ABSTRACT
Porcine reproductive and respiratory syndrome virus (PRRSV) nucleocapsid (N) protein is the main component of the viral capsid to encapsulate viral RNA, and it is also a multifunctional protein involved in the regulation of host cell processes. Nonstructural protein 9 (Nsp9) is the RNA-dependent RNA polymerase that plays a critical role in viral RNA transcription and replication. In this study, we demonstrate that PRRSV N protein is bound to Nsp9 by protein-protein interaction and that the contacting surface on Nsp9 is located in the two predicted α-helixes formed by 48 residues at the C-terminal end of the protein. Mutagenesis analyses identified E646, E608, and E611 on Nsp9 and Q85 on the N protein as the pivotal residues participating in the N-Nsp9 interaction. By overexpressing the N protein binding fragment of Nsp9 in infected Marc-145 cells, the synthesis of viral RNAs, as well as the production of infectious progeny viruses, was dramatically inhibited, suggesting that Nsp9-N protein association is involved in the process of viral RNA production. In addition, we show that PRRSV N interacts with cellular RNA helicase DHX9 and redistributes the protein into the cytoplasm. Knockdown of DHX9 increased the ratio of short subgenomic mRNAs (sgmRNAs); in contrast, DHX9 overexpression benefited the synthesis of longer sgmRNAs and the viral genomic RNA (gRNA). These results imply that DHX9 is recruited by the N protein in PRRSV infection to regulate viral RNA synthesis. We postulate that N and DHX9 may act as antiattenuation factors for the continuous elongation of nascent transcript during negative-strand RNA synthesis.
IMPORTANCE It is unclear whether the N protein of PRRSV is involved in regulation of the viral RNA production process. In this report, we demonstrate that the N protein of the arterivirus PRRSV participates in viral RNA replication and transcription through interacting with Nsp9 and its RdRp and recruiting cellular RNA helicase to promote the production of longer viral sgmRNAs and gRNA. Our data here provide some new insights into the discontinuous to continuous extension of PRRSV RNA synthesis and also offer a new potential anti-PRRSV strategy targeting the N-Nsp9 and/or N-DHX9 interaction.
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS) emerged in America in 1987 and later in Europe (1), and since then it has spread around the world and has brought great economic losses to the swine industry worldwide (2). The etiological agent, PRRS virus (PRRSV), is an enveloped positive-sense RNA virus that belongs to the family Arteriviridae in the order Nidovirales, along with equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV), and simian hemorrhagic fever virus (SHFV) (3).
The PRRSV genome is approximately 15 kb in length and includes at least 10 open reading frames (ORFs): ORF1a, ORF1b, ORF2a, ORF2b, ORF3 to -7, and the newly discovered ORF5a (4, 5). ORF1a and ORF1b encode polyproteins pp1a and pp1ab, and at least 14 viral nonstructural proteins (Nsps) are generated as a result of serial cleavage of pp1a and pp1ab. pp1a is processed to release Nsp1α, Nsp1β, Nsp2 to Nsp6, Nsp7α, Nsp7β, and Nsp8; Nsp9 to Nsp12, which have been shown to be involved in viral genome transcription and replication, are generated from pp1ab (6). Two more Nsps, nsp2TF and nsp2N, have been found recently to be produced from an alternative reading frame overlapping the viral replicase gene by programmed ribosomal frameshifting (7). ORF2a and ORF3 to ORF5 encode four N-glycosylated envelope proteins (GP2a, GP3, GP4, and GP5); ORF2b and ORF6 encode two other membrane-associated nonglycosylated proteins (E and M) (8). The nucleocapsid (N) protein is translated from the last ORF (ORF7) to encapsidate the viral RNA genome (9).
The ORF1b-encoded Nsp9 protein contains a putative RNA-dependent RNA-polymerase (RdRp) domain in the C-terminal part, and the N-terminal part of Nsp9 has a newly identified nidovirus RdRp-associated nucleotidyltransferase domain (6, 10). PRRSV RdRp is considered to be a core component of the viral replication and transcription complex (RTC) that is crucial for virus replication (11). PRRSV Nsp9 not only possesses polymerase activity, but also interacts with other cellular proteins that affect viral replication, including annexin A2, retinoblastoma protein, and DEAD box RNA helicase 5 (12–14).
The N protein of North American PRRSV contains 123 amino acids with a molecular mass of 15 kDa. This phosphorylated protein is the most abundant viral structural protein and exhibits multiple functions throughout the viral life cycle (15). The fundamental function of the N protein is to interact with itself through covalent and noncovalent interactions to form the viral capsid, which packages the viral genome (16). In addition, the N protein localizes in both the cytoplasm and the nucleus/nucleolus of the host cell and interacts with a number of host factors, and it can affect cellular gene expression involved in viral pathogenesis (16–18).
Nucleocapsid proteins of minus-strand RNA viruses have long been known as part of the helical ribonucleoprotein structure, which is the template for viral RNA transcription and replication (19, 20). Recently, the nucleocapsid protein of bunyavirus was reported to interact with its L polymerase and to mediate the switch from capped RNA-primed transcription to unprimed viral RNA replication (21). The nucleocapsid protein of hantavirus was also shown to interact with its RdRp, and their association was required for viral RNA synthesis (22), consistent with the requirement for N protein expression for the function of RdRp in two Bunyaviridae reverse-genetic systems (23, 24).
In plus-strand RNA viruses, the core protein of classic swine fever virus (CSFV) and the capsid protein of norovirus have been reported to enhance their RdRp activities, and their viral RNA syntheses are regulated by the interaction between their RdRps and core/capsid proteins (25, 26). A role in viral RNA synthesis has also been postulated for the N protein of coronavirus, as the expression of the nucleocapsid protein is required for the initiation of RNA synthesis in some reverse-genetic systems (27, 28). Recently, phosphorylated coronavirus N protein has been found to recruit RNA helicase DDX1, and this can facilitate the synthesis of longer viral subgenomic mRNAs (sgmRNAs) (29).
In arteriviruses, the N protein has been reported to colocalize with the viral replicases, including Nsp9, in early infection (11, 28). DHX9, a member of the DExH/D box superfamily of RNA helicases that plays critical roles in transcriptional regulation and translation, has been identified by proteomic analyses as a potential cellular partner of PRRSV nucleocapsid protein (17, 18). Why is N protein present in the replication complex? Does DHX9 play a role in PRRSV replication similar to that of DDX1 in coronavirus infection? These interesting questions remain to be investigated.
In this study, we provide evidence demonstrating that the PRRSV nucleocapsid protein interacts with Nsp9 and its RdRp and also recruits the cellular helicase DHX9 during virus infection to facilitate viral RNA synthesis and virus production. Our data suggest that these protein interactions play an important role in the regulation of arterivirus PRRSV RNA synthesis to balance the production of viral sgmRNAs and gRNA.
MATERIALS AND METHODS
Cells and virus.
Human embryonic kidney (HEK) 293T cells and Marc-145 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (HyClone) in a 5% CO2 atmosphere at 37°C. Growth media were supplemented with 10% fetal bovine serum (FBS) (HyClone), 0.1 mM sodium pyruvate, and penicillin (100 U/ml)-streptomycin (100 μg/ml) (Bio Basic, Canada). A highly pathogenic PRRSV isolate, TA-12 (GenBank accession no. HQ416720.1), and a recombinant PRRSV expressing enhanced green fluorescent protein (eGFP) reporter (30) were used in virus infection studies. Genes of the North American strain NSVL 97-7895 (GenBank accession no. AY545985.1) were amplified by PCR using replicon FL-12 as the template (31), and genes of the low-pathogenic European strain Olot/91 (GenBank accession no. KF203132.1) were obtained by reverse transcription (RT)-PCR as described previously (32).
Bioinformatics analysis.
The spatial structures of full-length Nsp9 and N proteins from PRRSV strain NVSL 97-7895 were predicted by the I-TASSER server online tool (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) as previously described (33). The N-Nsp9 interaction model was predicted by PRISM 2.0 (http://cosbi.ku.edu.tr/prism/index.php) (34). The structure files were analyzed using PyMOL software (PyMOL v1.0).
DNA transfection.
For pulldown or immunoprecipitation assays, HEK 293T cells grown in 100-mm plates were transfected with 10 μg of plasmids using calcium phosphate (35). Transfections of Marc-145 cells in 12-well plates were performed with 1 μg of plasmids per well using Lipofectamine 2000 reagent (Invitrogen, USA) according to the manufacturer's instructions.
Colocalization assay.
Cells were cotransfected with constructs expressing green fluorescent protein (GFP)- and mCherry-tagged proteins. At 36 h posttransfection, the cells were fixed and permeabilized with precooled isometric methyl alcohol and acetone for 10 min. After three washes with phosphate-buffered saline (PBS), the cells were dyed with DAPI (4′,6-diamidino-2-phenylindole) for 20 min in a darkroom at room temperature. The expression of fluorescent proteins was observed using a confocal microscope (Nikon, Japan).
Immunofluorescence assay.
Transfected HEK 293T cells or PRRSV-infected Marc-145 cells were immobilized using 4% paraformaldehyde and permeabilized using 0.2% Triton X-100 for 10 min. After being blocked with 5% skimmed milk, the cells were incubated with rabbit anti-DHX9 polyclonal antibodies (Proteintech, USA) at room temperature for 1 h. After three washes with PBS, the cells were incubated with tetramethyl rhodamine isocyanate (TRITC)-conjugated anti-rabbit IgG (CWBIO, China) for 1 h and then stained with DAPI for 20 min. The fluorescence signals were recorded by confocal microscopy.
Yeast two-hybrid assay.
For the yeast two-hybrid (Y2H) assay, the Nsp9 and N genes of strain NVSL 97-78975 were amplified by PCR using the corresponding primers listed in Table S1 in the supplemental material and then inserted into pGBKT7 and pGADT7, respectively. The truncated fragments and site-directed mutants of Nsp9 were cloned into plasmid pGBKT7, the same vector as for the full-length Nsp9, and the mutants of N were cloned into pGADT7. The inserts in the constructs used in this study were verified by DNA sequencing.
The pGADT7-N and pGBKT7-Nsp9 constructs were cotransformed into Saccharomyces cerevisiae (yeast) strain Gold using the Yeastmaker Yeast Transformation System kit (Clontech, USA) as described by the manufacturer. Each interaction assay was repeated in at least three independent experiments.
Bimolecular fluorescence complementation (BiFC) assay.
The Nsp9 and N fragments of NVSL strain 97-7895 were amplified using primer pairs HindIIINsp9-F/SalINsp9-R and EcoRIN-F/XhoIN-R, and the PCR products were digested with the indicated restriction enzymes and cloned into vector pBiFC-VN173 or pBiFC-VC155, respectively, resulting in plasmids pBiFC-Nsp9-VN173 and pBiFC-N-VC155 for the expression of fusion proteins Nsp9-EYFPN and N-EYFPC. HEK 293T cells were cotransfected with the two constructs using calcium phosphate. The supernatant was replaced with fresh medium at 8 h posttransfection, and the fluorescence was examined 24 h later using a DM5000 B microscope (Leica, Germany).
