ABSTRACT
Herpes simplex virus (HSV) replicates in the skin and mucous membranes, and initiates lytic or latent infections in sensory neurons. Assembly of progeny virions depends on the essential large tegument protein pUL36 of 3,164 amino acid residues that links the capsids to the tegument proteins pUL37 and VP16. Of the 32 tryptophans of HSV-1-pUL36, the tryptophan-acidic motifs 1766WD1767 and 1862WE1863 are conserved in all HSV-1 and HSV-2 isolates. Here, we characterized the role of these motifs in the HSV life cycle since the rare tryptophans often have unique roles in protein function due to their large hydrophobic surface. The infectivity of the mutants HSV-1(17+)Lox-pUL36-WD/AA-WE/AA and HSV-1(17+)Lox-CheVP26-pUL36-WD/AA-WE/AA, in which the capsid has been tagged with the fluorescent protein Cherry, was significantly reduced. Quantitative electron microscopy shows that there were a larger number of cytosolic capsids and fewer enveloped virions compared to their respective parental strains, indicating a severe impairment in secondary capsid envelopment. The capsids of the mutant viruses accumulated in the perinuclear region around the microtubule-organizing center and were not dispersed to the cell periphery but still acquired the inner tegument proteins pUL36 and pUL37. Furthermore, cytoplasmic capsids colocalized with tegument protein VP16 and, to some extent, with tegument protein VP22 but not with the envelope glycoprotein gD. These results indicate that the unique conserved tryptophan-acidic motifs in the central region of pUL36 are required for efficient targeting of progeny capsids to the membranes of secondary capsid envelopment and for efficient virion assembly.
IMPORTANCE Herpesvirus infections give rise to severe animal and human diseases, especially in young, immunocompromised, and elderly individuals. The structural hallmark of herpesvirus virions is the tegument, which contains evolutionarily conserved proteins that are essential for several stages of the herpesvirus life cycle. Here we characterized two conserved tryptophan-acidic motifs in the central region of the large tegument protein pUL36 of herpes simplex virus. When we mutated these motifs, secondary envelopment of cytosolic capsids and the production of infectious particles were severely impaired. Our data suggest that pUL36 and its homologs in other herpesviruses, and in particular such tryptophan-acidic motifs, could provide attractive targets for the development of novel drugs to prevent herpesvirus assembly and spread.
INTRODUCTION
The diversity of clinical herpesvirus isolates is shaped to a large extent by the long coevolution between virus and host as well as by homologous recombination among different strains (1–4). The subfamilies of the Herpesviridae are characterized by similar properties: for example, fast replication of members of the Alphaherpesvirinae in keratinocytes, epithelial cells, or fibroblasts and latency in sensory neurons. Infections with the herpes simplex viruses (HSV-1 or HSV-2) or varicella-zoster virus occur at young age, and besides the latent genomes in the neuronal nuclei, the viruses are mostly cleared from the host. Primary infections as well as reactivations from latency cause many diseases, such as herpes encephalitis, herpes neonatorum, or postherpetic neuralgia, particularly in immunocompromised patients (5–7).
Herpesvirus virions are enveloped, and their large DNA genomes are enclosed in capsids with a diameter of 125 nm. The tegument constitutes their most complex structure, and in HSV-1 occupies about two-thirds of the virion volume in an asymmetric crescent shape between the capsid and the envelope (8). Tegument proteins modulate nuclear and cytoplasmic viral and host functions and are incorporated into virions in a complex repertoire and with various stoichiometries (9, 10). Assembly of herpesviruses commences with nuclear procapsids that package viral genomes and mature into icosahedral capsids that diffuse to the inner nuclear membrane for primary budding (9, 11, 12). The primary envelopes fuse with the outer nuclear membrane and release the capsids into the cytosol, where they associate with inner tegument proteins: e.g., pUL36 and pUL37 of the Alphaherpesvirinae (10, 13, 14). Cytosolic capsids are transported along microtubules to the cytoplasmic organelles of secondary envelopment (15, 16; K. Döhner, A. Buch, L. Ivanova, A. Binz, A. Pohlmannn, M. Capucci, M. Sandbaumhüter, B. Sodeik, and R. Bauerfeind, submitted for publication).
The viral envelope and outer tegument proteins accumulate on cytoplasmic membranes which are accessible to endocytic tracers and harbor marker proteins of the trans-Golgi-network (10, 17–20). Here, the membranes with the envelope proteins and associated host and viral proteins wrap around cytosolic capsids in a process called secondary envelopment which results in the formation of transport vesicles that in the end fuse with the plasma membrane to release virions (9–11). Neighboring cells are infected by fusion of the viral envelopes with the plasma membrane or the limiting membranes of macropinosomes or endosomes (21–24). While at least some inner tegument proteins (e.g., pUL36 and pUL37), remain associated with the incoming capsids, envelope and outer tegument proteins dissociate (15, 25–28). The capsids again utilize microtubules for transport to the nucleus, where they dock at the nuclear pores to release the viral genomes into the nucleoplasm for viral transcription and replication (29–32).
The evolutionarily conserved large tegument protein HSV-1 pUL36 and its orthologs are expressed mainly after genome replication and contribute to various steps of the viral life cycle (33–35). The full-length version of HSV-1 pUL36 consists of 3,164 amino acid residues (c.f. Figure 1A). In cells infected with mutants of HSV-1 or of pseudorabies virus (PRV), a porcine alphaherpesvirus, that lack pUL36 or express severely truncated versions of pUL36, capsid assembly and nuclear egress into the cytosol proceed normally, but there is no secondary envelopment, and no infectious virions are formed (26, 28, 36–38). In addition to its function in targeting the capsids to cytoplasmic membranes for assembly, pUL36 is also crucial for capsid binding to the nuclear pore complexes during cell entry (29, 39, 40). An HSV-1(17+) mutant lacking only the C-terminal 167 residues of pUL36 assembles virions of regular morphology that also enter neighboring cells, but the incoming capsids are not targeted to the nuclear envelopes (26).
FIG 1.
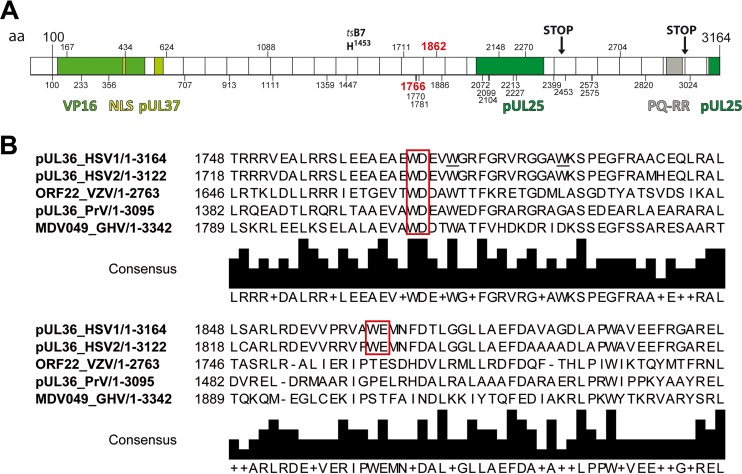
Primary structure of HSV-1-pUL36. (A) Primary structure of pUL36 and location of important point mutations, truncations, interaction domains, and tryptophans. The N-terminal third of pUL36 contains an ubiquitin-specific cysteine protease (USP) with a conserved catalytic residue cysteine at position 65, a nuclear localization signal spanning residues 426 to 432, binding sites for the transcriptional activator VP16, and the inner tegument protein pUL37 (35, 41, 42, 44–46, 48, 49, 81, 109). The tsB7 point mutation Y1453H prevents genome uncoating of incoming capsids at the nuclear pores at the nonpermissive temperature (29, 110, 111). The C-terminal region of pUL36 contains two binding sites for the capsid protein pUL25 and a PQ repeat-rich region (26, 40, 112). HSV-1-pUL36 truncation mutants lacking the C-terminal 735 residues (STOP at position 2430) do not associate with capsids, whereas the 167 C-terminal residues are required during early steps of infection (STOP at position 2998) (26). The positions of 32 tryptophan residues in HSV-1-pUL36 are indicated by vertical bars and the respective numbers. W1766 and W1862 (red) were mutated in the present study. (B) Alignments around conserved W motifs. HSV-1-pUL36 was aligned with its orthologs HSV2-pUL36, VZV-ORF22, PRV-pUL36 and GHV-MDV049 using UniProtKB and Clustal Omega. The tryptophan-acidic motifs are indicated by red boxes.
The N-terminal 533 residues of HSV-1-pUL36 and its orthologs encode a ubiquitin-specific protease (41, 42). Furthermore, a fragment of residues 1 to 767 interacts with tumor susceptibility gene 101, a component of the ESCRT complexes which regulate endosomal sorting (43). Residues 124 to 511 of HSV-1-pUL36 are required for interaction with the transcriptional activator VP16 and residues 548 to 572 for binding to the inner tegument protein pUL37 (35, 37, 44–47). The interaction with VP16 contributes to efficient virion assembly but is not essential per se, while deletion of the pUL37-binding site abrogates virus formation (37, 44, 45). HSV-1-pUL36 contains a conserved nuclear localization signal at residues 426 to 432 that is essential for targeting incoming HSV-1 capsids to the nuclear pores but not for virion formation (48, 49).
While Scrima et al. have solved the first crystal structure for HSV-1 pUL36, comprising residues 1625 to 1758, which consists mainly of α-helices (50), no functional domains have yet been assigned to the central third of the large tegument proteins (c.f. Fig. 1A). The most C-terminal 60 residues of HSV-1 pUL36 and PRV pUL36 encode a binding domain for pUL25 that forms a complex with pUL17 on the capsid surface around the vertices, and the pUL36-pUL25-pUL17 structures contribute to the capsid vertex-specific components (40, 51, 52). However, an HSV-1 pUL36 of residues 1 to 2998 but lacking the C-terminal 167 residues still associates with capsids during assembly, whereas one with a larger C-terminal truncation of 735 residues does not (26). This observation may be related to a second pUL25 binding site that has been identified among residues 2037 to 2353 (40). Interestingly, capsids recruit pUL36 of residues 1 to 2894 that includes this pUL25 binding site during assembly but do not hold onto it during nuclear targeting upon cell entry (26). Superresolution fluorescence microscopy of extracellular HSV-1 virions localizes pUL36 in close proximity to the capsids, whereas pUL37 and VP16 are shifted toward the envelope consistent with pUL36's proposed structural linker function (53).
Hot spots on protein surfaces are specific amino acid peptide motifs that contribute more than others to the binding affinity of protein-protein interaction interfaces (54–56). The composition of hot spots is distinctive and not random, with tryptophans (W) having the highest frequency; when tryptophan with its large hydrophobic surface is mutated to alanine (A), the size difference of these residues generates a cavity that creates a local destabilization (54, 57, 58). There are 32 tryptophans among the 3,164 residues of HSV-1 pUL36. To begin to understand their potential functional roles, we have replaced conserved central tryptophan-acidic motifs by alanine, namely, 1766WD/AA1767 or 1862WE/AA1863. Our data show that we have identified novel viral determinants that are required for efficient secondary envelopment and virion formation. These motifs could either contribute to pUL36 self-interactions required for proper folding of the central part of pUL36, mediate important interactions with other structural HSV-1 proteins, or bind to host factors facilitating the secondary envelopment of tegumented cytosolic capsids with cytoplasmic membranes harboring the HSV-1 envelope proteins.
MATERIALS AND METHODS
Alignments.
We used the website Universal Protein Knowledgebase curated by the public research organization UniProtKB (http://www.uniprot.org/ [accessed October 2015]) and the entries for HSV-1-pUL36 (P10220), HSV2-pUL36 (P89459), VZV-ORF22 (P09278), PrV-pUL36 (G3G960), GHV-MDV049 (Q9E6N3), HCMV-pUL48 (P16785), HHV6-pU31 (P52340), HHV7-pU31 (P52362), EBV-BPLF1 (P03186), KSHV-ORF64 (Q2HR64), and MHV-68-ORF64 (O41965) to align the primary sequence of HSV-1-pUL36 and its homologs among the Alpha-, Beta-, and Gammaherpesvirinae.
