Abstract
Background
Mitochondria play fundamental roles in cellular metabolism, signaling, and viability. Disruption of mitochondria not only leads to dysfunction of the organelles but also activates mechanisms of cell injury and death, contributing to the pathogenesis of various diseases.
Summary
Removal of damaged mitochondria is therefore crucial for cellular homeostasis and survival. Mitophagy, the selective elimination of mitochondria via autophagy, is an important mechanism of mitochondrial quality control in physiological and pathological conditions. Defects in mitophagy have been implicated in a variety of human disorders, including both acute and chronic kidney diseases. However, the role and regulatory mechanisms of mitophagy in kidney cells and tissues remain largely unknown.
Key Message
This review provides updated information on mitophagy and suggests a potential role of mitophagy in renal pathophysiology.
Key Words: Kidney diseases, Mitochondria, Mitophagy, Cell death
Introduction
Mitochondria are organelles of eukaryotic cells playing fundamental roles in cellular metabolism, especially in ATP production through oxidative phosphorylation. Mitochondria are also a major intracellular source of reactive oxygen species (ROS) in a variety of pathologic conditions including hypoxia [1], ischemia and reperfusion [2,3], aging [4], and chemical inhibition of mitochondrial respiration [5]. Excessive mitochondrial ROS production leads to oxidative damage to cellular components that include mitochondrial proteins, lipids and DNA, which in turn leads to enhanced ROS production and further damage to mitochondria, forming a vicious cycle. Mitochondrial damage leads to the release of proapoptotic proteins, such as cytochrome c (cyt c), resulting in cell death. In addition, mitochondrial ROS production can activate the mitochondrial permeability transition (MTP) pore for cell death [6]. To ensure cellular homeostasis for viability, several mitochondrial quality control mechanisms have evolved [7,8]. Mitophagy, the selective autophagic elimination of damaged or dysfunctional mitochondria serves as an important mitochondrial quality control mechanism. Defects in mitophagy have been associated with a variety of human disorders including cancer, heart diseases and neurodegenerative diseases [9].
As a high metabolic demanding organ, the kidney is rich in mitochondria. Mitochondrial pathology has been implicated in the pathogenesis of both acute and chronic kidney diseases [10,11]. As such, mitophagy may be critical to the maintenance of homeostasis in kidney cells, and disruption of mitophagy has been implicated in several kidney diseases. However, the role and regulation of mitophagy in renal physiology and pathogenesis remain largely unknown. This review aims at providing updated information on understanding mitophagy and the relevance of mitophagy in kidney diseases.
Overview of Autophagy
Autophagy is an evolutionarily conserved intracellular system for the degradation of organelles and long-life proteins. There are three types of autophagy, including macroautophagy, chaperon-mediated autophagy and microautophagy. In microautophagy, lysosomal membrane directly engulfs a portion of the cytoplasm [12]. In chaperon-mediated autophagy, cargo proteins containing a consensus KFERQ-like motif are recognized by HSPA8, and then the substrates bind to the lysosomal protein LAMP-2A followed by translocating across the lysosomal membrane for degradation [13]. Macroautophagy (hereafter referred to as autophagy) is characterized by the formation of a unique double-membrane organelle called autophagosome.
Autophagy is initiated by the nucleation of an isolation membrane, and then the isolation membrane elongates and closes to form an autophagosome. The origin of autophagosome membranes still remains controversial, but in mammalian cells, the endoplasmic reticulum seems to be the major source of autophagosome membrane [14]. Autophagosome formation is regulated by a complex molecular machinery. Among them, two ubiquitin-like conjugation systems, Atg12-Atg5 and Atg8-PE are involved in the expansion of autophagosome [15]. Autophagosome will fuse with lysosome to form a hybrid organelle named autolysosome. The cellular contents sequestered by autophagosome are degraded in autolysosome. Autophagy was considered as a nonselective bulk degradative process where the autophagosomes randomly engulf contents in the cytosol. However, it has been recently demonstrated that autophagy can specifically or selectively recognize and degrade protein aggregates, organelles or invading microbes [12]. To date, several selective types of autophagy have been verified, including the cytoplasm-to-vacuole targeting, pexophagy, ribophagy, xenophagy and mitophagy [16]. Autophagy plays critical roles in adaptive responses to starvation and other forms of cellular stress, homeostasis, and differentiation and development [17,18,19]. Defects in autophagy have been implicated in a range of diseases including cancer, neurodegenerative diseases, infectious diseases, and metabolic diseases [20,21].