Protein expression and purification.
For protein expression, primer pairs NcoINsp9-F/XhoINsp9-R and EcoRIN-F/BamHIN-R were used to amplify Nsp9 and N fragments from North American strains NVSL 97-78975 and TA-12 and European strain Olot/91, respectively. The Nsp9 genes were inserted into pTriEx-1.1 to make pTriEx-Nsp9 constructs, and the N genes were cloned into pGEX-2T. Recombinant glutathione S-transferase (GST)-N proteins were expressed in Escherichia coli BL21(DE3) at 37°C, while the expression of Nsp9-His proteins was performed at 16°C overnight, except for Olot/91 at 4°C for 2 days. The cells were disrupted by sonication. GST-N was purified with GST-Bind Sepharose (CWBIO) in phosphate buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 Mm KH2PO4 [pH 8.0]). For Nsp9-His, the purification was preformed using His-Bind Ni-nitrilotriacetic acid (NTA) Sepharose (CWBIO), and the protein was eluted in 20 mM Tris (pH 7.9), 0.5 M NaCl, and 200 mM imidazole.
Pulldown assay.
Protein mixtures of Nsp9-His and GST-N were incubated at 4°C for 2 h. In the reaction mixtures treated with RNase, RNase was added to the mixtures to a final concentration of 10 μg/ml. GST was incubated with Nsp9-His as the negative control. The solutions were then incubated with PureProteome nickel magnetic beads (Millipore, USA) in the tubes. After washing with 50 mM sodium phosphate, 300 mM NaCl, and 10 mM imidazole (pH 8.0), proteins bound with the beads were eluted with elution buffer (50 mM sodium phosphate, 300 mM NaCl, and 300 mM imidazole [pH 8.0]). The eluates were separated by SDS-PAGE and stained with Coomassie brilliant blue or detected by Western blotting.
Coimmunoprecipitation (co-IP) assay.
DNA-transfected or PRRSV-infected cells were washed with PBS three times and lysed with a One Step Animal Cell Active Protein Exaction kit (Sangon Biotech, China). The cell lysates were collected by centrifugation at 14,000 × g for 15 min at 4°C. Aliquots of the cell extract (about 1 × 107 cells) were immunoprecipitated using the indicated antibodies and applied to PureProteome Protein A Mix magnetic beads (Millipore, USA) following the supplier's protocol. The eluates were analyzed by Western blotting.
Western blotting.
Protein samples were separated by SDS-PAGE or Tricine–SDS-PAGE gels for the detection of protein fragments smaller than 10 kDa (36) and subsequently transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad, USA). The membranes were blocked for 1 h in 5% skimmed milk and then incubated with the indicated primary antibodies at 4°C overnight. The primary antibodies used in this study included mouse anti-His (1:5,000; CWBIO), anti-GST monoclonal antibodies (MAbs) (1:4,000; BioSino, China), rabbit anti-GFP polyclonal antibody (1:5,000; CWBIO), and rabbit anti-DHX9 polyclonal antibody (1:5,000; Proteintech). After three washes with washing buffer (0.1% Tween 20 in Tris-buffered saline [TBS]), the membranes were probed with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit IgG (1:10,000; CWBIO) at 37°C for 2 h. Immunodetection was performed using enhanced chemiluminescence (ECL) reagents (CWBIO) according to the supplier's instructions, and protein bands were quantified by densitometry using Image Lab software (Bio-Rad).
Competitive-inhibition assay.
Marc-145 cells grown in 12-well plates were transfected with 1 μg of plasmid per well expressing the wild-type (WT) Nsp9599–646 fragment or its indicated mutant. At 16 h posttransfection, the cells were infected with PRRSV at a multiplicity of infection (MOI) of 1. To boost the expression of Nsp9599–646, the cells were transfected again at 8 h postinfection (hpi). The infected cells were harvested at 0, 24, and 48 hpi. The total RNAs were extracted, and the mRNA levels for N and Nsp1 were quantified by real-time RT-PCR. The expression level of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene was detected as an internal control. The quantities of Nsp1 and N represent the levels of genomic RNA and subgenomic RNAs, respectively.
ShRNA knockdown and overexpression assays.
For short hairpin RNA (shRNA)-mediated knockdown of DHX9, the target sequence of DHX9 was selected according to the method of a previous study (37). The forward and reverse oligonucleotides (shDHX9-F, GGTTAATGAACGTATGCTGATCTCTTGAATCAGCATACGTTCATTAACTTTTTTC; shDHX9-R, TCGAGAAAAAAGTTAATGAACGTATGCTGATTCAAGAGATCAGCATACGTTCATTAACCTGCA) contain complementary sequences: the sense and antisense sequences that encode a hairpin structure within a 19-nucleotide (nt) stem and a 9-nt loop (5′-TCTCTTGAA-3′) in the middle. The PstI and XhoI restriction enzyme sites (underlined sequences) were added to the 5′ ends of the forward and reverse primers, respectively, to form overhangs. The annealed double-stranded oligonucleotides were cloned into vector pTriEx-U6 (modified in our laboratory) to generate pTriEx-shDHX9. Sequence from the luciferase reporter gene was also cloned as a control (pTriEx-shCT), using primers shCT-F (GCTTACGCTGAGTCTTCGATCTCTTGAATCGAAGACTCAGCGTAAGTTTTTTC) and shCT-R (TCGAGAAAAAACTTACGCTGAGTCTTCGATTCAAGAGATCGAAGACTCAGCGTAAGCTGCA).
For DHX9 overexpression, a construct of pTriEx-DHX9 was generated in two steps due to the limited available restriction sites in the vector. First, the 3′ part of DHX9 was amplified using the primer pair EcoRIDHX9r2-F/HindIIIDHX9r2-R and cloned into the pTriEx-1.1 plasmid between the EcoRI and HindIII sites. Second, the 5′ part of DHX9 was amplified using the primer pair BamHIDHX9r1-F/EcoRIDHX9r1-R and cloned into the construct between the BamHI and EcoRI sites.
To detect the effects of DHX9 knockdown and overexpression on PRRSV transcription, Marc-145 cells in 12-well plates were transfected with pTriEx-shDHX9 or pTriEx-DHX9. Empty vector and pTriEx-shCT were transfected as negative controls. At 16 h posttransfection, the cells were infected with PRRSV strain TA-12 at an MOI of 1, and cells were harvested at 48 hpi for subsequent analyses.
Quantitative real-time PCR.
Infected Marc-145 cells were washed three times with PBS, and the total RNAs were extracted with TRIzon Reagent (CWBIO). Reverse transcriptions were completed using a 5× All-in-One RT MasterMix kit (ABM, Canada). Quantitative real-time PCR (qRT-PCR) was performed in a CFX96 Real-Time PCR detection system (Bio-Rad), using EvaGreen qPCR MasterMix (ABM) according to the manufacturer's instructions. The following cycling conditions were used for real-time PCRs: 95°C for 30 s and 40 cycles of 95°C for 5 s and 60°C for 30 s. To measure viral total RNA, a pair of internal primers in the N gene were used to amplify all sgmRNAs and gRNA. To measure gRNA and each viral sgmRNA, a forward primer in the 5′ leader and a reverse primer in the corresponding ORF were used for the PCR. The cellular GAPDH was quantified as the internal control to normalize the cDNA amounts. All primers used for qRT-PCR are listed in Table S2 in the supplemental material. The 2−ΔΔCT method was used to calculate the relative levels of viral genomic and subgenomic RNAs.
Virus titration.
Marc-145 cells grown in 96-well plates were infected with PRRSV strain TA-12 at serial dilutions. After 1 h incubation at 37°C, the supernatants were replaced with fresh medium containing 2% FBS. Viral titers were determined by endpoint dilution analyses. The Spearman-Karber method was used to determine the 50% tissue culture infected dose (TCID50) 5 days postinfection (38).
Statistical analysis.
Statistical analyses of quantitative real-time PCR were performed with paired two-tailed Student's t tests, and analysis of the virus titer was carried out using one-way analysis of variance (ANOVA). The data were calculated as means and standard deviations (SD) of three independent experiments. Statistical analyses were completed using GraphPad Prism 5 (GraphPad Software, USA).
RESULTS
Nsp9 of PRRSV strain NVSL 97-7895 interacts with its nucleocapsid protein.
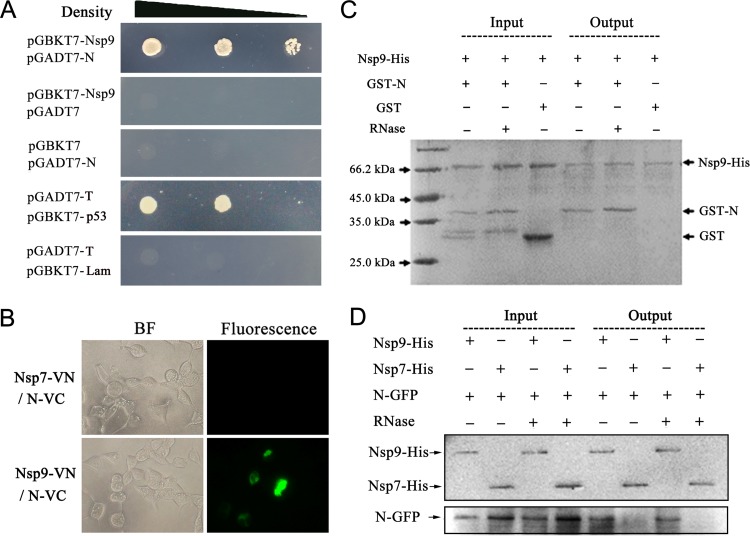
To investigate the interaction between Nsp9 and N, their gene fragments were amplified from a highly pathogenic strain, NSVL 97-7895, and inserted into the vectors pGBKT7 and pGADT7, respectively, for Y2H analysis. The constructs pGBKT7-Nsp9 and pGADT7-N containing Nsp9 and N genes were cotransformed into the yeast strain Gold, and empty vectors pGADT7 and pGBKT7 were cotransformed with pGBKT7-Nsp9 and pGADT7-N, respectively, as negative controls. Protein interactions were checked by growing on selective medium, synthetic dropout minimal base (SD) medium without Ade, His, Leu, and Trp (SD −Ade/−His/−Leu/−Trp). The results showed that neither Nsp9 fused with the DNA binding domain (BD) of the transcription factor nor N protein fused with the activation domain (AD) could activate the expression of the reporter, which was essential for the growth of the yeast cells on the selective medium, whereas the cells cotransformed with pGBKT7-Nsp9 and pGADT7-N were alive on the selective medium, indicating that N-AD bound to Nsp9-BD and activated the reporter in yeast cells (Fig. 1A).
FIG 1.