Cells.
Vero cells (ATCC CCL-81) and Vero-HS30 cells were cultured in minimal essential medium (MEM) Eagle (CytoGen, Wetzlar, Germany) supplemented with 7.5% (vol/vol) fetal calf serum (FCS) (Life Technologies, Darmstadt, Germany). Vero-HS30 cells provide HSV-1 pUL36 in trans for complementation and contain the UL36 gene under the control of its authentic promoter together with 2,100 kb upstream and 474 kb downstream flanking sequences and a neomycin resistance gene (36); 500 μg/ml G418 was added to their culture medium in every 4th passage (PAA Laboratories GmbH, Pasching, Austria).
Antibodies.
We used mouse monoclonal antibody (MAb) 3B6 (Virusys Corporation, Taneytown, MD) for detection of capsid protein VP5, MAb DL6 for glycoprotein D (59), MAb 1501 for actin (Millipore, Billerica, MA), MAb 5F8 for mCherry (ChromoTek, Hauppauge, NY), and rabbit polyclonal antibodies (pAbs) raised against residues 95 to 112 of VP26 (60), against a pUL36 fragment of residues 1408 to 2112 (pAb 147 [26]), or a peptide of residues 936 to 951 (pAb 3; Eurogentec, Seraing, Belgium), against full-length glutathione S-transferase (GST)-tagged pUL37 (61), against VP16 (631209; BD Biosciences, NJ), pAb AGV30 against a GST-VP22 fusion protein (62), and pAb Romulus V raised against numerous HSV-1 structural proteins (63, 64). Secondary goat anti-rabbit and anti-mouse antibodies for fluorescence microscopy were cross-adsorbed against other species and coupled to Alexa Fluor 488 or Alexa Fluor 546 (Life Technologies, Darmstadt, Germany) and for immunoblotting coupled to infrared fluorescent dyes with emission wavelengths between 680 and 800 nm (LI-COR Biosciences, Lincoln, NE).
Ethics statement.
Sera of healthy, HSV-1-seronegative volunteers were obtained after written informed consent from blood donors. Permission was granted by the Internal Review Board of Hannover Medical School (approval no. 893).
HSV-1 strains and construction of mutants.
The strains HSV-1(17+)Lox and HSV-1(17+)Lox-ΔUL36 as well as HSV-1(17+)Lox-CheVP26 and HSV-1(17+)Lox-CheVP26-ΔUL36, in which the first 7 N-terminal residues of the small capsid protein VP26 have been replaced by the fluorescent protein monomeric Cherry (mCherry), have already been characterized (15, 26, 65). All pHSV-1(17+)Lox bacterial artificial chromosome (BAC) plasmids contain a floxed BAC cassette, inserted between UL22 and UL23, and containing bacterial genes for replication and maintenance in Escherichia coli, a chloramphenicol resistance gene and a Cre recombinase gene (15, 65–67). In eukaryotic cells, Cre recombinase catalyzes the recombination between LoxP sites, and thus excises the BAC cassette; the resulting HSV-1(17+)Lox strains contain one LoxP site between UL22 and UL23 and lack the OriL (67). To construct mutants harboring point mutations in the tryptophan-acidic motifs, the kanamycin resistance cassette was amplified from pEP-Kan-S2 and used as positive selection marker for two recombination rounds to perform a traceless replacement of specific amino acid residues (68). For quality control, we used seven different restriction enzymes to obtain fragments distributed uniformly over the viral genome and sequencing. To reconstitute the novel HSV-1(17+)Lox mutants, we transfected BAC DNA purified using the NucleoBond BAC 100 kit according to the manufacturer's instructions (Macherey & Nagel, Düren, Germany) into eukaryotic cells (67). Parental Vero or Vero-HS30 cells were cultured to 80 to 85% confluence, transfected with 10 μg/60-mm dish of BAC DNA (MBS Mammalian transfection kit, Stratagene, CA), and cultured until cytopathic effects developed. To obtain virus of passage one, the cells underwent three cycles of freeze-thawing, and the released virus was used to generate the next passage. The mutant viruses were propagated and titrated in the complementing Vero-HS30 and the parental strains in Vero cells (26). The parental strains HSV-1(17+)Lox and HSV-1(17+)Lox-CheVP26 had genome-to-PFU ratios of about 30, and the mutants HSV-1(17+)Lox-pUL36-WD/AA-WE/AA, HSV-1(17+)Lox-CheVP26-pUL36-WD/AA-WE/AA, and HSV-1(17+)Lox-CheVP26-ΔUL36 had genome-to-PFU ratios of below 100, after DNase treatment, indicating a low concentration of defective particles (64). All experiments were conducted with virus particles sedimented from the culture supernatant of infected cells (15, 26).
Plaque assays and growth curves.
Confluent Vero or Vero-HS30 cells were inoculated with serial dilutions of the respective virus preparations for 1 h at room temperature and then cultured for 2 days in medium containing 20 μg/ml human pooled IgGs containing neutralizing antibodies (Sigma-Aldrich, Schnelldorf, Germany). The cells were fixed in water-free methanol at −20°C, and the number of plaques was determined (69). The plaque sizes were measured (software ImageJ version 1.49k; Wayne Rasband, NIH) after labeling with pAb Romulus V and secondary antibodies conjugated to Alexa Fluor 488 and documentation with an inverted microscope (AxioObserver Z1, ZEISS Göttingen, Germany) equipped with a digital camera (Axiocam HRm) and controlled by the Axiovision software (version 4.8.2). Subconfluent Vero cells were inoculated at a multiplicity of infection (MOI) of 5 PFU/cell, corresponding to 1 × 107 PFU/ml, and incubated for 1 h on a rocking platform at room temperature. After 6, 12, 18, 24, or 30 hours postinfection (hpi), the supernatant and the cells were harvested. The intracellular virus was released by three cycles of freeze-thawing. Intra- and extracellular virus yields were titrated in duplicates.
Synchronous HSV-1 infection.
Vero or Vero-HS30 cells were infected synchronously as previously described (15, 26, 64, 70). Briefly, cells were seeded at a concentration of 5 × 104/cm2 for about 14 h, inoculated with an MOI of 10 PFU/cell corresponding to 5 × 106 PFU/ml, and incubated for 2 h on ice on a rocking platform to facilitate virus binding. Virus internalization was initiated by adding growth medium and shifting to 37°C. After 1 h, residual extracellular but not internalized virions were inactivated by a short treatment in 40 mM citrate at pH 3.0 with 135 mM NaCl and 10 mM KCl (71, 72), and the cells were further incubated at 37°C and 5% CO2.
Immunoblotting.
Vero and Vero-HS30 cells were infected synchronously for 8, 10, or 12 h and lysed in a sample buffer (50 mM Tris-HCl [pH 6.8], 1% [wt/vol] SDS, 1% [vol/vol] β-mercaptoethanol, 5% [vol/vol] glycerol, bromphenol blue) at 95°C and containing protease inhibitors AEL (aprotinin, E-64, and leupeptin), ABP (antipain, bestatin, pepstatin), and phenylmethylsulfonyl fluoride (PMSF). The cell lysates were maintained at 95°C for 5 min and loaded onto linear 5 to 15% gradient SDS-PAGE gels. The proteins were transferred to nitrocellulose membranes that were blocked in 5% (wt/vol) low-fat milk in phosphate-buffered saline (PBS) and probed overnight with primary antibodies at 4°C. After incubation with secondary antibodies (IRDye 800CW or IRDye 680RD) at room temperature (RT) for 1 h, the membranes were imaged with a digital camera (Odyssey infrared imaging system; LI-COR Biosciences, Lincoln, NE).
Electron microscopy.
Vero cells were seeded onto coverslips, infected synchronously, and fixed at 10 or 14 hpi with 2% (wt/vol) glutaraldehyde (Polysciences Inc., Warrington, PA) in 130 mM cacodylate buffer at pH 7.4 supplemented with 2 mM CaCl2 and 10 mM MgCl2 as previously described (15, 26, 64, 70). The specimens were incubated for 1 h with 1% (wt/vol) OsO4 (Polysciences) in 165 mM cacodylate buffer containing 1.5% (wt/vol) K3Fe(CN)6 followed by 0.5% (wt/vol) uranyl acetate (Agar Scientific, Essex, United Kingdom) overnight at 4°C (73) and embedded in situ in Epon stubs (Serva Electrophoresis GmbH, Heidelberg, Germany). After removal of the coverslips, ultrathin sections of 50 nm were cut parallel to the substrate (30, 74) and further contrasted with lead citrate (75) and uranyl acetate (76). Images were taken with Morgani or Tecnai G2 T20 electron microscopes (FEI, Eindhoven, The Netherlands) and further processed using ImageJ (version 1.49k) as reported before (15, 26, 31, 44, 77). For quantitation, random images of nonselected cells from the respective samples were taken at a low magnification (6,000×). To image an entire nucleoplasm cross section or an entire cytoplasm cross section of one cell, the corresponding images were digitally stitched together using Adobe Photoshop CS (version 6.0, Adobe Systems, CA). All capsids were counted, and the respective nuclear and cellular areas that had been sampled were measured using an ImageJ plugin.
Immunofluorescence microscopy.
Vero cells were cultured on coverslips, infected synchronously, and at the indicated time points, fixed and permeabilized with PHEMO fix (3.7% [wt/vol] paraformaldehyde, 0.05% [wt/vol] glutaraldehyde, 0.5% [vol/vol] Triton X-100 in PHEMO buffer of 68 mM PIPES, 25 mM HEPES [pH 6.9], 15 mM EGTA, 3 mM MgCl2, 10% [vol/vol] dimethyl sulfoxide [DMSO]) for 10 min and subsequently washed twice with PHEMO buffer (69). The residual paraformaldehyde was quenched with 50 mM NH4Cl for 10 min. To block unspecific protein interactions, the samples were incubated with 10% (vol/vol) serum from a healthy HSV-1-seronegative volunteer and 0.5% (wt/vol) bovine serum albumin (BSA) in PBS for 1 h at RT (69) and then probed with primary and secondary antibodies. DNA was stained by 0.05 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) in PBS with 0.05% (vol/vol) DMSO, 0.0005% (vol/vol) NP-40, 0.025% (wt/vol) BSA, 0.05 mM Tris-HCl, pH 7.4, 0.73 mM NaCl, 0.01 mM CaCl2, and 0.11 mM MgCl2. Images were acquired by confocal laser scanning microscopy (LSM 510 Meta [Zeiss] or TCS SP8 [Leica Microsystems, Wetzlar, Germany]) with plan-apochromat 63×/1.40 oil immersion objectives, and 405-, 488-, 561-, and 633-nm lasers. The images were processed using either Adobe Photoshop CS or ImageJ. Fluorescence intensities were measured in the 8-bit images along a 1-pixel-thick line of different lengths using the “plot profile” tool of ImageJ. The line profiles representing the plotted gray values were created using the software GraphPad Prism (Version 5, GraphPad Software, CA) (44).
RESULTS
Mutation of conserved tryptophan-acidic motifs of HSV-1 pUL36.
To identify tryptophans in conserved neighborhoods, we aligned the amino acid sequences of HSV-1-pUL36 (c.f. Fig. 1 for Alphaherpesvirinae) with 10 orthologs of other herpesviruses. Of the 32 tryptophans of the clinical isolate HSV-1 strain 17+, whose sequence was the first to be determined (78), 4 are evolutionarily maintained in all 11 human herpesviruses (W1711, W2072, W2213, and W2270), 5 more in Alpha- and Gammaherpesvirinae (W913, W1359, W1447, and W2227) or Alpha- and Betaherpesvirinae (W2399), 12 more within the Alphaherpesvirinae (W100, W167, W233, W356, W434, W1111, W1766, W1770, W1886, W2453, W2573, and W3024), and 9 more among HSV-1 and HSV-2 strains (W707, W1088, W1781, W1862, W2099, W2104, W2148, W2575, and W2704). To summarize, 30 tryptophans are conserved among HSV-1 and HSV-2 (Fig. 1A), but only 5 of these are flanked by conserved acidic residues; namely 1765WD1767, 1862WE1863, and 2212DW2213 in all Alphaherpesvirinae and 706EW707 and 2453WE2454 in HSV-1 and HSV-2 strains. Furthermore, 26 other clinical isolates of HSV-1, including the well-characterized strains F, KOS, H129, and McKrae, contain these motifs (3; http://szparalab.psu.edu/hsv-diversity/data).
