SUMMARY
Transcription elongation regulates the expression of many genes, including oncogenes. Histone deacetylase (HDAC) inhibitors (HDACIs) block elongation, suggesting HDACs are involved in gene activation. To understand this, we analyzed nascent transcription and elongation factor binding genome-wide after perturbation of elongation with small molecule inhibitors. We found that HDACI-mediated repression requires heat shock protein 90 (HSP90) activity. HDACIs promote the association of RNA polymerase II (RNAP2) and negative elongation factor (NELF), a complex stabilized by HSP90, at the same genomic sites. Additionally, HDACIs redistribute bromodomain-containing protein 4 (BRD4), a key elongation factor involved in enhancer activity: BRD4 binds to newly acetylated sites, and its occupancy at promoters and enhancers is reduced. Furthermore, HDACI reduce enhancer activity as measured by enhancer RNA production. Thus, HDACs are required for limiting acetylation in gene bodies and intergenic regions. This facilitates the binding of elongation factors to properly acetylated promoters and enhancers for efficient elongation.
Graphical abstract
INTRODUCTION
Transcription elongation is a critical step in regulating many human genes (Adelman and Lis, 2012; Gilchrist et al., 2010). We previously reported that inhibition of histone deacetylase (HDAC) activity results in a dramatic decrease in transcription elongation efficiency at multiple genes using global run-on sequencing (GRO-seq) (Core et al., 2008) to analyze RNA polymerase II (RNAP2) activity across the genome. We found that elongation repression occurs in several cell lines derived from both non-cancerous tissue and tumors, suggesting that this is a general effect of inhibiting HDACs in human cells (Kim et al., 2013). As a pivotal determinant of transcript level for many oncogenes, elongation is being investigated for cancer therapy because it is regulated by many factors targetable by small molecule inhibitors (Delmore et al., 2011; Zhai et al., 2002; Zuber et al., 2011). HDAC inhibitors (HDACIs) are used clinically in tumor treatment, and inhibit the zinc-dependent HDAC isoforms, which are often components of complexes associated with transcriptional silencing.
Transcription of protein-coding genes by RNAP2 can be regulated at initiation and elongation steps (Adelman and Lis, 2012). Initiation of transcription is catalyzed by the assembly of the preinitiation complex at the promoter (Thomas and Chiang, 2006) followed by the incorporation of the first several nucleotides downstream from the promoter (Core et al., 2008). Transcription through the gene body by the RNAP2 is prevented by factors that block elongation, such as negative elongation factor (NELF) and DRB-sensitivity inducing factor (DSIF) (Kwak and Lis, 2013). In order for RNAP2 to transition into the productive elongation phase and synthesize full-length pre-mRNA, elongation-inducing factors are recruited. Positive transcription elongation factor b (P-TEFb), which modifies RNAP2 and other factors required for overcoming the elongation block, is recruited by BRD4, an acetyl-lysine binding protein (Jang et al., 2005; Yang et al., 2005). P-TEFb contains cyclin dependent kinase 9 (CDK9) that phosphorylates DSIF, NELF, and serine 2 of the heptad repeats in the C-terminal domain (CTD) of the largest subunit of RNAP2 (Fujinaga et al., 2004). NELF can interact with nascent RNAs and is evicted when elongation is induced (Yamaguchi et al., 1999), whereas DSIF travels along with the elongating RNAP2 upon phosphorylation by P-TEFb (Wu et al., 2003).
It was surprising that HDACIs are capable of directly repressing the transcription of many genes (Chou et al., 2011; Kim et al., 2013; Scott et al., 2002), given that classical HDACs are components of complexes known to silence transcription. The two inhibitors used here, trichostatin A (TSA) and suberanilohydroxamic acid (SAHA, known clinically as vorinostat), inhibit the 11 classical HDAC isoforms (Bolden et al., 2006). They are found in the Sin3, nucleosome remodeling deacetylase (NuRD), and nuclear receptor corepressor 2/silencing mediator for retinoid or thyroid-hormone receptors (NCOR2/SMRT) complexes (Glass and Rosenfeld, 2000). Lysine acetylation is a well-known mark of transcriptionally active open chromatin (Eberharter and Becker, 2002), and acetylation of many transcription factors activates their function and deacetylation represses their function (Sterner and Berger, 2000). However, in support of a role for HDACs in active transcription, prior research shows that HDAC complexes are involved in both repression and activation of transcription in yeast (Vidal and Gaber, 1991; Vidal et al., 1991; Wang et al., 2002), and some transcription factors are activated when deacetylated (Chen et al., 1999; Wolf et al., 2002; Xu et al., 2003). These proteins appear to facilitate similar opposing functions in higher eukaryotes as well, because HDACs are associated strongly with actively transcribed genes in human cells (Wang et al., 2009).