The N protein of PRRSV strain NVSL 97-7895 interacts with its Nsp9. (A) Yeast two-hybrid analysis. Yeast cells were cotransformed with the indicated constructs. The group transformed with pGADT7-T/pGBKT7-p53 was set up as a positive control, and pGADT7-T/pGBKT7-Lam was the negative control. The yeast cells were grown for 3 days at 30°C on SD medium without Ade, His, Leu, and Trp. The three spots in each group were diluted 101-, 102-, or 103-fold. (B) BiFC assay. Constructs expressing Nsp9-EYFPN and N-EYFPC were cotransfected into HEK 293T cells, and the fluorescence was detected at 24 h posttransfection. The Nsp7-VN/N-VC group was set as a negative control. (C) Pulldown assay using purified proteins. The purified fusion proteins Nsp9-His and GST-N were incubated at 4°C for 2 h, RNase was added to a final concentration of 10 μg/ml in the RNase “+” groups. GST was purified and used as the control protein for the binding of GST-N to Nsp9-His. The protein mixtures were applied to nickel magnetic beads, and the bound proteins were eluted and then detected by SDS-PAGE and Coomassie brilliant blue R250 staining. (D) Pulldown assay to analyze the interaction between Nsp9-His purified from E. coli and N-GFP expressed in mammalian cells. A vector expressing N-GFP was transfected into HEK 293T cells, the cell lysate was incubated with purified Nsp9-His, and the proteins bound with Nsp9-His were purified using Ni-NTA beads. The samples were examined by Western blotting using anti-GFP antibody. Nsp7-His was used as a negative control.
To confirm the interaction of Nsp9 with the N protein, a bimolecular fluorescence complementation assay was then applied. As shown in Fig. 1B, Nsp9 and N coexpressed in 293T cells dragged their fused N fragments and C fragments of enhanced yellow fluorescent protein (EYFP) together and yielded observable fluorescence under a microscope, suggesting that the PRRSV N protein was able to get close to and contact Nsp9.
To investigate PRRSV Nsp9-N protein interaction in vitro, we expressed recombinant GST-N and 6×His-tagged Nsp9 in E. coli and purified the proteins for pulldown assays. As both Nsp9 and N protein are RNA-binding proteins, RNase was also added to the protein mixture to a final concentration of 10 μg/ml to rule out the involvement of RNAs in the interaction assay. The results showed that the N protein clearly bound to Nsp9 with or without RNase treatment (Fig. 1C), indicating that the N protein directly recognized Nsp9 through protein-protein interaction. To validate this result, HEK 293T cells were transfected with the vector expressing N-GFP, and then the cell lysate was harvested and incubated with purified Nsp9-His protein and nickel magnetic beads. After washing, N-GFP was successfully pulled out with Nsp9-His, but not with the control Nsp7-His from the RNase-treated and untreated cell lysates (Fig. 1D), confirming that N interacted with Nsp9 and that the interaction did not need the presence of RNA.
N protein interacts with Nsp9 by binding to the C-terminal end of Nsp9.
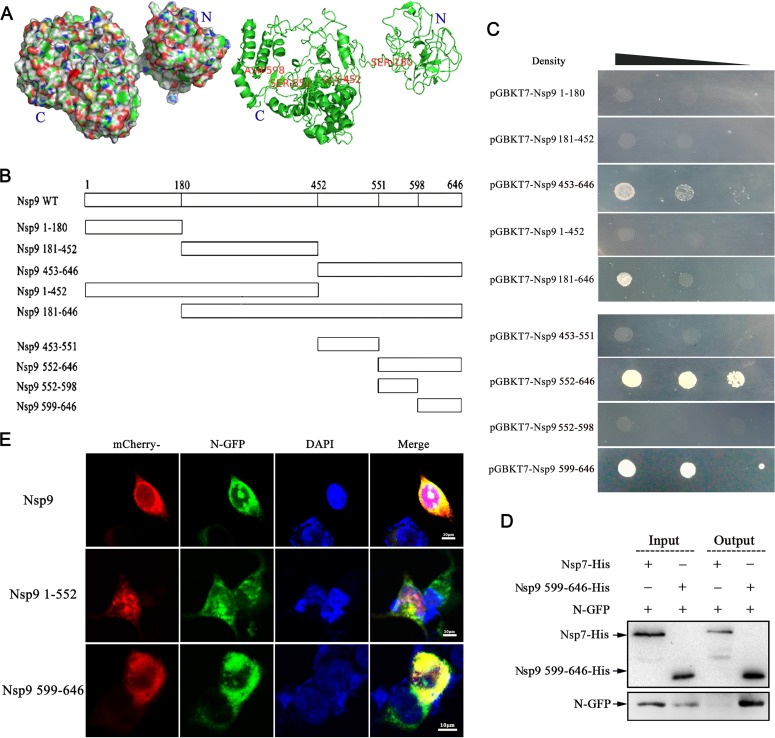
The full-length Nsp9 of PRRSV is a large protein that harbors 646 amino acids. To narrow down the Nsp9-N interaction region on Nsp9, the three-dimensional (3D) structure of Nsp9 from strain NSVL 97-7895 was predicted using the I-TASSER server online tool (Fig. 2A), and based on this structure, the protein was expressed as five BD-fused fragments containing amino acid residues 1 to 180, 181 to 452, 453 to 646, 1 to 452, and 181 to 646 (illustrated in Fig. 2B) for yeast two-hybrid assays. When these fragments were coexpressed with N-AD, BD-fused fragments 453 to 646 and 181 to 646 activated reporter expression (Fig. 2C), indicating that the N protein interaction site was located within the 453-to-646 region. Subsequently, fragment 453 to 646 was split in two, 453 to 551 and 552 to 646 (Fig. 2B), and it was shown that the N protein bound to the 552-to-646 polypeptide (Fig. 2C). Further truncation of the fragment 552 to 646 (552 to 598 and 599 to 646 in Fig. 2B) positioned the N protein binding site in the 599-to-646 region (Fig. 2C), which consisted of only 48 amino acid residues and was predicted to form two α-helixes at the C-terminal end of Nsp9.
FIG 2.
The Nsp9-N interaction domain in Nsp9 is located at the C-terminal end. (A) Predicted 3D structure of Nsp9 of PRRSV strain NVSL 97-7895. The structure was predicted using the I-TASSER tool and is presented in surface (left) and cartoon (right) formats. (B) Nsp9 fragments used in this study for identification of the Nsp9-N interaction domain. Full-length Nsp9 was truncated into five fragments in the first stage: Nsp91–180, Nsp9181–452, Nsp9453–646, Nsp91–452, and Nsp9181–646. The C-terminal fragment, Nsp9453–646, was subsequently split into fragments Nsp9453–551 and Nsp9552–646, and fragment Nsp9552–646 was finally split into fragments Nsp9552–598 and Nsp9599–646. (C) Vectors expressing the N protein and the indicated Nsp9 fragments were cotransformed into yeast cells. The yeast cells were grown on selective medium in decreasing densities. (D) Vectors expressing N-GFP and Nsp9599–646-His were cotransfected into HEK 293T cells. Thirty-six hours after transfection, the cell lysate was harvested and purified using Ni-NTA beads, and the eluate was examined by Western blotting using anti-GFP antibody. As a negative control, Nsp7-His was coexpressed with N-GFP. (E) HEK 293T cells were cotransfected with plasmids expressing N-GFP (green), along with the construct expressing mCherry-tagged full-length Nsp9 or the indicated fragment of Nsp9 (red). The nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) (blue).
To validate the binding of N to the C-terminal end of Nsp9, constructs expressing N-GFP and His-tagged Nsp9599–646 were cotransfected into 293T cells. Nsp7-His was coexpressed with N-GFP as a negative control. The cell lysates were then incubated with Ni-NTA beads, the eluted His-tagged proteins were examined by Western blotting using anti-His antibody, and N-GFP was detected using anti-GFP antibody. As shown in Fig. 2D, N-GFP was pulled down by Nsp9599–646-His, but not by the control protein, Nsp7-His. This result verified that the N protein binding domain of Nsp9 was located in the C-terminal 48 amino acids.
To directly visualize the interaction between the N protein and Nsp9599–646, we generated constructs expressing mCherry fused to full-length Nsp9 and the Nsp9599–646 and Nsp91–551 segments. After cotransfection with the plasmid expressing N-GFP, the fluorescence signals were examined by confocal microscopy. The results showed that Nsp9599–646, as well as the full-length Nsp9, colocalized with the N protein in the cytoplasm, although the N protein could enter the nucleolus while Nsp9 resided only in the cytoplasm (Fig. 2E, top and bottom). In contrast, no obvious colocalization between the N protein and Nsp91–551 was observed (Fig. 2E, middle). These results were consistent with the results of Y2H and pulldown assays and confirmed that the C-terminal 599-to-646 fragment of Nsp9 constituted the N protein binding domain.
Nsp9-N association exists in both types of PRRSV strains.
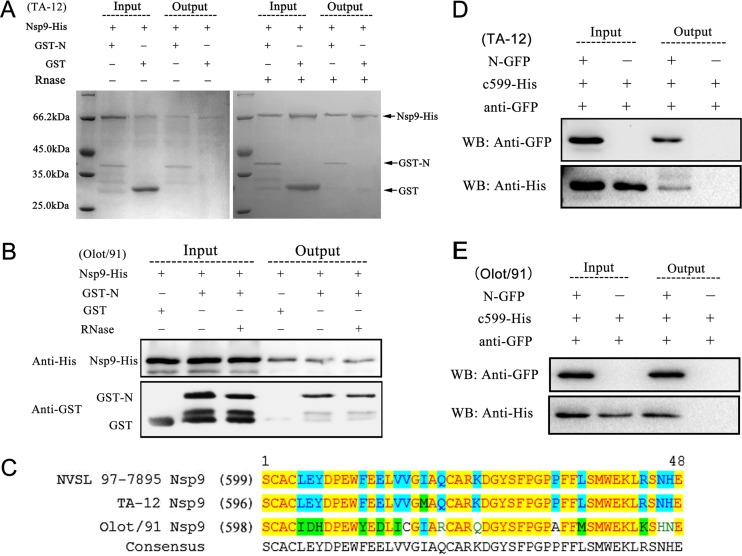
There are two PRRSV genotypes, represented by the North American prototype strain VR-2332 and the European prototype strain Lelystad virus (LV), which display extensive variation from one another (39). The above-mentioned data demonstrated the interaction between the Nsp9 and N proteins of PRRSV isolate NSVL97-7895, a highly virulent North American strain isolated in the United States in 1997 (31). To investigate whether N-Nsp9 interaction exists in other PRRSV strains, TA-12, another highly virulent North American strain isolated in China in 2008, and a low-virulence European strain, Olot/91, were used in this study. Sequence comparison (the sequence alignment is shown in Fig. S1 in the supplemental material) showed that strain TA-12 was close to NSVL97-7895, and the amino acid identity between them reached 98.5% for Nsp9 and 92.7% for the N protein. In comparison, the sequence similarity between the low-virulence European strain Olot/91 and the two American strains was only about 60% for the N protein and 70% for Nsp9.