Mitophagy-Autophagic Clearance of Mitochondria
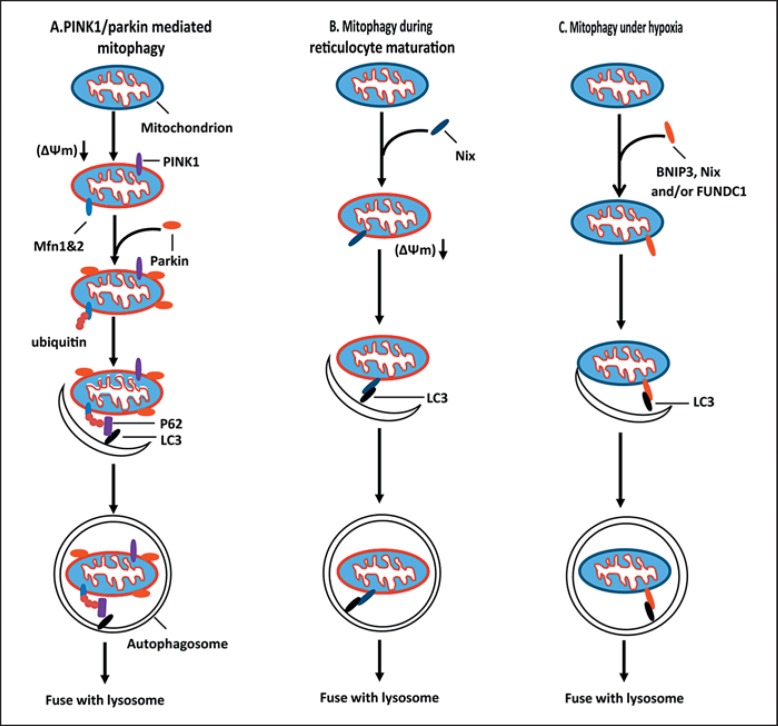
Mitophagy is the selective degradation of mitochondria by autophagy. In this process, mitochondria are sequestered in autophagosomes and delivered to lysosomes for hydrolytic degradation. Mitophagy occurs physiologically and pathologically. Physiologically, Mitophagy plays essential roles in development, including the complete removal of mitochondria during erythrocyte maturation and the selective destruction of sperm-derived mitochondria after oocyte fertilization [22]. Defects in mitophagy have been implicated in a variety of pathological conditions including neurodegenerative diseases, myopathies, aging, cardiac diseases, and autoimmune diseases [9,23]. Mitophagy shares the core molecular machinery with autophagy, and the induction and regulation of mitophagy involves a coordination of the mitochondrial dynamics machinery, specific mitophagy-initiating pathways, and the autophagy system. The work in the last few years has delineated two major mitophagy pathways that are centered on PINK1/Parkin and BNIP3/Nix [24,25] (fig. 1).
Fig. 1.
Mitophagy mechanisms. A PINK1/Parkin-mediated mitophagy. In healthy mitochondria, PINK1 is constitutively imported into mitochondria and degraded by the inner mitochondrial membrane protease PARL. In response to mitochondrial stress, mitochondrial membrane potential (Δψm) is dissipated to prevent PINK1 import, leading to the accumulation of PINK1 on the mitochondrial outer membrane. PINK1 then recruits Parkin to mitochondria. Upon recruitment, Parkin ubiquitinates mitochondrial membrane proteins such as mitofusins. P62 was recruited to ubiquitinated mitochondria to promote the delivery of ubiquitinated mitochondria to autophagosome via the binding to LC3. B Mitophagy during reticulocyte maturation. During reticulocyte maturation, upregulation of Nix induces mitochondrial depolarization and promotes the delivery of mitochondria to autophagosome by binding to LC3. C Mitophagy under hypoxia. In response to hypoxia, Nix and BNIP3 are induced. Nix and BNIP3 act as autophagy receptors to deliver mitochondria to autophagosome by binding to LC3. FUNDC1 may also act as an autophagy receptor to deliver mitochondria to autophagosome.
PINK1-Parkin Pathway of Mitophagy
The most recognized mitophagy pathway in mammalian cells is mediated by two Parkinson's disease-related proteins, PINK1 and Parkin [26,27]. PINK1 is serine/threonine kinase with a mitochondrial target sequence [26], and Parkin is a cytosol ubiquitin E3 ligase [28]. In healthy mitochondria, PINK1 is constitutively imported to the inner membrane where it is cleaved by the inner membrane presenilin-associated rhomboid-like protease PARL, and ultimately proteolytically degraded [29]. Mitochondrial depolarization prevents the import of PINK1 to the inner membrane, thereby stabilizing the full length PINK1 on the mitochondrial outer membrane [30,31]. In this manner, PINK1 acts as a sensor of mitochondrial damage. Accumulation of full-length PINK1 on the mitochondrial surface recruits Parkin from the cytosol to damaged mitochondria. Following the recruitment to mitochondria, Parkin ubiquitinates mitochondrial outer membrane proteins including mitofusin 1 and mitofusin 2 [32] to induce and promote the autophagic removal of mitochondria. PINK1 is necessary for Parkin-mediated mitophagy. PINK1-phosphorylated mitofusin 2 has been implicated as a Parkin receptor to attract Parkin to damaged mitochondria [33]. PINK1 is essential for activating Parkin E3 ligase activity by phosphorylating Parkin and ubiquitin [34,35]. On the other hand, the mitochondrial deuibiquitinase USP30 opposes Parkin-mediated mitophagy [36]. In addition, some proteins such as HSPA1L, BAG4 and SIAH3 have been shown to modulate the translocation of Parkin to damaged mitochondria [37].
