ABSTRACT
Although the mechanism is not well understood, growing evidence indicates that the nonenveloped parvovirus minute virus of mice (MVM) may actively egress before passive release through cell lysis. We have dissected the late maturation steps of the intranuclear progeny with the aims of confirming the existence of active prelytic egress and identifying critical capsid rearrangements required to initiate the process. By performing anion-exchange chromatography (AEX), we separated intranuclear progeny particles by their net surface charges. Apart from empty capsids (EC), two distinct populations of full capsids (FC) arose in the nuclei of infected cells. The earliest population of FC to appear was infectious but, like EC, could not be actively exported from the nucleus. Further maturation of this early population, involving the phosphorylation of surface residues, gave rise to a second, late population with nuclear export potential. While capsid surface phosphorylation was strictly associated with nuclear export capacity, mutational analysis revealed that the phosphoserine-rich N terminus of VP2 (N-VP2) was dispensable, although it contributed to passive release. The reverse situation was observed for the incoming particles, which were dephosphorylated in the endosomes. Our results confirm the existence of active prelytic egress and reveal a late phosphorylation event occurring in the nucleus as a selective factor for initiating the process.
IMPORTANCE In general, the process of egress of enveloped viruses is active and involves host cell membranes. However, the release of nonenveloped viruses seems to rely more on cell lysis. At least for some nonenveloped viruses, an active process before passive release by cell lysis has been reported, although the underlying mechanism remains poorly understood. By using the nonenveloped model parvovirus minute virus of mice, we could confirm the existence of an active process of nuclear export and further characterize the associated capsid maturation steps. Following DNA packaging in the nucleus, capsids required further modifications, involving the phosphorylation of surface residues, to acquire nuclear export potential. Inversely, those surface residues were dephosphorylated on entering capsids. These spatially controlled phosphorylation-dephosphorylation events concurred with the nuclear export-import potential required to complete the infectious cycle.
INTRODUCTION
The egress of enveloped viruses is well characterized and involves budding through host cell membranes (1, 2). The release of nonenveloped viruses is less well understood. In general, the release of nonenveloped viruses is associated with cellular lysis and thus is considered a passive process (3–5). However, accumulating data suggest that active egress precedes virus-induced cell lysis and subsequent passive release. For instance, bluetongue virus has been demonstrated to usurp the ESCRT machinery for egress by means of its L-domains (6, 7). Similarly, the release of hepatitis A virus requires ESCRT-associated proteins (8). Furthermore, drug-induced stimulation of the autophagy pathway increases the nonlytic spread of poliovirus, and progeny virions have been shown to accumulate unilaterally on the apical surfaces of polarized and productively infected epithelial cells (9, 10). Equally, simian vacuolating virus 40 and simian rotavirus have been recovered almost exclusively from the apical culture fluid of polarized epithelial cells prior to cell lysis. Electron microscopy studies and specific inhibition of vesicular transport pathways indicate a vesicle-associated release of progeny virions (11, 12).
An active egress process has also been suggested for parvoviruses (PV), a group of small nonenveloped viruses (13–15). Autonomous rodent PV, including minute virus of mice (MVM), display a T=1 icosahedral capsid containing a single-stranded DNA (ssDNA) genome of about 5 kb (16). Due to their simplicity, PV depend strongly on their host cells. Following entry, they are imported into the nucleus, where they profit from the replication machinery of the host for their own replication. Subsequently, assembly and genome packaging occur in the nucleus and give rise to infectious progeny. Productive PV infection causes dramatic morphological and physiological changes in host cells, culminating in cell death and the passive release of progeny virions. The cytotoxicity of PV is mediated mainly by the large nonstructural protein NS1 (3, 17, 18).
Besides passive egress by cell lysis, the existence of active, prelytic egress for MVM has been suggested (13–15). Several viral and cellular factors involved in PV egress have been identified. The highly stable interaction of the viral nonstructural protein NS2 with CRM1 has been proposed to play a role in egress (19, 20). Classical nuclear export signals (NES) exhibit low affinity for CRM1, preventing the formation of stable CRM1/cargo complexes in the cytoplasm, where RanGTP is absent (21). Surprisingly, the NES of NS2 belongs to the supraphysiological NES, which bind tightly to CRM1 regardless of the presence of RanGTP. Therefore, NS2 competitively inhibits CRM1 function by sequestering endogenous nuclear export receptors (22). MVM mutants with disabled CRM1 interaction were compromised in viral nuclear export and productive infection (23, 24). The exact role of NS2 in virus egress was not elucidated, and attempts to demonstrate an interaction of NS2 with viral capsid proteins were unsuccessful. Since NS2 has multiple functions, abrogation of the tight NS2–CRM1 interaction might interfere with early functions during a productive infection, which may indirectly affect the maturation of progeny and their export from the nucleus. In transformed human cells, NS2 was dispensable for infection (25), and progeny export was not affected by treatment with the antifungal antibiotic leptomycin B, a potent inhibitor of CRM1-dependent nuclear export. For these cells, an alternative export mechanism, involving the N terminus of VP2 (N-VP2) and the phosphorylation of its serines, has been proposed (15). Site-directed mutagenesis of the three distal serine residues at positions 2, 6, and 10 of N-VP2 revealed an important role for these phosphorylations in the CRM1-independent nuclear export of MVM in permissive transformed human cells (15).
It has been suggested that following nuclear export, MVM is released actively through a vesicle-associated, gelsolin-dependent mechanism involving major rearrangement of the cytoskeleton. Progeny virions have been shown to colocalize with exocytic, endosomal, and lysosomal markers in immunofluorescent experiments. Cell fractionation experiments have confirmed this observation by demonstrating comigration of viral particles with cytosolic vesicles (14). Progeny virions would become engulfed by COPII vesicles at the perinuclear endoplasmic reticulum (ER), where they accumulate with dynamin. Accordingly, dramatic retention of virions in the perinuclear area and inhibition of virion release into the medium were observed in cells lacking functional effectors of the secretory pathway (13). In addition, members of the ezrin-radixin-moesin (ERM) family have been shown to play a role in virus maturation and spreading capacity, as judged by their impact on MVM plaque morphology (26). Dominant negative radixin or moesin mutants failed to wrap progeny virions into transport vesicles, resulting in a marked reduction in virus egress.
Documentation of active egress by nonenveloped viruses requires accurate demonstration that no cell lysis occurred during the experiment. However, it is challenging to exclude the possibility that lysis of a few cells may passively release progeny virions, which could additionally contribute to uncontrolled second rounds of infection. Parvoviruses, particularly MVM, are highly robust and can persist as intact particles in the lysosomes of infected cells (27). The entry route and the proposed egress route largely overlap in the nucleus and in the dynamic endosomal pathway. Accordingly, distinguishing consistently between incoming and progeny virions represents a major challenge and requires exhaustive methodologies capable of identifying and isolating viruses at specific steps of the infection.
Through anion-exchange chromatography (AEX) in combination with cell fractionation and quantitative PCR (qPCR), viral particles were isolated by their particular capsid surface configurations. This approach allowed the detection of a previously unrecognized population of progeny virus and revealed a selective process of nuclear export and egress preceding passive release by cell lysis. Only DNA-containing capsids with an externalized N-VP2 and additional surface phosphorylations were able to egress actively. Although the phosphoserine-rich N-VP2 was found to be dispensable, critical phosphorylation of capsid surface residues was strictly associated with nuclear export potential. In agreement with a role in export, those residues were efficiently dephosphorylated on incoming capsids.
MATERIALS AND METHODS
Cells and viruses.
A9 mouse fibroblasts (28) and NB324K cells (29) were routinely propagated with a minimal number of passages in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal calf serum (FCS) at 37°C under a 5% CO2 atmosphere. Stocks of MVM were propagated on A9 cells. As soon as the cytopathic effect was complete, the supernatant was collected and was precleared of cell debris by low-speed centrifugation, and the virus was pelleted through a 20% sucrose cushion. The virus pellet was washed and was resuspended in phosphate-buffered saline (PBS). Titers were determined by quantitative PCR (qPCR) and are expressed as the number of DNA-containing particles per microliter. DNA-containing capsids (full capsids [FC]) and empty capsids (EC) were separated by CsCl gradient centrifugation as described previously (30). CsCl was removed by size exclusion chromatography through PD-10 desalting columns (GE Healthcare, Chalfont St Giles, United Kingdom), and when required, the capsids were concentrated in Amicon centrifugal filter devices (Merck Millipore, Billerica, MA).
Antibodies, chemicals, and enzymes.
