ABSTRACT
Antiviral immunity in the model organism Drosophila melanogaster involves the broadly active intrinsic mechanism of RNA interference (RNAi) and virus-specific inducible responses. Here, using a panel of six viruses, we investigated the role of hemocytes and autophagy in the control of viral infections. Injection of latex beads to saturate phagocytosis, or genetic depletion of hemocytes, resulted in decreased survival and increased viral titers following infection with Cricket paralysis virus (CrPV), Flock House virus (FHV), and vesicular stomatitis virus (VSV) but had no impact on Drosophila C virus (DCV), Sindbis virus (SINV), and Invertebrate iridescent virus 6 (IIV6) infection. In the cases of CrPV and FHV, apoptosis was induced in infected cells, which were phagocytosed by hemocytes. In contrast, VSV did not trigger any significant apoptosis but we confirmed that the autophagy gene Atg7 was required for full virus resistance, suggesting that hemocytes use autophagy to recognize the virus. However, this recognition does not depend on the Toll-7 receptor. Autophagy had no impact on DCV, CrPV, SINV, or IIV6 infection and was required for replication of the sixth virus, FHV. Even in the case of VSV, the increases in titers were modest in Atg7 mutant flies, suggesting that autophagy does not play a major role in antiviral immunity in Drosophila. Altogether, our results indicate that, while autophagy plays a minor role, phagocytosis contributes to virus-specific immune responses in insects.
IMPORTANCE Phagocytosis and autophagy are two cellular processes that involve lysosomal degradation and participate in Drosophila immunity. Using a panel of RNA and DNA viruses, we have addressed the contribution of phagocytosis and autophagy in the control of viral infections in this model organism. We show that, while autophagy plays a minor role, phagocytosis contributes to virus-specific immune responses in Drosophila. This work brings to the front a novel facet of antiviral host defense in insects, which may have relevance in the control of virus transmission by vector insects or in the resistance of beneficial insects to viral pathogens.
INTRODUCTION
Experiments in the model organism Drosophila melanogaster have shown that RNA interference (RNAi) plays a major role in antiviral immunity in insects: (i) flies with mutations for the three key genes of the small interfering RNA (siRNA) pathway, Dicer-2, Argonaute 2, and r2d2, show increased sensitivity to infection by RNA and DNA viruses (1–6); (ii) Dicer-2-dependent 21-nucleotide siRNAs of viral origin accumulate in virus-infected flies (1, 3, 4, 7–9); (iii) several insect viruses express viral suppressors of RNAi (5, 10–12). The importance of this pathway in the control of viral infections has been confirmed in other insects, in particular, the vector mosquito genera Aedes and Culex, which transmit important human pathogens such as dengue virus, West Nile virus, and other arthropod-borne viruses (13–18).
Inducible responses also contribute to the antiviral host defense in Drosophila, although they remain poorly characterized and involve virus-specific mechanisms (reviewed in references 19 and 20). We previously reported that a number of genes are induced following viral infection via the Jak/STAT pathway (21). Accordingly, flies with mutations for the Jak kinase Hopscotch are susceptible to infection by Drosophila C virus (DCV) and Cricket paralysis virus (CrPV), two members of the Dicistroviridae family, although they are as resistant as wild-type controls to other viruses (e.g., the alphavirus Sindbis virus [SINV] or the rhabdovirus vesicular stomatitis virus [VSV]) (3). Virus-induced autophagy and apoptosis have also been associated with antiviral immunity in Drosophila (22–24) and other insects (reviewed in reference 25).
Insects also mount cellular responses to fight infections mediated by blood cells called hemocytes. In Drosophila, macrophage-like plasmatocytes and two other nonphagocytic cells, the crystal cells and lamellocytes, have been described (26, 27). Plasmatocytes form the majority of differentiated blood cells (90 to 95% of hemocytes in Drosophila larvae). Commonly referred to as macrophages, they can engulf and degrade dead cells, debris, and invading pathogens (28, 29). Crystal cells (5% of larval hemocytes in Drosophila) are round cells with a 10- to 12-μm diameter and characteristic paracrystalline cytoplasmic inclusions. They play a key role in host defense through melanization and they participate in wound healing. Lamellocytes are large (15 to 40 μm in diameter), flat adherent cells which encapsulate and neutralize objects too large to be phagocytosed by plasmatocytes (30–32). Of note, the question of the involvement of hemocytes in antiviral immunity has not been a focus of interest so far for Drosophila.
Here, we performed a comparative analysis of the contribution of cellular immunity and autophagy to antiviral host defense, using a panel of 6 different viruses. We show that hemocytes participate in antiviral host defense against CrPV, Flock House virus (FHV), and VSV, but not against the three other viruses tested. FHV- and CrPV-infected cells undergo apoptosis and can be cleared by hemocytes. In addition, we confirmed that autophagy participates in the host defense against VSV infection, although its contribution is modest compared to that of RNAi. However, Drosophila strains with mutations of the essential autophagy gene Atg7 are more resistant to FHV infection, indicating that autophagy has a pro- rather than antiviral function in this context. Our results indicate that blood cells and autophagy display virus-specific functions in Drosophila and are not general antiviral pathways, in contrast to RNAi.
MATERIALS AND METHODS
Drosophila strains.
The fly stocks were raised on standard cornmeal agar medium at 25°C. Canton-S, w1118, y1 w1, Df(2R)BSC22/SM6a (stock number 7441), w*; P{UAS-mCherry.NLS}3 (stock 38424), Df(2R)BSC45, w+mC/SM6a (stock 7441), UAS-Mito::GFP (stock 8443), and actin-GAL4 (stock 25374) genotype flies were obtained from the Bloomington Fly Stock Center (Bloomington, IN). Atg7d14/Cyo-GFP, Atg7d77/Cyo-GFP, CG5335d30/Cyo-GFP (33), Dcr-2L811fsX (3), UAS-bax/CyO-actin-GFP, hml(Δ)-GAL4, UAS-eGFP (34), Toll-7g1-1 (35), and Toll-7p8 and Toll-7p114 (36) stocks have been described previously. A genomic rescue of the Toll-7 gene was established with the fosmid FlyFos 030116 (http://transgeneome.mpi-cbg.de) inserted at the landing site attP40 (3L), and the transgenic chromosome was associated with the deficiency Df(2R)BSC22, which uncovers the Toll-7 locus. For the rescue experiments, Toll-7g1-1 mutants were crossed with the Df(2R)BSC22–gToll-7 rescue line. All fly lines were tested and cleared of any Wolbachia spp. infection.
Phagocyte ablation experiments.
Surfactant-free, red, 0.3-μm-diameter carboxylate modified latex beads (Interfacial Dynamics Corp.) were washed and resuspended at a 4× concentration in 1× phosphate-buffered saline (PBS) (corresponding to 5 to 10% solids). Flies were injected with 69 nl of this solution 24 h prior to virus infection.
Infections.
Adult flies (half males and half females) 4 to 6 days old were used in infection experiments. VSV and VSV with a green fluorescent protein inserted (VSV-GFP) were grown and titers were determined on Vero cells. Supernatants of infected cells were centrifuged at 1,000 × g to pellet cell debris. The resulting virus suspensions were used to infect flies. A supernatant from uninfected cells was used as a control. For all other viruses, stocks were prepared, titers were determined as described previously (3), and the stocks were resuspended in 10 mM Tris-HCl (pH 7.5). Infections were done by intrathoracic injection (Nanoject II apparatus; Drummond Scientific) of 4.6 nl of a viral suspension (500 PFU/fly for DCV and FHV, 5 PFU/fly for CrPV, 2,500 PFU/fly for SINV, 5,000 PFU/fly for Invertebrate iridescent virus 6 [IIV6], and 10,000 PFU/fly for VSV and VSV-GFP). The size of the inoculum was chosen to take into account the kinetics of replication and colonization of Drosophila by the different viruses (3). Injection of the same volume of 10 mM Tris-HCl (pH 7.5) or mock-infected Vero cell culture supernatant for VSV and VSV-GFP experiments was used for controls. Infected flies were incubated at 25°C and monitored daily for survival or frozen for RNA or DNA isolation at the indicated time points.