How are ubiquitinated mitochondria delivered to the autophagy machinery? Accumulating evidence has demonstrated that P62 (also known as SQSTM1) is essential for delivering ubiquitinated mitochondria to autophagosome via binding to LC3 [30,38]. However, the role of p62 in Parkin-induced mitophagy remains controversial. Mitochondrial elimination has been reported to occur in the absence of p62-mediated clustering [39]. A very recent study by Wong and Holzbaur [40] showed that instead of p62, optineurin was required for autophagosome formation around damaged mitochondria in Parkin-mediated mitophagy by binding to LC3 via its LC3 interaction region (LIR) domain, and the recruitment of optineurin is independent on P62. These findings suggest Parkin can active mitophagy through recruiting preexisting phagophores via ubiquitination of outer mitochondrial membrane proteins and recruitment of receptors such as p62 and optineurin.
Parkin can also interact directly with autophagy-regulating proteins to facilitate mitophagy. For example, Van Humbeeck et al. [41] revealed that Parkin interacted with Ambra1, an activator of the Beclin1/Vps34 complex, to induce mitophagy. They showed that the interaction between Parkin and Ambra1 increased in depolarized mitochondria. Ambra1 was not required for the mitochondrial translocation of Parkin, but was critical for mitochondrial clearance through stimulating perimitochondrial nucleation of new phagophores via activating Vps34 in their immediate vicinity. These data suggest that the interaction of Parkin with Ambra1 is a key mechanism for inducing the final clearance step of Parkin-mediated mitophagy. However, it is worth noting that Ambra1 was also implicated to induce Parkin- and p62-independent but LC3-dependent mitophagy [42].
In addition to Parkin, some other ubiquitin E3 ligases, including cytoplasmic E3 ligase SMURF1 and Gp78, and the mitochondrial E3 ubiquitin protein ligase 1 (Mul1) have also been implicated in mitophagy. SMURF1 [43] and Gp78 [44] participate in Parkin-dependent and -independent mitophagy, respectively. Mul1 was shown to induce mitophagy in skeletal muscles [45]. A recent study in Drosophila and mammalian cells revealed that Mul1 acted in parallel to the PINK1/parkin pathway in regulating mitofusin degradation and compensated for the loss of PINK1/parkin, and Mul1 did not affect Parkin-mediated mitophagy [46].
Nix- and BNIP3-Mediated Mitophagy
BCL2 and adenovirus E1B 19-kDa-interacting protein 3 (BNIP3) and BNIP3-like (BNIP3L or Nix) are Bcl-2 family proteins with an atypical BH3 domain [47]. BNIP3 and Nix are mitochondrial outer membrane proteins and initially identified as pro-death proteins. Recent studies indicated that BNIP3 and Nix also mediate mitophagy in specific conditions. Nix seems indispensable for the complete mitochondrial elimination during reticulocyte maturation. In Nix-deficient mice, mitochondrial clearance in reticulocytes was dramatically inhibited or retarded [48,49,50]. Several studies also revealed that Nix deficiency inhibited mitochondrial depolarization, and treatment with uncoupling chemicals (e.g., CCCP) or a BH3 mimetic (e.g., ABT-737) was able to induce mitochondrial depolarization and then restored the sequestration of mitochondria into autophagosomes in Nix-deficient erythroid cells, suggesting that one mechanism for Nix induced-mitophagy is inducing mitochondrial depolarization [50]. Nix was also shown as a selective autophagy receptor for targeting mitochondria to autophagosomes via directly interacting with LC3 and GABARAP with its LIR domain [51].
Nix and BNIP3 have also been implicated in hypoxia-induced mitophagy. Under hypoxia conditions, Nix and BNIP3 are highly induced to activate mitophagy to prevent increased ROS production through elimination of damaged mitochondria, and this in turn mitigates cell death [52,53]. Similar to Nix, BNIP3 was implicated to active mitophagy via inducing mitochondrial depolarization [54] and/or delivering mitochondria to autophagosomes as a receptor via directly interacting with LC3 with its LIR domain [55]. In addition, as BH3-only proteins, Nix and BNIP3 can compete with Beclin-1 for binding to BCL-2 or BCL-xl leading to the release of Beclin1 [56]. Once released, Beclin1 is free to nucleate preautophagosomal membranes and induce autophagy.
In addition to BNIP3 and Nix, the integral mitochondrial outer membrane protein FUNDC1 was implicated in hypoxia-induced mitophagy through interacting with LC3 with its typical LC3-binding mitof Y(18)xxL(21) [57]. Moreover, a very recent study showed that FUNDC1 regulated ULK1 recruitment to damaged mitochondria, where ULK1 interacted and phosphorylated FUNDC1 at serine 17 to enhance FUNDC1 binding to LC3 [58]. It will be interesting to know whether FUNDC1 cooperates with BNIP3 and/or Nix to induce mitophagy under hypoxia.