Rabbit polyclonal antibodies (pAbs) against MVM structural proteins, a rabbit polyclonal antibody against the N terminus of VP2 (N-VP2), and a mouse monoclonal antibody (MAb) against intact capsids (clone B7) have been described previously (15, 31). Fluorescently labeled secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and horseradish peroxidase-conjugated antibodies were purchased from DakoCytomation (Glostrup, Denmark). Bafilomycin A1 (BafA1), chymotrypsin (CHT), and chymostatin were obtained from Sigma-Aldrich (St. Louis, MO) and were reconstituted in ethanol at 0.1 mg/ml, in 1 mM HCl with 2 mM CaCl2, or in dimethyl sulfoxide DMSO at 10 mM, respectively. To avoid enzymatic digestion or dephosphorylation during the processing of cell extracts, the lysis buffer was supplemented with protease inhibitors (Roche, Basel, Switzerland), 1 mM sodium orthovanadate (Na3VO4), and 1 mM sodium fluoride (NaF) (Sigma-Aldrich).
Virus infection.
A9 or NB324K cells (8 × 103 cells for qPCR; 3 × 106 cells for AEX) were infected with MVM (5,000 DNA-containing particles per cell, corresponding to approximately 10 PFU/cell [32]) for 1 h at 4°C for binding. Unbound virus was removed by extensive washes, and the cells were incubated at 37°C to initiate the infection. At various times postinternalization, total cellular DNA was extracted for qPCR analysis, or cells were fractionated and subjected to AEX. Cell viability was accessed via trypan blue exclusion using the TC10 automated cell counter (Bio-Rad, Hercules, CA).
Cell fractionation.
A9 and NB324K cytoplasmic fractions were extracted in 50 mM Tris-HCl–150 mM NaCl–5 mM EDTA–1% NP-40–1 mM Na3VO4–1 mM NaF–protease inhibitor cocktail (Roche), pH 7.2, at 4°C for 30 min. After vortexing, intact nuclei and cell debris were removed by high-speed centrifugation at 4°C. Nuclei were isolated by using the Nuclei EZ Prep nucleus isolation kit (Sigma-Aldrich) according to the manufacturer's instructions. In order to obtain highly pure nuclear fractions, the isolated nuclei were further processed by centrifugation at 500 × g for 10 min through a sucrose gradient. The integrity of the isolated nuclei was examined by light microscopy after trypan blue staining. The purity of the nuclei and the absence of the outer nuclear membrane were evaluated with antibodies against lamins A and C (lamin A/C) (for the inner membrane) and SERCA2 ATPase (for the outer membrane). Purified nuclei were lysed in 50 mM Tris-HCl–150 mM NaCl–5 mM EDTA–1% Triton X-100–1 mM Na3VO4–1 mM NaF–protease inhibitor cocktail (Roche), pH 7.2, at 4°C for 30 min. After vortexing, the nuclear lysate was passed 10 times through a 27-gauge needle, and nuclear debris was removed by high-speed centrifugation at 4°C.
qPCR.
Template DNA was extracted by using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's guidelines. Amplification and real-time detection of PCR products were performed by using the CFX96 Real-Time system with iTaq Universal SYBR green Supermix (Bio-Rad). Primers for MVM DNA amplification were as follows: forward, 5′-GACGCACAGAAAGAGAGTAACCAA-3′ (nucleotides 231 to 254); reverse, 5′-CCAACCATCTGCTCCAGTAAACAT-3′ (nucleotides 709 to 732). The specificity of the amplification was determined by melting curve analysis. As an external standard, an infectious clone of MVM (33) was used in 10-fold serial dilutions.
Iodixanol density gradient centrifugation.
When FC devoid of EC and defective particles were needed, virus stocks were fractionated by centrifugation through iodixanol gradients. After extensive cytopathic effect (8 days postinfection [dpi]), intracellular viruses were released into the culture medium by repeated freeze-thaw cycles. Cell debris was removed by low-speed centrifugation. Full capsids were separated from EC and defective particles by centrifugation through discontinuous iodixanol (OptiPrep; Axis-Shield, Oslo, Norway) gradients, as described previously (34). Viral capsids and DNA-containing particles in the fractions were detected by dot blot analysis and qPCR, respectively.
Anion-exchange chromatography (AEX).
The Mono Q HR 5/5 column (inside diameter, 5 mm; length, 50 mm; Pharmacia, Uppsala, Sweden) was connected to the ÄKTApurifier 10/100 chromatography system with a UPC-900 monitor, operated by UNICORN control software (GE Healthcare). The Mono Q column was equilibrated with 5 column volumes (CV) of starting buffer (20 mM Tris-HCl, 1 mM EDTA [pH 7.2]). Viruses (at least 108 virus particles) diluted in 1 ml starting buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8]) were applied to the Mono Q column through a 2-ml injection loop, rinsed with 6 CV of starting buffer, and eluted by a linear salt gradient (0 to 2 M NaCl) in 20 mM Tris-HCl–1 mM EDTA (pH 7.2). The flow rate was constantly kept at 1.5 ml/min, and the salt concentration was monitored by electrical conductivity. Viruses in each fraction (185 μl) were quantified by qPCR.
Plaque assay.
A9 cells were seeded in a 60-mm petri dish. On the following day, the cells were rinsed and inoculated with serial dilutions of FC progeny 1 (FC-P1) or FC progeny 2 (FC-P2) in DMEM without FCS. The virus was allowed to internalize at 37°C for 1 h. The cells were washed with PBS and were covered with low-gelling-temperature agarose (Sigma) containing DMEM and 5% FCS. After 6 days, the cells were fixed with paraformaldehyde and were stained using crystal violet.
Immunofluorescence microscopy.
A total of 3 × 105 A9 cells were seeded onto coverslips in 12-well plates. After 24 h, the cells were infected with iodixanol and AEX-purified FC-P1 or FC-P2 devoid of EC and defective particles at 2,500 DNA-containing particles per cell, corresponding to approximately 5 PFU/cell (35), for 1 h at 4°C. Subsequently, the cells were washed to remove unbound virus and were incubated at 37°C. At different times, cells were washed and processed for immunofluorescence as described previously (36, 37) with fluorescently conjugated secondary antibodies. Cells were mounted with Mowiol (Calbiochem, Billerica, MA) containing 30 mg/ml of 1,4-diazabicyclo(2,2,2)octane (DABCO; Sigma) as an antifade agent and were examined by laser scanning microscopy (LSM880; 63× objective; Carl Zeiss, Germany).
Immunoprecipitation.
Viruses were incubated with specific antibodies in Eppendorf LoBind tubes preblocked with PBSA (PBS containing 1% bovine serum albumin [BSA]) for 1 h at 4°C. Subsequently, 20 μl of protein G agarose beads was added, and the samples were further incubated overnight at 4°C. The beads were washed with PBSA. To remove residual BSA, an additional washing step was carried out with PBS.
Enzymatic reactions.
All enzymatic reactions were performed in a 50-μl reaction volume. Viruses diluted in PBS (108 virus particles) were incubated for 1.5 h at 37°C with chymotrypsin (0.5 mg/ml), and the reaction was stopped by adding 100 μM chymostatin. Lambda phosphatase treatment (40,000 U/ml) was performed in 50 mM Tris-HCl–100 mM NaCl–2 mM MnCl2–5 mM dithiothreitol (DTT), pH 7.8, for 3 h at 37°C. Phosphatase was inactivated by adding 1 mM Na3VO4 and 1 mM NaF. Free DNA was digested using DNase I (50 U) at 37°C for 1.5 h. DNase I was inhibited by incubation at 75°C for 15 min.
RESULTS
Two distinct populations of progeny DNA-containing particles are detectable in the nuclei of MVM-infected cells.
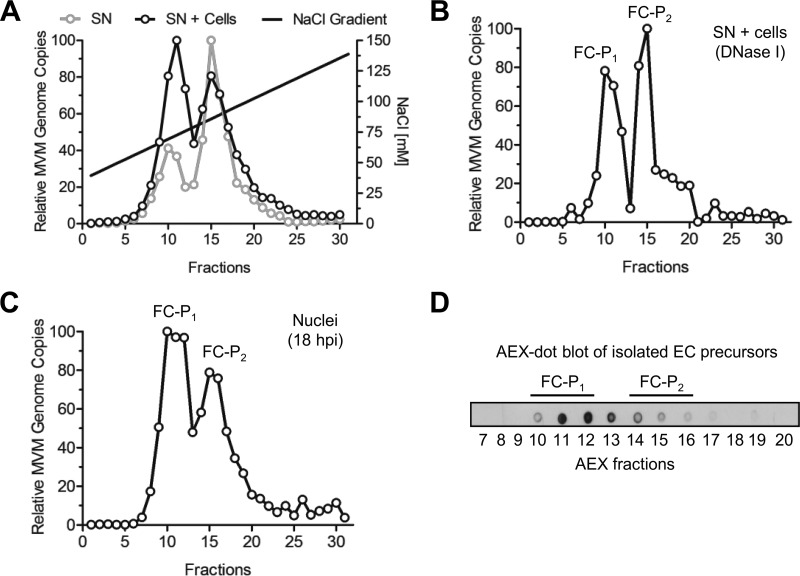
Progeny viral particles accumulating in the supernatants of infected A9 cells were collected 8 days postinfection (dpi), when cytopathic effect was complete. Unlysed cells and cell debris were excluded by centrifugation. The viral capsids collected were analyzed by anion-exchange chromatography (AEX), which can separate particles based on their net surface charges, followed by quantitative PCR (qPCR). Two distinct virus populations of DNA-containing particles (full capsids [FC]) were separated and their relative amounts determined by qPCR (Fig. 1A). A second AEX-qPCR analysis, which included both the medium and the cell lysate obtained by freeze-thaw cycles to release the remaining intracellular viruses, was performed in parallel. When these additional intracellular viruses were included, the same two populations were detected, but their proportions were different. The more positively charged population (referred to as FC progeny 1 [FC-P1]) was associated predominantly with cells; thus, it increased when additional intracellular viruses were included in the assay via freeze-thaw cycles. In contrast, the more negatively charged population (referred to as FC progeny 2 [FC-P2]) was the predominant population in the supernatant, from which many of the intracellular viruses were excluded.