Nsp9-His and GST-N from strain TA-12, like the proteins from strain NSVL 97-7895, were stable and could be abundantly purified and easily detected by Coomassie blue staining. In comparison, the expression level of Nsp9-His from strain Olot/91 was low, the protein degraded rapidly during the pulldown assay, and the eluate from the pulldown assay could be detected only by Western blotting. Nevertheless, the pulldown results showed that the N proteins from both strains TA-12 and Olot/91 bound to their Nsp9s, and the association was not mediated by RNA, consistent with the result from strain NSVL 97-7895 (Fig. 3A and B).
FIG 3.
Determination of Nsp9-N association in strains TA-12 and Olot/91. (A) Interaction of N with full-length Nsp9 from strain TA-12. Nsp9-His and GST-N were purified using Ni-NTA or GST-Bind Sepharose, respectively. The pulldown assays were carried out under the same conditions as for strain NVSL 97-7895 (Fig. 1C). The eluate was detected by SDS-PAGE with Coomassie brilliant blue staining. (B) Interaction of N with full-length Nsp9 from strain Olot/91. The pulldown assays were carried out as described in the legend to panel A. His-tagged proteins in the eluate were examined by Western blotting using anti-His antibodies, and the GST-tagged proteins were detected using anti-GST antibodies. (C) Sequence alignment of the 48 Nsp9 C-terminal residues of PRRSV isolates NVSL 97-7895, TA-12, and Olot/91. The residues shaded in yellow were conserved among all three strains, and those shaded in cyan were identical in two of the three strains. (D) Interaction of N with the Nsp9 C-terminal fragment from strain TA-12. (E) Interaction of N with the Nsp9 C-terminal fragment from strain Olot/91. (D and E) In coimmunoprecipitation assays, HEK 293T cells were cotransfected with vectors expressing N-GFP and His-tagged Nsp9599–646. Thirty-six hours posttransfection, the cell lysates were collected and incubated with anti-GFP antibody and protein A beads. The immunoprecipitated proteins were examined by Western blotting (WB) using anti-GFP and anti-His antibodies.
The C-terminal end of Nsp9, the region identified as the N binding domain in strain NSVL97-7895, was also highly conserved between the two highly pathogenic strains but diverged about 30% between the European and American strains (Fig. 3C). To validate the interaction domain on Nsp9 of strains TA-12 and Olot/91, a coimmunoprecipitation assay was carried out. The cell lysates coexpressing N-GFP and the His-tagged Nsp9 C-terminal-end 48 amino acid residues were harvested and immunoprecipitated with anti-GFP polyclonal antibody. The proteins eluted from the protein A column were subsequently detected by Western blotting. As the results shown in Fig. 3D and E illustrate, the Nsp9 C-terminal-end fragments from both strains TA-12 and Olot/91 were coimmunoprecipitated with their N proteins by the anti-GFP antibody. The data shown here demonstrated that PRRSV Nsp9 interacted with the N protein through its C-terminal domain, and this interaction existed in the low-pathogenic European strain, as well as in the highly pathogenic North American strains, in spite of their sequence divergence.
Determination of the crucial amino acids on Nsp9 and N protein participating in their interaction.
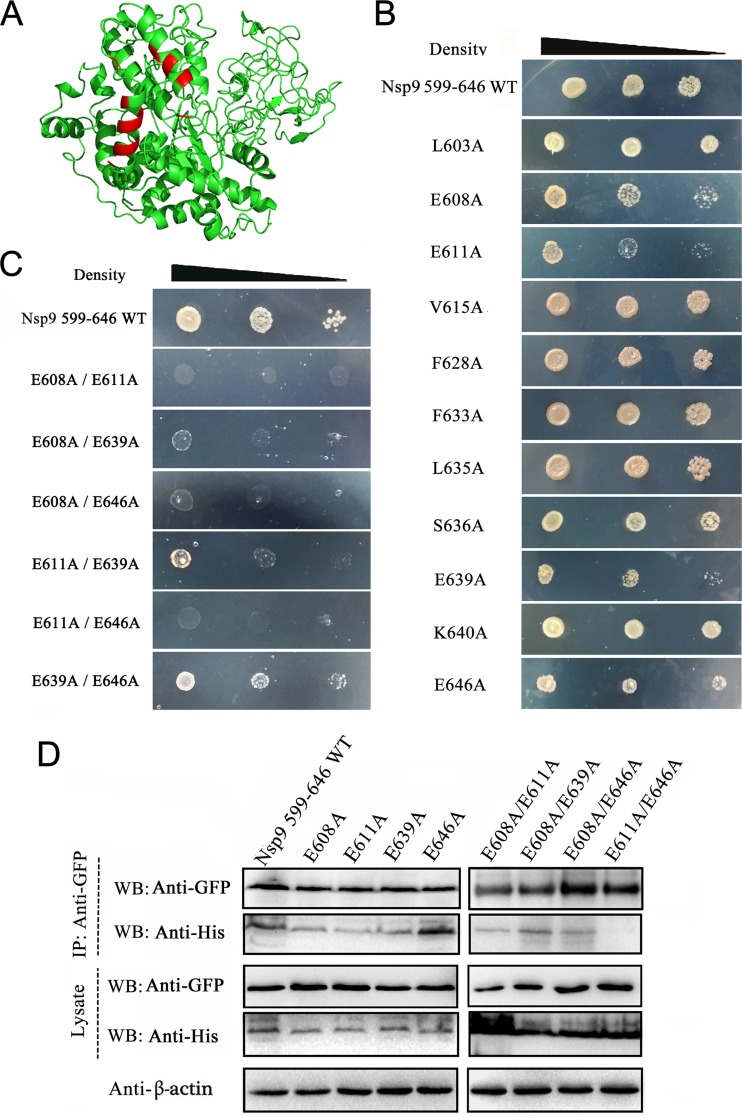
To identify the key amino acids involved in N-Nsp9 interaction, N-Nsp9 binding models for PRRSV strain NSVL 97-7895 were simulated by the online server PRISM 2.0 (34) (see Fig. S2 in the supplemental material). A series of residues exposed on the contacting surface of Nsp9 according to the binding models were chosen for mutagenesis analysis (Fig. 4A). The interactions of 11 single mutants of Nsp9599–646 with the N protein were assayed with the yeast two-hybrid system. As shown in Fig. 4B, compared with the WT Nsp9599–646, replacement of amino acids E608, E611, E639, and E646 with alanine effectively weakened but did not abrogate the association of the Nsp9 fragment with the N protein. Simultaneous mutation of two of the four residues showed that E608A/E611A, E608A/E646A, and E611A/E646A double mutants completely abolished the interaction between the N protein and the Nsp9599–646 fragment (Fig. 4C). The E608A/E639A double mutant significantly diminished the interaction, but E611A/E639A and E639A/E646A mutants showed growth ability on the selective media similar to that of the E611A and E646A single mutants (Fig. 4C).
FIG 4.
Identification of the amino acid residues on Nsp9 involved in N-Nsp9 binding. (A) Prediction of the probable amino acids on Nsp9 involved in N-Nsp9 interaction. Nsp9 and the N protein of strain NSVL 97-7895 were used for the structural prediction, and protein docking was carried out with a simulated-interaction online server (PRISM 2.0). The residues probably involved in the N-Nsp9 interaction, according to the predicted binding models (see Fig. S2 in the supplemental material), are shown in red. (B) Y2H assay of the single mutants of Nsp9599–646. BD-fused Nsp9 fragments containing the indicated mutations were coexpressed with N-AD in yeast cells. The yeast cells were grown on selective medium in decreasing concentrations. (C) Y2H assay of the double mutants of Nsp9599–646. (D) Analysis of the interaction between the N protein and the mutated Nsp9599–646 fragments by coimmunoprecipitation assay. HEK 293T cells were cotransfected with vectors expressing N-GFP and the indicated His-tagged Nsp9599–646 fragments. The precipitated proteins and the input cell lysates were examined by Western blotting using anti-GFP and anti-His antibodies. Cellular β-actin was detected as an internal control.
To verify the yeast two-hybrid results, coimmunoprecipitation assays were done using the mutated Nsp9599–646 fragments. N-GFP was coexpressed with either His-tagged WT or a single/double mutant of Nsp9599–646 in HEK 293T cells. The harvested cell lysates were incubated with anti-GFP antibodies, and the proteins eluted from protein A beads were analyzed by Western blotting using an anti-His MAb. The results showed that replacement of single amino acids had little effect on the N-Nsp9 interaction (Fig. 4D); however, the E611A/E646A double mutant lost the ability to interact with the N protein, and the E608A/E611A double mutant only weakly bound to the N protein. These results indicated that residues E608, E611, and E646 of Nsp9 could cooperatively participate in the Nsp9-N interaction.
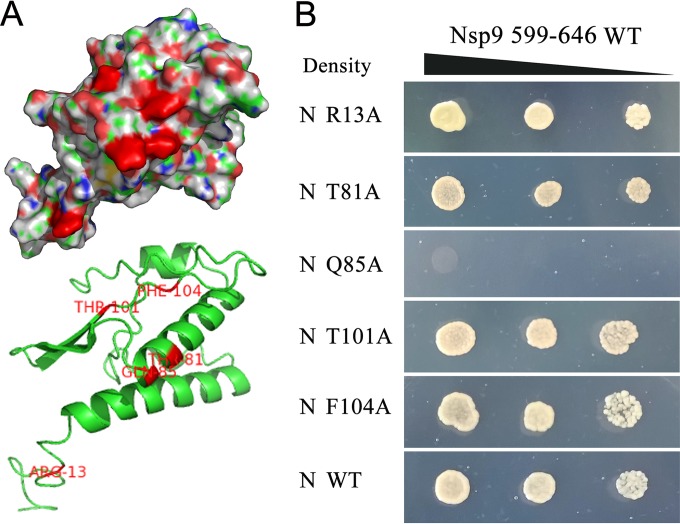
To identify the amino acid residues on the N protein that participated in the N-Nsp9 interaction, the three-dimensional structure for the N protein of strain NSVL 97-7895 was predicted (Fig. 5A) and displayed high similarity to the resolved structure for the N protein of strain VR-2332 (Protein Data Bank accession no. [PDB] 1P65), and a docking study on N-Nsp9 interaction was conducted. In the predicted binding models, amino acids R13, T81-Q85-F104, and T101 on the N protein could bind to the residues E608, E646, and E611, respectively, on Nsp9 (see Fig. S2 in the supplemental material). Here, constructs expressing the five site-directed mutants of the N protein were generated for Y2H assay. As shown in Fig. 5B, the Q85A mutant entirely abrogated the interaction with the WT Nsp9599–646 fragment, while the rest of the mutants displayed no difference from the WT N protein in the Y2H assay.
FIG 5.