Quantitative RT-PCR.
Analysis of RNA expression or viral DNA was performed by real-time quantitative reverse transcription-PCR (RT-PCR) as previously described (3). Primers used for quantitative PCR (qPCR) were as follows: RP49 (forward, 5′-GACGCTTCAAGGGACAGTATCTG-3′; reverse, 5′-AAACGCGGTTCTGCATGAG-3′), DCV (forward, 5′-TCATCGGTATGCACATTGCT-3′; reverse, 5′-CGCATAACCATGCTCTTCTG-3′), CrPV (forward, 5′-GCTGAAACGTTCAACGCATA-3′; reverse, 5′-CCACTTGCTCCATTTGGTTT-3′), FHV RNA1 (forward, 5′-TTTAGAAGCACATGCGTCCAG-3′; reverse, 5′-CGCTCACTTTCTTCGGGTTA-3′), VSV (forward, 5′-CATGATCCTGCTCTTCGTCA-3′; reverse, 5′-TGCAAGCCCGGTATCTTATC-3′), SINV (forward, 5′-CAAATGTGCCACAGATACCG-3′; reverse, 5′-ATACCCTGCCCTTTCAACAA-3′), Toll-7 (forward, 5′-GGGCGAGAATCAAATTCGTA-3′; reverse, 5′-CAGACCAGTCAGCTGGTGAA-3′), IIV6 (forward, 5′-TTGTTAGGAATTGGAACTGGAA-3′; reverse, 5′-GCCCTAGATGCTGCTTGTTC-3′).
Flow cytometry.
Cell death was assessed by Annexin-V–fluorescein isothiocyanate/7-aminoactinomycin D (7-AAD) double staining (catalog numbers 559925 and 556419; BD Biosciences) after infection at a multiplicity of infection (MOI) of 10 during 1 h at 4°C. After acquisition by a Gallios flow cytometry apparatus (Beckman Coulter), data were analyzed with FlowJo software (Tree Star) or imaged with a cell observer (spinning disk; Zeiss, Oberkochen, Germany) with adapted settings.
Live imaging.
Lab-Tek II chambered coverglasses (catalog number 155382; Thermo Scientific Nunc) were coated with a Cell-Tak solution (catalog number 354240; Corning) diluted in ultrapure water (1 to 5 μg/cm2). A total of 200,000 S2 cells were added per chamber during 30 min at 25°C. Cells were incubated with different viruses at an MOI of 10 during 1 h at 4°C. NucBlue was used for live DNA staining (catalog number R37605; Molecular Probes). Cells were then observed using adapted settings on a cell observer (spinning disk; Zeiss, Oberkochen, Germany).
Cocultured hemocytes/infected S2 cells.
A stable cell line derived from plasmatocyte-like S2 cells was established for the expression of cytoplasmic GFP and mCherry, using the construct Flag-mCherry-T2A-GFP-T2A-neo (catalog number 32426; Addgene) and G418 at a concentration of 2 mg/ml. Hemocytes were collected from actin-GAL4>UAS-Mito-GFP adult flies and added to Lab-Tek II chambered coverglasses with Schneider medium for 1 h at 25°C. Aliquots (100 μl) of infected S2 cells (at a concentration of 106 cells/ml) were added to each well. Cells were imaged for 12 h, with 1 picture every 20 min, under a confocal microscope (spinning disk cell observer; Zeiss, Oberkochen, Germany) using adapted settings.
Statistical analysis.
An unpaired two-tailed Student t test was used for statistical analysis of data within GraphPad Prism (GraphPad Software). Survival curves were plotted and analyzed by log-rank analysis (Kaplan-Meier method) using the Prism program (GraphPad Software). P values less than 0.05 were considered statistically significant.
RESULTS
Virus-specific role of plasmatocytes in antiviral host defense.
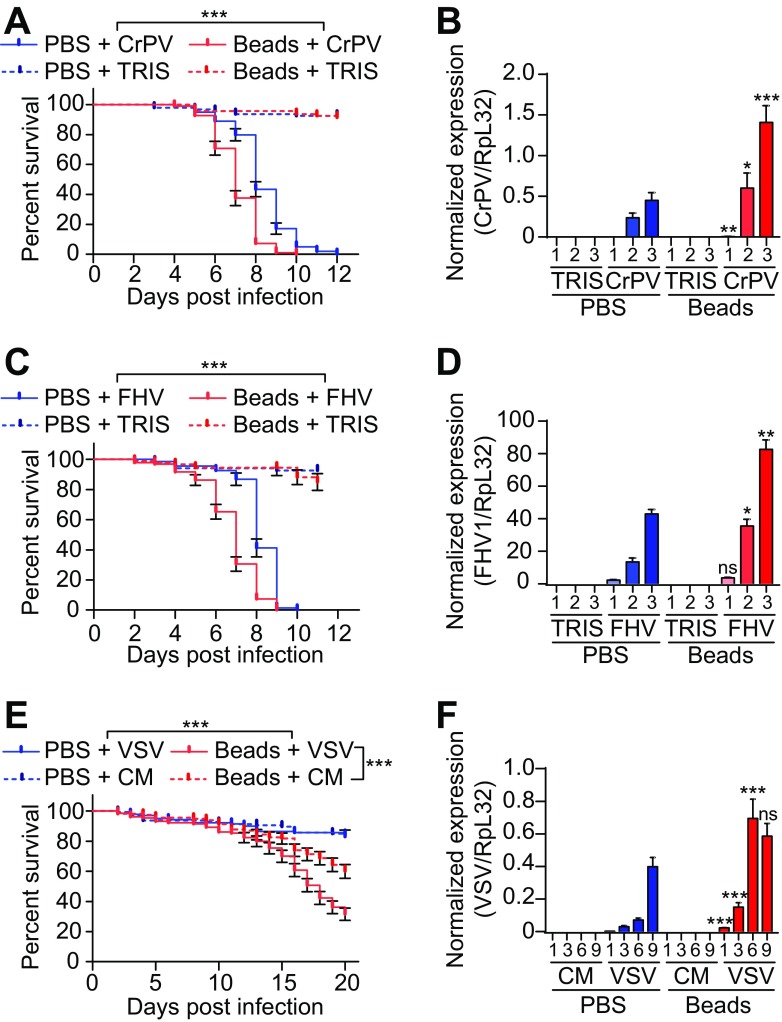