FIG 1.
Isolation of two distinct populations of progeny DNA-containing particles. (A) Progeny viruses (1010 DNA-containing particles) were collected from the culture media of infected A9 monolayers (supernatant [SN]) after extensive cytopathic effect (8 dpi). Alternatively, the medium was enriched with additional intracellular particles by repeated freeze-thaw cycles (SN + cells). AEX was performed, and fractions were collected. The DNA-containing particles in each fraction were quantified by qPCR. (B) Prior to AEX-qPCR, the virus progeny (SN + cells) was treated with DNase I. (C) A9 cells (3 × 106) were infected at a multiplicity of infection of 5,000 DNA-containing particles per cell for 1 h at 4°C and were then washed to remove unbound virus. The cells were further incubated at 37°C for 18 h. To avoid reinfection, neuraminidase and an anti-capsid MAb were added to the cells. Nuclei were purified, and the nuclear progeny was analyzed by AEX-qPCR. (D) EC isolated from infected A9 cells (8 dpi) were subjected to AEX followed by dot blot analysis using the anti-capsid MAb for detection.
In order to verify the integrity of the two DNA-containing viral populations, we collected supernatants and intracellular viruses and subjected them to nucleolytic digestion. As shown in Fig. 1B, both virus populations were resistant to nuclease digestion, and their AEX profiles did not change, indicating that both particle types represent fully assembled DNA-containing particles.
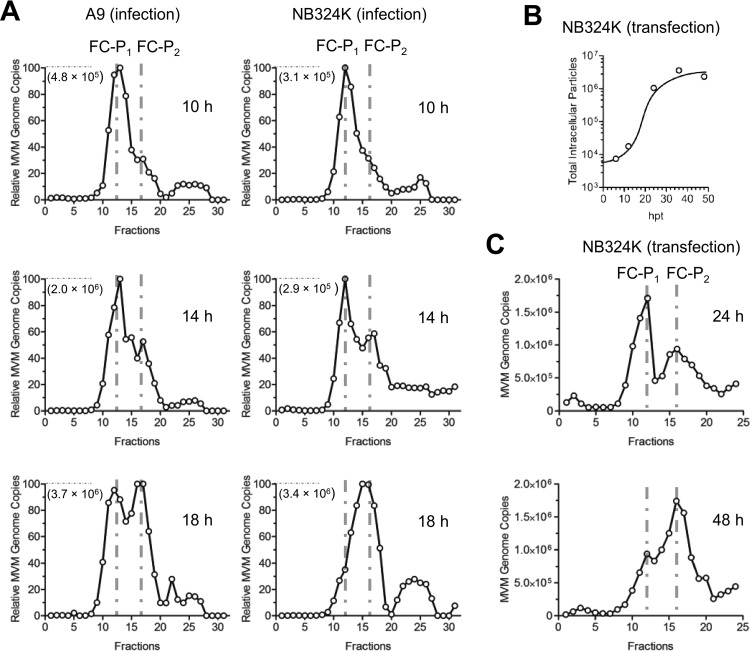
MVM capsid assembly occurs in the nucleus. It was therefore of interest to verify the presence of both virus populations in the nucleus early, at the onset of assembly and packaging. Viruses were collected from isolated nuclei of infected murine fibroblasts early after infection and were subjected to AEX-qPCR. The integrity of the isolated nuclei was examined by trypan blue staining, and the absence of the outer nuclear membrane was evaluated with antibodies against lamin A/C (inner membrane) and SERCA2 ATPase (outer membrane). As shown in Fig. 1C, by 18 h postinfection (hpi), the two DNA-containing viral populations coexisted in the cell nucleus.
Besides DNA-containing capsids, MVM infection results in the accumulation of empty capsids (EC), which represent assembled capsid precursors that have not yet packaged viral genomes (38). To verify their AEX profiles, EC precursors were purified by CsCl centrifugation, subjected to AEX, and visualized by dot blot analysis using an antibody against assembled capsids (MAb B7) (31). As illustrated in Fig. 1D, EC precursors had an AEX profile resembling that of the FC-P1 population. This observation, along with the fact that FC-P1 particles are predominantly cell associated (Fig. 1A), suggests that FC-P1 represent immature particles without egress potential. In contrast, FC-P2 may represent particles displaying further maturation, enabling selective egress.
FC-P1 and FC-P2 are infectious and exhibit differences in density and N-VP2 conformation.
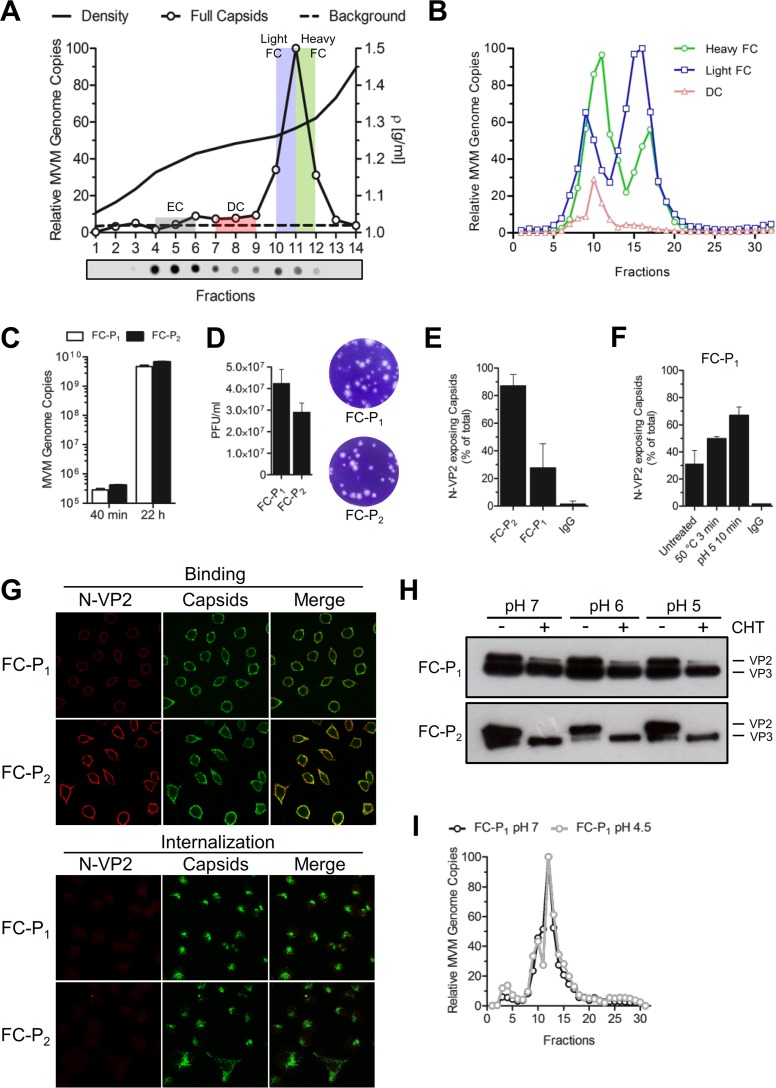
In order to further characterize the two FC populations, the virus progeny was fractionated by centrifugation through iodixanol gradients, as described previously (34). Fractions corresponding to defective particles (fractions 7 to 9), light FC (fraction 10 and half of fraction 11), and heavy FC (half of fraction 11 and fraction 12) (Fig. 2A) were pooled and were analyzed by AEX. The results showed that FC-P1 and FC-P2 originate from the FC fractions (fractions 10 to 12, corresponding to 1.26 to 1.30 g/ml). Interestingly, while FC-P2 particles were enriched in the light fraction (1.26 g/ml), FC-P1 particles were enriched in the heavier fraction (1.30 g/ml), demonstrating a slightly denser composition of FC-P1 (Fig. 2B). Like EC (Fig. 1D), the minor population of defective particles eluted in the same fractions as FC-P1 (Fig. 2B). The similarities of FC-P1 with the EC precursors and defective particles, and the unique AEX profile of FC-P2, further emphasize the differences in virus maturation between the two FC populations.