To reveal potential functions of tryptophan-acidic motifs in the middle third of pUL36, we replaced 1766WD1767 or/and 1862WE1863 with alanines in HSV-1(17+)Lox or HSV-1(17+)Lox-CheVP26 (Fig. 1B). In the latter strain, the small capsid protein VP26 has been tagged with mCherry (15, 26, 65; Döhner et al., submitted). We mutated the BAC plasmids pHSV-1(17+)Lox and pHSV-1(17+)Lox-CheVP26 to encode pUL36-WD/AA, pUL36-WE/AA, or pUL36-WD/AA-WE/AA. Restriction digest analyses with AscI showed the expected band shift from 3.1 kb to 4.1 kb upon a first recombination that had inserted a kanamycin resistance cassette together with the AA codons (not shown). A second recombination removed the cassette, as indicated by shifting back to 3.1 kb (Fig. 2A). Further analyses with BamHI, EcoRI, HindIII, NotI, SalI, and XhoI yielded the expected fragments (not shown). However, there were some heterogeneous AscI bands between 2 and 2.5 kb as reported before (70, 79; Döhner et al., submitted). The respective BAC plasmids were transfected into Vero-HS30 cells that provide pUL36 in trans and that complement the growth of HSV-1-ΔUL36 mutants (26, 36). While all HSV-1-pUL36 mutants formed plaques in Vero-HS30 cells within 3 days posttransfection (dpt), it took HSV-1-pUL36-WD/AA and HSV-1-pUL36-WE/AA 4 dpt, and HSV-1-pUL36-WD/AA-WE/AA 6 to 7 dpt to form plaques in the parental Vero cells. Sequencing of different HSV-1(17+)Lox stocks after the third passage in Vero-HS30 cells validated the maintenance of the WD/AA, WE/AA, or WD/AA-WE/AA mutations in pUL36 in the virus stocks (A. Pohlmann, L. Ivanova, E. Hage, and B. Sodeik, unpublished data).
FIG 2.
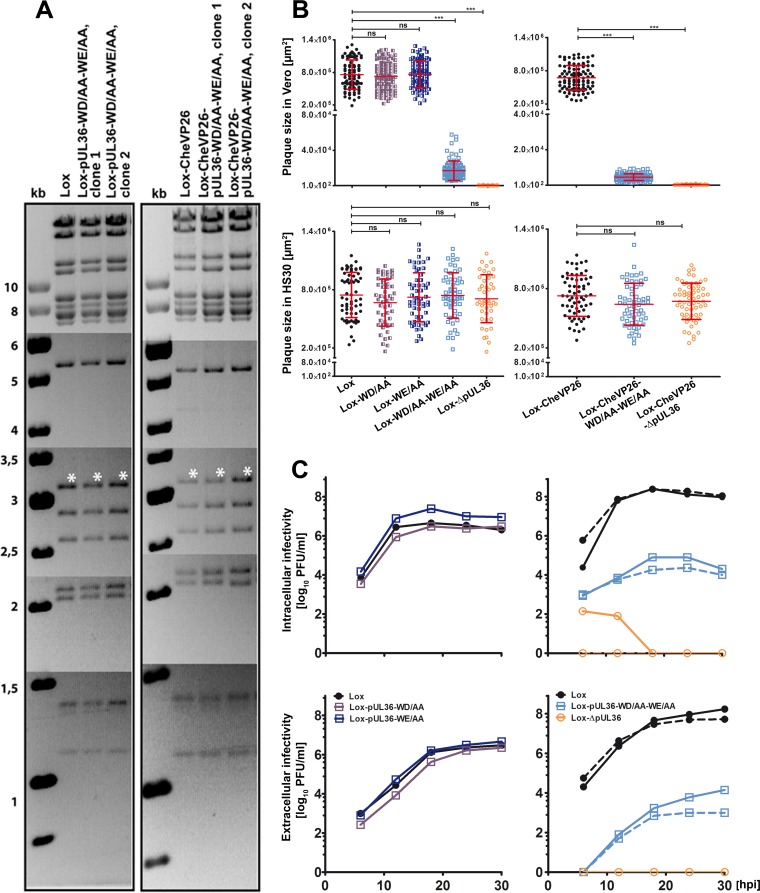
Characterization of HSV-1-pUL36 mutants. (A) Agarose gels of AscI restriction digests of the parental BACs pHSV-1(17+)Lox and pHSV-1(17+)Lox-mCheVP26 or the derived mutants pHSV-1-pUL36-WD/AA-WE/AA and pHSV-1-mCheVP26-pUL36-WD/AA-WE/AA. The sizes of the restriction fragments and molecular size markers are indicated in kilobase pairs. (B) Plaque formation in Vero and Vero-HS30 cells. The plaque sizes in Vero cells were determined in three independent experiments and those in Vero-HS30 cells in two independent experiments. Values are means ± standard deviations (SD) (***, P < 0.0001 as determined by Tukey's multiple comparison test). (C) Vero cells were infected with an MOI of 5 PFU/cell (1 × 107 PFU/ml), and the samples were harvested at the indicated time points. The extracellular and intracellular infectivities were determined in duplicates. The virus yields of the parental strains Lox (solid line) and Lox-CheVP26 (dashed line) are depicted by closed circles, those of Lox-WD/AA (solid dark blue line), Lox-WE/AA (solid blue line), Lox-WD/AA-WE/AA (solid light blue line), and Lox-CheVP26-WD/AA-WE/AA (dashed light blue line) by squares, and those of the deletion mutants HSV-1-ΔUL36 deletion mutants are represented by open circles with a solid or dashed line for Lox and Lox-CheVP26, respectively.
Characterization of HSV-1 strains with mutated pUL36 tryptophan-acidic motifs.
Virions of HSV-1-pUL36-WD/AA or -pUL36-WE/AA that had been complemented with pUL36 in Vero-HS30 cells formed in the noncomplementing Vero cells plaques of the same size as the parental HSV-1(17+)Lox (Fig. 2B). However, mutants with pUL36-WD/AA-WE/AA formed only small plaques, and mutants lacking pUL36 did not replicate at all. In contrast, in the complementing Vero-HS30, the plaque sizes of all strains were indistinguishable (Fig. 2B). Mutation of the motifs in pUL36 changed the fraction of infectious particles of an inoculum to a minor extent as the ratios of genomes to PFU were only about 2-fold higher for HSV-1(17+)Lox-pUL36-WD/AA-WE/AA strains than for their respective parental strains.
Next, we determined the amount of infectious virions after inoculating Vero cells with an MOI of 5 PFU/cell (Fig. 2C). The growth curves of HSV-1(17+)Lox, Lox-pUL36-WD/AA, and Lox-pUL36-WE/AA were indistinguishable, and maximal amounts of intracellular and extracellular infectivity had been produced within 18 h postinfection (hpi). The titers of both, HSV-1(17+)Lox-pUL36-WD/AA-WE/AA and -Lox-mCheVP26-pUL36-WD/AA-WE/AA, were about 3 logs lower for cell-associated and extracellular virions, but there was a significant amplification. In contrast, there was no virus production after inoculation with HSV-1(17+)Lox-pUL36 or -CheVP26-ΔUL36. Their low intracellular infectivity at 6 and 12 hpi was most likely derived from the inoculum that had been trans-complemented with wild-type pUL36. Thus, pUL36-WD/AA-WE/AA significantly reduced plaque sizes as well as production of infectious intracellular and extracellular virions. There were no differences in the growth properties of the respective HSV-1(17+)Lox and HSV-1(17+)Lox-mCheVP26 strains indicating that tagging VP26 did not aggravate the pUL36-WD/AA-WE/AA phenotype.
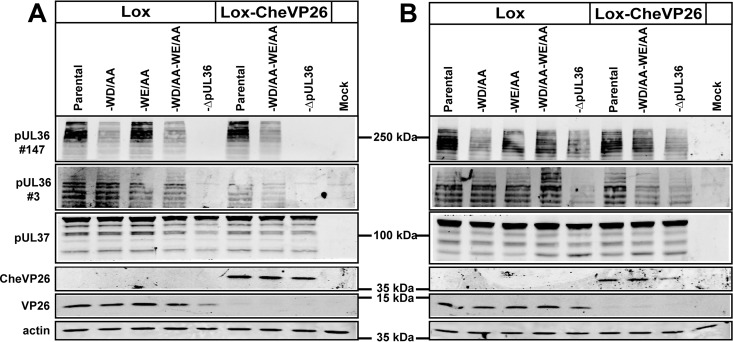
To analyze the expression patterns of the different HSV-1-pUL36 versions, Vero cells were inoculated with HSV-1(17+)Lox or HSV-1(17+)Lox-mCheVP26 and the respective mutants at 10 PFU/cell for 8, 10 (not shown), or 12 h (Fig. 3A). In addition to full-length HSV-1-pUL36 with an apparent molecular mass of more than 250 kDa, the cells had synthesized several pUL36 forms of lower molecular mass as reported before (26, 28, 44, 80). HSV-1(17+)Lox-pUL36-WD/AA, -pUL36-WE/AA, and -pUL36-WD/AA-WE/AA and the corresponding Lox-CheVP26 strains expressed the same size pattern of pUL36 proteins (Fig. 3A). Similar results were also obtained for the HSV-1-ΔUL36 mutants upon infection of Vero-HS30 cells that complemented pUL36 in trans (Fig. 3B). Using a polyclonal antiserum generated against residues 1408 to 2112 (pAb 147), it appeared that less pUL36-WD/AA and pUL36WD/AA-WE/AA had been synthesized compared to the respective parental strains. Since these mutations are contained in the protein fragment that had been used to generate pAb 147, we generated another antibody against pUL36 residues 936 to 951 (pAb 3), which detected similar amounts of pUL36 in all strains. Furthermore, mutation of 1766WD1767 and 1862WE1863 had no influence on the expression of pUL37 or VP26 from the neighboring genes, UL37 and UL35. Thus, the codon changes apparently had not influenced the stability of UL36 mRNAs, and these tryptophan acidic motifs did not contribute to the overall protein stability, e.g., by influencing their rate of synthesis or their susceptibility to proteases in a noncompensating manner.
FIG 3.
HSV-1-pUL36-WD/AA-WE/AA does not affect the expression pattern of pUL36 proteins. Vero cells (A) and Vero-HS30 cells (B) were mock infected or infected synchronously with the indicated HSV-1 strains and harvested at 12 hpi. The proteins were separated in gradient polyacrylamide gels and probed with antibodies directed against pUL36 (pAb 147; residues 1408 to 2112), pUL36 (pAb 3; residues 936 to 951), pUL37 (pAb anti-pUL37), VP26 (pAb anti-VP26), CheVP26 (MAb anti-red fluorescent protein), or as loading control actin (MAb 1501).
HSV-1-pUL36 tryptophan-acidic motifs are required for efficient secondary envelopment.