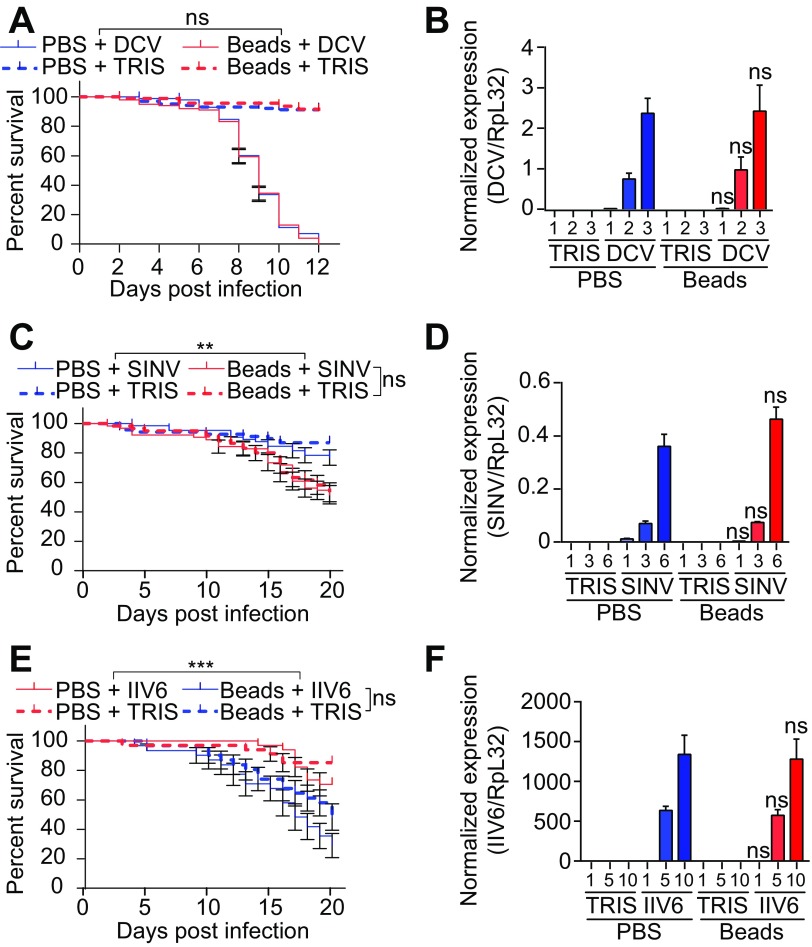
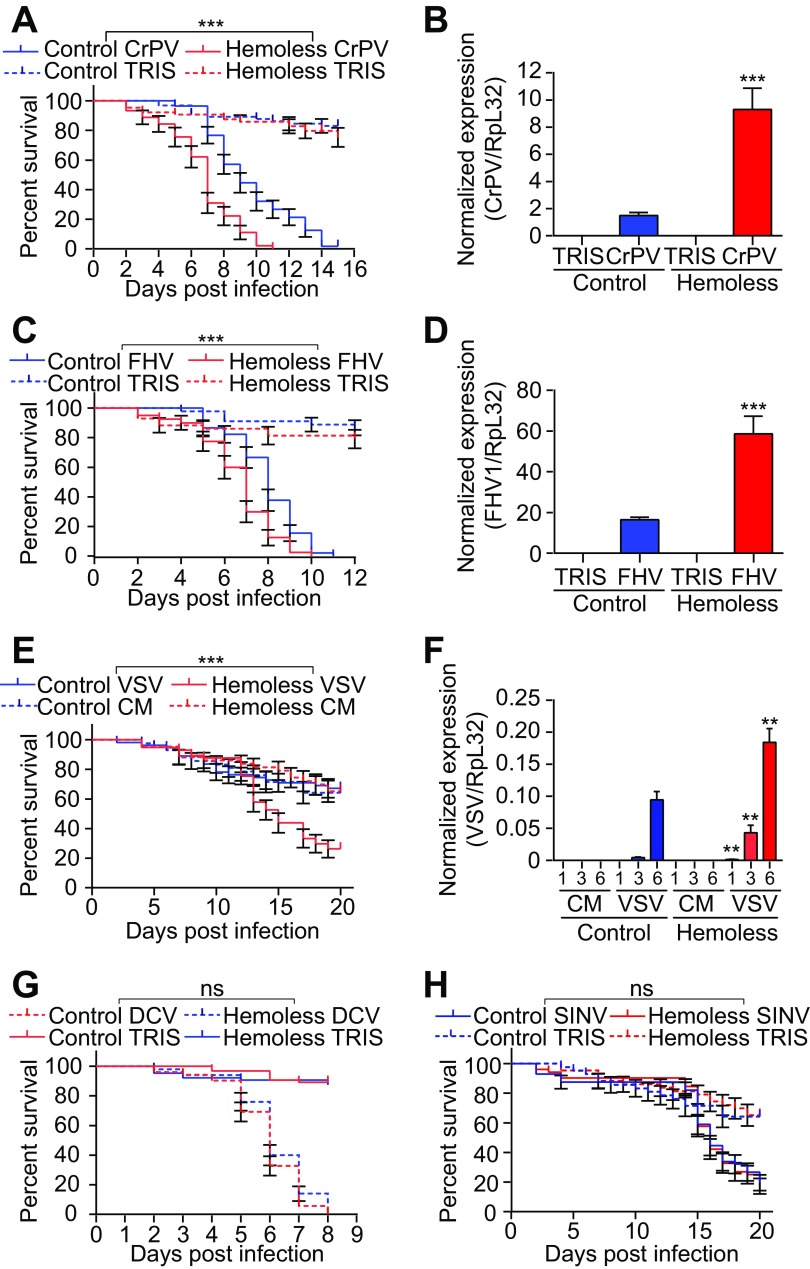
Injection of latex beads into wild-type flies blocks phagocytosis and provides a convenient way to address the contribution of plasmatocytes in host defense (37, 38). As observed previously (39), flies injected with latex beads showed decreased survival upon infection with the dicistrovirus CrPV (Fig. 1A). In addition, bead injection led to a significant increase in the CrPV titer, suggesting that phagocytes are important to control viral replication (Fig. 1B). Resistance of wild-type flies to the nodavirus FHV and the rhabdovirus VSV was also significantly reduced when phagocytosis was impaired (Fig. 1C to F). In contrast, injection of latex beads did not affect survival or virus load following challenge with the alphavirus SINV, the iridovirus IIV6, or DCV, which belongs to the same family as CrPV (Fig. 2). Similar results were obtained using transgenic flies genetically depleted of hemocytes, referred to as hemoless (34). Hemoless flies were more sensitive to infection by CrPV, FHV, and VSV than were the wild-type controls [hml(Δ)-GAL4, UAS-eGFP/+] (Fig. 3). In agreement with results using latex beads, no differences were observed for the other viruses. These findings uncover the involvement of hemocytes in the control of viral infections, thus revealing a novel arm of the antiviral defense in insects. Significantly, however, they also showed that this host defense reaction is limited to some viral species.
FIG 1.
Inhibition of phagocytosis affects resistance to CrPV, FHV, and VSV. (A, C, and E) Survival of Canton-S wild-type flies injected with latex beads or PBS 1 day before challenge with CrPV (A), FHV (C), or VSV (E). (B, D, and F) Quantitative RT-PCR analysis of the accumulation of viral RNA at the indicated days postinfection in Canton-S flies injected with latex beads or PBS 1 day before challenge with CrPV (B), FHV (D), or VSV (F). Tris or conditioned medium (CM) was used as a control. Data represent the means ± standard errors (A, C, and E) or SD (B, D, and F) of 3 independent experiments, each containing three groups of 10 (A, C, and E) or 6 (B, D, and F) flies. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 (log rank [A, C, and E] or unpaired t test [B, D, and F]).
FIG 2.
Phagocytosis is not required during DCV, SINV, and IIV6 infection. (A, C, and E) Survival of Canton-S wild-type flies injected with latex beads or PBS 1 day before challenge with DCV (A), SINV (C), or IIV6 (E). (B, D, and F) Quantitative RT-PCR analysis of the accumulation of viral RNA at the indicated days postinfection in Canton-S flies injected with latex beads or PBS 1 day before challenge with DCV (B), SINV (D), or IIV6 (F). Tris was used as a control. Data represent the means ± standard errors (A, C, and E) or SD (B, D, and F) of 3 independent experiments, each containing three groups of 10 (A, C, and E) or 6 (B, D, and F) flies. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 (log rank [A, C, and E] or unpaired t test [B, D, and F]).