FIG 2.
Density, infectivity, and N-VP2 conformation of the FC progeny. (A) Viruses were collected 8 dpi and were purified by iodixanol density gradient centrifugation. The gradient was fractionated, and the capsids in each fraction were detected by dot blot analysis using an anti-capsid MAb. FC were detected by qPCR analysis, and relative MVM genome copies are indicated for each fraction. DC, defective capsids. (B) DC (fractions 7 to 9), light FC (fraction 10 and half of fraction 11), and heavy FC (fraction 12 and half of fraction 11) were pooled and were analyzed by AEX-qPCR. The AEX profile for each capsid type is shown. (C) A9 cells (8 × 103) were infected with purified FC-P1 or FC-P2 particles at a multiplicity of infection of 2,500 DNA-containing particles per cell for 1 h at 4°C, followed by washing to remove unbound virus. The cells were further incubated at 37°C for 40 min or 22 h. Total DNA was extracted and quantified as described in Materials and Methods. (D) Representative results illustrating the ratio and morphology of plaques from iodixanol-purified FC-P1 and FC-P2. (E) Immunoprecipitation of 108 iodixanol-purified FC-P1 or FC-P2 particles with an anti-capsid MAb or a rabbit pAb against N-VP2. The specificity of the antibodies was confirmed using nonspecific rabbit IgG. (F) Iodixanol-purified FC-P1 particles (108) were incubated at 50°C for 3 min or at pH 5 for 10 min. Immunoprecipitation was performed as explained above. (G) A9 cells (3 × 105) were infected with purified FC-P1 or FC-P2 virions as indicated above. At different intervals p.i., the proteolytic processing of N-VP2 was examined by immunofluorescence with an anti-capsid MAb (green) and a pAb against N-VP2 (red). (H) Iodixanol-purified FC-P1 or FC-P2 particles (108) were incubated at pH 7, 6, or 5 and were either treated with CHT (+) or left untreated (−). Proteolytic N-VP2 processing was analyzed by 10% SDS-PAGE followed by Western blotting. (I) AEX of FC-P1 with different amounts of exposed N-VP2. Purified FC-P1 particles (1010) were treated at pH 7 or pH 4.5 (to externalize additional N-VP2 subunits), followed by dilution in Tris-EDTA buffer (pH 8) and AEX-qPCR analysis.
The AEX fractions corresponding to FC-P1 and FC-P2 (Fig. 2B) were pooled and were used for further studies. The abilities of the purified viral populations to initiate infection in A9 cells were investigated. As demonstrated in Fig. 2C, both virus populations were able to reach the nucleus, and their genomes were replicated without significant differences. This finding was confirmed by a plaque assay, where the two populations of FC displayed equivalent infectivities. Plaque morphology and size were also similar (Fig. 2D). The particle-to-PFU ratio was 59 ± 8 for FC-P1 and 86 ± 12 for FC-P2.
The N-terminal region of VP2 (N-VP2) occupies an external positon in the virion. However, during entry, N-VP2 is cleaved by endosomal proteases to render a shorter protein named VP3 (39, 40). The function of N-VP2 cleavage is not fully understood, but it is required to allow the exposure of the N-terminal region of VP1 (N-VP1) (41, 42), which harbors important functional motifs essential for infection (43), particularly endosomal escape (44) and nuclear targeting (36) motifs. We analyzed the surface conformations of N-VP2 in the two populations of FC, devoid of EC and defective particles, by immunoprecipitation with a specific antibody raised against this region (15), followed by quantitative PCR. In the majority of FC-P2 particles, N-VP2 was accessible, and the particles were efficiently immunoprecipitated. In contrast, in a significant proportion of FC-P1 particles, N-VP2 was not accessible to antibody binding (Fig. 2E). Exposure of FC-P1 to 50°C or acidic conditions (pH 5) increased the accessibility of N-VP2 (Fig. 2F). The poor accessibility of N-VP2 in FC-P1 particles was further investigated by immunofluorescence microscopy. When incubated with A9 cells, the two virion species bound similarly to cells. However, while N-VP2 from FC-P2 was detectable on the surfaces of cells, N-VP2 from FC-P1 was barely visible. Following internalization, N-VP2 was fully processed in the acidic endosomes and became undetectable in both populations (Fig. 2G).
The conformation of N-VP2 was also analyzed by proteolytic digestion with chymotrypsin (27). To this end, the purified FC populations, devoid of EC and defective particles, were analyzed by SDS-PAGE. As shown in Fig. 2H, a significant amount of VP2 was already processed to VP3 in both FC populations. This process is known to occur in acidic endosomes during cell entry (27). However, while the remaining N-VP2 from the FC-P2 progeny could be fully cleaved by chymotrypsin, a proportion of N-VP2 subunits from the FC-P1 population was inaccessible at a neutral pH, and cleavage increased marginally under acidic conditions.
We next investigated whether the particular N-VP2 conformation in FC-P1 is responsible for its distinct AEX profile. To this end, FC-P1 virions were exposed to acidic conditions in order to increase the externalization of N-VP2 (as shown in Fig. 2F) and were analyzed by AEX-qPCR. The results indicated that despite more-prominent externalization of N-VP2, the AEX profile remained unchanged (Fig. 2I). Accordingly, the difference in the N-VP2 conformation is not responsible for the different AEX profiles. These results also confirm that genome packaging is required to externalize N-VP2; however, further maturation is necessary to mediate the prominent N-VP2 externalization that characterizes mature particles.
The surface phosphorylation status of the capsid, other than the phosphoserine-rich N-VP2, is the key determinant of the different AEX profiles.
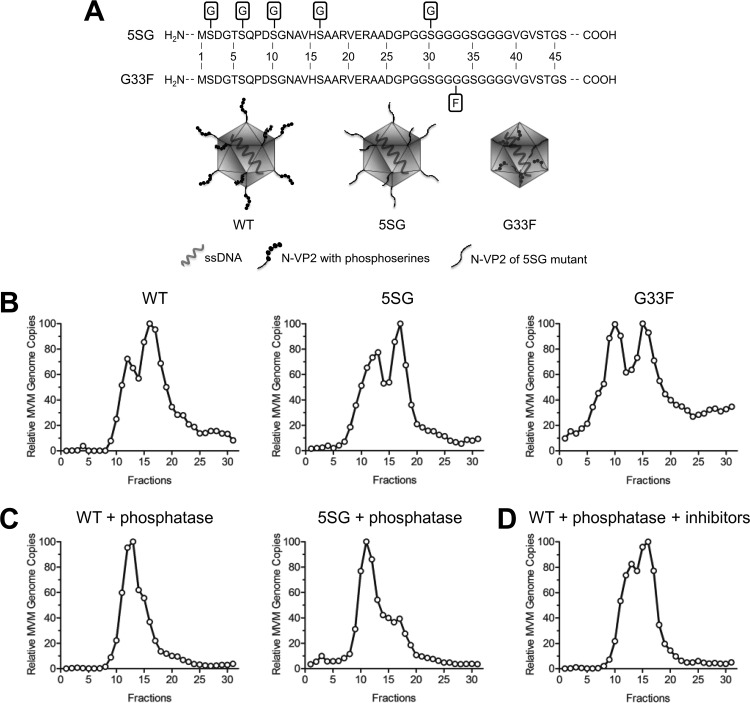
In order to further confirm that the phosphoserine-rich N-VP2 is not responsible for the distinct AEX profiles, we used two N-VP2 mutants (schematically illustrated in Fig. 3A). In one mutant, the four distal serines on N-VP2 and an additional serine in the polyglycine region were replaced by glycine (referred to as the 5SG mutant). Additionally, we used an MVM mutant containing a bulky phenylalanine residue at position 33 within the flexible polyglycine stretch (referred to as the G33F mutant). Due to this substitution, the mutant progeny particles were unable to externalize N-VP2 following DNA packaging. Accordingly, transfection with this mutant generated DNA-containing particles that were not infectious, due to failure to process N-VP2 and, consequently, failure to externalize N-VP1 during entry (45). Like transfection with the wild type (WT), transfection with the 5SG or G33F mutant generated FC-P1 and FC-P2 particles (Fig. 3B). We next treated WT and 5SG progeny particles with lambda phosphatase and subsequently analyzed their AEX profiles. When dephosphorylated, FC-P2 changed its AEX profile to that of FC-P1, which remained unchanged (Fig. 3C). In contrast, the AEX profile did not change in the presence of phosphatase inhibitors (Fig. 3D). These results confirm that phosphorylation of a critical capsid surface residue(s), other than the distal phosphoserines in N-VP2, is present exclusively in the FC-P2 population and is responsible for its particular AEX profile.