Enhancers are transcriptional regulatory elements that promote the transcription of a gene or genes (Moreau et al., 1981; Shlyueva et al., 2014). From a linear perspective of DNA sequences, they are often located far away from the transcription start sites (TSSs) of genes, but folding and looping of the chromosome can bring these elements within close proximity of the genes whose transcription they affect (Jin et al., 2013). Recently, active enhancers have been found to be sites of bidirectional transcription, and create unstable transcripts called enhancer RNA (eRNA). eRNAs are reliable markers of active enhancers (Danko et al., 2015), and the amount of eRNA produced relates to the activity of the enhancer. Knockdown of eRNAs reduces the transcription of the target genes of an enhancer (Banerjee et al., 2014; Hsieh et al., 2014; Melo et al., 2013). BRD4 is strongly associated with enhancers, and repression of BRD4 results in a block of elongation in target genes of enhancers (Loven et al., 2013). NELF may be involved in the link between enhancers and the promoters they induce as well because it has an RNA interacting domain that binds paused transcripts, and potentially eRNAs as well (Yamaguchi et al., 2002; Yamaguchi et al., 1999). It is postulated that the NELF-induced elongation block is overcome when the RNA interacting domain exchanges the paused transcript for an eRNA (Schaukowitch et al., 2014).
In this study, we set out to investigate the positive effect of HDACs on transcription elongation. Our data show that HDAC-regulated transcription elongation requires heat shock protein 90 (HSP90) activity. In contrast, the elongation block that results from CDK9 inhibition does not, suggesting CDK9 functions downstream of HDACs and HSP90. Treatment with HDACIs causes redistribution of other elongation factors across the genome. Particularly, colocalization of RNAP2 with the NELF complex across the genome, whose stability is regulated by the HSP90 chaperone, is strongly increased after HDACI treatment. These inhibitors induce global acetylation changes, which redistributes BRD4 binding, which is an important factor that is involved in promoting enhancer activity, and affects the regulatory organization of the genome. Because BRD4 and NELF are associated with enhancers, we looked at enhancer activity after HDACI treatment. We found that HDACIs reduce eRNA synthesis at high eRNA-producing enhancer sites. This is associated with corresponding changes in the expression of neighboring genes. Overall, we show that HDACs are important regulators of elongation and play an essential role in active gene transcription for many genes.
RESULTS
HDAC inhibition blocks elongation of RNAP2
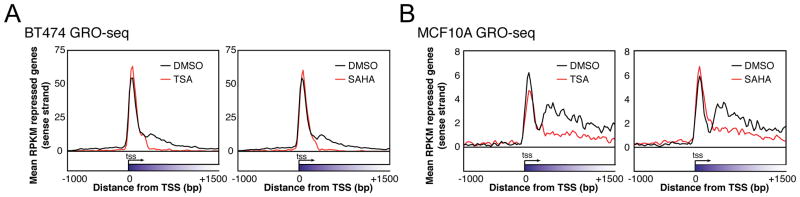
HDACIs repress transcription by blocking elongation, as we have shown previously in human breast cancer (BT474) and non-cancerous breast epithelial (MCF10A) cell lines using GRO-seq (Kim et al., 2013). Analysis of expression within different gene regions by RPKM normalization (reads per kilobase of annotated region per million mapped sequence reads) indicates that repressed genes have an impediment in the transcription of gene bodies, but the transcription near the start of genes is not significantly changed, or is increased after a 4 hr. treatment with either of two pan-specific HDACIs, TSA and SAHA, in BT474 (Figure 1A and S1A) and MCF10A (Figure 1B and S1A). GRO-seq gene body RPKM was used to classify genes into three groups based on expression changes in response to HDACIs: genes whose expression goes down after SAHA treatment were defined as repressed, genes whose expression is unchanged were identified as not changed, and genes whose expression is increased were defined as activated. There is a high percentage of overlap between genes in the three expression change groupings seen in this data compared to previously generated GRO-seq data (Kim et al., 2013) in these cell lines (Table S1). ChIP-seq of RNAP2 was conducted to validate the three groupings, and we found that SAHA does not significantly change the density of RNAP2 in the promoters of repressed genes (Figure S1B, Table S2). In the gene body, RNAP2 binding decreases in GRO-seq repressed genes, stays the same in not changed genes, and significantly increases in activated genes (Figure S1B, Table S3).