FIG 3.
Hemocyte-depleted flies are susceptible to some, but not all, viral infections. (A, C, E, G, and H) Survival of hemoless [hml(Δ)-GAL4, UAS-eGFP, or UAS-Bax] and control flies [hml(Δ)-GAL4, UAS-eGFP,+] after CrPV (A), FHV (C), VSV (E), DCV (G), or SINV (H) infection. (B, D, and F) Quantitative RT-PCR analysis of the accumulation of viral RNA in hemoless and control flies 3 days postinfection with CrPV (B) or FHV (D) and at the indicated days postinfection following VSV infection (F). Tris or conditioned medium (CM) was used as the injection control. Data represent the means ± standard errors (A, C, E, G, and H) or SD (B, D, and F) of 3 independent experiments, each containing three groups of 10 (A, C, E, G, and H) or 6 (B, D, and F) flies. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 (log rank [A, C, and E] or unpaired t test [B, D, and F]).
FHV and CrPV, but not VSV, induce apoptosis of infected cells.
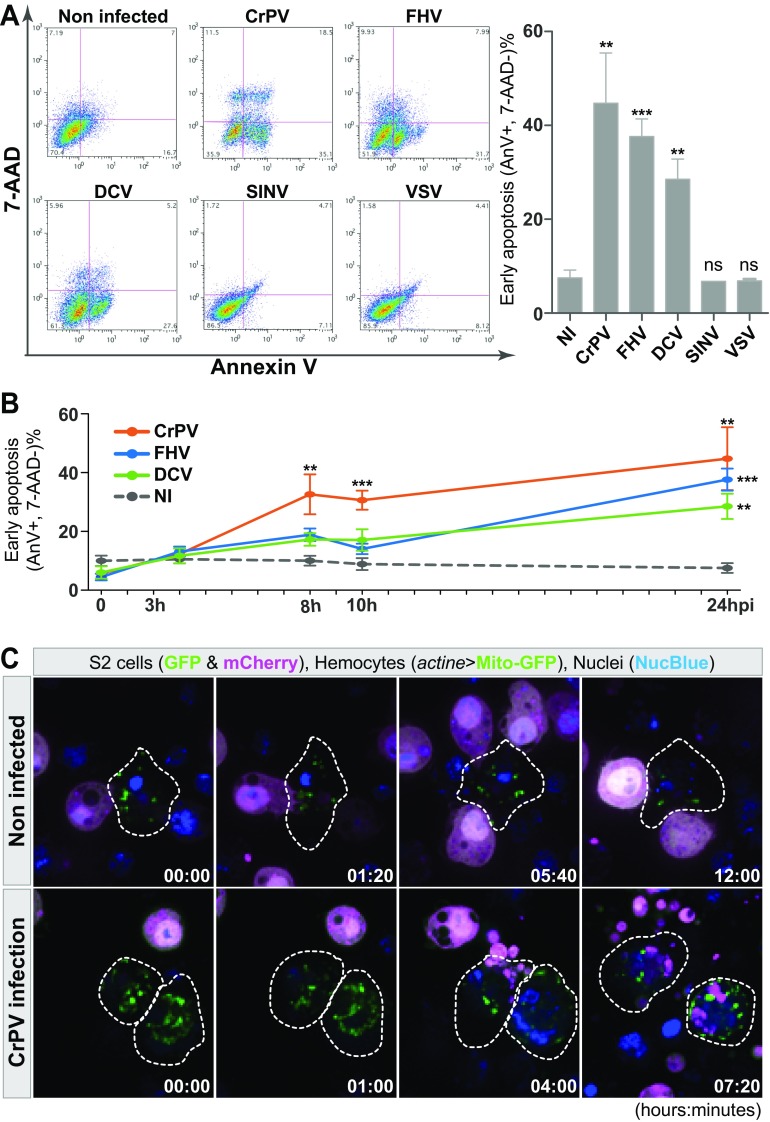
The requirement for phagocytosis in antiviral immunity raises the question of the recognition of virus-infected cells by plasmatocytes. FHV was one of the viruses impacted by hemocytes and is known to induce apoptosis (23, 40), raising the possibility that plasmatocytes are involved in the clearance of infected apoptotic cells. One hallmark of apoptosis is the early surface exposure of phosphatidylserine, which can readily be detected by Annexin-V staining. As expected, Drosophila S2 cells (a hemocyte-like cell line) infected with FHV had a significant increase in Annexin-V staining 24 h postinfection (Fig. 4A). CrPV-infected cells, and to a lesser extent DCV-infected cells, also showed increased Annexin-V staining compared to controls, suggesting that Dicistroviridae trigger apoptosis as well, although we did not detect a contribution of hemocytes in the control of DCV (Fig. 4A) (see Discussion). Notably, CrPV-infected cells showed a significant increase in Annexin-V as early as 8 h postinfection (Fig. 4B). Ex vivo, plasmatocytes isolated from adult flies engulfed apoptotic bodies derived from CrPV-infected cells, supporting the hypothesis that plasmatocytes clear apoptotic cells and prevent the release of infectious particles from dead or dying cells (Fig. 4C; see also Movies S1 and S2 in the supplemental material). Intriguingly, we did not detect Annexin-V staining or cell death of VSV-infected S2 cells (Fig. 4A), despite the importance of phagocytosis and plasmatocytes in the control of this virus in vivo. This suggests that blood cells participate in the control of VSV by a mechanism different from clearance of apoptotic cells.
FIG 4.
Apoptosis-dependent phagocytosis of CrPV-infected cells. (A) Annexin-V/7-AAD double staining to quantify viable S2 cells after infection by the indicated viruses to quantify viable cells (Annexin-V−, 7-AAD−), early apoptotic cells (Annexin-V+, 7-AAD−), and late apoptotic/necrotic cells (AnnexinV+, 7-AAD+) 24 h postinfection. The right panel represents the percentage of early apoptotic cells (Annexin-V+, 7-AAD−) after 24 h of infection with different viruses. (B) Kinetics of appearance of early apoptotic cells (Annexin-V+, 7-AAD−) during infection of S2 cells with CrPV, FHV, or DCV. Measurements were made at 0, 3, 8, 10, and 24 h postinfection. NI, noninfected. For panels A and B, graphs represent means ± standard errors of the means. ***, P < 0.001; **, P < 0.01; *, P < 0.05 (Student's t test). (C) Phagocytosis of apoptotic cells after CrPV infection. Hemocytes (actine>mito-GFP cells), surrounded by the dotted lines, were incubated with mock- or CrPV-infected S2 cells (cytoplasmic GFP and mCherry markers) for 12 h. NucBlue was used to stain DNA in live cells. One picture was taken every 20 min. The corresponding movies can be found in the supplemental material.
Autophagy has opposite effects on VSV and FHV in Drosophila.