FIG 3.
The phosphorylation status of the capsid surface, aside from the phosphoserine-rich N-VP2, determines the distinct AEX profiles of FC-P1 and FC-P2. (A) Schematic representations of the wild-type (WT) virus and N-VP2 mutants. (B) Intracellular virus progenies from the WT and from the 5SG and G33F mutants were analyzed by AEX-qPCR. (C) Intracellular virus progenies from the WT and the 5SG mutant were treated with lambda phosphatase and were subsequently analyzed by AEX-qPCR. (D) AEX and phosphatase treatment in the presence of sodium orthovanadate and sodium fluoride.
Only FC-P2 has nuclear export potential and can actively egress from the infected host cell.
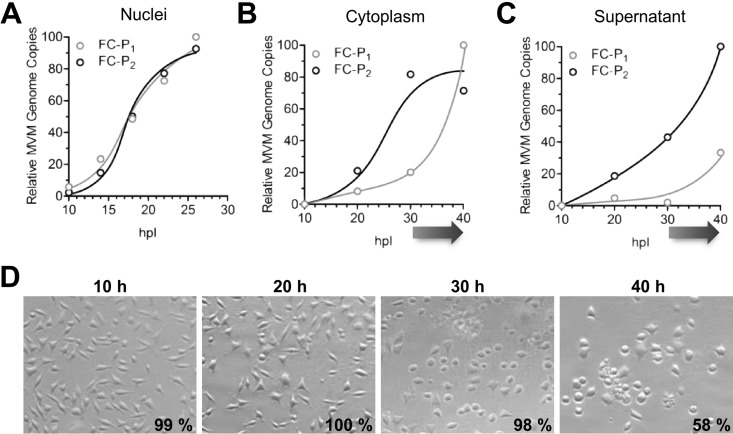
The subcellular distribution of the two full-capsid populations was examined. A9 cells were infected with MVM, and at various times postinfection (p.i.), progeny viruses were collected from nuclear, cytoplasmic, and supernatant fractions and were subjected to AEX-qPCR. While the two FC populations accumulated with similar kinetics in the nucleus, the accumulation of FC-P2 preceded that of FC-P1 in the cytoplasm and in the supernatant (Fig. 4A to C). FC-P2 egress started before the onset of cell lysis, which occurred from 30 hpi, as judged from the trypan blue exclusion assay (Fig. 4D), and resulted in the appearance of FC-P1 in the cytoplasmic and supernatant fractions. The compartment-dependent segregation of the two full-capsid entities at increasing times p.i. confirms the existence of an active mechanism of nuclear export and egress involving FC-P2 particles exclusively and preceding the passive release of FC-P1 and EC through late virus-induced cell lysis.
FIG 4.
Time- and compartment-dependent segregation of the MVM progeny. The FC-P2 progeny actively egresses from the infected host cell. A9 cells (3 × 106) were infected with 5,000 DNA-containing particles per cell at 4°C. After washing to remove unbound viruses, the cells were incubated at 37°C for the indicated times in the presence of neuraminidase and an anti-capsid MAb. Cells were fractionated as described in Materials and Methods and were subjected to AEX-qPCR. The relative amounts of FC-P1 and FC-P2 virions were calculated and plotted. (A) Progeny in the nuclei. (B) Progeny in the cytoplasm. (C) Progeny in the culture medium. An arrow under the x axis indicates the onset of cell lysis. (D) Cell viability was calculated via trypan blue exclusion, and the averages of three independent measurements are given. Phase-contrast pictures of the infected cells were taken using a Zeiss Axiovert 35 microscope with a 20× objective.
FC-P1 is the precursor of FC-P2.
During infection in the presence of neuraminidase and an anti-capsid antibody to prevent reinfections, FC-P1 was the first population to appear in the nuclei of murine A9 cells and transformed human NB324K cells. While in NB324K cells FC-P1 disappeared progressively to give rise to FC-P2, in A9 cells this transfer was less efficient, leading to the accumulation of both populations (Fig. 4A and 5A). In order to further confirm that FC-P1 is the precursor of FC-P2, NB324K cells were transfected in the presence of neuraminidase and an anti-capsid antibody. FC-P1 and FC-P2 virions were quantitatively analyzed by AEX-qPCR from 24 to 48 h posttransfection (hpt), when no significant de novo production or degradation of progeny virions was observed (Fig. 5B). As shown in Fig. 5C, FC-P1 was the predominant virus population at 24 hpt. However, at 48 hpt, the total quantity of FC-P1 virions declined significantly, representing only one-third of the whole virus progeny and giving rise to a significant increase in the quantity of FC-P2 particles. Collectively, these results indicate that FC-P1 particles are the precursors of FC-P2 virions. The maturation of FC-P1 into FC-P2, which involves surface phosphorylation, would be more efficient in the transformed human cells than in the A9 murine fibroblasts.
FIG 5.
Dynamics of FC-P1 and FC-P2 in infection and transfection. (A) A9 and NB324K cells (3 × 106) were infected with 5,000 DNA-containing particles per cell at 4°C. After washing to remove unbound viruses, the cells were incubated at 37°C for the indicated times in the presence of neuraminidase and an anti-capsid MAb. Nuclei were isolated, and AEX-qPCR analysis was performed, as described in Materials and Methods. The genome copy number that corresponds to 100% is shown in each graph. (B) NB324K cells (106) were transfected in the presence of neuraminidase and an anti-capsid MAb. The intracellular progeny was immunoprecipitated with an anti-capsid MAb and was quantified by PCR at the indicated time points posttransfection. (C) NB324K cells were transfected as described above. Intracellular viruses were analyzed by AEX-qPCR at the indicated time points.
The phosphoserine-rich N-VP2 contributes to passive release but is dispensable for active egress.
In sharp contrast to FC-P1 and empty particles, FC-P2 capsids are selectively exported from the nucleus and are subsequently released from the host cell prior to cell lysis. The more-prominent external conformation of N-VP2 in FC-P2 and the additional surface phosphorylations represent a late maturation step and might confer nuclear export potential on FC-P2. N-VP2 is heavily phosphorylated at serine residues, which have been suggested previously to assist nuclear export (15).
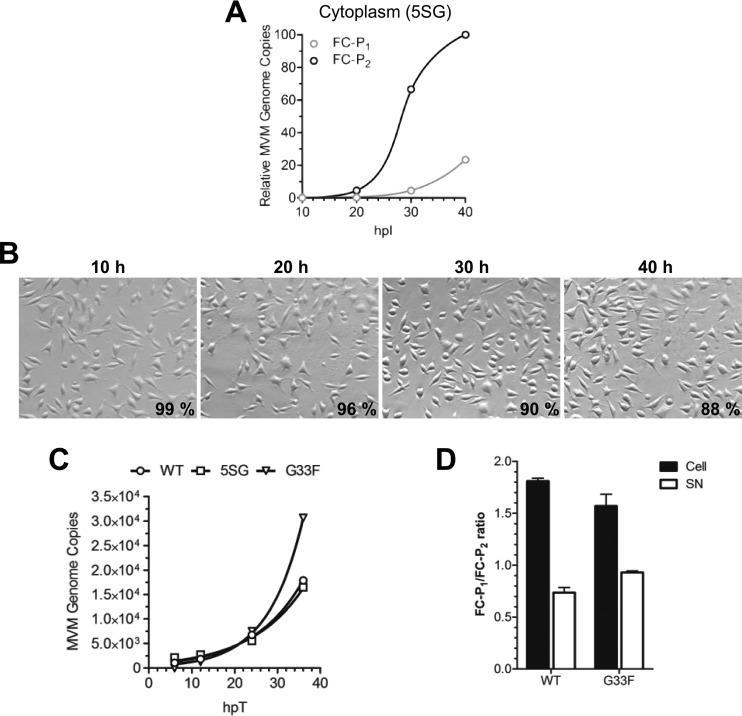
To investigate the role of N-VP2 in nuclear export, we used the 5SG mutant, which lacks all N-VP2 distal phosphorylations (Fig. 3A). The nuclear export of the two full-capsid populations generated by the 5SG mutant (Fig. 3B) was examined. Cells were infected with the 5SG mutant, and at various times p.i., progeny viruses were collected from the cytoplasmic fraction and were subjected to AEX-qPCR. The results showed that as with the WT (Fig. 4B), the accumulation of FC-P2 in the cytoplasm preceded that of FC-P1 (Fig. 6A), revealing that like that of the WT, the FC-P2 population of the 5SG mutant was selectively exported. The selective nuclear export of FC-P2 started largely before the onset of cell lysis, which was barely detectable at 40 hpi, as judged from the trypan blue exclusion assay (Fig. 6B), and resulted in the appearance of a few FC-P1 capsids in the cytoplasmic fraction. Interestingly, cell rounding, which was induced by the WT, was not detected for cells infected by the 5SG mutant, and cell lysis was significantly delayed. The delayed cell killing resulted in retarded passive release of FC-P1. This observation suggests a possible role of N-VP2 phosphoserines in MVM-mediated cytotoxicity, which would accelerate passive release, contributing to virus spread.