Mitochondrial Fragmentation in Mitophagy
Mitochondria are highly dynamic organelles that are engaged in repeated cycles of fusion and fission. Mitochondrial fission and fusion are regulated by members of a family of conserved large GTPases. In mammals, mitochondrial fission is mediated by the cytoplasmic dynamin-related protein 1 (DRP1) [59] and the outer mitochondrial membrane protein FIS1 [60]. Mitochondrial fusion is regulated by Mfn1, Mfn2 and OPA1. Mfn1 and Mfn2 mediate outer membrane fusion, and OPA1 mediates inner membrane fusion [61,62]. Mitochondria fuse and divide in response to cell demands and environmental changes. Under cell stress, mitochondrial fission is activated via Drp1 and fusion is arrested through the interaction of Bak, a proapoptotic Bcl-2 family protein on mitochondria, with mitofusins, culminating in mitochondrial fragmentation [63,64]. Mitochondrial fragmentation has been identified in various human diseases, including cancer as well as neurologic and cardiovascular diseases [65,66].
Recently, accumulating evidence has demonstrated that mitochondrial fragmentation is a prerequisite for mitophagy. The study by Twig et al. [67] revealed that fusion and fission were paired and occurred most commonly as fusion quickly followed by fission. Often, a fission event yields two types of daughter mitochondria, including hyperpolarized mitochondria and depolarized mitochondria. The depolarized daughter mitochondria have decreased levels of OPA1 and a reduced probability to fuse and will be removed by autophagy or mitophagy. Inhibition of fission reduces mitophagy, resulting in the accumulation of damaged mitochondria. Depolarization also causes the loss of Mfn1 and Mfn1 on mitochondria and thereby promotes mitochondrial fission, leading to Parkin-and/or Mul1-mediated mitophagy [46,68]. Another study by Lee et al. [69] showed that inhibition of Drp1 or overexpression of Mfn1 prevented BNIP3-mediated mitophagy. However, although mitochondrial fragmentation promotes mitophagy, fission itself cannot induce mitophagy, for which a concomitant mitochondrial damage such as depolarization and increased ROS or other unclear signals are required [70].
MTP and Mitophagy
MTP is the result of opening mitochondria permeability transition pores in the inner membrane, which conduct all solutes less than about 1.5 KD [71]. Mitochondrial injuries from calcium overloading, ROS, and thiol cross-linking agents trigger MTP [72]. MTP causes mitochondrial depolarization, mitochondrial swelling, outer membrane rupture and consequent release of apoptogenic factors. MPT is recognized as a key event in cell death, especially necrosis.
MPT has recently been implicated in the activation of mitophagy, probably through inducing mitochondrial depolarization, a well-documented trigger of mitophagy in mammalian cells. For example, a recent study by Elmore et al. [73] showed that cyclosporin A, an immunosuppressant that blocks MPT, prevented serum deprivation and glucagon stimulated-mitochondrial depolarization and the subsequent sequestration of mitochondria into autophagosomes in hepatocytes [73,74]. In another study, Cui et al. [75] revealed that silence of PINK1-induced mitophagy was mediated MTP in dopaminergic MN9D cells. However, no direct measurement of mitophagy was performed when MPT was inhibited in these studies. Also, it is worth noting that not all mitochondrial depolarizations are mediated by MPT.
Mitophagy and Cell Death
In general, mitophagy preserve viability by removing damaged mitochondria. However, the interplay between mitophagy and cell death is complicated [76,77]. Firstly, many stimuli can trigger both mitophagy and cell death in the same cell. The outcome (life or death) is dependent on the degree and duration of the stress. If the stress is mild and only a subset of mitochondria is damaged, the cell can remove those mitochondria via mitophagy to preserve viability. On the other hand, when cellular stress is severe so that the number of damaged mitochondria overwhelms the capacity of mitophagy, or if mitophagy is defective, then the cell dies.
Secondly, several proteins have the capacity to regulate both mitophagy and apoptosis. For instance, Bcl-2 family proteins are involved in regulating mitophagy. Hollville et al. [78] showed that prosurvival Bcl-2 family proteins (Bcl-xL, Mcl-1, and Bcl-W) inhibit Parkin-mediated mitophagy probably by interfering with the stable recruitment of Parkin to mitochondria via direct interaction with Parkin. In a sharp contrary, BH3-only proteins (Bad, Bim, Noxa, or Puma) or a BH3 mimetic enhanced the mitochondrial translocation of Parkin leading to mitophagy activation. It is noteworthy that the effect on mitophagy may be secondary to the effect of these proteins on mitochondrial integrity and depolarization. Bcl-2 family proteins Nix and BNIP3 are proapoptotic proteins. However, as mentioned above, they both also mediate mitophagy. Moreover, BNIP3 overexpression-induced mitophagy was shown to be independent of the intrinsic apoptotic cell death pathway [54]. However, what remains unknown is exactly how these proteins switch from promoting cell death to preserving viability by regulating mitophagy.
