID:23636174; http://dx.doi.org/ 10.1038/ki.2013.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wagner CA. When proton pumps go sour: Urinary acidification and kidney stones. Kidney Int 2008; 73:1103-5; PMID:18449176; http://dx.doi.org/ 10.1038/ki.2008.137 [DOI] [PubMed] [Google Scholar]
- [22].He Y, Chen X, Yu Z, Wu D, Lv Y, Shi S, Zhu H. Sodium dicarboxylate cotransporter-1 expression in renal tissues and its role in rat experimental nephrolithiasis. J Nephrol 2004; 17:34-42; PMID:15151257 [PubMed] [Google Scholar]
- [23].Ozgurtas T, Yakut G, Gulec M, Serdar M, Kutluay T. Role of urinary zinc and copper on calcium oxalate stone formation. Urologia Internationalis 2004; 72:233-6; PMID:15084769; http://dx.doi.org/ 10.1159/000077122 [DOI] [PubMed] [Google Scholar]
- [24].Chi T, Kim MS, Lang S, Bose N, Kahn A, Flechner L, Blaschko SD, Zee T, Muteliefu G, Bond N, et al.. A Drosophila model identifies a critical role for zinc in mineralization for kidney stone disease. PloS One 2015; 10:e0124150; PMID:25970330; http://dx.doi.org/ 10.1371/journal.pone.0124150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Friedman PA. Codependence of renal calcium and sodium transport. Annu Rev Physiol 1998; 60:179-97; PMID:9558460; http://dx.doi.org/ 10.1146/annurev.physiol.60.1.179 [DOI] [PubMed] [Google Scholar]
- [26].Friedman PA, Figueiredo JF, Maack T, Windhager EE. Sodium-calcium interactions in the renal proximal convoluted tubule of the rabbit. Am J Physiol 1981; 240:F558-68; PMID:6787932 [DOI] [PubMed] [Google Scholar]
- [27].Reusz G. [Idiopathic hypercalciuria in childhood]. Orvosi Hetilap 1998; 139:2957-62; PMID:9879200 [PubMed] [Google Scholar]
- [28].Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 1984; 138:141-3; PMID:6731838; http://dx.doi.org/ 10.1016/0003-2697(84)90782-6 [DOI] [PubMed] [Google Scholar]
- [29].Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics: MCP 2011; 10:M111.011015; PMID:21642640; http://dx.doi.org/ 10.1074/mcp.M111.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].dos Santos G, Schroeder AJ, Goodman JL, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res 2015; 43:D690-7; PMID:25398896; http://dx.doi.org/ 10.1093/nar/gku1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