Figure 1. HDACIs block elongation of RNAP2 transcription in the genes they repress.
(A) Metagene plots of GRO-seq RPKM in the sense direction for repressed genes in BT474. Repressed genes had significantly (log-likelihood ratio P < 10−20, one GRO-seq experiment) reduced RPKM in the gene body (300 bp downstream of TSSs to gene ends). 6354 repressed genes were analyzed for TSA, and 7389 for SAHA.
(B) Same as (A) for MCF10A cells. There were 3866 repressed genes for TSA, and 4643 for SAHA.
See Figure S1 for GRO-seq statistics, RNAP2 ChIP-seq, GRO-seq of a shorter treatment of SAHA in BT474, SAHA blocking elongation in another cell line, and validation that SAHA and TSA repress transcription through their effects on HDACs.
See Table S1 for the number of genes common to each expression change subgroupings between the GRO-seq shown here and previously published GRO-seq data.
See Table S2 for median fold changes in subclassifications of gene expression changes for different factors by ChIP-seq in promoter regions.
See Table S3 for median fold changes in subclassifications of gene expression changes for different factors by ChIP-seq in gene body regions.
In order to examine the kinetics by which HDACIs suppress transcription elongation, the effect of SAHA treatment for a shorter time was examined in BT474 with GRO-seq. SAHA represses genes via an elongation block even after a short 30 min. treatment, and resulted in a similar, though less intense, global pattern of elongation inhibition as the 4 hr. treatment (Figure S1C and S1D). In order to test whether the repressive effect of SAHA on transcription elongation is applicable to cells of a different origin, we applied SAHA to a neuroblastoma cell line, SK-N-SH and examined the transcription elongation pattern. SAHA blocked elongation in SK-N-SH (Figure S1E), underscoring the general applicability of HDACIs as a transcription elongation blocker. Overall, our data shows that HDAC inhibition blocks the transition of RNAP2 into productive elongation in HDACI-repressed genes in a short time window and in a broad cellular context.
We examined whether overexpression of the HDAC1 isoform can rescue the effect of SAHA to validate that HDACIs are inhibiting the intended target, and that HDACIs are causing repression through a block in the catalysis of deacetylation. We overexpressed recombinant HDAC1 wild-type (WT) and catalytically dead mutant (mut; Figure S1F), and examined the expression of two of well-characterized HDACI-repressed oncogenes regulated by elongation in BT474, ERBB2 and MYC (Kim et al., 2013). Overexpression of the WT HDAC1 isoform, but not mut, in BT474 antagonizes SAHA-mediated repression of these two oncogenes (Figure S1G). TSA is also antagonized by HDAC1 overexpression, and the effect of the overexpression could be overcome by increasing the dose of the drug (S1H). This demonstrates that HDACIs are repressing transcription by blocking the deacetylation catalyzed by HDAC1, a class I deacetylase, and possibly other isoforms. Furthermore, ChIP-seq analysis of HDAC1 shows that prior to drug treatment, this enzyme is more enriched in genes that can be repressed by SAHA treatment compared to genes that do not change their expression after SAHA (Figure S1I, comparison of DMSO samples). Together, this data suggests that genes that are repressed by HDACI treatment are regulated by deacetylation.
HDACI repression of transcription requires HSP90
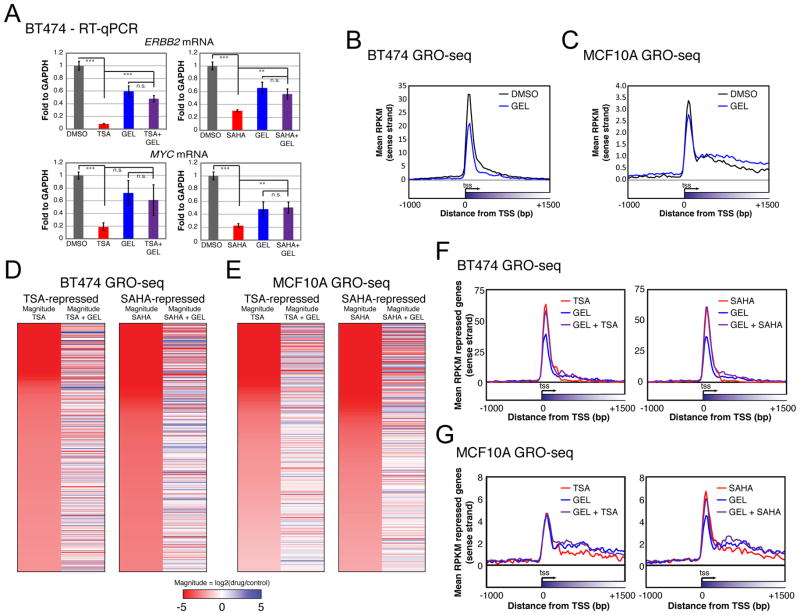