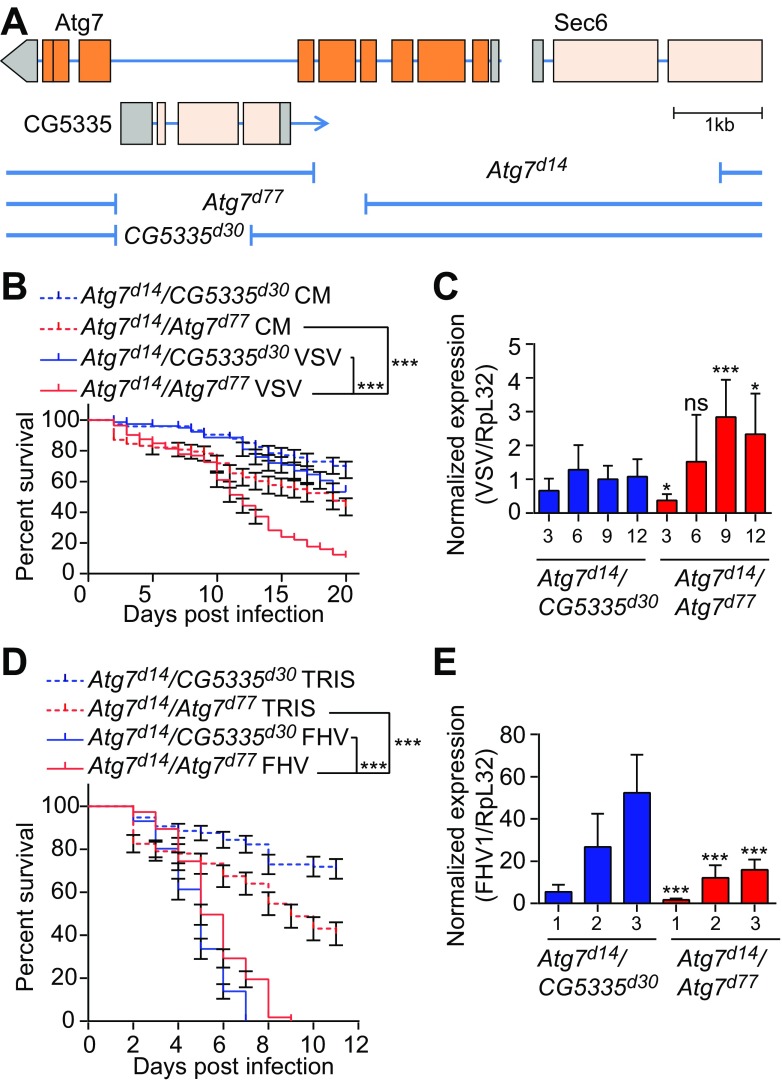
Plasmatocytes could be involved in the direct recognition of viruses. In mammals, VSV infection promotes autophagy in dendritic cells, leading to recognition of viral RNA by Toll-like receptor 7 (TLR-7) upon fusion of autophagosomes with lysosomes (41). Of note, autophagy and a Toll receptor, Toll-7, were reported to participate in the control of VSV infection in Drosophila (22, 42). To address the global role of autophagy in antiviral defenses in flies, we used transheterozygous adult flies carrying two null alleles of the Atg7 gene (Atg7d14/d77). These flies are viable but exhibit a strong impairment of the autophagy pathway and succumb rapidly to starvation (data not shown) (33). We compared these flies to controls carrying one of the null alleles in trans, with a shorter deletion affecting the gene CG53335 but not the coding sequence of Atg7 (Atg7d14/CG5335d30) (Fig. 5A). Of note, the three different deletion mutants are in the same genetic background (33). In agreement with their shorter life span and reported susceptibility to oxidative stress, Atg7 mutant flies (Atg7d14/d77) were more sensitive to an injection of buffer than were control flies (Atg7d14/CG5335d30) (Fig. 5B and D). In addition, Atg7 mutants exhibited reduced survival upon VSV infection (Fig. 5B), as previously described (22). A significant but modest increase (almost 3-fold) in the VSV viral titer was observed in Atg7 mutant flies at late time points of infection that coincided with a decrease in survival (9 and 12 days postinfection [dpi]) (Fig. 5C). In contrast, we did not observe significant differences in the resistance of Atg7 mutants versus control flies to infection with SINV, DCV, CrPV, or IIV6 (Fig. 6). Surprisingly, Atg7 mutant flies showed increased survival and decreased viral loads compared to controls infected with FHV, suggesting that autophagy plays a proviral role in this context (Fig. 5D and E). Altogether, these data indicate that autophagy is not a broad antiviral mechanism in Drosophila and can impact viral replication negatively or positively, depending on the virus.
FIG 5.
Atg7 can have pro- or antiviral activity in Drosophila. (A) Schematic representation of the Atg7 locus and the deletion mutants used. Exons are indicated by rectangles with the coding region highlighted in orange. Deletions are indicated by the gaps in the blue lines. (B and D) Survival of Atg7 mutant (Atg7d14/Atg7d77) and control (Atg7d14/CG5335d30) flies after VSV (B) or FHV (D) infection. Tris or conditioned medium (CM) was used as an injection control. (C and E) Quantitative RT-PCR analysis of the accumulation of viral RNA at the indicated day postinfection in control (Atg7d14/CG5335d30) and Atg7 mutant (Atg7d14/Atg7d77) flies injected with VSV (C) or FHV (E). Data represent the means ± standard errors (B and D) or SD (C and E) of three independent experiments, each containing three groups of 10 (B and D) or 6 (C and E) flies. ns, not significant; *, P < 0.05; ***, P < 0.001 (log rank [B and D] or unpaired t test [C and E]).
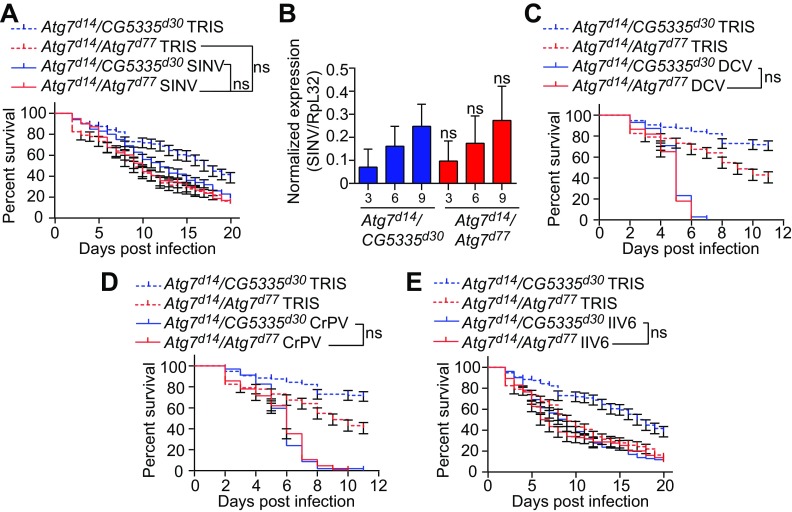
FIG 6.
Atg7 mutant flies show resistance similar to controls after infection by SINV, DCV, CrPV, or IIV6. (A, C, D, and E) Survival of Atg7 mutant (Atg7d14/Atg7d77) and control (Atg7d14/CG5335d30) flies after SINV (A), DCV (C), CrPV (D), or IIV6 (E) infection. Tris injection was used as a control. (B) Quantitative RT-PCR analysis of the accumulation of viral RNA at the indicated day postinfection in control (Atg7d14/CG5335d30) and Atg7 mutant (Atg7d14/Atg7d77) flies injected with SINV. Data represent means ± standard errors (A, C, D, and E) or SD (B) of 3 independent experiments, each containing three groups of 10 (A, C, D, and E) or 6 (B) flies. ns, not significant (log rank [A, C, D, and E] or unpaired t test [B]).
Toll-7 and resistance to VSV infection.