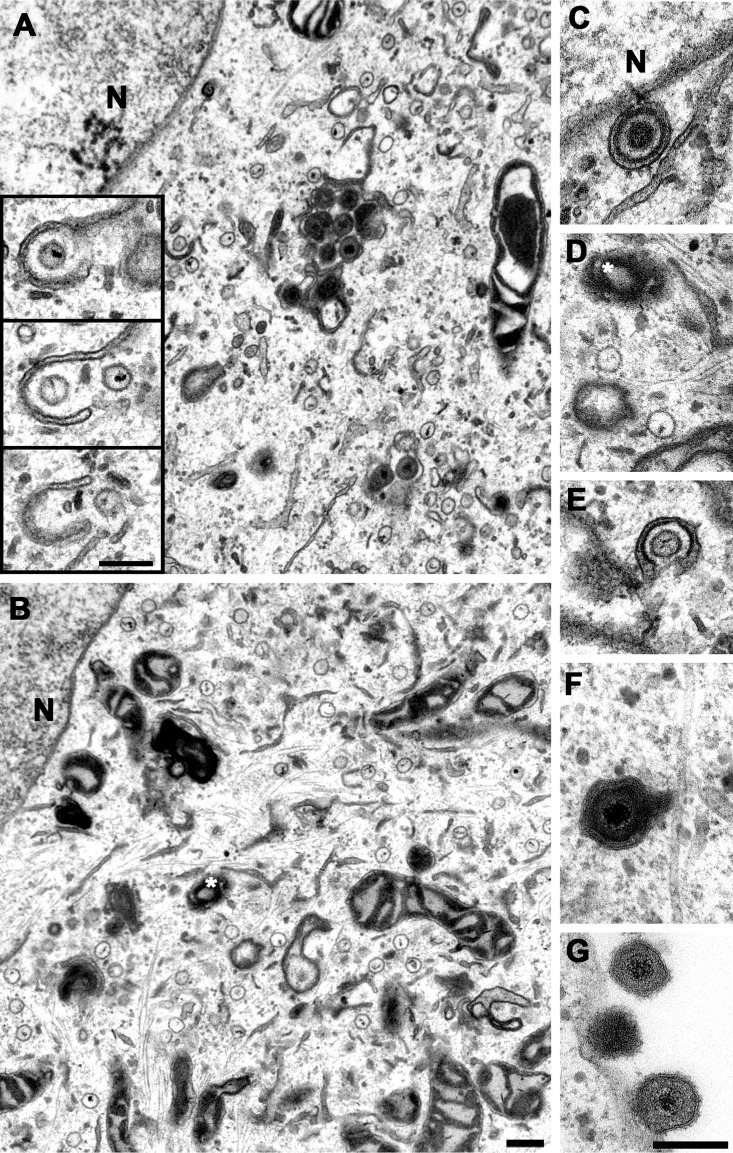
To characterize the morphogenesis of the novel mutants, Vero cells were infected synchronously with 10 PFU/cell, fixed at 10 or 14 hpi, and processed for electron microscopy. To achieve an optimal contrast for the HSV-1 capsids and to reveal their typical hexagonal cross section of about 125 nm, we used very thin sections of about 50-nm thickness in which the electron-dense HSV-1 DNA genome is often spread over two or three consecutive capsid cross sections (Fig. 4A, inset). In cells infected with HSV-1(17+)Lox (Fig. 4A) or HSV-1(17+)Lox-mCheVP26 (not shown), all known assembly intermediates had formed by 10 hpi (not shown) and could be easily detected by 14 hpi: nuclear A, B, and C capsids, primary virions between the inner and the outer nuclear envelope, cytosolic capsids, capsids in the process of secondary envelopment, intracellular vesicles harboring apparently intact virions (Fig. 4A), and extracellular progeny virions attached to the plasma membrane (not shown). In contrast, after infection with HSV-1(17+)Lox-pUL36-WD/AA-WE/AA (Fig. 4B to G) or -Lox-mCheVP26-pUL36-WD/AA-WE/AA (not shown), the cells contained a larger number of cytosolic capsids that clustered in a perinuclear region (Fig. 4B), few capsids in the process of being wrapped by cytoplasmic membranes (Fig. 4E), as well as rare vesicles harboring intact virions (Fig. 4F) and extracellular virions bound to the plasma membranes (Fig. 4G).
FIG 4.
Accumulation of cytosolic capsids and reduced secondary envelopment in cells infected with HSV-1(17+)Lox-pUL36-WD/AA-WE/AA. Vero cells were infected synchronously with HSV-1(17+)Lox (A) or HSV-1(17+)Lox-pUL36-WD/AA-WE/AA (B to G) PFU, fixed at 14 hpi, and analyzed by electron microscopy. The inset in panel A contains three consecutive sections showing viral genomes within cytoplasmic capsids. A white asterisk indicates the microtubule-organizing center, and N indicates the nucleus. The primary enveloped virion (C), cytosolic capsids (D), wrapping intermediate (E), enveloped virus particle (F), and extracellular virions (G) were assembled upon infection with HSV-1-pUL36-WD/AA-WE/AA. Scale bars, 200 nm.
To quantify the relative amounts of the different assembly intermediates (Table 1), we systematically evaluated single cross sections of 11 randomly imaged cells infected with HSV-1(17+)Lox and of 10 cells infected with HSV-1(17+)Lox-pUL36-WD/AA-WE/AA. Due to the small volume that is sampled in thin sections, there was a considerable heterogeneity among different cells. We therefore expanded the analysis to a second experiment and sampled more than 40 randomly imaged nuclei and more than 180 random snapshots from cytoplasmic regions for the parental strains and the mutants expressing pUL36-WD/AA-WE/AA. In reference to all cytoplasmic capsids, the percentage of cytosolic capsids whose cross section showed no membrane association had increased on average to 80% in the mutants compared to 49% in the parental strains. In accordance, the amount of wrapping intermediates was reduced from 28% in the parental strains to 13% and that of the enveloped virions from 23% to 7% in the mutants. The number of nuclear capsids was a bit lower in the mutants than in the respective parental strains. The abundance of the different HSV-1 assembly intermediates was not influenced by adding the mCherry tag to the small capsid protein VP26. The electron microscopy analysis shows that these tryptophan-acidic motifs in pUL36 are important for efficient membrane association and secondary envelopment of cytosolic capsids and subsequent virus formation and egress, but not for nuclear capsid formation and nuclear egress.
TABLE 1.
HSV-1(17+)-pUL36-WD/AA-WE/AA is impaired in secondary capsid envelopmenta
| Exp no. and HSV-1(17+) type | No. of cells or snapshotsb |
No. of capsids |
Area (1,000 μm2) |
No. of capsids/area (1/1,000 μm2) |
% of cytoplasmic capsids |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nuclear | Cytoplasmic | Nuclear | Cytoplasmic | Nuclear | Cytoplasmic | Nuclear | Cytoplasmic | Cytosolic | Wrapping intermediates | Enveloped virions | |
| Exp 1, Lox | 11* | 11* | 940 | 715 | 1.9 | 3.5 | 493 | 204 | 42 | 32 | 26 |
| Exp 2, Lox | 22* | 84# | 1,362 | 910 | 3.5 | 1.9 | 387 | 479 | 51 | 26 | 22 |
| Exp 2, Lox-CheVP26 | 20* | 101# | 1,541 | 908 | 4.3 | 2.0 | 356 | 454 | 54 | 26 | 20 |
| Total or avg | 53* | 11*; 185# | 3,843 | 2,533 | 9.7 | 7.4 | 412 | 379 | 49 | 28 | 23 |
| Exp 1, Lox-WD/AA-WE/AA | 10* | 10* | 920 | 552 | 2.1 | 3.2 | 446 | 173 | 70 | 17 | 13 |
| Exp 2, Lox-WD/AA-WE/AA | 22* | 92# | 905 | 1,867 | 3.2 | 1.9 | 279 | 983 | 84 | 12 | 4 |
| Exp 2, Lox-CheVP26-WD/AA-WE/AA | 20* | 100# | 1,262 | 1,720 | 3.5 | 2.2 | 362 | 782 | 86 | 9 | 4 |
| Total or avg | 52* | 10*; 192# | 3,087 | 4,139 | 8.8 | 7.3 | 362 | 646 | 80 | 13 | 7 |
Vero cells infected with HSV-1(17+)Lox, HSV-1(17+)Lox-pUL36-WD/AA-WE/AA, HSV-1(17+)Lox-mCheVP26, or HSV-1(17+)Lox-mCheVP26-pUL36-WD/AA-WE/AA were analyzed by electron microscopy, and the numbers of nuclear capsids, cytosolic capsids, wrapping intermediates, and enveloped virions were determined.
Shown are the number of cells of which the entire nucleus or the entire cytoplasm had been imaged (*) or the number of snapshots from a cytoplasmic region (#).
Tegument acquisition of HSV-1 capsids lacking tryptophan-acidic motifs in pUL36.
Next, we investigated by confocal fluorescence microscopy the subcellular localization of several structural HSV-1 proteins after inoculation with HSV-1(17+)Lox-pUL36-WD/AA-WE/AA (data not shown) or -Lox-CheVP26-pUL36-WD/AA-WE/AA. For parental and mutated HSV-1 strains, nuclear capsids were detected as early as 6 hpi by antibodies against the major capsid protein VP5 or by CheVP26 expression (data not shown). As the infection progressed, increasing amounts of VP5 or CheVP26 puncta appeared in the cytoplasm indicating successful nuclear egress (15, 26, 31, 65, 70). There were also diffuse nuclear and cytoplasmic pUL36 signals in addition to the capsid-associated pUL36 as reported before (15, 26, 65, 81). Cytoplasmic capsids of HSV-1(17+)Lox were distributed over the entire cytoplasm and also targeted to the cell periphery as the infection proceeded (Fig. 5Ai).
FIG 5.
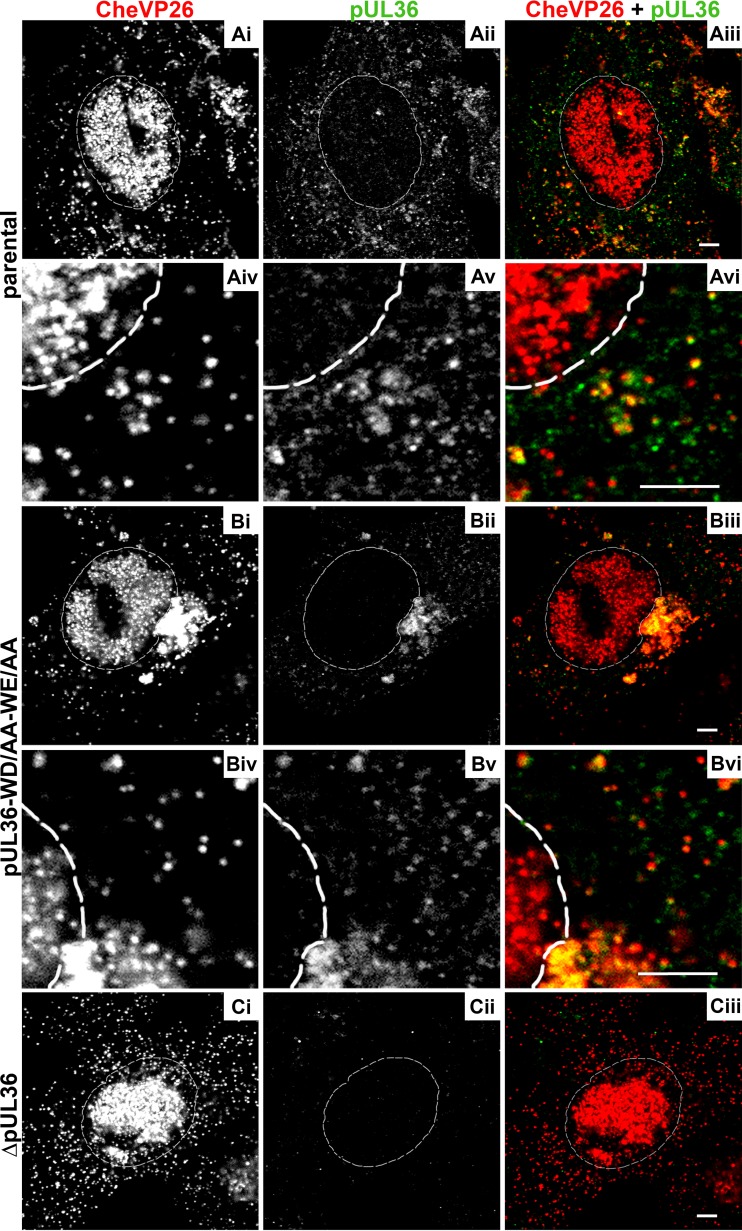
HSV-1-pUL36-WD/AA-WE/AA accumulates capsids in the perinuclear region and recruits pUL36. Vero cells were infected synchronously with HSV-1(17+)Lox-CheVP26 (A), HSV-1(17+)Lox-CheVP26-WD/AA-WE/AA (B), or HSV-1(17+)Lox-CheVP26-ΔUL36 (C) and fixed at 10 hpi. The specimens were labeled for pUL36 (pAb 147; white in panels ii and v and green in panels iii and vi) and analyzed by confocal microscopy. The capsids were visualized by fusion protein CheVP26 (white in panels i and iv and red in panels iii and vi). The nuclei were identified by DAPI labeling and are depicted by dashed white lines. Scale bars, 3 μm.