Thirdly, apoptosis may inhibit mitophagy by cleaving autophagy proteins. For instance, Beclin1 [79,80] and Atg5 [81] were reported to be cleaved by caspase and calpain, respectively. AMBRA1 cleavage by caspases and calpains has also been reported [82]. The cleavage of these autophagy proteins may result in the inactivation of autophagy. Moreover, some autophagy protein fragments resulting from caspase or calpain cleavage may translocate from cytosol to mitochondria, disrupting the interaction between Bcl-2 with Beclin1 and leading to augmented apoptosis [79,81]. These studies demonstrated that when there is vast mitochondrial damage, activation of apoptosis may shut down autophagy to ensure cell death. Based on these findings, enhancement of mitophagy may provide a therapeutic strategy to protect against cell death.
Mitochondrial Pathology in Kidney Diseases
Mitochondrial pathology occurs in and is known as an important pathogenic event of kidney diseases. Firstly, mitochondrial dynamics is disrupted during kidney injury, resulting in morphological and structural changes. For example, in cisplatin-induced acute kidney injury, disruption of mitochondrial cristae and extensive mitochondrial swelling in proximal tubular epithelium were observed [83]. Mitochondrial fragmentation was detected in kidney tubule cells under acute and chronic kidney injury [11,64,66,84,85]. Using an in vivo multiphoton imaging technique, Hall et al. [86] showed that, in response to ischemia, the mitochondrial membrane potential dissipated rapidly, and mitochondria became shortened and fragmented in proximal tubules but not in distal tubules. Secondly, mitochondrial biogenesis in the kidney is affected during kidney injury. In a cisplatin-induced AKI model, a decreased mitochondrial mass was observed, indicating impaired mitochondrial biogenesis [83]. In a rat model with chronic cyclosporin A (CsA) treatment, mitochondrial DNA copy number, along with the expression of nuclear and mitochondrial DNA-encoded oxidative phosphorylation (OXPHOS) proteins, peroxisome proliferator-activated receptor-γ coactivator (PGC)-1α, and mitochondrial transcription factor-A (Tfam) was dramatically decreased [87]. Interestingly, in a mouse model with 5/6 nephrectomy, decreases of mitochondrial content also happened in muscles, which resulted in impaired exercise endurance, a well-documented feature in chronic kidney diseases [88]. Mitochondrial biogenesis increases the mitochondrial mass and functional capacity or helps preserve it by replacing damaged mitochondria, so defects in mitochondrial biogenesis may deteriorate kidney injury. In addition, as will be discussed later, mitochondrial degradation in the kidney was also affected during kidney injury [89].
Mitochondrial structural changes are often associated with functional impairment evidenced by reduced ATP production, excessive generation of ROS, release of proapoptotic proteins, or disturbance in calcium homeostasis, etc. [10,90]. In the kidney, mitochondrial dysfunction is implicated in both acute and chronic kidney injury. For instance, in cisplatin-induced acute kidney injury, ATP production was affected by inhibiting complexes I-IV of the respiratory chain [91] and the electron transport chain enzyme cytochrome c oxidase [91]. Increasing evidence suggests that mitochondrial generation of ROS in response to pathogenic conditions, such as ischemia-reperfusion and hyperglycemia, plays a pathogenic role in kidney disease [92]. Release of proapoptotic proteins from mitochondrial intermembrane space resulting from permeabilization of mitochondrial outer and/or inner membrane was shown to contribute to tubular cell death that characterizes acute and chronic kidney disease [93,94]. Collectively, mitochondrial pathology plays a pathogenic role in a variety of kidney diseases, so timely removal of damaged mitochondria is imperative to keep the kidney healthy.
Mitophagy in Kidney Diseases
Mitophagy has been implicated in several kidney disease models, including both AKI and CKD. In response to ischemia-reperfusion injury, BNIP3 was upregulated in a HIF-1-dependent manner. Interestingly, BNIP3 overexpression was shown to induce mitophagy, and inhibition of BNIP3 reduced mitophagy, suggesting BNIP3-mediated mitophagy may happen in tubular cells in AKI [95]. In the cell and mouse models of metabolic acidosis, a common complication of CKD, mitophagy was shown to be induced in proximal tubular cells, which was indispensable for maintaining mitochondrial membrane potential, morphology and functions [96], suggesting a possible link between mitophagy and metabolic acidosis. In a diabetic kidney disease model, inhibition of mitophagy and altered expression of mitophagic proteins in the kidney were observed, indicating that a disruption of renal tubular mitochondrial quality control may contribute to the pathogenesis of diabetic kidney disease [84]. In the kidney of a FSGS mouse model with autophagy deficiency in nephron and human idiopathic FSGS kidney biopsy specimens, mitochondrial dysfunction was observed, suggesting an association between mitophagy and cell injury in FSGS [97]. During renal injury induced by high-calorie diet, mitophagy was dramatically reduced accompanied by abnormal mitochondrial morphology. However, calorie restriction markedly enhanced renal autophagy and ameliorated oxidative damage in kidneys [98]. Taken together, these studies suggest that mitophagy may play a renoprotective role in both acute and chronic kidney injury. However, the regulatory mechanisms of mitophagy under these conditions remain largely unknown.