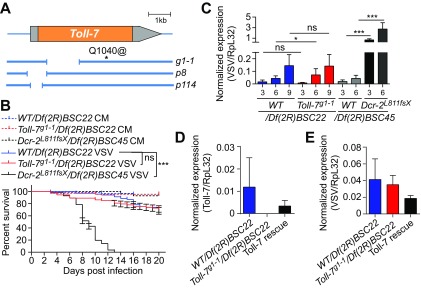
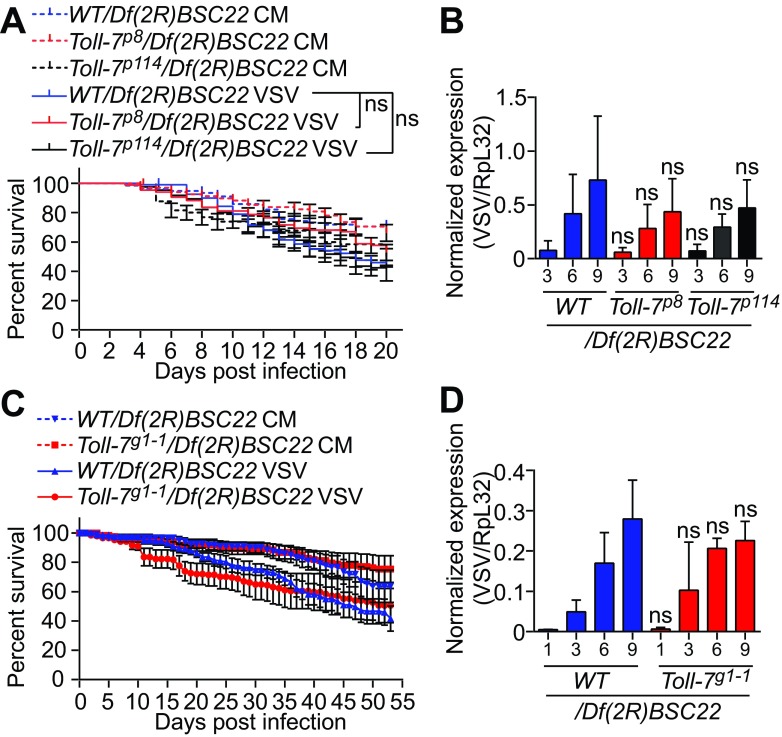
The VSV-specific antiviral role of autophagy led us to further investigate the previously described mechanisms. The Toll-7 receptor has been proposed to function as a direct VSV sensor to activate autophagy and control infection (42). However, we observed that Toll-7-deficient flies carrying a null allele over a deficiency that covers the Toll-7 locus [Toll-7g1-1/Df(2R)BSC22] (35) (Fig. 7A) survived VSV infection, similar to control flies carrying one wild-type allele of Toll-7 [WT/Df(2R)BSC22]. For comparison to Toll-7 mutants, we used Dcr-2 mutant flies [Dcr-2L811fsX/Df(2R)BSC45], which showed significantly increased VSV replication and rapidly succumbed to infection (Fig. 7B and C). VSV RNA levels remained similar in Toll-7 hemizygote mutants and in control flies, despite a 1.6-fold significant increase observed at only one time point. However, this mild phenotype was not reproducible when we rescued mutant flies with a transgene carrying a wild-type copy of Toll-7 (Fig. 7D and E). In addition, flies carrying two other null alleles of Toll-7 (p8 and p114) (36) also resisted VSV infection, similar to control flies (Fig. 8A and B). We noted that the original study showed increased sensitivity of Toll-7 mutants to a recombinant VSV-GFP virus, whereas we used a wild-type VSV (Indiana strain); this could help explain the differences we observed. However, we obtained similar results using a recombinant VSV-GFP in a separate set of experiments (Fig. 8C and D). We conclude that Toll-7 does not participate in the autophagy and hemocyte-mediated host defense against VSV infection.
FIG 7.
Toll7g1-1 mutant flies are not sensitive to VSV infection. (A) Schematic representation of the Toll-7 gene and the deletion mutants evaluated. The coding region is highlighted in orange, and deletions are indicated by the gaps in the blue lines. For the Toll-7g1-1 allele, a stop codon replaces a glutamine at position 1040. (B) Survival of Toll-7g1-1 null mutant and control flies after VSV infection. Flies carrying a null allele of Toll-7 or controls from the same genetic background (yw; Toll-7g1-1 and y1 w1) were crossed with a line containing a deficiency covering the Toll-7 gene [Df(2R)BSC22] and hemizygous flies from the progeny were injected with conditioned medium (CM) or VSV. Dcr-2 null mutant (Dcr-2L811fsX) and WT flies crossed with the deficient Df(2R)BSC45 flies were used as positive controls. (C) Quantitative RT-PCR analysis of the accumulation of VSV RNA on the indicated day postinfection in control flies [WT/Df(2R)BSC22 and WT/Df(2R)BSC45], Toll-7 null mutant flies [Toll-7g1-1/Df(2R)BSC22], and Dcr-2 mutant flies [Dcr-2L811fsX/Df(2R)BSC45]. (D and E) Quantitative RT-PCR analysis of Toll-7 mRNA expression (D) and VSV viral RNA accumulation (E) in control [WT/Df(2R)BSC22], Toll-7 null mutant [Toll-7g1-1/Df(2R)BSC22], and Toll-7 rescue [Toll-7g1-1/Df(2R)BSC22; +/gToll-7] flies at 6 dpi. Data represent means ± standard errors (B) or SD (C, D, and E) of three independent experiments, each containing three groups of 10 (B) or 6 (C, D, and E) flies. ns, not significant; *, P < 0.05; ***, P < 0.001 (log rank [B] or unpaired t test [C, D, and E]).
FIG 8.
Toll-7 is not required to control VSV infection. (A and C) Survival of Toll-7 mutant and control flies after VSV (A) or VSV-GFP (C) infection. Flies carrying a null allele of Toll-7 or controls from the same genetic background (w1118; Toll-7p8 or Toll-7p114 and w1118 [A] and yw; Toll-7g1-1 and y1 w1 [C] were crossed with a line containing a deficiency covering the Toll-7 gene [Df(2R)BSC22] and hemizygous flies from the progeny were injected with conditioned medium (CM) or VSV (A) or VSV-GFP (C). (B and D) Quantitative RT-PCR analysis of the accumulation of VSV RNA in control and the indicated Toll-7 mutant flies injected with VSV (B) or VSV-GFP (D) at the indicated day postinfection. Data represent means ± standard errors (A and C) or SD (B and D) of three independent experiments, each containing three groups of 10 (A and C), 6 (B), or 5 (D) individual flies. ns, not significant (log rank [A] or unpaired t test [B and D]).
DISCUSSION
We investigated the involvement of hemocytes and autophagy in the resistance to a panel of six viruses representative of different families of RNA and DNA viruses. Our data revealed that (i) unlike RNAi, which acts via a broad antiviral pathway, blood cells play a critical role only in defense against certain viruses, such as CrPV, FHV, and VSV; (ii) CrPV and FHV induce apoptosis of infected cells that likely act in concert with hemocytes to control these viruses; (iii) autophagy contributes to the containment of VSV infection together with hemocytes, but it does not seem to be important for the other viruses tested.
How can hemocytes sense infected cells and control viral infections in flies? FHV, CrPV, and VSV belong to different virus families and do not share obvious features that would explain why hemocytes are required to control them. However, FHV and CrPV trigger surface exposure of phosphatidylserine, suggesting that plasmatocytes clear apoptotic cells containing infectious particles. Recently, Nakanishi and colleagues reported a role for hemocytes in the control of DCV infection. They demonstrated that DCV infection triggers activation of effector caspases, phosphatidylserine exposure, and efferocytosis of the dying cells that is mediated by the receptors integrin βν and Draper (24). Curiously, in our hands blood cells were not required to control DCV infection in vivo. This virus induced exposure of phosphatidylserine and cell death in tissue culture S2 cells, but at lower rates than FHV and CrPV. Of note, DCV is less virulent than the two other viruses, possibly because its suppressor of RNAi is less potent (43). This could account for a threshold difference for the induction of apoptosis. In this regard, the difference between the two studies may have been due to the high dose of DCV used by Nainu et al. (up to 80,000 50% tissue culture infective doses [TCID50] per fly, versus a dose of ∼800 TCID50 in our study).