In contrast, capsids of HSV-1(17+)Lox-pUL36-WD/AA-WE/AA decorated with pUL36 had accumulated in a perinuclear region of the cytoplasm (Fig. 5B). Such capsid clusters had already formed at earlier time points and increased in size as the infection progressed. They most likely corresponded to the cytosolic capsids detected by electron microscopy in the neighborhood of the nuclear envelopes. In addition, there were few capsids with pUL36 in the more peripheral cytoplasm. Parallel infections with HSV-1(17+)Lox-ΔUL36 yielded no signals by the anti-pUL36 antibodies (Fig. 5Cii), confirming their specificity as reported before (15, 26). Both wild-type pUL36 (yellow in Fig. 5Aiii) and pUL36-WD/AA-WE/AA (yellow in Fig. 5Biii), were recruited onto cytosolic but not onto nuclear capsids. However, the signal on the capsids of HSV-1(17+)Lox-pUL36-WD/AA-WE/AA was lower, possibly because the polyclonal antibodies had been raised against a recombinant fragment from aa 1408 to 2112 (pAb 147) that comprises the mutated residues. The subcellular localization of pUL36-WD/AA-WE/AA suggested that its C-terminal region had maintained its affinity for pUL25 since it had been recruited to cytosolic capsids. A labeling for γ-tubulin showed that the capsids of HSV-1(17+)Lox-pUL36-WD/AA-WE/AA had clustered around the microtubule-organizing center (data not shown).
We next analyzed the subcellular distribution of pUL37, another inner tegument protein that depends on pUL36 to be recruited onto progeny cytosolic capsids, and of the tegument protein VP16, which connects pUL36 to the outer tegument (10, 15, 44, 46, 53). pUL37 colocalized to a similar extent with cytoplasmic capsids of HSV-1(17+)Lox (Fig. 6A) and -Lox-pUL36-WD/AA-WE/AA (Fig. 6B)—both in the cell periphery and on cytosolic capsids of Lox-pUL36-WD/AA-WE/AA that had accumulated in a perinuclear region (yellow in Fig. 6Biii). pUL37 did not colocalize with nuclear capsids, neither in the parental nor in the mutant strains, as reported before (15, 28, 82, 83). After inoculation with HSV-1(17+)Lox-ΔUL36, there was no pUL37 detected on the cytoplasmic capsids (Fig. 6Cii, green in Fig. 6Ciii) as reported before (15, 26).
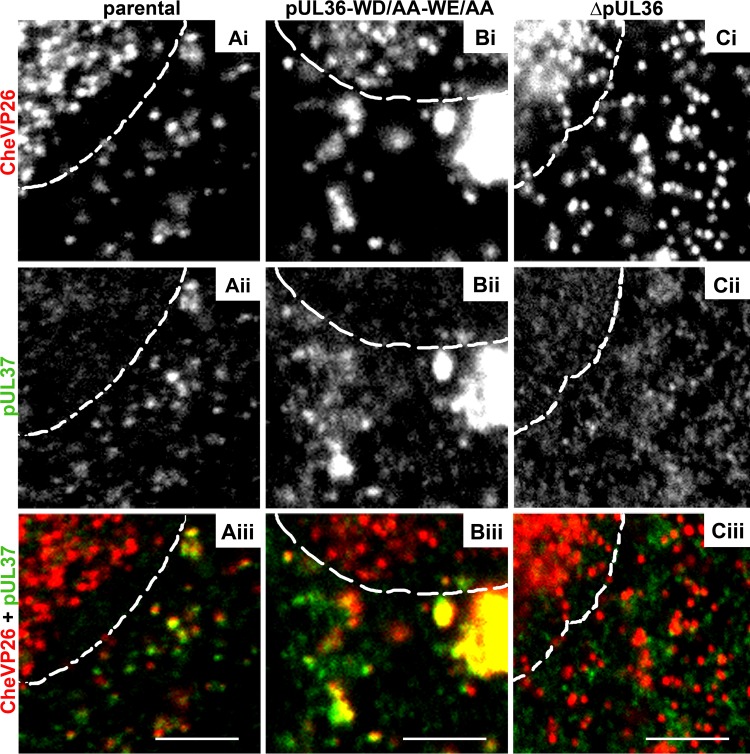
FIG 6.
The cytoplasmic capsids of HSV-1-CheVP26-pUL36-WD/AA-WE/AA recruit pUL37. Vero cells were infected synchronously with HSV-1(17+)Lox-CheVP26 (A), HSV-1(17+)Lox-CheVP26-WD/AA-WE/AA (B), or HSV-1(17+)Lox-CheVP26-ΔUL36 (C) and fixed at 10 hpi. The specimens were labeled for pUL37 (pAb α-pUL37; white in panels ii and green in panels iii) and analyzed by confocal microscopy. The capsids were visualized by CheVP26 (white in panels i and red in panels iii). The nuclei were identified by DAPI labeling and are depicted by dashed white lines. Scale bars, 3 μm.
VP16 also colocalized to a similar extent with the cytoplasmic capsids of HSV-1(17+)Lox (Fig. 7A) and of -Lox-pUL36-WD/AA-WE/AA (Fig. 7B); both in the cell periphery and on cytosolic capsids of -Lox-pUL36-WD/AA-WE/AA that had accumulated in a perinuclear region (Fig. 7B). The colocalization of pUL36-WD/AA-WE/AA with pUL37 and VP16 on cytosolic capsids suggests that its N-terminal region had folded properly and maintained its affinity for other HSV-1 structural proteins. After inoculation with HSV-1(17+)Lox-ΔUL36, there was no VP16 detected on the cytoplasmic capsids (Fig. 7Cii; green in Fig. 7Ciii).
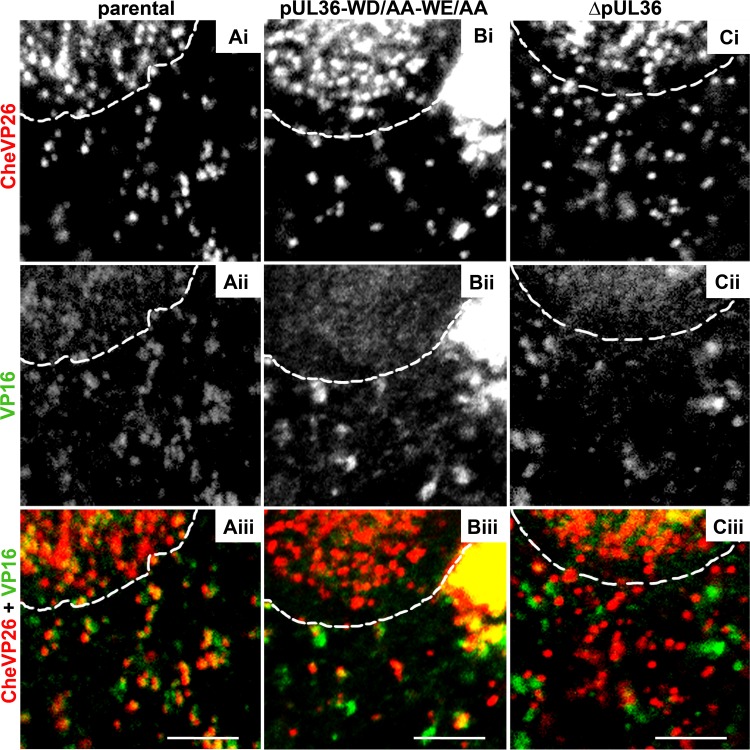
FIG 7.
The capsids of HSV-1-CheVP26-pUL36-WD/AA-WE/AA acquire VP16. Vero cells were infected synchronously with HSV-1(17+)Lox-CheVP26 (A), HSV-1(17+)Lox-CheVP26-WD/AA-WE/AA (B), or HSV-1(17+)Lox-CheVP26-ΔUL36 (C) and fixed at 10 hpi. The specimens were labeled for VP16 (pAb 631209; white in panels ii and green in panels iii) and analyzed by confocal microscopy. The capsids were visualized by CheVP26 (white in panels i and red in panels iii). The nuclei were identified by DAPI labeling and are depicted by dashed white lines. Scale bars, 3 μm.
The major outer tegument protein VP22 interacts with VP16 and links it to other tegument proteins as well as to envelope proteins gE and gM (10, 84–86). VP22 was highly enriched in the nucleus as reported before (87–89). Many but not all cytoplasmic capsids of HSV-1(17+)Lox colocalized with VP22 (Fig. 8Aii; yellow in Fig. 8Aiv). Similarly, some but not all capsids that had accumulated in close proximity to the nucleus after infection with HSV-1(17+)Lox-pUL36-WD/AA-WE/AA colocalized with VP22 (Fig. 8Bii; yellow in Fig. 8Biv). After inoculation with HSV-1(17+)Lox-ΔUL36, there was no VP22 detected on the cytoplasmic capsids (Fig. 8Cii; red in Fig. 8Civ).
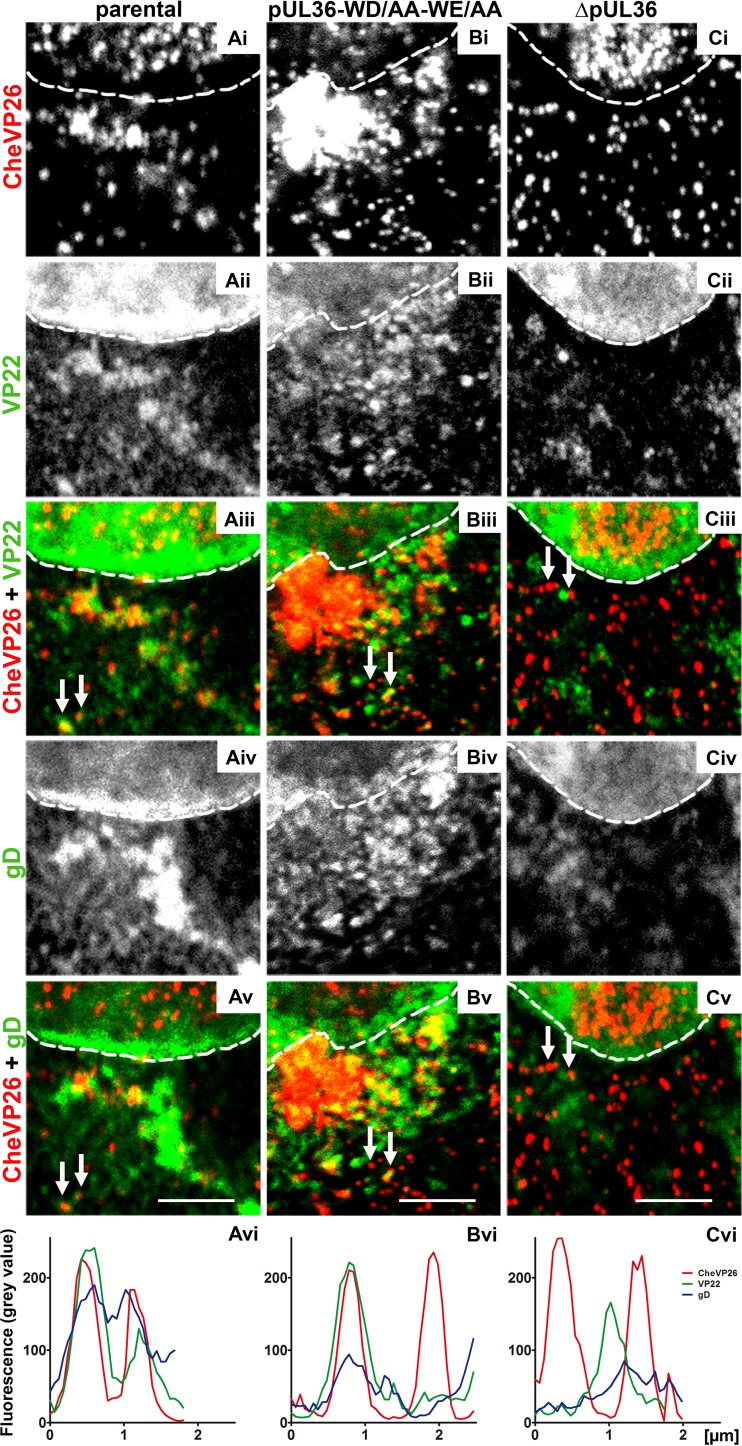
FIG 8.