Identification of the amino acids on the N protein involved in N-Nsp9 binding. (A) Prediction of the probable amino acids on the N protein involved in the N-Nsp9 interaction. The predicted three-dimensional structure of the N protein of strain NSVL 97-7895 is shown in surface (top) and cartoon (bottom) formats. The residues probably involved in the N-Nsp9 interaction, according to the predicted binding models (see Fig. S2 in the supplemental material), are shown in red. (B) Analysis of the interaction between the mutated N protein and the Nsp9599–646 fragment by Y2H assay. Vectors expressing the Nsp9599–646 fragment and the indicated mutants of the N protein or WT N protein were cotransformed into yeast. The yeast cells were grown on selective medium in decreasing densities at dilutions of 10−1, 10−2, and 10−3.
Taken together, the site-directed mutagenesis data presented here showed that amino acid Q85 on the N protein plays a critical role in the interaction with Nsp9. Three key amino acids on Nsp9 (E608, E611, and E646) involved in the N-Nsp9 interaction were identified, but further studies are needed to investigate the actual roles of these amino acids in N-Nsp9 binding.
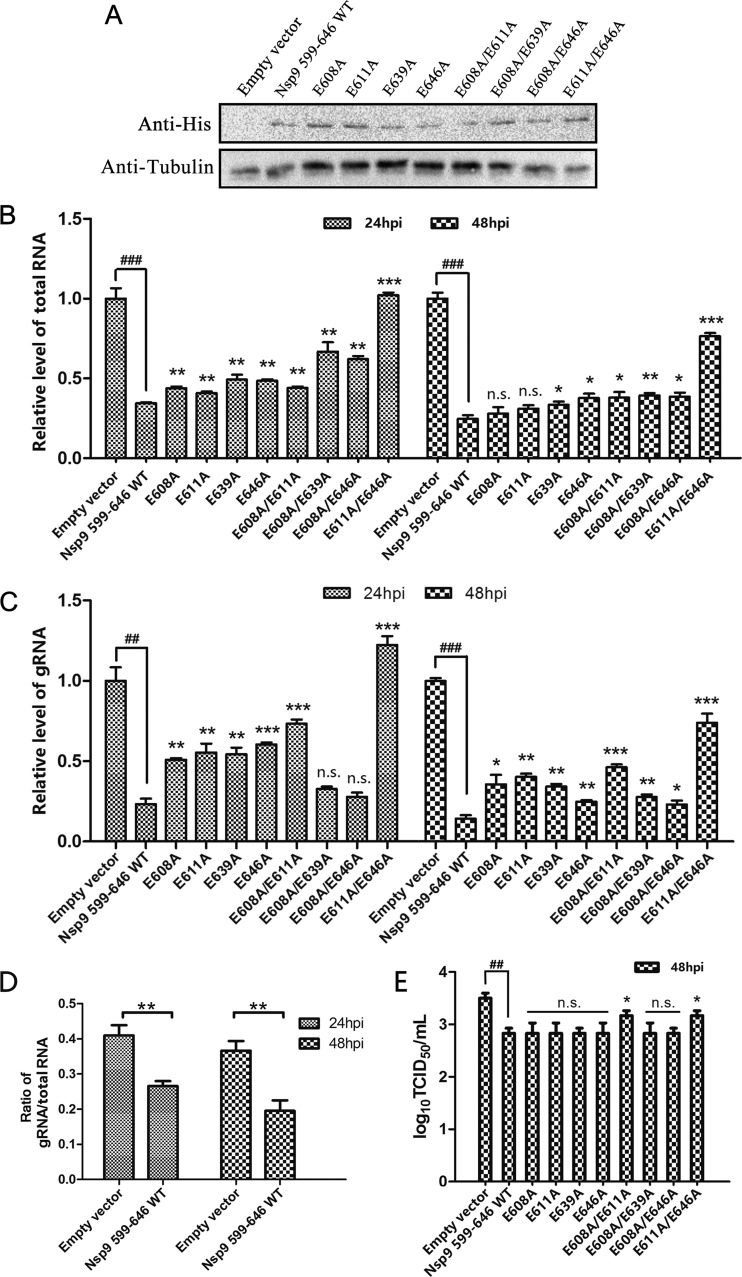
Competitive inhibition of N-Nsp9 interaction suppressed the synthesis of viral subgenomic and genomic RNAs.
In order to investigate whether the Nsp9-N interaction plays a role in PRRSV transcription, the vector expressing the polypeptide Nsp9599–646, which had been determined by pulldown and co-IP assays to be able to bind to the N protein, was transfected into Marc-145 cells. The transfected cells were then infected with PRRSV at an MOI of 1 at 16 h posttransfection, when the expression of the Nsp9599–646 fragment was detectable by Western blotting (Fig. 6A), and then viral total RNAs, including the gRNA, were quantified by real-time RT-PCR at 24 and 48 hpi. The qRT-PCR data showed that expression of Nsp9599–646 significantly inhibited the synthesis of both viral gRNA and viral total RNAs compared to the control transfected with an empty vector (Fig. 6B and C). Calculation of the gRNA/total RNA ratio revealed that the expression of Nsp9599–646 not only inhibited the synthesis of viral RNAs, but also reduced the proportion of the full-length gRNA in the total viral RNA (Fig. 6D), suggesting that the N-Nsp9 interaction participated in the regulation of PRRSV RNA replication and transcription.
FIG 6.
Competitive inhibition of the N-Nsp9 interaction by overexpression of Nsp9599–646 or its mutants and effects on the production of viral RNAs and progeny viruses. Mar-145 cells were transfected with constructs expressing the indicated fragments and then infected with PRRSV at an MOI of 1 at 16 h posttransfection. Empty vector was transfected as a control. (A) The expression of Nsp9 fragments in the transfected cells at 16 h posttransfection was detected by Western blotting using anti-His monoclonal antibody. Tubulin was detected as an internal control. (B and C) At 24 and 48 hpi, viral total RNA (B) and gRNA (C) were quantified by real-time PCR using primers targeting the N and Nsp1 genes, and the relative levels of viral RNAs versus those at 0 hpi are shown. The GAPDH level was monitored as an internal control for qRT-PCR. (D) The ratios of viral gRNA to total RNA levels in cells overexpressing WT Nsp9599–646 and in the control group were calculated. (E) Virus titers at 48 hpi were determined by limiting-dilution analysis. The data from three independent experiments were averaged and used to calculate the standard deviations, shown as error bars. Statistical analyses were performed with Student's t test. (B, C, D, and E) Significance was calculated relative to the WT Nsp9599–646 group, and the significance between empty vector and WT Nsp9599–646 is indicated: ### and ***, P < 0.001; ## and **, P < 0.01; *, P < 0.05; n.s., not significant.
In the N-Nsp9 interaction inhibition assay, expression of Nsp9599–646 mutants in parallel with WT Nsp9599–646 showed that most of the mutants displayed weaker inhibition ability than WT Nsp9599–646. The E611A/E646A double mutant, which lost the ability to bind to N protein in Y2H and co-IP assays, exhibited no activity inhibitive to the synthesis of viral RNAs compared to the empty vector. These results illustrated that interfering with the N-Nsp9 interaction suppressed the synthesis of viral RNAs and that replacing critical residues in the interfering fragments to relieve the inhibition of the N-Nsp9 interaction could also relieve the suppression effect on viral RNA synthesis, confirming the involvement of the N-Nsp9 interaction in viral RNA production.
Examination of the yield of viral progeny showed that expression of WT Nsp9599–646 significantly reduced the production of infectious viruses at 48 hpi. The E608A/E611A and E611A/E646A double mutants, which suppressed less viral gRNA synthesis, also exhibited less inhibition of viral progeny production than the other mutants (Fig. 6E).
PRRSV N protein interacts with the cellular RNA helicase DHX9 and redistributes DHX9 into the cytoplasm.
The observation of the involvement of the N-Nsp9 interaction in viral RNA synthesis raised the interesting question of how the N-Nsp9 interaction influenced the gRNA/sgmRNA product ratio. In coronaviruses, the phosphorylated N protein has been found to recruit the RNA helicase DDX1, and this can facilitate the synthesis of longer viral sgmRNAs (29). Mass spectrometry coupled with pulldown or coimmunoprecipitation assays have also found a panel of cellular RNA helicases associated with the PRRSV N protein (17, 18), and both of the studies have identified DHX9 as a potential cellular partner for the N protein, suggesting a possibility that DHX9 could play a role in PRRSV infection similar to that of DDX1 in coronavirus infection.
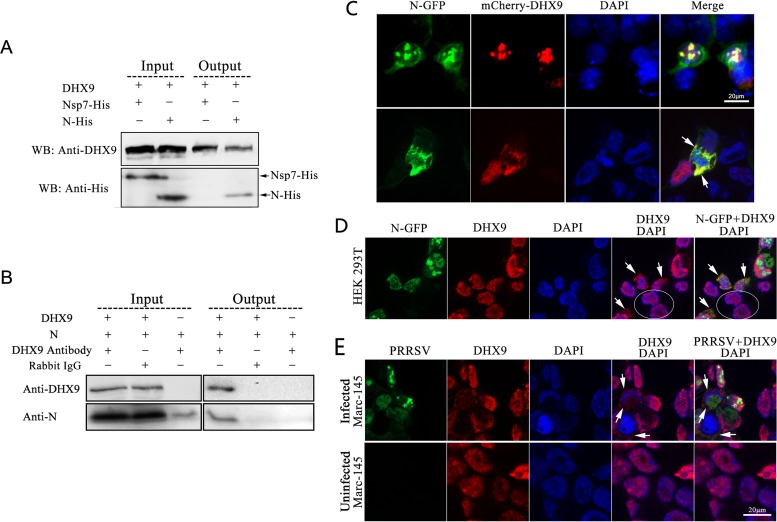
To verify the interaction of the PRRSV N protein with cellular DHX9, His-tagged N protein was coexpressed with DHX9 in HEK 293T cells. The coimmunoprecipitation assay results showed that the N protein was efficiently coimmunoprecipitated with DHX9 by anti-DHX9 antibodies (Fig. 7A), confirming the interaction between DHX9 and the PRRSV N protein. To investigate whether N interacted with DHX9 during PRRSV infection, Marc-145 cells were infected with PRRSV TA-12, and the cell extract was used for coimmunoprecipitation assays. As shown in Fig. 7B, anti-DHX9 antibody specifically coimmunoprecipitated the N protein with DHX9 from the cell extract. In the control assays, anti-DHX9 antibody failed to precipitate purified N protein without the presence of DHX9, and neither the N protein nor DHX9 was precipitated by the irrelevant rabbit IgG.
FIG 7.