FIG 6.
The phosphoserine-rich N-VP2 is dispensable for active egress. (A) A9 cells (3 × 106) were infected with the 5SG mutant (5,000 DNA-containing particles per cell) at 4°C. After washing to remove unbound viruses, the cells were incubated at 37°C for the indicated times in the presence of neuraminidase and an anti-capsid MAb. Progeny from the cytoplasmic fraction was analyzed by AEX-qPCR. The relative numbers of FC-P1 and FC-P2 virions were calculated and plotted. (B) Phase-contrast pictures of cells infected with the 5SG mutant were taken using a Zeiss Axiovert 35 microscope with a 20× objective. Cell viability was calculated via trypan blue exclusion, and the average of three independent measurements is given. (C) NB324K cells (106) were transfected with the WT infectious clone or the indicated mutant in the presence of neuraminidase and an anti-capsid MAb. The cells were incubated in 1.5 ml of medium. The virus progeny in the supernatant (200 μl) was treated with DNase I and was quantified. (D) AEX-qPCR analysis of intracellular (cell) and released (SN) virions was performed 24 hpt, and the FC-P1/FC-P2 ratio was calculated.
Because the G33F mutant cannot expose N-VP2, it is defective for virus entry (45). In order to examine the egress potential of the G33F mutant and to bypass the entry step, the mutant was tested in transfection experiments, and the results were compared to those for the egress-competent WT and the 5SG mutant. Upon transfection, DNA-containing progeny particles of the 5SG and G33F mutants accumulated progressively in the cell culture medium to quantities similar to those of WT virions and with similar kinetics (Fig. 6C). Transfection leads to increased cell lysis due to cell damage and therefore is also expected to increase passive release. To examine whether the observed extracellular accumulation of particles resulted mostly from active egress, we analyzed the intra- and extracellular FC-P1/FC-P2 ratios 24 h after transfection with the WT or the G33F mutant. At this time, the FC-P1 population exceeded the FC-P2 population in the nucleus; however, in the extracellular milieu, the ratio was inversed (Fig. 6D). The inverted ratios can be explained only by active egress of FC-P2 particles, despite the presence of increased passive release due to transfection-mediated cell damage. These results further emphasize that N-VP2 and its distal phosphorylations do not play a direct role in the nuclear export and egress of MVM.
During entry, FC-P2 particles are dephosphorylated, acquiring the AEX profile of FC-P1.
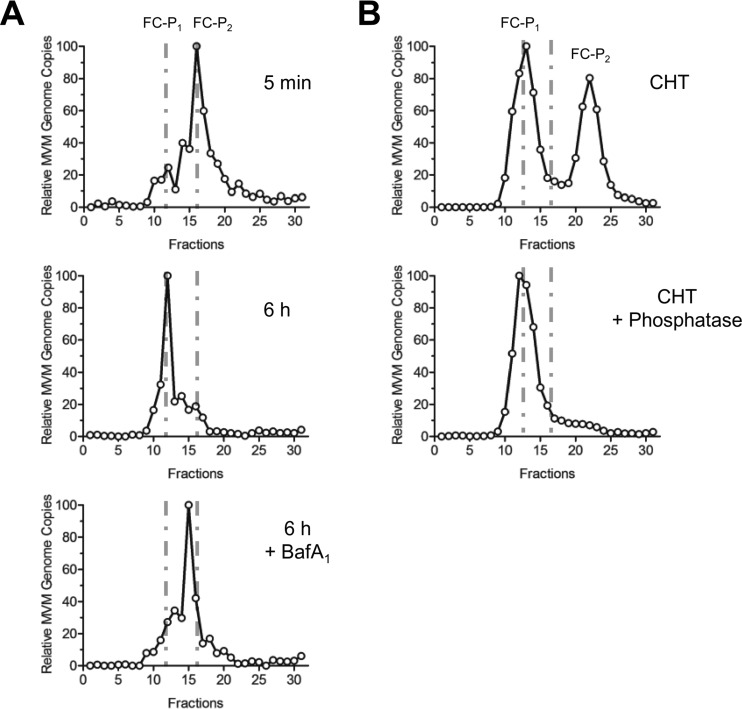
In the nucleus, FC-P1 precursors mature through surface phosphorylations to generate FC-P2, which are particles with nuclear export potential. During entry, the reverse situation was observed: incoming viruses composed mainly of FC-P2 particles were processed to generate FC-P1-like particles (Fig. 7A). The N-VP2 of incoming capsids became cleaved by endosomal proteases (Fig. 2G). However, as shown above (Fig. 2I and 3), the presence or absence of N-VP2 with its distal phosphoserines has no influence on the AEX profile, which is mediated by additional phosphorylation of capsid surface residues. These critical residues on the surfaces of incoming FC-P2 particles are dephosphorylated by endosomal acid phosphatases early in the entry process. In order to confirm the involvement of acid phosphatases in the processing of incoming particles, bafilomycin A1 (BafA1), which raises the endosomal pH and inhibits acid phosphatases, was applied to A9 cells. As shown in Fig. 7A, BafA1 totally abrogated the dephosphorylation of the incoming FC-P2 population.
FIG 7.
During entry, acid phosphatases dephosphorylate the surface residues associated with nuclear export potential. (A) A9 mouse fibroblasts (3 × 106) were infected with purified FC-P2 (5,000 particles per cell) at 4°C. After the removal of unbound viruses, cells were incubated at 37°C for the indicated times and were subjected to AEX-qPCR. In order to inhibit acid phosphatases, 150 nM BafA1 was added 15 min prior to virus internalization at 37°C. (B) Virus progenies with equivalent amounts of FC-P1 and FC-P2 were treated with CHT alone or with CHT plus phosphatase and were analyzed by AEX-qPCR.
The results obtained in vivo could be mimicked in vitro only by a dephosphorylation reaction, not by the proteolytic cleavage of N-VP2. When virions with equal proportions of FC-P1 and FC-P2 were treated with chymotrypsin alone to cleave N-VP2, only FC-P2 exhibited a change in its AEX profile and became even more negatively charged, most probably due to conformational changes consecutive to the proteolytic modification. In contrast, when the progeny was treated first with CHT and then with lambda phosphatase, the resulting particles had the same AEX profile as the incoming particles in vivo (Fig. 7B).
DISCUSSION
The active egress of enveloped viruses is well documented and involves budding through host cell membranes. The egress of nonenveloped viruses is generally assumed to result from the lytic burst occurring at the end of the infection. However, growing evidence indicates that egress may not be a mere consequence of the passive release induced by cell lysis but may involve active prelytic transport of the mature progeny (8, 9, 11, 13). Proving active prelytic egress for nonenveloped viruses is challenging, since the lysis of a few heavily infected cells can passively release virions, complicating the task of distinguishing between active and passive release. Therefore, it is not sufficient to detect progeny virions in the culture medium prior to significant cell lysis. In addition, an active process must be recognized. By means of anion-exchange chromatography (AEX) and cell fractionation, we observed a selective process of nuclear export and egress for the model parvovirus minute virus of mice (MVM) prior to passive release by cell lysis. Additionally, we identified late capsid maturation steps occurring in the cell nucleus before nuclear export.
The current model of MVM morphogenesis and egress suggests that EC precursors are first assembled in the nucleus and are subsequently filled with viral ssDNA to generate full capsids (FC) (46). As a consequence of packaging, the phosphoserine-rich N-VP2 becomes exposed outside the shell through the 5-fold axis of symmetry (16, 39). The exposed N-VP2 has been suggested to mediate the export of the FC progeny from the nucleus (15), followed by virus egress, which has been proposed to occur by vesicular transport through the endoplasmic reticulum and Golgi complex (13).
By use of AEX, proteins can be separated on the basis of their net surface charges. We performed AEX to separate and characterize parvovirus progeny particles displaying different protein surface configurations. Apart from EC precursors, the AEX profile of intranuclear MVM progeny revealed not one but two well-defined DNA-containing progeny populations, here named FC-P1 and FC-P2. FC-P1 shares many characteristics with the EC precursors. The particles appear early and have a similar surface phosphorylation pattern; N-VP2 is not fully accessible; and the particles cannot be exported from the nucleus. FC-P2 virions appear later and feature additional surface phosphorylations; N-VP2 is more prominently exposed; and the virions exhibit nuclear export potential. FC-P1 virions represent a previously unrecognized stage in MVM morphogenesis, intermediate between EC precursors and the late FC-P2 virions. The nuclear export-competent FC-P2 virions represent the fully mature infectious progeny. The infectivities and AEX profiles of the FC-P2 progeny isolated from the nucleus (preegress) and the FC-P2 progeny actively released from the cells (postegress) were identical (data not shown). Hence, complete virus progeny maturation is achieved in the nucleus, and no further maturation steps are required during the active process of egress for the acquisition of full infectivity.