Few capsids of HSV-1(17+)Lox-mCheVP26-pUL36-WD/AA-WE/AA undergo secondary envelopment. Vero cells were infected synchronously with HSV-1(17+)Lox-mCheVP26 (A), HSV-1(17+)Lox-mCheVP26-pUL36-WD/AA-WE/AA (B), or HSV-1(17+)Lox-CheVP26-ΔUL36 (C), fixed at 10 hpi and labeled for VP22 (pAb AGV30; white in panels ii and green in panels iii) and glycoprotein D (MAb DL-6; white in panels iv and green in panels v) and analyzed by confocal fluorescence microscopy. The capsids were detected by mCheVP26 (white in panels i and red in panels iv and v). The nuclei were identified by DAPI labeling and are depicted by dashed white lines. The fluorescence intensity of randomly selected capsids (white arrows) was measured in 8-bit images along a 1-pixel-thick and 2- to 2.5-μm-long line. The histograms represent the plotted gray values of CheVP26 (red), VP22 (green), and gD (blue) against the length of the line in micrometers. Scale bars, 3 μm.
Further labeling experiments for the envelope proteins gB (not shown) or gD (Fig. 8Biv) indicated that the perinuclear region in which capsids had clustered after inoculation with HSV-1(17+)Lox-pUL36-WD/AA-WE/AA was not enriched for cytoplasmic membranes (Fig. 8Biii; red in Fig. 8Bv), consistent with the electron microscopy data. For a more quantitative evaluation, we obtained line profiles from triple-labeling experiments of cytoplasmic viral structures. While there was a high degree of colocalization as indicated by a copeaking of the CheVP26, VP22 and gD signals after infection with HSV-1(17+)Lox-mCheVP26 (Fig. 8Avi), cytoplasmic capsids colocalized to a lower extent with gD (Fig. 8Bvi), gB (not shown), or VP22 (Fig. 8Bvi) after infection with HSV-1(17+)Lox-CheVP26-pUL36-WD/AA-WE/AA. After infection with HSV-1(17+)Lox-ΔUL36, there were several instances in which VP22 copeaked with gD in the absence of a capsid signal (Fig. 8Cvi). These structures most likely represented membrane vesicles decorated by tegument on their limiting membrane or L-particles within transport vesicles.
Thus, the N-terminal binding sites on pUL36 for pUL37 and VP16 and the C-terminal binding site for capsids seemed to operate normally in the mutated protein pUL36-WD/AA-WE/AA. pUL36 and pUL37, which are both essential for secondary envelopment, were associated with cytosolic capsids, and the capsids had also gathered in a perinuclear region around the microtubule-organizing center, but the electron and confocal fluorescence microscopy data show that they did not associate well with cytoplasmic membranes and were not enveloped efficiently.
DISCUSSION
The rare amino acid residue tryptophan contributes in a disproportionally high frequency to short peptide motifs that mediate protein-protein interactions (54–56). In the large tegument proteins of the Alphaherpesvirinae, 21 of the 32 tryptophans are conserved. To identify further functional domains in these essential proteins, we replaced the two most central of the conserved tryptophan-acidic motifs by alanines using traceless BAC mutagenesis and generated the HSV-1(17+)Lox and HSV-1(17+)Lox-CheVP26 strains in which either one or both motifs in pUL36 had been changed. HSV-1(17+)Lox mutants encoding pUL36-1766WD/AA1767-1862WE/AA1863 were severely impaired in plaque formation, secondary envelopment, and virion formation. Our study therefore identified a novel important functional determinant among residues 1700 to 1900 of HSV-1-pUL36. This region has been suggested to be of functional importance since deletion of a larger region of PRV-pUL36 of residues 1294 to 2025 including the homologous residues cannot complement virus replication (90). The HSV-1 lysine at position 1799 located between our tryptophan-acidic motifs is ubiquitinated and targeted by the autocatalytic USP activity of pUL36; however it is not conserved among the alphaherpesviruses (91). The novel HSV-1 mutants were amplified to the same titers as their parental strains in the complementing Vero-HS30 cells. This indicated that pUL36-WD/AA-WE/AA did not operate as a dominant-negative competitor of the authentic pUL36.
HSV-1 strains with mutated tryptophan-acidic motifs in pUL36 are impaired in membrane association and secondary envelopment of cytosolic capsids.
Our quantitative electron microscopy analysis showed that the first steps impaired in the viral cycle were the targeting to cytoplasmic membranes and secondary envelopment of progeny capsids, while capsid assembly and their nuclear egress into the cytosol seemed to proceed normally (c.f. Table 1). The most prominent phenotype of the HSV-1-pUL36-WD/AA-WE/AA strains was a strong accumulation of progeny cytosolic capsids in a perinuclear region around the microtubule-organizing center, with fewer capsids in the peripheral regions of the cytoplasm. These perinuclear capsids had still recruited pUL36-WD/AA-WE/AA and its partner pUL37, as in the parental strains. The association of these two inner tegument proteins is essential for secondary envelopment (15, 26, 28, 44). Furthermore, the capsids could still associate with the tegument protein VP16 and to some extent with the tegument protein VP22. VP16 provides a major link to important abundant outer tegument proteins such as VP22 (10, 45, 46). This is in contrast to the cytosolic capsids of the complete deletion mutant HSV-1(17+)-CheVP26-ΔUL36 that did not recruit any of these tegument proteins. However, although the known interaction partners of pUL36 still associated with cytoplasmic capsids, envelopment and virion formation were inefficient. These results indicate that events contributing to targeting progeny capsids to cytoplasmic membranes during assembly were impaired by mutating the central 1766WD1767 and 1862WE1863 motifs to alanine.
Besides crystal structures of an N-terminal fragment of the residues 1 to 236 of the MCMV homolog comprising the deubiquitinating and deneddylating activity (41, 92, 93), and a fragment of HSV-1 residues 1625 to 1758 (50), there is yet no further information on the structure of HSV-1-pUL36 available. The algorithm PredictProtein (94) suggests that 1766WD1767 may be part of an α-helix and that 1862WE1863 might be located at a coiled-coil region. Polyclonal antibodies that have been raised against residues 1408 to 2112 that include the mutated region detected several pUL36 forms after infection with parental strains (15, 26, 95). This complicated expression pattern of different versions of pUL36 was unchanged upon infection with HSV-1-pUL36-WD/AA-WE/AA strains indicating a similar usage of multiple translation start sites, premature translation termination, and/or susceptibility to proteases. The association of HSV-1-pUL36-WD/AA-WE/AA, pUL37, and VP16 with cytosolic capsids implies that the C-terminal binding domains for pUL25 on the capsid vertices and the N-terminal regions had folded properly. These observations suggest that the phenotype of the HSV-1-pUL36-WD/AA-WE/AA strains is mainly due to conformational changes in its central region but that the N- and C-terminal domains still fulfilled their bona fide functions.
Potential functions of the HSV-1-pUL36 tryptophan motifs.
One scenario to rationalize the phenotype of the novel HSV-1-pUL36-WD/AA-WE/AA strains is to surmise that 1766WD/AA1767 and 1862WE/AA1863 constitute small peptide binding motifs that may allow pUL36 to fold into a more rigid and compact conformation by internal pUL36 interactions and/or to interact with further as yet unknown host or viral factors. The inner tegument proteins pUL36 and pUL37 of the Alphaherpesvirinae are considered to be likely receptors or regulators of microtubule motors on the surface of cytosolic capsids, on the wrapping membrane mediating secondary capsid envelopment, and on the transport vesicles harboring fully enveloped virions (14, 15, 26, 95–103).
It has not escaped our notice that the 1766WD1767 and 1862WE1863 motifs in pUL36 bear some resemblance to tryptophan motifs mediating binding to the C-terminal tetratricopeptide repeats of kinesin light chains; these repeats are required to recruit kinesin-1 to host cargoes or to intracellular mature particles of vaccinia virus via the vaccinia virus A36 envelope protein (104–106). Based on several experimentally validated kinesin cargo interactions, Dodding et al. defined mono- and bipartite kinesin light chain binding motifs with conserved tryptophans embedded by one or two hydrophilic (asparagine, N; glutamine, Q) or acidic residues (104, 106). None of the HSV-1 tryptophans is accompanied by such hydrophilic residues, but 9 are flanked by conserved acidic aspartic acid (D) or glutamic acid (E). The linker within a bipartite kinesin light chain binding motif should have a length of 20 to 100 residues but lack any additional tryptophans (104). However, there are two further tryptophans W1770 and W1781 located between 1766WD/AA1767 and 1862WE/AA1863, while other tryptophans are, at least in the primary sequence, too far away. An inability to recruit kinesin-1 would be consistent with the accumulation of HSV-1 particles around the microtubule-organizing center during assembly after infection with HSV-1-pUL36-WD/AA-WE/AA. Transport along microtubules is required to ensure efficient meeting of tegumented cytosolic capsids with the cytoplasmic membranes mediating secondary envelopment and thus virion formation; if microtubule transport is impaired during assembly, HSV-1 particle formation is not completely blocked but severely reduced (15, 16, 107; Döhner et al., submitted). Furthermore, kinesin-1 can bind to isolated capsids exposing pUL36 and pUL37 on their surfaces (96), and kinesin-1 colocalizes with transport vesicles harboring fully assembled virions in epithelial cells (108).
Biochemical and live cell imaging experiments are required to elucidate potential differences in binding of host and viral factors to the central region of pUL36-WD/AA-WE/AA in comparison to authentic pUL36 and to unravel further functional consequences of these mutations. It may furthermore be possible to obtain structural data on these central domains of pUL36 or other large tegument proteins of the Herpesviridae. Our analysis of the HSV-1-pUL36-WD/AA-WE/AA mutants has identified a novel determinant in the central region of pUL36 that fulfils important functions in intracellular targeting of the capsids during assembly. It is tempting to speculate that the phenotype of HSV-1-pUL36-WD/AA-WE/AA might be explained by a reduced affinity of kinesin-1 for viral particles and that kinesin-1-mediated transport is required for efficient transport of capsids and possibly other HSV-1 particles along microtubules. However, structural rearrangements that impair other functions of pUL36 may also contribute to the phenotype of the HSV-1-pUL36-WD/AA-WE/AA mutants.
ACKNOWLEDGMENTS
We are grateful to Russel Diefenbach (Centre for Virus Research, University of Sydney, Australia) and Abel Viejo-Borbolla, Thomas Krey, Fenja Anderson, and Angelika Hinz (Institute of Virology, Hannover Medical School) for fruitful discussions and critical feedback on the manuscript and to the MHH Research Core Unit for Laser Microscopy (RECOLA) and the MHH Research Core Unit for Electron Microscopy for continuous support. We thank Prashant Desai (Johns Hopkins University, Baltimore, MD), Greg Smith (Northwestern University, Chicago, IL), Karsten Tischer and Klaus Osterrieder (Freie Universität Berlin, Germany), and Eva Borst and Martin Messerle (Institute of Virology, MHH) for their generous donation of invaluable cell lines, viruses, and plasmids. Antibodies were generously provided by Helena Browne and Tony Minson (University of Cambridge, United Kingdom), Gary Cohen and Roselyn Eisenberg (University of Pennsylvania, Philadelphia), Richard Courtney (Pennsylvania State University, Hershey), Prashant Desai (Johns Hopkins University), Gillian Elliott and Peter O'Hare (Imperial College, London, United Kingdom), Ari Helenius (ETH, Zurich, Switzerland), and Bill Newcomb and Jay Brown (University of Virginia, Charlottesville).