Autophagy may be used to combat infection by intracellular pathogens, such as viruses, by isolating them from the cytosol through an isolation membrane and targeting them to lysosomes, where they can be degraded. The role of autophagy in the control of viral infection is complex, since many RNA viruses hijack the autophagy machinery to generate the network of intracellular membranes that will nest their replication sites (44). The complexity of the relationship between viruses and autophagy is well illustrated by the case of measles virus, in which a first wave of autophagy, triggered during viral entry, is antiviral, whereas a second wave of autophagy is proviral (45, 46). Our data indicate that only two of the six viruses tested are affected by mutation of the essential autophagy gene Atg7. The first is FHV, for which autophagy is proviral, as shown by the reduced viral titer and increased survival of Atg7 mutant flies. Interestingly, FHV replicates on the outer membrane of mitochondria, and an important and well-characterized role of autophagy is the regulation of the turnover of mitochondria (mitophagy) (47). We propose that efficient removal of damaged mitochondria, which may not fully support the activity of the viral polymerase, contributes to the success of FHV replication. The second virus affected by autophagy is VSV, for which we observed an increase in replication by Atg7 mutants, as previously reported (22). This increase is, however, modest (3-fold increase), especially compared to that in RNAi-deficient flies (>50-fold increase). Therefore, we conclude that autophagy does not represent a major pathway of antiviral defense in Drosophila.
A previous study reported the provocative finding that one of the nine Toll receptors encoded in the Drosophila genome, Toll-7, senses VSV infection and triggers antiviral autophagy (42). However, our genetic studies using three different well-characterized alleles of Toll-7 do not support this initial observation. Although the sizes of the families of Toll receptors in Drosophila and mammals are similar, with about 10 members, phylogenetic analysis has clearly indicated that Toll receptors evolved independently in different animal phyla (48, 49). In Drosophila, only Toll itself has so far been shown to participate in the induction of an immune response in flies, although Toll-8 acts as a negative regulator of antimicrobial defenses in the respiratory tract (50–53). Drosophila Toll receptors are highly expressed during embryogenesis, pointing to developmental functions (49, 54). Indeed, a subset of Toll receptors, including Toll-7, function as neurotrophin receptors and are activated by members of the Spaetzle family in the developing nervous system (36, 55). Drosophila Toll receptors were also recently shown to function as adhesion molecules during elongation of the antero-posterior axis of the embryo (56). In contrast, experimental evidence that members of the Toll family other than Toll itself participate in the activation of innate immunity in Drosophila is still lacking.
Supplementary Material
ACKNOWLEDGMENTS
We thank François Leulier for Hml(Δ)-gal4, uas-Bax flies, Thomas P. Neufeld for Atg7d14, Atg7d77 and CG5335d30 mutant flies, Tony Ip for Toll7g1-1 mutant flies, and Alicia Hidalgo for Toll7p8 and Toll7p114 mutant flies. Expert technical assistance from Estelle Santiago and Stefanie Pietsch is gratefully acknowledged.
O.L., C.M., J.T.M., and J.-L.I. designed the experiments; O.L., J.A., I.J.D.S.D.F., and R.P.O. performed the experiments; O.L., J.A., I.J.D.S.D.F., R.P.O., F.B., C.M., J.A.H., J.T.M., and J.-L.I. analyzed the data; and O.L., J.A.H., J.T.M., and J.-L.I. wrote the manuscript.
This work was supported by CNRS, the University of Strasbourg Institute for Advanced Study, and grants from the NIH (PO1 AI070167) and ANR (ANR-11-ISV3-002 and ANR-13-BSV3-0009) and Investissements d'Avenir programs (NetRNA ANR-10-LABX-36 and I2MC ANR-11-EQPX-0022). I.J.D.S.D.F. and R.P.O. were supported by fellowships from CNPq and CAPES. J.T.M. was supported by grants from CAPES, CNPq, and FAPEMIG.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00238-16.
REFERENCES
- 1.Bronkhorst AW, van Cleef KWR, Vodovar N, Ince IA, Blanc H, Vlak JM, Saleh M-C, van Rij RP. 2012. The DNA virus Invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc Natl Acad Sci U S A 109:E3604–E3613. doi: 10.1073/pnas.1207213109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler J-L. 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat Immunol 7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 3.Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, Dostert C, Troxler L, Hetru C, Meignin C, Pfeffer S, Hoffmann JA, Imler J-L. 2013. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol 190:650–658. doi: 10.4049/jimmunol.1102486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller S, Gausson V, Vodovar N, Deddouche S, Troxler L, Perot J, Pfeffer S, Hoffmann JA, Saleh M-C, Imler J-L. 2010. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc Natl Acad Sci U S A 107:19390–19395. doi: 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Rij RP, Saleh M-C, Berry B, Foo C, Houk A, Antoniewski C, Andino R. 2006. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X-H, Aliyari R, Li W-X, Li H-W, Kim K, Carthew R, Atkinson P, Ding S-W. 2006. RNA interference directs innate immunity against viruses in adult Drosophila. Science 312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aliyari R, Wu Q, Li H-W, Wang X-H, Li F, Green LD, Han CS, Li W-X, Ding S-W. 2008. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marques JT, Wang J-P, Wang X, de Oliveira KPV, Gao C, Aguiar ERGR, Jafari N, Carthew RW. 2013. Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS Pathog 9:e1003579. doi: 10.1371/journal.ppat.1003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguiar ERGR, Olmo RP, Paro S, Ferreira FV, de Faria IJ DS, Todjro YMH, Lobo FP, Kroon EG, Meignin C, Gatherer D, Imler J-L, Marques JT. 2015. Sequence-independent characterization of viruses based on the pattern of viral small RNAs produced by the host. Nucleic Acids Res 43:6191–6206. doi: 10.1093/nar/gkv587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Mierlo JT, Overheul GJ, Obadia B, van Cleef KWR, Webster CL, Saleh M-C, Obbard DJ, van Rij RP. 2014. Novel Drosophila viruses encode host-specific suppressors of RNAi. PLoS Pathog 10:e1004256. doi: 10.1371/journal.ppat.1004256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Y-H, Luo Y-J, Wu Q, Jovel J, Wang X-H, Aliyari R, Han C, Li W-X, Ding S-W. 2011. RNA-based immunity terminates viral infection in adult Drosophila in the absence of viral suppression of RNA interference: characterization of viral small interfering RNA populations in wild-type and mutant flies. J Virol 85:13153–13163. doi: 10.1128/JVI.05518-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bronkhorst AW, van Rij RP. 2014. The long and short of antiviral defense: small RNA-based immunity in insects. Curr Opin Virol 7:19–28. doi: 10.1016/j.coviro.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. 2008. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc Natl Acad Sci U S A 105:19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sánchez-Vargas I, Scott JC, Poole-Smith BK, Franz AWE, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. 2009. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito's RNA interference pathway. PLoS Pathog 5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. 2012. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog 8:e1002470. doi: 10.1371/journal.ppat.1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vodovar N, Bronkhorst AW, van Cleef KWR, Miesen P, Blanc H, van Rij RP, Saleh M-C. 2012. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One 7:e30861. doi: 10.1371/journal.pone.0030861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Léger P, Lara E, Jagla B, Sismeiro O, Mansuroglu Z, Coppée JY, Bonnefoy E, Bouloy M. 2013. Dicer-2- and Piwi-mediated RNA interference in Rift Valley fever virus-infected mosquito cells. J Virol 87:1631–1648. doi: 10.1128/JVI.02795-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siu RWC, Fragkoudis R, Simmonds P, Donald CL, Chase-Topping ME, Barry G, Attarzadeh-Yazdi G, Rodriguez-Andres J, Nash AA, Merits A, Fazakerley JK, Kohl A. 2011. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J Virol 85:2907–2917. doi: 10.1128/JVI.02052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingsolver MB, Huang Z, Hardy RW. 2013. Insect antiviral innate immunity: pathways, effectors, and connections. J Mol Biol 425:4921–4936. doi: 10.1016/j.jmb.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamiable O, Imler J-L. 2014. Induced antiviral innate immunity in Drosophila. Curr Opin Microbiol 20C:62–68. doi: 10.1016/j.mib.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler J-L. 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol 6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 22.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. 2009. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B, Behura SK, Clem RJ, Schneemann A, Becnel J, Severson DW, Zhou L. 2013. P53-mediated rapid induction of apoptosis conveys resistance to viral infection in Drosophila melanogaster. PLoS Pathog 9:e1003137. doi: 10.1371/journal.ppat.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nainu F, Tanaka Y, Shiratsuchi A, Nakanishi Y. 2015. Protection of insects against viral infection by apoptosis-dependent phagocytosis. J Immunol 195:5696–5706. doi: 10.4049/jimmunol.1500613. [DOI] [PubMed] [Google Scholar]
- 25.Clem RJ. 12 February 2016. Arboviruses and apoptosis: the role of cell death in determining vector competence. J Gen Virol doi: 10.1099/jgv.0.000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanot R, Zachary D, Holder F, Meister M. 2001. Postembryonic hematopoiesis in Drosophila. Dev Biol 230:243–257. doi: 10.1006/dbio.2000.0123. [DOI] [PubMed] [Google Scholar]
- 27.Evans CJ, Hartenstein V, Banerjee U. 2003. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev Cell 5:673–690. doi: 10.1016/S1534-5807(03)00335-6. [DOI] [PubMed] [Google Scholar]
- 28.Franc NC, Heitzler P, Ezekowitz RA, White K. 1999. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science 284:1991–1994. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- 29.Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, Strom C, Conto SL, Hetru C, Stuart LM, Stehle T, Hoffmann JA, Reichhart J-M, Ferrandon D, Rämet M, Ezekowitz RAB. 2005. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123:335–346. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 30.Binggeli O, Neyen C, Poidevin M, Lemaitre B. 2014. Prophenoloxidase activation is required for survival to microbial infections in Drosophila. PLoS Pathog 10:e1004067. doi: 10.1371/journal.ppat.1004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh S, Singh A, Mandal S, Mandal L. 2015. Active hematopoietic hubs in Drosophila adults generate hemocytes and contribute to immune response. Dev Cell 33:478–488. doi: 10.1016/j.devcel.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Kronhamn J, Ekström J-O, Korkut GG, Hultmark D. 2015. JAK/STAT signaling in Drosophila muscles controls the cellular immune response against parasitoid infection. EMBO Rep 16:1664–1672. doi: 10.15252/embr.201540277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juhász G, Erdi B, Sass M, Neufeld TP. 2007. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev 21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Defaye A, Evans I, Crozatier M, Wood W, Lemaitre B, Leulier F. 2009. Genetic ablation of Drosophila phagocytes reveals their contribution to both development and resistance to bacterial infection. J Innate Immun 1:322–334. doi: 10.1159/000210264. [DOI] [PubMed] [Google Scholar]
- 35.Yagi Y, Nishida Y, Ip YT. 2010. Functional analysis of Toll-related genes in Drosophila. Dev Growth Differ 52:771–783. doi: 10.1111/j.1440-169X.2010.01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McIlroy G, Foldi I, Aurikko J, Wentzell JS, Lim MA, Fenton JC, Gay NJ, Hidalgo A. 2013. Toll-6 and Toll-7 function as neurotrophin receptors in the Drosophila melanogaster CNS. Nat Neurosci 16:1248–1256. doi: 10.1038/nn.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elrod-Erickson M, Mishra S, Schneider D. 2000. Interactions between the cellular and humoral immune responses in Drosophila. Curr Biol 10:781–784. doi: 10.1016/S0960-9822(00)00569-8. [DOI] [PubMed] [Google Scholar]
- 38.Rutschmann S, Kilinc A, Ferrandon D. 2002. Cutting edge: the toll pathway is required for resistance to gram-positive bacterial infections in Drosophila. J Immunol 168:1542–1546. doi: 10.4049/jimmunol.168.4.1542. [DOI] [PubMed] [Google Scholar]
- 39.Costa A, Jan E, Sarnow P, Schneider D. 2009. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS One 4:e7436. doi: 10.1371/journal.pone.0007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Settles EW, Friesen PD. 2008. Flock house virus induces apoptosis by depletion of Drosophila inhibitor-of-apoptosis protein DIAP1. J Virol 82:1378–1388. doi: 10.1128/JVI.01941-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. 2007. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 42.Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, Gold B, Cherry S. 2012. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity 36:658–667. doi: 10.1016/j.immuni.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nayak A, Berry B, Tassetto M, Kunitomi M, Acevedo A, Deng C, Kruchinsky A, Gross J, Antoniewski C, Andino R. 2010. Cricket paralysis virus (CrPV) antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol 17:547–554. doi: 10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deretic V, Levine B. 2009. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joubert P-E, Meiffren G, Grégoire IP, Pontini G, Richetta C, Flacher M, Azocar O, Vidalain P-O, Vidal M, Lotteau V, Codogno P, Rabourdin-Combe C, Faure M. 2009. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe 6:354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Grégoire IP, Richetta C, Meyniel-Schicklin L, Borel S, Pradezynski F, Diaz O, Deloire A, Azocar O, Baguet J, Le Breton M, Mangeot PE, Navratil V, Joubert P-E, Flacher M, Vidalain P-O, André P, Lotteau V, Biard-Piechaczyk M, Rabourdin-Combe C, Faure M. 2011. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog 7:e1002422. doi: 10.1371/journal.ppat.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol 5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Imler J-L, Zheng L. 2004. Biology of Toll receptors: lessons from insects and mammals. J Leukoc Biol 75:18–26. doi: 10.1189/jlb.0403160. [DOI] [PubMed] [Google Scholar]
- 49.Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. 2000. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci U S A 97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. 1996. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86:973–983. doi: 10.1016/S0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 51.Narbonne-Reveau K, Charroux B, Royet J. 2011. Lack of an antibacterial response defect in Drosophila Toll-9 mutant. PLoS One 6:e17470. doi: 10.1371/journal.pone.0017470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akhouayri I, Turc C, Royet J, Charroux B. 2011. Toll-8/Tollo negatively regulates antimicrobial response in the Drosophila respiratory epithelium. PLoS Pathog 7:e1002319. doi: 10.1371/journal.ppat.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ligoxygakis P, Bulet P, Reichhart J-M. 2002. Critical evaluation of the role of the Toll-like receptor 18-Wheeler in the host defense of Drosophila. EMBO Rep 3:666–673. doi: 10.1093/embo-reports/kvf130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kambris Z, Hoffmann JA, Imler J-L, Capovilla M. 2002. Tissue and stage-specific expression of the Tolls in Drosophila embryos. Gene Expr Patterns 2:311–317. doi: 10.1016/S1567-133X(02)00020-0. [DOI] [PubMed] [Google Scholar]
- 55.Ballard SL, Miller DL, Ganetzky B. 2014. Retrograde neurotrophin signaling through Tollo regulates synaptic growth in Drosophila. J Cell Biol 204:1157–1172. doi: 10.1083/jcb.201308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paré AC, Vichas A, Fincher CT, Mirman Z, Farrell DL, Mainieri A, Zallen JA. 2014. A positional Toll receptor code directs convergent extension in Drosophila. Nature 515:523–527. doi: 10.1038/nature13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.