The clear surface conformation of N-VP2 in FC-P2 contrasts with the less prominent exposure in FC-P1. This observation indicates that DNA packaging, although required, is not sufficient to trigger the complete external N-VP2 conformation found in the mature particles. However, in contrast to those of EC, the internal N-VP2 subunits in FC-P1 were unstable, and exposure to mild heat, a low pH (Fig. 2F), or cesium chloride (data not shown) was sufficient to trigger further externalization. The presence of internal N-VP2 subunits in FC-P1 may explain the slightly higher density of FC-P1 than of FC-P2. Interestingly, the specific AEX profile was not determined by the particular conformation of the phosphoserine-rich N-VP2 but by the phosphorylation of additional capsid surface residues (Fig. 2I and 3).
N-VP2, particularly the phosphorylation of its distal serine residues, has been suggested previously to play a crucial role in the nuclear export of de novo-synthesized virion progeny (15). These data are in line with our findings demonstrating that FC-P1 particles, which exhibit incomplete N-VP2 exposure, did not have nuclear export capacity. In order to challenge a possible involvement of N-VP2 and its prominent distal phosphorylations in the export of the late FC-P2 population, we used two mutants with dysfunctional N-VP2. The 5SG mutant lacks the distal serine phosphorylations within N-VP2. The G33F mutant is unable to externalize the N-VP2 sequence on the surface of the capsid due to the insertion of a bulky phenylalanine residue at position 33 of its polyglycine stretch within the VP2 protein sequence (45) (Fig. 3A). Confirming our observations, dephosphorylation of the distal serine residues of N-VP2 or prevention of its externalization did not influence the different AEX profiles, which were exclusively defined by additional surface phosphorylations (Fig. 3B and C). Despite their disabled N-VP2, both mutants were able to egress with kinetics and quantities similar to those observed for the WT (Fig. 6). These results confirm that N-VP2 and the phosphorylation of its distal serine residues are not key players in virus egress.
Parvoviruses display a high mutation rate, similar to that of RNA viruses (47, 48). Accordingly, genetic substitutions that interfere with crucial stages of the viral life cycle result in reversions after only a few rounds of infection. The distal S-to-G substitutions in N-VP2 were highly stable, and no genetic reversions were observed following several passages (data not shown). In infection experiments, the 5SG mutant displayed inefficient nuclear entry (data not shown), resulting in moderately delayed egress (Fig. 4B and 6A). In contrast to WT infection, infection of A9 cells with the 5SG mutant did not cause cell rounding; the cells remained mostly intact as late as 40 hpi despite the active egress of the progeny (Fig. 6A and B). In agreement with our findings, deletion of 7 amino acids within the sequence of N-VP2 in order to disturb its function did not affect the egress of progeny particles. However, this truncation caused retarded entry, leading to delayed progeny egress. Additionally, this mutant showed a lower cytotoxic effect (40). MVM infection is reported to induce dramatic changes in the cytoskeleton of the host cell, resulting in cell rounding and detachment and culminating in lysis and passive progeny release (49). These observations suggest the involvement of the phosphoserine-rich N-VP2 in the MVM-induced remodeling of actin filaments observed during virus egress and confirm previous observations that actin remodeling is not a mere side effect of virus replication (13, 49). Therefore, N-VP2 might play a role in the passive release of particles by accelerating cell lysis and contributing to virus spread.
Apart from the N-VP2 conformation, the surface phosphorylation pattern is the most prominent difference between FC-P1 and FC-P2 (Fig. 3). Therefore, it is tempting to speculate that the phosphorylation of surface residues other than those of N-VP2 confers nuclear export potential on the late progeny population. The efficiency of this late phosphorylation step was cell type dependent; the process was more efficient in transformed human NB324K cells than in murine A9 cells (Fig. 5). The more efficient phosphorylation in NB324K cells is in agreement with previous studies reporting lower overall capsid phosphorylation levels in murine A9 cells than in transformed human cells (50).
In agreement with a role in nuclear export, the late phosphorylation of capsid residues in the nucleus was efficiently reversed on the incoming capsids by acidic endosomal phosphatases, resulting in a complete reversion to nuclear FC-P1 particles (Fig. 7). Together with N-VP2 cleavage and N-VP1 externalization, the dephosphorylation of surface residues appears to represent a novel processing step during parvovirus cell entry that could be critical for avoiding interference with nuclear localization signals by stabilizing the incoming particles in the nuclei of infected cells. In line with this concept, it has been shown previously that the endocytic route is required for the nuclear targeting of canine parvovirus and adeno-associated virus. Particles microinjected into the cytoplasm to bypass the endocytic route failed to target the nucleus, even when pretreated under acidic conditions (51, 52). MVM that had cleaved N-VP2 and was exposed to acidic conditions remained sensitive to BafA1 (unpublished data), suggesting that further intraendosomal capsid modifications are required for the infection.
The requirement for NS2 in progeny egress has already been demonstrated for murine A9 cells, but NS2 is not a prerequisite in transformed NB324K cells. NS2 harbors a supraphysiological NES and interacts tightly with CRM1. Prevention of the NS2–CRM1 interaction has been demonstrated previously to impede the nuclear export of progeny virions in restrictive mouse fibroblasts (23, 24). These results explain the previously observed cell type-specific inhibition of nuclear export by the CRM1 inhibitor leptomycin B (15). We were able to demonstrate that the NS2–CRM1 interaction is not required for the late nuclear maturation of the virion progeny. A mutant containing amino acid substitutions within the consensus NES sequence produced WT levels of FC-P2 progeny (data not shown). Efforts to demonstrate a direct or indirect interaction between N-VP2 or other capsid regions and CRM1 have been unsuccessful. Therefore, the dependence of progeny egress on NS2 and the CRM1 export pathway may be indirect, via the supraphysiological interaction between NS2 and CRM1 (22).
In summary, we have identified a DNA-containing progeny population that accumulates early in the nucleus. This precursor requires further maturation, involving N-VP2 exposure and the phosphorylation of capsid surface residues, in order to acquire nuclear export potential. In contrast to N-VP2, which was dispensable for active egress, the phosphorylation of capsid surface residues was consistently associated with export capacity and was cell type dependent. Those surface residues were efficiently dephosphorylated during entry by acidic endosomal phosphatases. The spatially and temporally controlled changes in capsid surface phosphorylation may play a pivotal role by providing the nuclear import and export potential required to complete the infectious cycle of the karyophilic virus. Further studies are required to identify the critical capsid surface phosphorylations and to reveal their specific role in selective nuclear export. Additionally, it will be important to elucidate the mechanism of the phosphoserine-rich N-VP2 in cytotoxicity.