Lyudmila Ivanova and Anna Buch were Ph.D. Fellows of the Hannover Biomedical Research School (HBRS), Hannover Medical School, Centre of Infection Biology (ZIB), and this work is part of Lyudmila Ivanova's thesis.
Funding Statement
This study was supported by the Niedersachsen Research Network on Neuroinfectiology (N-RENNT) of the Ministry of Science and Culture of Lower Saxony, the Deutsche Forschungsgemeinschaft (DFG; German Research Council) (program project grant SFB 900 TP C2 and Cluster of Excellence Regenerative Biology and Reconstructive Therapies [REBIRTH] award EXC 62/1 to Beate Sodeik), and the Deutsches Zentrum für Infektionsforschung (DZIF; German Center for Infection Research) (TTU Infections of the Immunocompromised Host).
REFERENCES
- 1.Kolb AW, Larsen IV, Cuellar JA, Brandt CR. 2015. Genomic, phylogenetic, and recombinational characterization of herpes simplex virus 2 strains. J Virol 89:6427–6434. doi: 10.1128/JVI.00416-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee K, Kolb AW, Sverchkov Y, Cuellar JA, Craven M, Brandt CR. 2015. Recombination analysis of herpes simplex virus 1 reveals a bias toward GC content and the inverted repeat regions. J Virol 89:7214–7223. doi: 10.1128/JVI.00880-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szpara ML, Gatherer D, Ochoa A, Greenbaum B, Dolan A, Bowden RJ, Enquist LW, Legendre M, Davison AJ. 2014. Evolution and diversity in human herpes simplex virus genomes. J Virol 88:1209–1227. doi: 10.1128/JVI.01987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davison AJ. 2011. Evolution of sexually transmitted and sexually transmissible human herpesviruses. Ann N Y Acad Sci 1230:E37–E49. doi: 10.1111/j.1749-6632.2011.06358.x. [DOI] [PubMed] [Google Scholar]
- 5.Corey L, Wald A. 2009. Maternal and neonatal herpes simplex virus infections. N Engl J Med 361:1376–1385. doi: 10.1056/NEJMra0807633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinchington PR, Leger AJ, Guedon JM, Hendricks RL. 2012. Herpes simplex virus and varicella zoster virus, the house guests who never leave. Herpesviridae 3:5. doi: 10.1186/2042-4280-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson C, Whitley R. 2011. Neonatal herpes simplex virus infections: where are we now? Adv Exp Med Biol 697:221–230. doi: 10.1007/978-1-4419-7185-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grünewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 9.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res 143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Owen DJ, Crump CM, Graham SC. 2015. Tegument sssembly and secondary envelopment of alphaherpesviruses. Viruses 7:5084–5114. doi: 10.3390/v7092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 12.Bosse JB, Hogue IB, Feric M, Thiberge SY, Sodeik B, Brangwynne CP, Enquist LW. 2015. Remodeling nuclear architecture allows efficient transport of herpesvirus capsids by diffusion. Proc Natl Acad Sci U S A 112:E5725–E5733. doi: 10.1073/pnas.1513876112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henaff D, Remillard-Labrosse G, Loret S, Lippé R. 2013. Analysis of the early steps of herpes simplex virus 1 capsid tegumentation. J Virol 87:4895–4906. doi: 10.1128/JVI.03292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith G. 2012. Herpesvirus transport to the nervous system and back again. Annu Rev Microbiol 66:153–176. doi: 10.1146/annurev-micro-092611-150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandbaumhüter M, Döhner K, Schipke J, Binz A, Pohlmann A, Sodeik B, Bauerfeind R. 2013. Cytosolic herpes simplex virus capsids not only require binding inner tegument protein pUL36 but also pUL37 for active transport prior to secondary envelopment. Cell Microbiol 15:248–269. doi: 10.1111/cmi.12075. [DOI] [PubMed] [Google Scholar]
- 16.Pasdeloup D, McElwee M, Beilstein F, Labetoulle M, Rixon FJ. 2013. Herpesvirus tegument protein pUL37 interacts with dystonin/BPAG1 to promote capsid transport on microtubules during egress. J Virol 87:2857–2867. doi: 10.1128/JVI.02676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollinshead M, Johns HL, Sayers CL, Gonzalez-Lopez C, Smith GL, Elliott G. 2012. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. EMBO J 31:4204–4220. doi: 10.1038/emboj.2012.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibiricu I, Huiskonen JT, Döhner K, Bradke F, Sodeik B, Grünewald K. 2011. Cryo electron tomography of herpes simplex virus during axonal transport and secondary envelopment in primary neurons. PLoS Pathog 7:e1002406. doi: 10.1371/journal.ppat.1002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turcotte S, Letellier J, Lippé R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J Virol 79:8847–8860. doi: 10.1128/JVI.79.14.8847-8860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albecka A, Laine RF, Janssen AF, Kaminski CF, Crump CM. 2016. HSV-1 glycoproteins are delivered to virus assembly sites through dynamin-dependent endocytosis. Traffic 17:21–39. doi: 10.1111/tra.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devadas D, Koithan T, Diestel R, Prank U, Sodeik B, Döhner K. 2014. Herpes simplex virus internalization into epithelial cells requires Na+/H+ exchangers and p21-activated kinases but neither clathrin- nor caveolin-mediated endocytosis. J Virol 88:13378–13395. doi: 10.1128/JVI.03631-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. 2012. Herpes virus fusion and entry: a story with many characters. Viruses 4:800–832. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campadelli-Fiume G, Menotti L, Avitabile E, Gianni T. 2012. Viral and cellular contributions to herpes simplex virus entry into the cell. Curr Opin Virol 2:28–36. doi: 10.1016/j.coviro.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Nicola AV, Hou J, Major EO, Straus SE. 2005. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J Virol 79:7609–7616. doi: 10.1128/JVI.79.12.7609-7616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurer UE, Sodeik B, Grünewald K. 2008. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc Natl Acad Sci U S A 105:10559–10564. doi: 10.1073/pnas.0801674105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schipke J, Pohlmann A, Diestel R, Binz A, Rudolph K, Nagel CH, Bauerfeind R, Sodeik B. 2012. The C terminus of the large tegument protein pUL36 contains multiple capsid binding sites that function differently during assembly and cell entry of herpes simplex virus. J Virol 86:3682–3700. doi: 10.1128/JVI.06432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luxton GW, Haverlock S, Coller KE, Antinone SE, Pincetic A, Smith GA. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc Natl Acad Sci U S A 102:5832–5837. doi: 10.1073/pnas.0500803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts AP, Abaitua F, O'Hare P, McNab D, Rixon FJ, Pasdeloup D. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J Virol 83:105–116. doi: 10.1128/JVI.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batterson W, Furlong D, Roizman B. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol 45:397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sodeik B, Ebersold MW, Helenius A. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol 136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rode K, Döhner K, Binz A, Glass M, Strive T, Bauerfeind R, Sodeik B. 2011. Uncoupling uncoating of herpes simplex virus genomes from their nuclear import and gene expression. J Virol 85:4271–4283. doi: 10.1128/JVI.02067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McElwee M, Beilstein F, Labetoulle M, Rixon FJ, Pasdeloup D. 2013. Dystonin/BPAG1 promotes plus-end-directed transport of herpes simplex virus 1 capsids on microtubules during entry. J Virol 87:11008–11018. doi: 10.1128/JVI.01633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spear PG, Roizman B. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol 9:143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNabb DS, Courtney RJ. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221–232. doi: 10.1016/0042-6822(92)91208-C. [DOI] [PubMed] [Google Scholar]
- 35.Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J Virol 76:3065–3071. doi: 10.1128/JVI.76.6.3065-3071.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desai PJ. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol 74:11608–11618. doi: 10.1128/JVI.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuchs W, Klupp BG, Granzow H, Mettenleiter TC. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J Virol 78:11879–11889. doi: 10.1128/JVI.78.21.11879-11889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kharkwal H, Furgiuele SS, Smith CG, Wilson DW. 2015. Herpes simplex virus capsid-organelle association in the absence of the large tegument protein UL36p. J Virol 89:11372–11382. doi: 10.1128/JVI.01893-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Copeland AM, Newcomb WW, Brown JC. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J Virol 83:1660–1668. doi: 10.1128/JVI.01139-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J Virol 83:6610–6623. doi: 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell 19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Bolstad M, Abaitua F, Crump CM, O'Hare P. 2011. Autocatalytic activity of the ubiquitin-specific protease domain of herpes simplex virus 1 VP1-2. J Virol 85:8738–8751. doi: 10.1128/JVI.00798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calistri A, Munegato D, Toffoletto M, Celestino M, Franchin E, Comin A, Sartori E, Salata C, Parolin C, Palu G. 2015. Functional interaction between the ESCRT-I component TSG101 and the HSV-1 tegument ubiquitin specific protease. J Cell Physiol 230:1794–1806. doi: 10.1002/jcp.24890. [DOI] [PubMed] [Google Scholar]
- 44.Kelly BJ, Bauerfeind R, Binz A, Sodeik B, Laimbacher AS, Fraefel C, Diefenbach RJ. 2014. The interaction of the HSV-1 tegument proteins pUL36 and pUL37 is essential for secondary envelopment during viral egress. Virology 454-455:67–77. doi: 10.1016/j.virol.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Svobodova S, Bell S, Crump CM. 2012. Analysis of the interaction between the essential herpes simplex virus 1 tegument proteins VP16 and VP1/2. J Virol 86:473–483. doi: 10.1128/JVI.05981-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ko DH, Cunningham AL, Diefenbach RJ. 2010. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2). J Virol 84:1397–1405. doi: 10.1128/JVI.01721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stellberger T, Hauser R, Baiker A, Pothineni VR, Haas J, Uetz P. 2010. Improving the yeast two-hybrid system with permutated fusions proteins: the varicella zoster virus interactome. Proteome Sci 8:8. doi: 10.1186/1477-5956-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hennig T, Abaitua F, O'Hare P. 2014. Functional analysis of nuclear localization signals in VP1-2 homologues from all herpesvirus subfamilies. J Virol 88:5391–5405. doi: 10.1128/JVI.03797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abaitua F, Hollinshead M, Bolstad M, Crump CM, O'Hare P. 2012. A nuclear localization signal in herpesvirus protein VP1-2 is essential for infection via capsid routing to the nuclear pore. J Virol 86:8998–9014. doi: 10.1128/JVI.01209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scrima N, Lepault J, Boulard Y, Pasdeloup D, Bressanelli S, Roche S. 2015. Insights into herpesvirus tegument organization from structural analyses of the 970 central residues of HSV-1 UL36 protein. J Biol Chem 290:8820–8833. doi: 10.1074/jbc.M114.612838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cardone G, Newcomb WW, Cheng NQ, Wingfield PT, Trus BL, Brown JC, Steven AC. 2012. The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J Virol 86:4058–4064. doi: 10.1128/JVI.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan WH, Roberts AP, McElwee M, Bhella D, Rixon FJ, Lauder R. 2015. The large tegument protein pUL36 is essential for formation of the capsid vertex-specific component at the capsid-tegument interface of herpes simplex virus 1. J Virol 89:1502–1511. doi: 10.1128/JVI.02887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laine RF, Albecka A, van de Linde S, Rees EJ, Crump CM, Kaminski CF. 2015. Structural analysis of herpes simplex virus by optical super-resolution imaging. Nat Commun 6:5980. doi: 10.1038/ncomms6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bogan AA, Thorn KS. 1998. Anatomy of hot spots in protein interfaces. J Mol Biol 280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 55.Morrow JK, Zhang S. 2012. Computational prediction of protein hot spot residues. Curr Pharm Des 18:1255–1265. doi: 10.2174/138161212799436412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tompa P, Davey NE, Gibson TJ, Babu MM. 2014. A million peptide motifs for the molecular biologist. Mol Cell 55:161–169. doi: 10.1016/j.molcel.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 57.Moreira IS, Fernandes PA, Ramos MJ. 2007. Hot spots—a review of the protein-protein interface determinant amino-acid residues. Proteins 68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 58.Samanta U, Pal D, Chakrabarti P. 2000. Environment of tryptophan side chains in proteins. Proteins 38:288–300. [PubMed] [Google Scholar]