Conclusions and Future Directions
Significant progress has been made in our understanding of the roles and regulation of mitophagy in the last decade. However, many questions still remain unanswered. For example, mounting evidence supports the fact that mitophagy plays cytoprotective roles in pathological conditions, but emerging evidence also implicates that mitophagy is a possible effector of cell death programs [99]. The reason for the opposing role of mitophagy in different pathological conditions remains to be verified and delineated. Several mitophagy mechanisms have been identified, but which mechanism dominates and how they interconnect to modulate the level of mitophagy in a specific pathological condition remains largely unclear. In addition, the molecular mechanism underlying the switch of Nix and BNIP3 from promoting cell death to preserving viability by regulating mitophagy is unclear. Mitochondrial fragmentation not only promotes mitophagy but also apoptosis [63,100]. The determinants of the outcome (apoptosis vs. mitophagy) of mitochondrial fragmentation need to be further investigated. As to the kidney, although mitophagy has been implicated in model systems of kidney diseases, the exact role and regulatory mechanism of mitophagy under these conditions of kidney pathology remains unclear. Therefore, future studies using animal models with the deletion of mitophagy-related genes specifically in kidney tissues will be needed to further our understanding of mitophagy in kidney diseases.
Disclosure Statement
The authors have no conflicts of interest to declare.
Acknowledgements
This study was supported in part by grants from the National Basic Research Program of China 973 (program No. 2012CB517601), the National Natural Science Foundation of China (81430017) and the National Institutes of Health and Department of Veterans Administration of USA.
References
- 1.Chandel NS, McClintock DS, Feliciano CE, et al. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Jang HS, Park KM. Reactive oxygen species generated by renal ischemia and reperfusion trigger protection against subsequent renal ischemia and reperfusion injury in mice. Am J Physiol Renal Physiol. 2010;298:F158–F166. doi: 10.1152/ajprenal.00474.2009. [DOI] [PubMed] [Google Scholar]
- 3.Dobashi K, Ghosh B, Orak JK, Singh I, Singh AK. Kidney ischemia-reperfusion: modulation of antioxidant defenses. Mol Cell Biochem. 2000;205:1–11. doi: 10.1023/a:1007047505107. [DOI] [PubMed] [Google Scholar]
- 4.Liochev SI. Reactive oxygen species and the free radical theory of aging. Free Radic Biol Med. 2013;60:1–4. doi: 10.1016/j.freeradbiomed.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Watabe M, Nakaki T. Mitochondrial complex I inhibitor rotenone inhibits and redistributes vesicular monoamine transporter 2 via nitration in human dopaminergic SH-SY5Y cells. Mol Pharmacol. 2008;74:933–940. doi: 10.1124/mol.108.048546. [DOI] [PubMed] [Google Scholar]
- 6.Brenner C, Moulin M. Physiological roles of the permeability transition pore. Circ Res. 2012;111:1237–1247. doi: 10.1161/CIRCRESAHA.112.265942. [DOI] [PubMed] [Google Scholar]
- 7.Kotiadis VN, Duchen MR, Osellame LD. Mitochondrial quality control and communications with the nucleus are important in maintaining mitochondrial function and cell health. Biochim Biophys Acta. 2014;1840:1254–1265. doi: 10.1016/j.bbagen.2013.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer F, Hamann A, Osiewacz HD. Mitochondrial quality control: an integrated network of pathways. Trends Biochem Sci. 2012;37:284–292. doi: 10.1016/j.tibs.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Redmann M, Dodson M, Boyer-Guittaut M, Darley-Usmar V, Zhang J. Mitophagy mechanisms and role in human diseases. Int J Biochem Cell Biol. 2014;53:127–133. doi: 10.1016/j.biocel.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Che R, Yuan Y, Huang S, Zhang A. Mitochondrial dysfunction in the pathophysiology of renal diseases. Am J Physiol Renal Physiol. 2014;306:F367–F378. doi: 10.1152/ajprenal.00571.2013. [DOI] [PubMed] [Google Scholar]
- 11.Zhan M, Brooks C, Liu F, Sun L, Dong Z. Mitochondrial dynamics: regulatory mechanisms and emerging role in renal pathophysiology. Kidney Int. 2013;83:568–581. doi: 10.1038/ki.2012.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 15.Nakatogawa H. Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy. Essays Biochem. 2013;55:39–50. doi: 10.1042/bse0550039. [DOI] [PubMed] [Google Scholar]
- 16.Jin M, Liu X, Klionsky DJ. SnapShot: Selective autophagy. Cell. 2013;152:368–368. doi: 10.1016/j.cell.2013.01.004. e362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 21.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu H, Li G, Liu L, Feng L, Wang X, Jin H. Regulation and function of mitophagy in development and cancer. Autophagy. 2013;9:1720–1736. doi: 10.4161/auto.26550. [DOI] [PubMed] [Google Scholar]