ACKNOWLEDGMENTS
We thank J. M. Almendral for critical reading of the manuscript and for kindly providing the G33F mutant and antibodies against MVM. We thank N. Salomé for the MVM-NES22 infectious clone. We acknowledge A. Stocker and W. Aeschimann for competent support and excellent technical assistance with the ÄKTA chromatography system.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Martin-Serrano J, Neil SJ. 2011. Host factors involved in retroviral budding and release. Nat Rev Microbiol 9:519–531. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- 2.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daeffler L, Horlein R, Rommelaere J, Nüesch JP. 2003. Modulation of minute virus of mice cytotoxic activities through site-directed mutagenesis within the NS coding region. J Virol 77:12466–12478. doi: 10.1128/JVI.77.23.12466-12478.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maul GG. 1976. Fibrils attached to the nuclear pore prevent egress of SV40 particles from the infected nucleus. J Cell Biol 70:714–719. doi: 10.1083/jcb.70.3.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tucker SP, Compans RW. 1993. Virus infection of polarized epithelial cells. Adv Virus Res 42:187–247. doi: 10.1016/S0065-3527(08)60086-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohl BP, Roy P. 2014. Bluetongue virus capsid assembly and maturation. Viruses 6:3250–3270. doi: 10.3390/v6083250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wirblich C, Bhattacharya B, Roy P. 2006. Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101. J Virol 80:460–473. doi: 10.1128/JVI.80.1.460-473.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird SW, Maynard ND, Covert MW, Kirkegaard K. 2014. Nonlytic viral spread enhanced by autophagy components. Proc Natl Acad Sci U S A 111:13081–13086. doi: 10.1073/pnas.1401437111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tucker SP, Thornton CL, Wimmer E, Compans RW. 1993. Vectorial release of poliovirus from polarized human intestinal epithelial cells. J Virol 67:4274–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clayson ET, Brando LV, Compans RW. 1989. Release of simian virus 40 virions from epithelial cells is polarized and occurs without cell lysis. J Virol 63:2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jourdan N, Maurice M, Delautier D, Quero AM, Servin AL, Trugnan G. 1997. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J Virol 71:8268–8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bär S, Rommelaere J, Nüesch JP. 2013. Vesicular transport of progeny parvovirus particles through ER and Golgi regulates maturation and cytolysis. PLoS Pathog 9:e1003605. doi: 10.1371/journal.ppat.1003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bär S, Daeffler L, Rommelaere J, Nüesch JP. 2008. Vesicular egress of non-enveloped lytic parvoviruses depends on gelsolin functioning. PLoS Pathog 4:e1000126. doi: 10.1371/journal.ppat.1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maroto B, Valle N, Saffrich R, Almendral JM. 2004. Nuclear export of the nonenveloped parvovirus virion is directed by an unordered protein signal exposed on the capsid surface. J Virol 78:10685–10694. doi: 10.1128/JVI.78.19.10685-10694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore SF, Tattersall P. 1987. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res 33:91–174. doi: 10.1016/S0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 17.Caillet-Fauquet P, Perros M, Brandenburger A, Spegelaere P, Rommelaere J. 1990. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J 9:2989–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero Y, Calle M, Cornelis JJ, Herold-Mende C, Rommelaere J, Schlehofer JR, Geletneky K. 2004. Parvovirus H-1 infection of human glioma cells leads to complete viral replication and efficient cell killing. Int J Cancer 109:76–84. doi: 10.1002/ijc.11626. [DOI] [PubMed] [Google Scholar]
- 19.Bodendorf U, Cziepluch C, Jauniaux JC, Rommelaere J, Salomé N. 1999. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J Virol 73:7769–7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohshima T, Nakajima T, Oishi T, Imamoto N, Yoneda Y, Fukamizu A, Yagami K. 1999. CRM1 mediates nuclear export of nonstructural protein 2 from parvovirus minute virus of mice. Biochem Biophys Res Commun 264:144–150. doi: 10.1006/bbrc.1999.1478. [DOI] [PubMed] [Google Scholar]
- 21.Nachury MV, Weis K. 1999. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci U S A 96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelsma D, Valle N, Fish A, Salomé N, Almendral JM, Fornerod M. 2008. A supraphysiological nuclear export signal is required for parvovirus nuclear export. Mol Biol Cell 19:2544–2552. doi: 10.1091/mbc.E08-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichwald V, Daeffler L, Klein M, Rommelaere J, Salomé N. 2002. The NS2 proteins of parvovirus minute virus of mice are required for efficient nuclear egress of progeny virions in mouse cells. J Virol 76:10307–10319. doi: 10.1128/JVI.76.20.10307-10319.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller CL, Pintel DJ. 2002. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J Virol 76:3257–3266. doi: 10.1128/JVI.76.7.3257-3266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naeger LK, Cater J, Pintel DJ. 1990. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J Virol 64:6166–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nüesch JP, Bär S, Lachmann S, Rommelaere J. 2009. Ezrin-radixin-moesin family proteins are involved in parvovirus replication and spreading. J Virol 83:5854–5863. doi: 10.1128/JVI.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mani B, Baltzer C, Valle N, Almendral JM, Kempf C, Ros C. 2006. Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J Virol 80:1015–1024. doi: 10.1128/JVI.80.2.1015-1024.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tattersall P, Bratton J. 1983. Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J Virol 46:944–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shein HM, Enders JF. 1962. Multiplication and cytopathogenicity of simian vacuolating virus 40 in cultures of human tissues. Proc Soc Exp Biol Med 109:495–500. doi: 10.3181/00379727-109-27246. [DOI] [PubMed] [Google Scholar]
- 30.Segovia JC, Real A, Bueren JA, Almendral JM. 1991. In vitro myelosuppressive effects of the parvovirus minute virus of mice (MVMi) on hematopoietic stem and committed progenitor cells. Blood 77:980–988. [PubMed] [Google Scholar]
- 31.López-Bueno A, Mateu MG, Almendral JM. 2003. High mutant frequency in populations of a DNA virus allows evasion from antibody therapy in an immunodeficient host. J Virol 77:2701–2708. doi: 10.1128/JVI.77.4.2701-2708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tattersall P. 1972. Replication of the parvovirus MVM. I. Dependence of virus multiplication and plaque formation on cell growth. J Virol 10:586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merchlinsky MJ, Tattersall P, Leary JJ, Cotmore SF, Gardiner EM, Ward DC. 1983. Construction of an infectious molecular clone of the autonomous parvovirus minute virus of mice. J Virol 47:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Abramo AM, Ali AA, Wang F, Cotmore SF, Tattersall P. 2005. Host range mutants of Minute Virus of Mice with a single VP2 amino acid change require additional silent mutations that regulate NS2 accumulation. Virology 340:143–154. doi: 10.1016/j.virol.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 35.Rubio MP, López-Bueno A, Almendral JM. 2005. Virulent variants emerging in mice infected with the apathogenic prototype strain of the parvovirus minute virus of mice exhibit a capsid with low avidity for a primary receptor. J Virol 79:11280–11290. doi: 10.1128/JVI.79.17.11280-11290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lombardo E, Ramírez JC, García J, Almendral JM. 2002. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J Virol 76:7049–7059. doi: 10.1128/JVI.76.14.7049-7059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lombardo E, Ramírez JC, Agbandje-McKenna M, Almendral JM. 2000. A β-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J Virol 74:3804–3814. doi: 10.1128/JVI.74.8.3804-3814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller DE, Siegl G. 1983. Maturation of parvovirus LuIII in a subcellular system. II. Isolation and characterization of nucleoprotein intermediates. J Gen Virol 64:1055–1067. [DOI] [PubMed] [Google Scholar]
- 39.Tattersall P, Shatkin AJ, Ward DC. 1977. Sequence homology between structural polypeptides of minute virus of mice. J Mol Biol 111:375–394. doi: 10.1016/S0022-2836(77)80060-0. [DOI] [PubMed] [Google Scholar]
- 40.Tullis GE, Burger LR, Pintel DJ. 1992. The trypsin-sensitive RVER domain in the capsid proteins of minute virus of mice is required for efficient cell binding and viral infection but not for proteolytic processing in vivo. Virology 191:846–857. doi: 10.1016/0042-6822(92)90260-V. [DOI] [PubMed] [Google Scholar]
- 41.Cotmore SF, D'Abramo AM, Ticknor CM, Tattersall P. 1999. Controlled conformational transitions in the MVM virion expose the VP1 N-terminus and viral genome without particle disassembly. Virology 254:169–181. doi: 10.1006/viro.1998.9520. [DOI] [PubMed] [Google Scholar]
- 42.Farr GA, Cotmore SF, Tattersall P. 2006. VP2 cleavage and the leucine ring at the base of the fivefold cylinder control pH-dependent externalization of both the VP1 N terminus and the genome of minute virus of mice. J Virol 80:161–171. doi: 10.1128/JVI.80.1.161-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tullis GE, Burger LR, Pintel DJ. 1993. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J Virol 67:131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farr GA, Zhang LG, Tattersall P. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc Natl Acad Sci U S A 102:17148–17153. doi: 10.1073/pnas.0508477102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castellanos M, Pérez R, Rodríguez-Huete A, Grueso E, Almendral JM, Mateu MG. 2013. A slender tract of glycine residues is required for translocation of the VP2 protein N-terminal domain through the parvovirus MVM capsid channel to initiate infection. Biochem J 455:87–94. doi: 10.1042/BJ20130503. [DOI] [PubMed] [Google Scholar]
- 46.King JA, Dubielzig R, Grimm D, Kleinschmidt JA. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J 20:3282–3291. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shackelton LA, Holmes EC. 2006. Phylogenetic evidence for the rapid evolution of human B19 erythrovirus. J Virol 80:3666–3669. doi: 10.1128/JVI.80.7.3666-3669.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shackelton LA, Parrish CR, Truyen U, Holmes EC. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci U S A 102:379–384. doi: 10.1073/pnas.0406765102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nüesch JP, Lachmann S, Rommelaere J. 2005. Selective alterations of the host cell architecture upon infection with parvovirus minute virus of mice. Virology 331:159–174. doi: 10.1016/j.virol.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 50.Maroto B, Ramírez JC, Almendral JM. 2000. Phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J Virol 74:10892–10902. doi: 10.1128/JVI.74.23.10892-10902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vihinen-Ranta M, Kalela A, Mäkinen P, Kakkola L, Marjomäki V, Vuento M. 1998. Intracellular route of canine parvovirus entry. J Virol 72:802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonntag F, Bleker S, Leuchs B, Fischer R, Kleinschmidt JA. 2006. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J Virol 80:11040–11054. doi: 10.1128/JVI.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]