The HSP90 chaperone complex promotes RNAP2 pausing in Drosophila (Sawarkar et al., 2012). Therefore, we tested if the mechanism of HDACI suppression of transcription elongation is dependent on HSP90. A potent HSP90 inhibitor, geldanamycin (GEL), was applied to reduce pausing and was used in combination with HDACIs in BT474 cells to see how these small molecules interact to affect elongation. RT-qPCR shows that ERBB2 and MYC repression by HDACIs is antagonized by GEL treatment (Figure 2A), whereas RPS10 and ACTG1, which are not repressed by HDACIs, do not show a significant increase in expression after combination treatment (Figure S2A). To look at the global elongation changes brought on by the drug combinations, we conducted GRO-seq with single and combined treatment with the inhibitors. As expected, treating with GEL alone reduces the amount of promoter proximal transcripts in both BT474 and MCF10A cell lines (Figure 2B, 2C, and S2B), validating this treatment as a repressor of pausing. GEL antagonizes the repression of the majority of the top 1000 HDACI-repressed genes (Figure 2D and 2E) by more than 5-fold (Figure S2C). Combination treatment with GEL antagonizes the gene body repression induced by HDACIs (Figure 2F, 2G, and S3D). These results show that HDACI-mediated repression of elongation is dependent on HSP90 activity.
Figure 2. Inhibition of HSP90 antagonizes HDACI repression.
(A) mRNA level of HDACI-repressed genes in combination with GEL. Quantitation of ERBB2 and MYC mRNA relative to GAPDH in BT474 after single and combined treatments is shown. Statistical significance was determined with a two-tailed t-test where *** = p < 0.001, ** = p < 0.01, and n.s. = not significant (p > 0.05). Bars represent standard error. n = 11 to 14 from at least 4 biological replicates. GEL was administered at 20 μM.
(B) GRO-seq metagenes of GEL treatment for all genes in BT474. One GRO-seq experiment conducted. 37467 genes are represented in the metaplot.
(C) Same as (B) for MCF10A.
(D) Heatmaps of the top 1000 TSA and SAHA repressed genes in BT474 as determined by GRO-seq and those same genes when GEL is also added. Magnitude of expression for HDACIs was calculated relative to DMSO, and HDACI+GEL was relative to GEL. Colors represent magnitude expression change in log scale, with red representing repression and blue activation.
(E) Same as (D) for MCF10A.
(F) Metagenes of HDACI-repressed genes in BT474 when GEL is also added. Same genes as Figure 1A.
(G) Same as (F) for MCF10A. Same genes as Figure 1B.
See Figure S2 for RT-qPCR of genes not repressed by HDACIs after combination treatment, GRO-seq statistics, and percentages of genes antagonized by GEL.
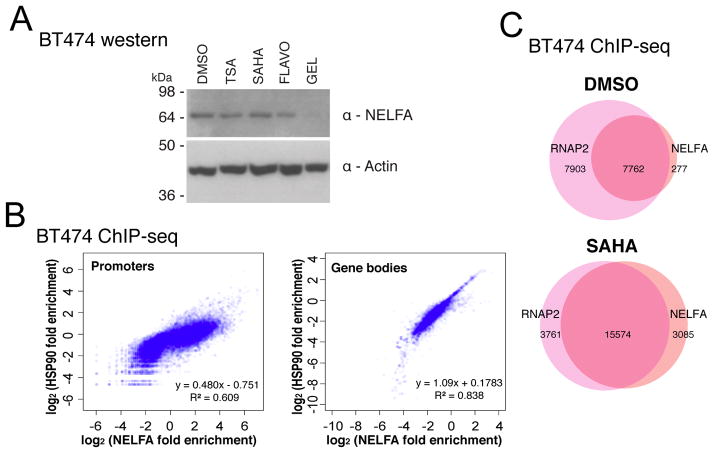
NELFE, a subunit of the NELF complex, is destabilized upon HSP90 inhibition in cells from several organisms (Sawarkar et al., 2012). Destabilization of just one subunit of the four-subunit NELF complex leads to the degradation of the entire complex (Narita et al., 2007; Sun and Li, 2010; Sun et al., 2008). We tested whether NELF is a downstream effector of HSP90 that could be mediating transcriptional elongation repression by HDACIs. In BT474, GEL destabilizes the NELFA subunit (Figure 3A), which likely leads to the destabilization of the entire NELF complex. In order to examine the change in binding of NELF upon HDACI treatment, we performed ChIP-seq of NELFA and HSP90 in DMSO- and SAHA-treated BT474. NELFA and HSP90 density are correlated in promoters and gene bodies, with a stronger correlation in gene bodies (Figure 3B). More than half of RNAP2 binding peaks are not colocalized with NELFA in the DMSO treatment control. In contrast, SAHA treatment dramatically increases the number of NELFA peaks, and these peaks predominantly overlap with RNAP2 (Figure 3C). These results show that global distribution of NELF binding is affected by SAHA, indicating that HDACI treatment may result in transcription pausing through NELF.