Association of RNA helicase DHX9 with PRRSV N protein and virus infection. (A) Verification of the interaction between DHX9 and N by coimmunoprecipitation assay. DHX9 and N-His were coexpressed in HEK 293T cells, and Nsp7-His was coexpressed with DHX9 as a control. At 36 h posttransfection, the cells were harvested, and the cell lysates were incubated with anti-DHX9 antibody and protein A beads. Western blotting of the input lysates and precipitated proteins was performed using anti-His (WB: Anti-His) and anti-DHX9 (WB: Anti-DHX9) antibodies. (B) Identification of the interaction between DHX9 and PRRSV N protein in infected cells. Marc-145 cells were infected with PRRSV at an MOI of 1, and the cells were lysed at 48 hpi. The coimmunoprecipitation assays were carried out using anti-DHX9 antibody, with rabbit anti-goat IgG (CWBIO; CW0105) as a control. Purified N protein was also immunoprecipitated with rabbit anti-DHX9 antibody in parallel as a negative control. The eluted proteins were examined by Western blotting. (C) Colocalization of overexpressed N-GFP and mCherry-DHX9. HEK 293T cells were cotransfected with plasmids expressing N-GFP and mCherry-DHX9. The cells were fixed at 36 h posttransfection using prechilled methyl alcohol and acetone and stained with DAPI. Fluorescence was detected by confocal microscopy. Arrows indicate the redistribution of DHX9 to the cytoplasm. (D) Redistribution of endogenous cellular DHX9 in cells expressing N-GFP. HEK 293T cells were transfected with vector expressing N-GFP for 48 h and then stained with rabbit anti-DHX9 antibody after immobilization. The arrows indicate the redistribution of DHX9 to the cytoplasm in transfected cells, and the untransfected cells are circled. (E) Redistribution of cellular DHX9 in PRRSV-infected cells. Marc-145 cells were infected with PRRSV SD16-eGFP, fixed at 48 hpi, and incubated sequentially with rabbit anti-DHX9 antibody and TRITC-conjugated anti-rabbit IgG. The arrows indicate the redistribution of DHX9 to the cytoplasm in infected cells.
To visually observe the DHX9-N association, fusion proteins N-GFP and mCherry-DHX9 were coexpressed in HEK 293T cells. In transfected cells, the majority of the overexpressed mCherry-DHX9 entered cell nuclei, and N-GFP localized in both the cytoplasm and cell nuclei. In most of the cotransfected cells, the red fluorescence superimposed well on the green fluorescence in cell nuclei (Fig. 7C, top), but colocalization of DHX9 with the N protein in the cytoplasm was also observed in a small number of cotransfected cells (Fig. 7C, bottom).
The localization of overexpressed DHX9 in cell nuclei is consistent with the function of the protein in transcription regulation. However, the transcription and replication of PRRSV RNAs, like those of other nidoviruses, are carried out in the cytoplasm of the host cells. If DHX9 is recruited by the PRRSV N protein and is involved in virus replication, it will require the redistribution of the protein into the cytoplasm. To investigate whether the N protein could redistribute the localization of cellular DHX9, HEK 293T cells were transfected with the construct expressing N-GFP, and the cells' endogenous DHX9 was visualized by immunofluorescence assay. As shown in Fig. 7D, the endogenous DHX9 localized exclusively in the nuclei in the untransfected cells, while in the transfected cells expressing N-GFP, some of the DHX9 proteins were obviously redistributed from the nucleus into the cytoplasm, where they colocalized with N-GFP.
To observe the localization of DHX9 in PRRSV-infected cells, Marc-145 cells were infected at an MOI of 1 with a recombinant PRRSV strain (SD16) expressing eGFP (30), and then the localization of DHX9 was examined by immunofluorescence analysis using anti-DHX9 antibodies at 48 hpi. Consistent with the subcellular localization of the protein in untransfected 293T cells, DHX9 predominantly localized to the nucleus in uninfected Marc-145 cells (Fig. 7E, bottom). In PRRSV-infected cells, part of the endogenous DHX9 proteins were clearly redistributed into the cytoplasm (Fig. 7E, top), implying that DHX9 might be recruited into the virus RTCs and involved in the regulation of virus replication.
N protein recruits DHX9 to benefit the synthesis of viral gRNA and long sgmRNAs.
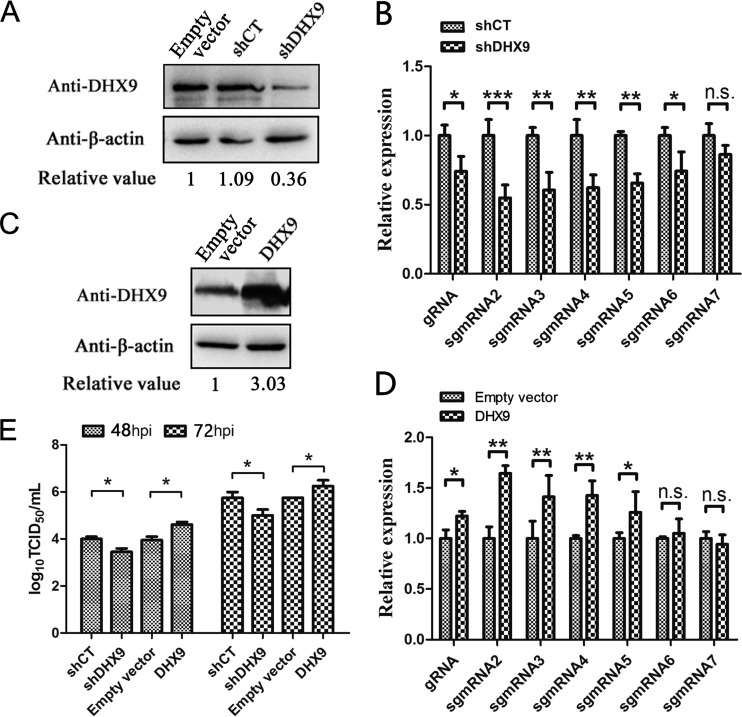
To investigate the role of DHX9 in PRRSV replication, DHX9 in Marc-145 cells was knocked down by transfection of an RNA interference vector expressing a short hairpin RNA targeting the DHX9 gene (shDHX9). Western blot analysis showed that expression of shDHX9 reduced the level of DHX9 protein by about 60% compared to the controls transfected with the empty vector or plasmid expressing interfering RNA of the luciferase reporter gene (shCT) (Fig. 8A). After the infection of the Marc-145 cells with PRRSV, synthesized viral RNAs were quantified by real-time RT-PCR. As shown in Fig. 8B, the relative levels of viral gRNA and sgmRNAs, including sgmRNA2 to sgmRNA6, apparently declined in DHX9 knockdown cells compared to the control, but the relative levels of the shortest viral sgmRNA, sgmRNA7, had no significant differences. This result suggested the positive role of DHX9 in the process of synthesizing long subgenomic mRNAs and genomic RNA.
FIG 8.
Effects of knockdown and overexpression of DHX9 on PRRSV infection. (A) Interference with DHX9 expression by shRNA. MARC-145 cells were transfected with the empty vector or vectors expressing short hairpin RNA targeting the DHX9 gene (shDHX9) or shCT targeting the luciferase gene. DHX9 protein was detected using anti-DHX9 polyclonal antibodies, and β-actin was detected as an internal control. The relative value was calculated using β-actin to normalize the amount of protein loaded. (B) Effects of DHX9 knockdown on PRRSV RNA synthesis. MARC-145 cells were infected with PRRSV at an MOI of 1 at 16 h posttransfection of the indicated vectors. The levels of viral RNAs were monitored at 48 hpi by quantitative RT-PCR, and the levels of shCT treatment were normalized to a value of 1. (C) Detection of overexpression of DHX9 protein by Western blotting. β-Actin was examined as an internal control. The relative value is the DHX9/β-actin ratio in each group. (D) Effects of DHX9 overexpression on PRRSV RNA synthesis. Cell transfection and RNA quantification were performed as described for panel B. MARC-145 cells transfected with empty vector were infected with PRRSV as a control, and the viral RNA levels in control cells were normalized to a value of 1. (E) Progeny virus titers produced in DHX9 knockdown and overexpressing cells. Virus titers at 48 and 72 hpi were determined by endpoint dilution assay. The data shown are averages of the results of three independent experiments. The error bars indicate standard deviations. Statistical analyses were performed with Student's t test. ***, P < 0.001; **, P < 0.01; *, P < 0.05; n.s., not significant.
Subsequently, Marc-145 cells were transfected with a construct overexpressing DHX9 before PRRSV infection. The overexpression of DHX9 in the transfected cells was confirmed by Western blotting (Fig. 8C). Quantification of the viral RNAs showed that overexpression of DHX9 markedly increased the relative levels of long viral RNAs compared with the control group, while no significant changes were found in the synthesis of short sgmRNA6 and sgmRNA7 (Fig. 8D). These data confirmed the beneficial role of the cellular RNA helicase DHX9 in the synthesis of long viral RNAs.
Titration of PRRSV at 48 and 72 hpi revealed that knockdown of DHX9 expression inhibited the production of progeny viruses. On the other hand, overexpression of DHX9 prominently increased virus production compared with the control group (Fig. 8E). These results demonstrated that DHX9 played a positive role in PRRSV infection.
DISCUSSION
The N protein of arteriviruses has been known as a multifunctional protein more than a structural component of the viral capsid to encapsulate the viral genomic RNA (16, 40). It has been reported to be involved in other aspects of viral biology, such as modulating host cell function during infection and recruiting cellular factors to promote viral replication (41, 42). The N protein of arterivirus EAV colocalizes with the viral replication complex in early infection, probably due to the the proximity of the sites of encapsidation and genome replication (11, 28), as it has been shown that EAV genome replication and sgmRNA transcription require only the expression of the replicase gene (28). PRRSV can be successfully rescued by reverse genetics without the preexisting N protein (30, 31), suggesting that N protein is also dispensable for the initiation of PRRSV RNA synthesis. However, whether N protein of PRRSV is involved in the regulation of the viral RNA synthesis process has remained obscure.
Here, we showed that the N protein of the arterivirus PRRSV bound with Nsp9, its RdRp, by protein-protein interaction. The N-Nsp9 interaction existed not only in the highly pathogenic North American strains NSVL 97-7895 and TA-12, but also in a low-virulence European strain, Olot/91, indicating that the protein-protein interaction could be common in PRRSV strains and may play some important roles in virus infection. In this study, we detected only the interaction of tagged N protein with Nsp9 due to the lack of appropriate antibodies for co-IP assays. Further studies are needed to examine whether N actually interacts with Nsp9 in PRRSV-infected cells.
To investigate how the N protein interacted with Nsp9, truncated Nsp9 fragments were used in Y2H, pulldown/co-IP, and immunofluorescence colocalization assays, and the C-terminal-end fragment consisting of amino acid residues 599 to 646 was identified as the N protein binding region. This region is close to but does not overlap the predicted RdRp catalytic domain, which spans amino acid residues 334 to 583 (43). Site-directed mutagenesis analyses identified the negatively charged residues E646, E608, and E611 on Nsp9 and a polar residue, Q85, on the N protein as the key residues participating in the N-Nsp9 interaction.
Competitive inhibition of the N-Nsp9 interaction with overexpressed Nsp9599–646 resulted in a dramatic decline in viral RNA synthesis, especially gRNA production, and consequently inhibited the production of infectious progeny viruses. In contrast, the mutated Nsp9599–646, with reduced ability to bind the N protein, displayed less inhibition activity than WT Nsp9599–646, and the E611A/E646A double mutant, which lost the ability to bind N protein, had the least inhibition effect. These data demonstrated that the interaction of N protein with Nsp9 could benefit the synthesis of PRRSV RNAs.