- 23.de Vries RL, Przedborski S. Mitophagy and Parkinson's disease: be eaten to stay healthy. Mol Cell Neurosci. 2013;55:37–43. doi: 10.1016/j.mcn.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 27.Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 28.Riley BE, Lougheed JC, Callaway K, et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deas E, Plun-Favreau H, Gandhi S, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisler S, Holmstrom KM, Skujat D, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature cell biology. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 31.Matsuda N, Sato S, Shiba K, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kane LA, Lazarou M, Fogel AI, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koyano F, Okatsu K, Kosako H, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 36.Bingol B, Tea JS, Phu L, et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 37.Hasson SA, Kane LA, Yamano K, et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013;504:291–295. doi: 10.1038/nature12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu CT, Ji J, Dagda RK, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci USA. 2014;111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Humbeeck C, Cornelissen T, Vandenberghe W. Ambra1:a Parkin-binding protein involved in mitophagy. Autophagy. 2011;7:1555–1556. doi: 10.4161/auto.7.12.17893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strappazzon F, Nazio F, Corrado M, et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015;22:419–432. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orvedahl A, Sumpter R, Jr., Xiao G, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu M, St-Pierre P, Shankar J, Wang PT, Joshi B, Nabi IR. Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Mol Biol Cell. 2013;24:1153–1162. doi: 10.1091/mbc.E12-08-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lokireddy S, Wijesoma IW, Teng S, et al. The ubiquitin ligase Mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab. 2012;16:613–624. doi: 10.1016/j.cmet.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 46.Yun J, Puri R, Yang H, et al. MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. Elife. 2014;3:e01958. doi: 10.7554/eLife.01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen G, Cizeau J, Vande Velde C, et al. Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J Biol Chem. 1999;274:7–10. doi: 10.1074/jbc.274.1.7. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J, Ney PA. NIX induces mitochondrial autophagy in reticulocytes. Autophagy. 2008;4:354–356. doi: 10.4161/auto.5552. [DOI] [PubMed] [Google Scholar]
- 49.Chen M, Sandoval H, Wang J. Selective mitochondrial autophagy during erythroid maturation. Autophagy. 2008;4:926–928. doi: 10.4161/auto.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bellot G, Garcia-Medina R, Gounon P, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Bosch-Marce M, Shimoda LA, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Thomas RL, Kubli DA, Gustafsson AB. Bnip3-mediated defects in oxidative phosphorylation promote mitophagy. Autophagy. 2011;7:775–777. doi: 10.4161/auto.7.7.15536. [DOI] [PubMed] [Google Scholar]
- 55.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu L, Feng D, Chen G, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 58.Wu W, Tian W, Hu Z, et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J Cell Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- 61.Meeusen S, DeVay R, Block J, et al. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 62.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brooks C, Dong Z. Regulation of mitochondrial morphological dynamics during apoptosis by Bcl-2 family proteins: a key in Bak? Cell Cycle. 2007;6:3043–3047. doi: 10.4161/cc.6.24.5115. [DOI] [PubMed] [Google Scholar]
- 64.Wei Q, Dong G, Chen JK, Ramesh G, Dong Z. Bax and Bak have critical roles in ischemic acute kidney injury in global and proximal tubule-specific knockout mouse models. Kidney Int. 2013;84:138–148. doi: 10.1038/ki.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Archer SL. Mitochondrial dynamics-mitochondrial fission and fusion in human diseases. N Engl J Med. 2013;369:2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 66.Brooks C, Wei Q, Cho SG, Dong Z. Regulation of mitochondrial dynamics in acute kidney injury in cell culture and rodent models. J Clin Investig. 2009;119:1275–1285. doi: 10.1172/JCI37829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Twig G, Elorza A, Molina AJ, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee Y, Lee HY, Hanna RA, Gustafsson AB. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am J Physiol Heart Circ Physiol. 2011;301:H1924–H1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gomes LC, Scorrano L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim Biophys Acta. 2008;1777:860–866. doi: 10.1016/j.bbabio.2008.05.442. [DOI] [PubMed] [Google Scholar]
- 71.Bernardi P. The mitochondrial permeability transition pore: a mystery solved? Front Physiol. 2013;4:95. doi: 10.3389/fphys.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong Z, Saikumar P, Weinberg JM, Venkatachalam MA. Calcium in cell injury and death. Annu Rev Pathol. 2006;1:405–434. doi: 10.1146/annurev.pathol.1.110304.100218. [DOI] [PubMed] [Google Scholar]
- 73.Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 74.Rodriguez-Enriquez S, Kai Y, Maldonado E, Currin RT, Lemasters JJ. Roles of mitophagy and the mitochondrial permeability transition in remodeling of cultured rat hepatocytes. Autophagy. 2009;5:1099–1106. doi: 10.4161/auto.5.8.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cui T, Fan C, Gu L, et al. Silencing of PINK1 induces mitophagy via mitochondrial permeability transition in dopaminergic MN9D cells. Brain Res. 2011;1394:1–13. doi: 10.1016/j.brainres.2011.01.035. [DOI] [PubMed] [Google Scholar]