- 59.Eisenberg RJ, Ponce de Leon M, Friedman HM, Fries LF, Frank MM, Hastings JC, Cohen GH. 1987. Complement component C3b binds directly to purified glycoprotein C of herpes simplex virus types 1 and 2. Microb Pathog 3:423–435. doi: 10.1016/0882-4010(87)90012-X. [DOI] [PubMed] [Google Scholar]
- 60.Desai P, DeLuca NA, Person S. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 61.Leege T, Granzow H, Fuchs W, Klupp BG, Mettenleiter TC. 2009. Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type 1. J Gen Virol 90:1560–1568. doi: 10.1099/vir.0.010322-0. [DOI] [PubMed] [Google Scholar]
- 62.Elliott G, O'Hare P. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223–233. doi: 10.1016/S0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 63.Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol Cell Biol 20:4922–4931. doi: 10.1128/MCB.20.13.4922-4931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Döhner K, Radtke K, Schmidt S, Sodeik B. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated capsid transport without the small capsid protein VP26. J Virol 80:8211–8224. doi: 10.1128/JVI.02528-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagel CH, Döhner K, Binz A, Bauerfeind R, Sodeik B. 2012. Improper tagging of the non-essential small capsid protein VP26 impairs nuclear capsid egress of herpes simplex virus. PLoS One 7:e44177. doi: 10.1371/journal.pone.0044177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nygardas M, Paavilainen H, Müther N, Nagel CH, Roytta M, Sodeik B, Hukkanen V. 2013. A herpes simplex virus-derived replicative vector expressing LIF limits experimental demyelinating disease and modulates autoimmunity. PLoS One 8:e64200. doi: 10.1371/journal.pone.0064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagel CH, Pohlmann A, Sodeik B. 2014. Construction and characterization of bacterial artificial chromosomes (BACs) containing herpes simplex virus full-length genomes. Methods Mol Biol 1144:43–62. doi: 10.1007/978-1-4939-0428-0_4. [DOI] [PubMed] [Google Scholar]
- 68.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 69.Döhner K, Wolfstein A, Prank U, Echeverri C, Dujardin D, Vallee R, Sodeik B. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol Biol Cell 13:2795–2809. doi: 10.1091/mbc.01-07-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nagel CH, Döhner K, Fathollahy M, Strive T, Borst EM, Messerle M, Sodeik B. 2008. Nuclear egress and envelopment of herpes simplex virus capsids analyzed with dual-color fluorescence HSV1(17+). J Virol 82:3109–3124. doi: 10.1128/JVI.02124-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson DW, Davis-Poynter N, Minson AC. 1994. Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J Virol 68:6985–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Farnham AE, Newton AA. 1959. The effect of some environmental factors on herpes virus grown in HeLa cells. Virology 7:449–461. doi: 10.1016/0042-6822(59)90073-X. [DOI] [PubMed] [Google Scholar]
- 73.McDonald K. 1984. Osmium ferricyanide fixation improves microfilament preservation and membrane visualization in a variety of animal cell types. J Ultrastruct Res 86:107–118. doi: 10.1016/S0022-5320(84)80051-9. [DOI] [PubMed] [Google Scholar]
- 74.Steiner M, Schofer C, Mosgoeller W. 1994. In situ flat embedding of monolayers and cell relocation in the acrylic resin LR white for comparative light and electron microscopy studies. Histochem J 26:934–938. doi: 10.1007/BF02388570. [DOI] [PubMed] [Google Scholar]
- 75.Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watson ML. 1958. Staining of tissue sections for electron microscopy with heavy metals. J Biophys Biochem Cytol 4:475–478. doi: 10.1083/jcb.4.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Funk C, Ott M, Raschbichler V, Nagel CH, Binz A, Sodeik B, Bauerfeind R, Bailer SM. 2015. The herpes simplex virus protein pUL31 escorts nucleocapsids to sites of nuclear egress, a process coordinated by its N-terminal domain. PLoS Pathog 11:e1004957. doi: 10.1371/journal.ppat.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGeoch DJ, Dalrymple MA, Davison AJ, Dolan A, Frame MC, McNab D, Perry LJ, Scott JE, Taylor P. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol 69:1531–1574. [DOI] [PubMed] [Google Scholar]
- 79.Umene K, Sakaoka H. 1991. Homogeneity and diversity of genome polymorphism in a set of herpes simplex virus type 1 strains classified as the same genotypic group. Arch Virol 119:53–65. doi: 10.1007/BF01314323. [DOI] [PubMed] [Google Scholar]
- 80.Jovasevic V, Liang L, Roizman B. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J Virol 82:3311–3319. doi: 10.1128/JVI.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abaitua F, O'Hare P. 2008. Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein. J Virol 82:5234–5244. doi: 10.1128/JVI.02497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Granzow H, Klupp BG, Fuchs W, Veits J, Osterrieder N, Mettenleiter TC. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J Virol 75:3675–3684. doi: 10.1128/JVI.75.8.3675-3684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mettenleiter TC. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet Microbiol 113:163–169. doi: 10.1016/j.vetmic.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 84.Maringer K, Stylianou J, Elliott G. 2012. A network of protein interactions around the herpes simplex virus tegument protein VP22. J Virol 86:12971–12982. doi: 10.1128/JVI.01913-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tanaka M, Kato A, Satoh Y, Ide T, Sagou K, Kimura K, Hasegawa H, Kawaguchi Y. 2012. Herpes simplex virus 1 VP22 regulates translocation of multiple viral and cellular proteins and promotes neurovirulence. J Virol 86:5264–5277. doi: 10.1128/JVI.06913-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Starkey JL, Han J, Chadha P, Marsh JA, Wills JW. 2014. Elucidation of the block to herpes simplex virus egress in the absence of tegument protein UL16 reveals a novel interaction with VP22. J Virol 88:110–119. doi: 10.1128/JVI.02555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kotsakis A, Pomeranz LE, Blouin A, Blaho JA. 2001. Microtubule reorganization during herpes simplex virus type 1 infection facilitates the nuclear localization of VP22, a major virion tegument protein. J Virol 75:8697–8711. doi: 10.1128/JVI.75.18.8697-8711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pomeranz LE, Blaho JA. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J Virol 73:6769–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Leeuwen H, Okuwaki M, Hong R, Chakravarti D, Nagata K, O'Hare P. 2003. Herpes simplex virus type 1 tegument protein VP22 interacts with TAF-I proteins and inhibits nucleosome assembly but not regulation of histone acetylation by INHAT. J Gen Virol 84:2501–2510. doi: 10.1099/vir.0.19326-0. [DOI] [PubMed] [Google Scholar]
- 90.Lee JI, Luxton GW, Smith GA. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J Virol 80:12086–12094. doi: 10.1128/JVI.01184-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huffmaster NJ, Sollars PJ, Richards AL, Pickard GE, Smith GA. 2015. Dynamic ubiquitination drives herpesvirus neuroinvasion. Proc Natl Acad Sci U S A 112:12818–12823. doi: 10.1073/pnas.1512559112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gastaldello S, Hildebrand S, Faridani O, Callegari S, Palmkvist M, Di Guglielmo C, Masucci MG. 2010. A deneddylase encoded by Epstein-Barr virus promotes viral DNA replication by regulating the activity of cullin-RING ligases. Nat Cell Biol 12:351–361. doi: 10.1038/ncb2035. [DOI] [PubMed] [Google Scholar]
- 93.Schlieker C, Weihofen WA, Frijns E, Kattenhorn LM, Gaudet R, Ploegh HL. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol Cell 25:677–687. doi: 10.1016/j.molcel.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, Honigschmid P, Schafferhans A, Roos M, Bernhofer M, Richter L, Ashkenazy H, Punta M, Schlessinger A, Bromberg Y, Schneider R, Vriend G, Sander C, Ben-Tal N, Rost B. 2014. PredictProtein—an open resource for online prediction of protein structural and functional features. Nucleic Acids Res 42:W337–343. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Radtke K, Döhner K, Sodeik B. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol 8:387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 96.Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog 6:e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sodeik B. 2000. Mechanisms of viral transport in the cytoplasm. Trends Microbiol 8:465–472. doi: 10.1016/S0966-842X(00)01824-2. [DOI] [PubMed] [Google Scholar]
- 98.Wolfstein A, Nagel CH, Radtke K, Döhner K, Allan VJ, Sodeik B. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227–237. doi: 10.1111/j.1600-0854.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- 99.Luxton GW, Lee JI, Haverlock-Moyns S, Schober JM, Smith GA. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J Virol 80:201–209. doi: 10.1128/JVI.80.1.201-209.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shanda SK, Wilson DW. 2008. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J Virol 82:7388–7394. doi: 10.1128/JVI.00225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zaichick SV, Bohannon KP, Hughes A, Sollars PJ, Pickard GE, Smith GA. 2013. The herpesvirus VP1/2 protein is an effector of dynein-mediated capsid transport and neuroinvasion. Cell Host Microbe 13:193–203. doi: 10.1016/j.chom.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kramer T, Enquist LW. 2013. Directional spread of alphaherpesviruses in the nervous system. Viruses 5:678–707. doi: 10.3390/v5020678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Diefenbach RJ, Miranda-Saksena M, Douglas MW, Cunningham AL. 2008. Transport and egress of herpes simplex virus in neurons. Rev Med Virol 18:35–51. doi: 10.1002/rmv.560. [DOI] [PubMed] [Google Scholar]
- 104.Dodding MP, Mitter R, Humphries AC, Way M. 2011. A kinesin-1 binding motif in vaccinia virus that is widespread throughout the human genome. EMBO J 30:4523–4538. doi: 10.1038/emboj.2011.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dodding MP, Way M. 2011. Coupling viruses to dynein and kinesin-1. EMBO J 30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pernigo S, Lamprecht A, Steiner RA, Dodding MP. 2013. Structural basis for kinesin-1:cargo recognition. Science 340:356–359. doi: 10.1126/science.1234264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Avitabile E, Di Gaeta S, Torrisi MR, Ward PL, Roizman B, Campadelli-Fiume G. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J Virol 69:7472–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bearer EL, Breakefield XO, Schuback D, Reese TS, LaVail JH. 2000. Retrograde axonal transport of herpes simplex virus: evidence for a single mechanism and a role for tegument. Proc Natl Acad Sci U S A 97:8146–8150. doi: 10.1073/pnas.97.14.8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee JH, Vittone V, Diefenbach E, Cunningham AL, Diefenbach RJ. 2008. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378:347–354. doi: 10.1016/j.virol.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 110.Abaitua F, Daikoku T, Crump CM, Bolstad M, O'Hare P. 2011. A single mutation responsible for temperature-sensitive entry and assembly defects in the VP1-2 protein of herpes simplex virus. J Virol 85:2024–2036. doi: 10.1128/JVI.01895-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abaitua F, Souto RN, Browne H, Daikoku T, O'Hare P. 2009. Characterization of the herpes simplex virus (HSV)-1 tegument protein VP1-2 during infection with the HSV temperature-sensitive mutant tsB7. J Gen Virol 90:2353–2363. doi: 10.1099/vir.0.012492-0. [DOI] [PubMed] [Google Scholar]
- 112.Coller KE, Lee JI, Ueda A, Smith GA. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol 81:11790–11797. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]