In minus-strand RNA viruses, the N protein has been well known to play important roles in viral RNA transcription and replication (19, 20). Recently, the N protein of bunyavirus was found to interact with its RdRp, and the interaction regulated the switch from capped RNA-primed transcription to unprimed viral RNA replication (21). In hantavirus, the interaction of N with its RdRp has been reported to be required for viral RNA synthesis, and it is postulated to be involved in cap-snatching or RdRp-recruiting processes (22).
In the plus-strand viruses CSFV and norovirus, the core/capsid proteins have been reported to interact with the RdRps and to be involved in viral RNA synthesis (25, 26). Here, we found that the PRRSV N protein also interacted with its RdRp, Nsp9. To our knowledge, this is the first study to demonstrate the interaction of the N protein with its viral RdRp in nidoviruses and the involvement of the N protein in arterivirus RNA synthesis. It is noteworthy that the production of PRRSV genomic RNA was reduced more severely than that of the total viral RNAs by inhibition of the N-Nsp9 interaction. In coronaviruses, the N protein has been reported to be required for efficient replication of the viral genome (44, 45). Whether the N and RdRp interaction also exists in coronaviruses and participates in RNA synthesis regulation remains to be determined.
In our previous studies, a panel of cellular RNA helicases interacting with the PRRSV N protein were identified by proteomic assays (17, 18). Among them, DHX9 was detected in both studies and scored high in the list of proteins pulled down by the His-tagged N protein. Here, we showed that the N protein could be coimmunoprecipitated with DHX9 by anti-DHX9 antibodies, confirming the interaction of PRRSV N with DHX9. The redistribution of endogenous cellular DHX9 from the nucleus to the cytoplasm, where PRRSV replication compartments are located, was observed in both N-transfected and PRRSV-infected cells, suggesting that the N protein may recruit this RNA helicase into the RTC to regulate viral RNA synthesis. Furthermore, we showed that overexpression of DHX9 increased the production of viral gRNA and sgmRNAs longer than sgmRNA6, while knockdown of the expression of DHX9 using shRNA inhibited the synthesis of viral gRNA and longer sgmRNAs. These data demonstrated that DHX9 was recruited by the N protein to benefit the replication of viral gRNA and the transcription of long sgmRNAs.
DHX9 is also called RNA helicase A, and its function has been known to be unwinding double-stranded RNA (46). Nakajima et al. (47) have reported that DHX9 acts as a bridging factor between CREB-binding protein (CBP) and RNA polymerase II and cooperates with CBP to activate transcription. In virus infections, DHX9 was first identified interacting with HIV gag protein and HIV RNA, and it was also found to be incorporated into HIV virions (48). During foot-and-mouth disease virus infection, it was reported to associate with the 5′-terminal viral genome and viral proteins 2C and 3A and also complexed with the cellular protein PABP (49). Recently, DHX9 was shown to be required for the replication and transcription of influenza virus RNAs through interaction with viral NS1 protein in an RNA-dependent manner (50).
All nidoviruses contain a positive-strand long genomic RNA. During viral RNA synthesis, the viruses first produce a set of coterminal negative-strand RNAs, which are generated through a unique discontinuous minus-strand RNA synthesis mechanism, and then use these RNAs as the template to synthesize their positive-strand gRNA and sgmRNAs (51, 52). The discontinuous process for the production of negative-strand RNAs is controlled by a conserved transcription-regulating sequence (TRS), which is located after the leader sequence (leader TRS) and in front of each gene (body TRS) (51, 53). In nidovirus infections, shorter sgmRNAs are produced more abundantly than longer viral RNAs in general, suggesting that transcription may stop upon reaching the 3′ end of the body TRS to produce shorter sgmRNAs (51, 54). To synthesize longer sgmRNAs and gRNA, which are essential to the viral life cycle, the transcription has to pass the body TRS in an appropriate proportion. To illustrate how the nested gRNA and sgmRNA are generated, a “discontinuous extension of minus-strand RNA synthesis” model has been proposed (54). In this model, the synthesis of minus-strand RNA is initiated at the 3′ end of a genomic RNA; when a body TRS and/or its flanking sequences located upstream of each ORF is encountered, a proportion of the nascent RNA elongation stops and relocates to complete its synthesis of the 5′ end of the genome; some RTC will pass the TRS and continue to synthesize longer minus-strand RNAs (51, 54, 55). However, the model does not explain how the virus can control the balance between the synthesis of shorter and longer sgmRNAs and gRNA.
In coronaviruses, Wu et al. (29) reported that the phosphorylated N protein recruited RNA helicase DDX1 to facilitate template readthrough and enable synthesis of longer sgmRNAs, probably because DDX1 could unwind the hairpin loop structure of viral gRNA during the discontinuous minus-strand RNA synthesis process. In this study, we found that the arterivirus PRRSV N protein interacted with its RdRp and recruited the cellular helicase DHX9 to facilitate the synthesis of gRNA and longer sgmRNAs. This result is consistent with the coronavirus data showing that the N protein may recruit RNA helicase to control the switch from discontinuous to continuous RNA synthesis. It would be interesting to investigate whether the involvement of the N protein in coronavirus transcription regulation is also through the interaction with the RdRp and whether the phosphorylation of the N protein plays a role in PRRSV RNA synthesis. Considering that both the PRRSV N proteins produced in HEK 293T cells and those from E. coli interact with Nsp9 and cellular DHX9 (17, 18), the phosphorylation of the N protein is unlikely to be involved in the N-Nsp9 and N-DHX9 associations. As both the N proteins from HEK 293T and E. coli cells associate, not only with DHX9, but also with a panel of other cellular RNA helicases (17, 18), and Nsp9 has also been reported to be able to recruit host RNA helicases (14), it is possible that cellular RNA helicases other than DHX9 are involved in the regulation of the discontinuous minus-strand RNA synthesis process during PRRSV infection.
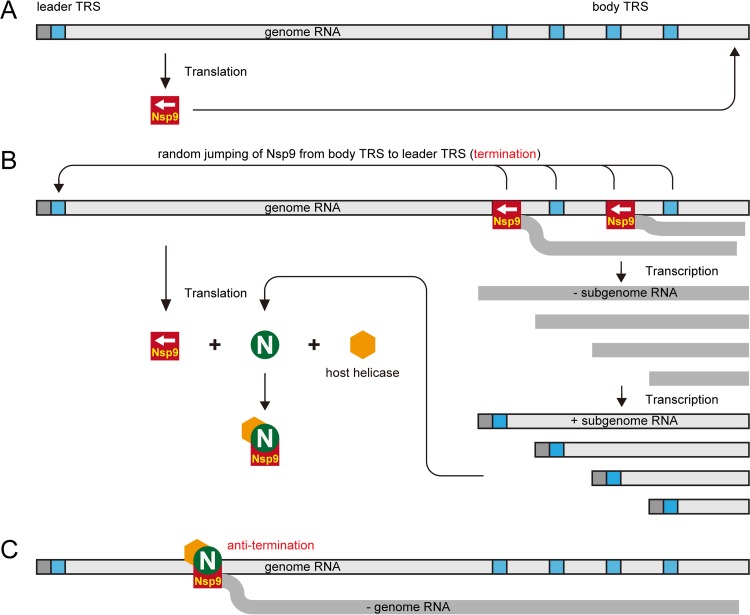
Based on the data obtained in this study and the previous discontinuous extension of minus-strand RNA synthesis model, we propose a modified model to illustrate the mechanism by which the arterivirus PRRSV controls the switch from discontinuous to continuous synthesis of viral RNAs (Fig. 9). Since the uncoated viral gRNA released in the infected cell can be used as the mRNA to synthesize the proteins encoded by ORF1, Nsp9, the viral RdRp, can be produced early in infection. After it is produced, the RdRp activity of Nsp9 initiates sgmRNA synthesis, especially the synthesis of short sgmRNAs. As the N protein is encoded by the shortest sgmRNA, it can then be abundantly produced. The newly synthesized N protein binds to Nsp9 and recruits cellular RNA helicases, including DHX9, to join the RTC. N protein binding with RNA may help stabilize the association of the RTC with the RNA template when the body TRSs are encountered and may act as an antitermination or antiattenuation factor for discontinuous RNA synthesis. The recruited RNA helicases unwind the RNA secondary structures and allow the RdRp to read through the template and generate longer viral RNAs.
FIG 9.
Proposed model to illustrate the involvement of N protein and cellular RNA helicase in the regulation of PRRSV viral production. (A) Following PRRSV entry and the release of viral genomic RNA, viral biosynthesis starts with the translation of polyproteins pp1a and pp1ab, using plus-strand gRNA as the mRNA. The nonstructural proteins, including Nsp9, are proteolytically processed by virus-encoded proteinases. (B) Nsp9 begins to transcribe nascent RNAs. The body TRSs can lead to the premature termination of negative-strand RNA synthesis and facilitate the accumulation of short minus-strand subgenomic RNAs (− sgRNA) and then their plus-strand subgenomic RNAs (+ sgRNA). The nucleocapsid protein is translated using subgenomic mRNA7 as the template, and the newly synthesized N proteins bind to Nsp9 and recruit cellular RNA helicases, such as DHX9, to form protein complexes. (C) Host RNA helicases in the protein complexes unwind the RNA secondary structures on gRNA. Consequently, Nsp9 reads through the body TRS and/or its flanking sequences, and longer − sgRNAs and − gRNA are generated.
In conclusion, we have demonstrated here that the N protein of the arterivirus PRRSV participates in viral RNA replication and transcription by interacting with Nsp9, its RdRp, and recruiting cellular RNA helicase to unwind the duplex RNA structure of gRNA and promote the production of longer viral sgmRNAs and gRNA. Our data from this study provide some new insights into the discontinuous to continuous synthesis of PRRSV RNA and offer a new potential anti-PRRSV strategy targeting the N-Nsp9 and/or N-DHX9 interaction. Further studies are ongoing to investigate by reverse genetics whether mutation of the N-Nsp9 binding sites can attenuate PRRSV.
Supplementary Material
ACKNOWLEDGMENT
This work was funded by Fundamental Research Funds for the Central Universities (QN2011065).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03216-15.