- 76.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kubli DA, Gustafsson AB. Mitochondria and mitophagy: the yin and yang of cell death control. Circ Res. 2012;111:1208–1221. doi: 10.1161/CIRCRESAHA.112.265819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hollville E, Carroll RG, Cullen SP, Martin SJ. Bcl-2 family proteins participate in mitochondrial quality control by regulating Parkin/PINK1-dependent mitophagy. Mol Cell. 2014;55:451–466. doi: 10.1016/j.molcel.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 79.Wirawan E, Vande Walle L, Kersse K, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cho DH, Jo YK, Hwang JJ, Lee YM, Roh SA, Kim JC. Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett. 2009;274:95–100. doi: 10.1016/j.canlet.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 81.Yousefi S, Perozzo R, Schmid I, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 82.Pagliarini V, Wirawan E, Romagnoli A, et al. Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ. 2012;19:1495–1504. doi: 10.1038/cdd.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zsengeller ZK, Ellezian L, Brown D, et al. Cisplatin nephrotoxicity involves mitochondrial injury with impaired tubular mitochondrial enzyme activity. J Histochem Cytochem. 2012;60:521–529. doi: 10.1369/0022155412446227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhan M, Usman IM, Sun L, Kanwar YS. Disruption of Renal Tubular Mitochondrial Quality Control by Myo-Inositol Oxygenase in Diabetic Kidney Disease. J Am Soc Nephrol. 2014 doi: 10.1681/ASN.2014050457. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang W, Wang Y, Long J, et al. Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell Metab. 2012;15:186–200. doi: 10.1016/j.cmet.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hall AM, Rhodes GJ, Sandoval RM, Corridon PR, Molitoris BA. In vivo multiphoton imaging of mitochondrial structure and function during acute kidney injury. Kidney Int. 2013;83:72–83. doi: 10.1038/ki.2012.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rehman H, Krishnasamy Y, Haque K, et al. Green tea polyphenols stimulate mitochondrial biogenesis and improve renal function after chronic cyclosporin a treatment in rats. PLoS One. 2014;8:e65029. doi: 10.1371/journal.pone.0065029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tamaki M, Miyashita K, Wakino S, Mitsuishi M, Hayashi K, Itoh H. Chronic kidney disease reduces muscle mitochondria and exercise endurance and its exacerbation by dietary protein through inactivation of pyruvate dehydrogenase. Kidney Int. 2014;85:1330–1339. doi: 10.1038/ki.2013.473. [DOI] [PubMed] [Google Scholar]
- 89.Higgins GC, Coughlan MT. Mitochondrial dysfunction and mitophagy: the beginning and end to diabetic nephropathy? Br J Pharmacol. 2014;171:1917–1942. doi: 10.1111/bph.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sivitz WI, Yorek MA. Mitochondrial dysfunction in diabetes: from molecular mechanisms to functional significance and therapeutic opportunities. Antioxid Redox Signal. 2010;12:537–577. doi: 10.1089/ars.2009.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kruidering M, Van de Water B, de Heer E, Mulder GJ, Nagelkerke JF. Cisplatin-induced nephrotoxicity in porcine proximal tubular cells: mitochondrial dysfunction by inhibition of complexes I to IV of the respiratory chain. J Pharmacol Exp Ther. 1997;280:638–649. [PubMed] [Google Scholar]
- 92.Forbes JM, Coughlan MT, Cooper ME. Oxidative stress as a major culprit in kidney disease in diabetes. Diabetes. 2008;57:1446–1454. doi: 10.2337/db08-0057. [DOI] [PubMed] [Google Scholar]
- 93.Sanz AB, Santamaria B, Ruiz-Ortega M, Egido J, Ortiz A. Mechanisms of renal apoptosis in health and disease. J Am Soc Nephrol. 2008;19:1634–1642. doi: 10.1681/ASN.2007121336. [DOI] [PubMed] [Google Scholar]
- 94.Linkermann A, Chen G, Dong G, Kunzendorf U, Krautwald S, Dong Z. Regulated cell death in AKI. J Am Soc Nephrol. 2014;25:2689–2701. doi: 10.1681/ASN.2014030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ishihara M, Urushido M, Hamada K, et al. Sestrin-2 and BNIP3 regulate autophagy and mitophagy in renal tubular cells in acute kidney injury. Am J Physiol Renal Physiol. 2013;305:F495–F509. doi: 10.1152/ajprenal.00642.2012. [DOI] [PubMed] [Google Scholar]
- 96.Namba T, Takabatake Y, Kimura T, et al. Autophagic clearance of mitochondria in the kidney copes with metabolic acidosis. J Am Soc Nephrol. 2014;25:2254–2266. doi: 10.1681/ASN.2013090986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kawakami T, Gomez IG, Ren S, et al. Deficient autophagy results in mitochondrial dysfunction and FSGS. J Am Soc Nephrol. 2014 doi: 10.1681/ASN.2013111202. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cui J, Shi S, Sun X, et al. Mitochondrial autophagy involving renal injury and aging is modulated by caloric intake in aged rat kidneys. PLoS One. 2013;8:e69720. doi: 10.1371/journal.pone.0069720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mizumura K, Cloonan SM, Nakahira K, et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J Clin Investig. 2014;124:3987–4003. doi: 10.1172/JCI74985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]