REFERENCES
- 1.Wensvoort G, Terpstra C, Pol J, Ter Laak E, Bloemraad M, De Kluyver E, Kragten C, Van Buiten LD, Den Besten A, Wagenaar F. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q 13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- 2.Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc 227:385–392. doi: 10.2460/javma.2005.227.385. [DOI] [PubMed] [Google Scholar]
- 3.Cavanagh D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol 142:629–633. [PubMed] [Google Scholar]
- 4.Firth AE, Zevenhoven-Dobbe JC, Wills NM, Go YY, Balasuriya UBR, Atkins JF, Snijder EJ, Posthuma CC. 2011. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J Gen Virol 92:1097–1106. doi: 10.1099/vir.0.029264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. 2011. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J Gen Virol 92:1107–1116. doi: 10.1099/vir.0.030213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Y, Snijder EJ. 2010. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res 154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Treffers EE, Napthine S, Tas A, Zhu L, Sun Z, Bell S, Mark BL, van Veelen PA, van Hemert MJ, Firth AE, Brierley I, Snijder EJ, Fang Y. 2014. Transactivation of programmed ribosomal frameshifting by a viral protein. Proc Natl Acad Sci U S A 111:E2172–E2181. doi: 10.1073/pnas.1321930111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D. 2000. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch Virol 145:659–688. doi: 10.1007/s007050050662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spilman MS, Welbon C, Nelson E, Dokland T. 2009. Cryo-electron tomography of porcine reproductive and respiratory syndrome virus: organization of the nucleocapsid. J Gen Virol 90:527–535. doi: 10.1099/vir.0.007674-0. [DOI] [PubMed] [Google Scholar]
- 10.Lehmann KC, Gulyaeva A, Zevenhoven-Dobbe JC, Janssen GMC, Ruben M, Overkleeft HS, van Veelen PA, Samborskiy DV, Kravchenko AA, Leontovich AM, Sidorov IA, Snijder EJ, Posthuma CC, Gorbalenya AE. 2015. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res 43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knoops K, Bárcena M, Limpens RW, Koster AJ, Mommaas AM, Snijder EJ. 2012. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol 86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong JG, Zhang N, Ge XN, Zhou L, Guo X, Yang HC. 2014. The interaction of nonstructural protein 9 with retinoblastoma protein benefits the replication of genotype 2 porcine reproductive and respiratory syndrome virus in vitro. Virology 464:432–440. doi: 10.1016/j.virol.2014.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li JN, Guo DW, Huang L, Yin MM, Liu QF, Wang Y, Yang CM, Liu YY, Zhang LJ, Tian ZJ, Cai XH, Yu LY, Weng CJ. 2014. The interaction between host Annexin A2 and viral Nsp9 is beneficial for replication of porcine reproductive and respiratory syndrome virus. Virus Res 189:106–113. doi: 10.1016/j.virusres.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 14.Zhao S, Ge X, Wang X, Liu A, Guo X, Zhou L, Yu K, Yang H. 2015. The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp9 in vitro. Virus Res 195:217–224. doi: 10.1016/j.virusres.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Music N, Gagnon CA. 2010. The role of porcine reproductive and respiratory syndrome (PRRS) virus structural and non-structural proteins in virus pathogenesis. Anim Health Res Rev 11:135–163. doi: 10.1017/S1466252310000034. [DOI] [PubMed] [Google Scholar]
- 16.Yoo D, Wootton SK, Li G, Song C, Rowland RR. 2003. Colocalization and interaction of the porcine arterivirus nucleocapsid protein with the small nucleolar RNA-associated protein fibrillarin. J Virol 77:12173–12183. doi: 10.1128/JVI.77.22.12173-12183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jourdan SS, Osorio F, Hiscox JA. 2012. An interactome map of the nucleocapsid protein from a highly pathogenic North American porcine reproductive and respiratory syndrome virus strain generated using SILAC-based quantitative proteomics. Proteomics 12:1015–1023. doi: 10.1002/pmic.201100469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Lear Z, Hughes DJ, Wu W, Zhou EM, Whitehouse A, Chen H, Hiscox JA. 2015. Resolution of the cellular proteome of the nucleocapsid protein from a highly pathogenic isolate of porcine reproductive and respiratory syndrome virus identifies PARP-1 as a cellular target whose interaction is critical for virus biology. Vet Microbiol 176:109–119. doi: 10.1016/j.vetmic.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb A. 1996. Paramyxoviridae: the virus and their replication, p 1177–1204. In Fields BN, Knipe DM, Howley PM (ed), Fields virology, 3rd ed Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 20.Lamb RA, Krug RM. 2001. Orthomyxoviridae: the viruses and their replication, p 1487–1531. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 21.Newcomb LL, Kuo RL, Ye Q, Jiang Y, Tao YJ, Krug RM. 2009. Interaction of the influenza a virus nucleocapsid protein with the viral RNA polymerase potentiates unprimed viral RNA replication. J Virol 83:29–36. doi: 10.1128/JVI.02293-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng E, Wang Z, Mir MA. 2014. Interaction between hantavirus nucleocapsid protein (N) and RNA-dependent RNA polymerase (RdRp) mutants reveals the requirement of an N-RdRp interaction for viral RNA synthesis. J Virol 88:8706–8712. doi: 10.1128/JVI.00405-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown K, Ebihara H, Feldmann H. 2012. Development of a minigenome system for Andes virus, a New World hantavirus. Arch Virol 157:2227–2233. doi: 10.1007/s00705-012-1401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikegami T, Peters CJ, Makino S. 2005. Rift Valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J Virol 79:5606–5615. doi: 10.1128/JVI.79.9.5606-5615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W, Zhang Y, Kao CC. 2014. The classic swine fever virus (CSFV) core protein can enhance de novo-initiated RNA synthesis by the CSFV polymerase NS5B. Virus Genes 49:106–115. doi: 10.1007/s11262-014-1080-x. [DOI] [PubMed] [Google Scholar]
- 26.Subba-Reddy CV, Yunus MA, Goodfellow IG, Kao CC. 2012. Norovirus RNA synthesis is modulated by an interaction between the viral RNA-dependent RNA polymerase and the major capsid protein, VP1. J Virol 86:10138–10149. doi: 10.1128/JVI.01208-12. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Casais R, Thiel V, Siddell SG, Cavanagh D, Britton P. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J Virol 75:12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molenkamp R, van Tol H, Rozier BC, van der Meer Y, Spaan WJ, Snijder EJ. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J Gen Virol 81:2491–2496. doi: 10.1099/0022-1317-81-10-2491. [DOI] [PubMed] [Google Scholar]
- 29.Wu CH, Chen PJ, Yeh SH. 2014. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe 16:462–472. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Huang B, Kong N, Li Q, Ma Y, Li Z, Gao J, Zhang C, Wang X, Liang C. 2013. A novel porcine reproductive and respiratory syndrome virus vector system that stably expresses enhanced green fluorescent protein as a separate transcription unit. Vet Res 44:104. doi: 10.1186/1297-9716-44-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Truong HM, Lu Z, Kutish GF, Galeota J, Osorio FA, Pattnaik AK. 2004. A highly pathogenic porcine reproductive and respiratory syndrome virus generated from an infectious cDNA clone retains the in vivo virulence and transmissibility properties of the parental virus. Virology 325:308–319. doi: 10.1016/j.virol.2004.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plana Duran J, Climent I, Sarraseca J, Urniza A, Cortés E, Vela C, Casal JI. 1997. Baculovirus expression of proteins of porcine reproductive and respiratory syndrome virus strain Olot/91. Involvement of ORF3 and ORF5 proteins in protection. Virus Genes 14:19–29. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Zhang Y. 2015. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuncbag N, Gursoy A, Nussinov R, Keskin O. 2011. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc 6:1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan M, Schallhorn A, Wurm FM. 1996. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res 24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schagger H. 2006. Tricine-SDS-PAGE. Nat Protoc 1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 37.Huang M, Mitchell BS. 2008. Guanine nucleotide depletion mediates translocation of nucleolar proteins, including RNA helicase A (DHX-9). Nucleosides Nucleotides Nucleic Acids 27:704–711. doi: 10.1080/15257770802145132. [DOI] [PubMed] [Google Scholar]
- 38.Kärber G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn-Schmiedebergs Arch Exp Pathol Pharmakol 162:480–483. doi: 10.1007/BF01863914. [DOI] [Google Scholar]
- 39.Nelsen CJ, Murtaugh MP, Faaberg KS. 1999. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol 73:270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowland R, Kervin R, Kuckleburg C, Sperlich A, Benfield DA. 1999. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res 64:1–12. doi: 10.1016/S0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- 41.Lee C, Hodgins D, Calvert JG, Welch S-KW, Jolie R, Yoo D. 2006. Mutations within the nuclear localization signal of the porcine reproductive and respiratory syndrome virus nucleocapsid protein attenuate virus replication. Virology 346:238–250. doi: 10.1016/j.virol.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoo D, Song C, Sun Y, Du Y, Kim O, Liu HC. 2010. Modulation of host cell responses and evasion strategies for porcine reproductive and respiratory syndrome virus. Virus Res 154:48–60. doi: 10.1016/j.virusres.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie J, Zhou H, Cui J, Chen Y, Zhang M, Deng S, Zhou P, Su S, Zhang G. 2014. Inhibition of porcine reproductive and respiratory syndrome virus by specific siRNA targeting Nsp9 gene. Infect Genet Evol 28:64–70. doi: 10.1016/j.meegid.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 44.Almazán F, Galán C, Enjuanes L. 2004. The nucleoprotein is required for efficient coronavirus genome replication. J Virol 78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schelle B, Karl N, Ludewig B, Siddell SG, Thiel V. 2005. Selective replication of coronavirus genomes that express nucleocapsid protein. J Virol 79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee C-G, Hurwitz J. 1992. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J Biol Chem 267:4398–4407. [PubMed] [Google Scholar]
- 47.Nakajima T, Uchida C, Anderson SF, Lee C-G, Hurwitz J, Parvin JD, Montminy M. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107–1112. doi: 10.1016/S0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 48.Roy BB, Hu J, Guo X, Russell RS, Guo F, Kleiman L, Liang C. 2006. Association of RNA helicase a with human immunodeficiency virus type 1 particles. J Biol Chem 281:12625–12635. doi: 10.1074/jbc.M510596200. [DOI] [PubMed] [Google Scholar]
- 49.Lawrence P, Rieder E. 2009. Identification of RNA helicase A as a new host factor in the replication cycle of foot-and-mouth disease virus. J Virol 83:11356–11366. doi: 10.1128/JVI.02677-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin L, Li Y, Pyo H-M, Lu X, Raman SNT, Liu Q, Brown EG, Zhou Y. 2012. Identification of RNA helicase A as a cellular factor that interacts with influenza A virus NS1 protein and its role in the virus life cycle. J Virol 86:1942–1954. doi: 10.1128/JVI.06362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasternak AO, Spaan WJ, Snijder EJ. 2006. Nidovirus transcription: how to make sense? J Gen Virol 87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 52.Snijder EJ, Siddell SG, Gorbalenya AE. 2005. The order Nidovirales, p 390–404. In Topley WWC. (ed), Topley and Wilson microbiology and microbial infections. ASM Press, Washington, DC. [Google Scholar]
- 53.Pasternak AO, van den Born E, Spaan WJM, Snijder EJ. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J 20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawicki SG, Sawicki DL, Siddell SG. 2007. A contemporary view of coronavirus transcription. J Virol 81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van den Born E, Gultyaev AP, Snijder EJ. 2004. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA 10:424–437. doi: 10.1261/rna.5174804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.