Figure 3. HSP90 stabilizes NELFA, and SAHA increases NELFA binding.
(A) NELFA stability after treatment with different elongation-affecting drugs, shown with western of BT474 cell lysates, representative image for one of two blots.
(B) Correlation of NELFA and HSP90 fold enrichments in promoter and gene body regions for 33119 annotated RefSeq genes. Two biological replicates for each ChIP-seq were conducted and average signal is plotted.
(C) Venn diagrams of RNAP2 and NELFA peak overlaps. Overlaps were within a 500 base pair window. Numbers of genes in each section are shown.
P-TEFb inhibition affects transcription elongation, but not through HSP90
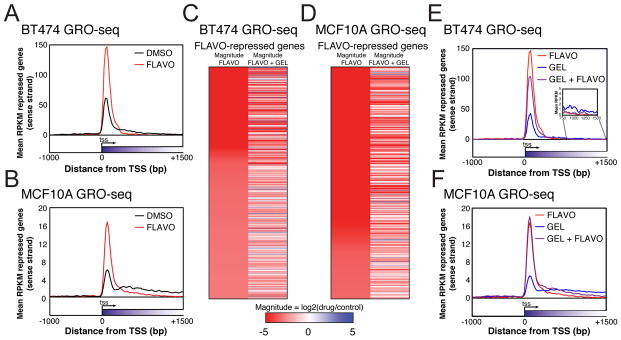
P-TEFb is an important regulator of elongation, so we sought to determine how P-TEFb might act in connection with HDACs and HSP90. In addition to the HSP90 inhibitor GEL, we used elongation inhibitor flavopiridol (FLAVO) that inhibits the CDK9 subunit of the P-TEFb complex. We aimed to compare its effects on elongation with HDACI and GEL combination treatment. Like HDACIs, FLAVO represses ERBB2 and MYC transcript levels in BT474 (Figure S3A). Globally, FLAVO-repressed genes show a decrease in gene body transcription in GRO-seq experiments. However, unlike HDACI-repressed genes, FLAVO-repressed genes displayed a dramatic increase in promoter proximal transcription (Figure 4A, 4B, and Figure S3B). Furthermore, transcription in the gene bodies of FLAVO-repressed genes in BT474 and MCF10A is still repressed in the presence of GEL (Figure 4C, 4D, and S3B), whereas the transcription of HDACI-repressed genes was recovered by GEL treatment (Figure 2D, 2E, and S2C). Elongation patterns observed in GRO-seq were examined after combined treatments with these drugs. The FLAVO-induced elongation repression pattern persists in the presence of GEL (Figure 4E, 4F, and S3B), suggesting its mechanism of repression is HSP90-independent. This shows that HDACI-mediated transcription suppression is through a different mechanism than FLAVO. Perhaps FLAVO suppresses elongation via more immediate effects on the phosphorylation of RNAP2 CTD, than HDACIs, which suppress genes under tight control of HSP90.
Figure 4. Elongation block caused by P-TEFb inhibition is not dependent on HSP90.
(A) FLAVO-repressed (log-likelihood ratio P < 10−20 for gene body RPKM changes, 9706 genes) metagenes from BT474. Data is from one GRO-seq experiment.
(B) Same as (A) for MCF10A, 12659 genes.
(C) Heatmaps of the expression changes in the top 1000 FLAVO repressed genes with the greatest magnitude of change in BT474 as determined by GRO-seq (log-likelihood ratio P < 10−20) and those same genes when GEL is also added. Colors indicate magnitude expression change in log scale. Red represents repression, blue activation.
(D) Same as (C) for MCF10A.
(E) Metagenes of all FLAVO-repressed genes when GEL is also added to BT474.
(F) Same as (E) for MCF10A.
See also Figure S3 for FLAVO repression of ERBB2 and MYC, and GRO-seq statistics.
Acetylation and BRD4 binding changes in gene bodies and intergenically after HDACI
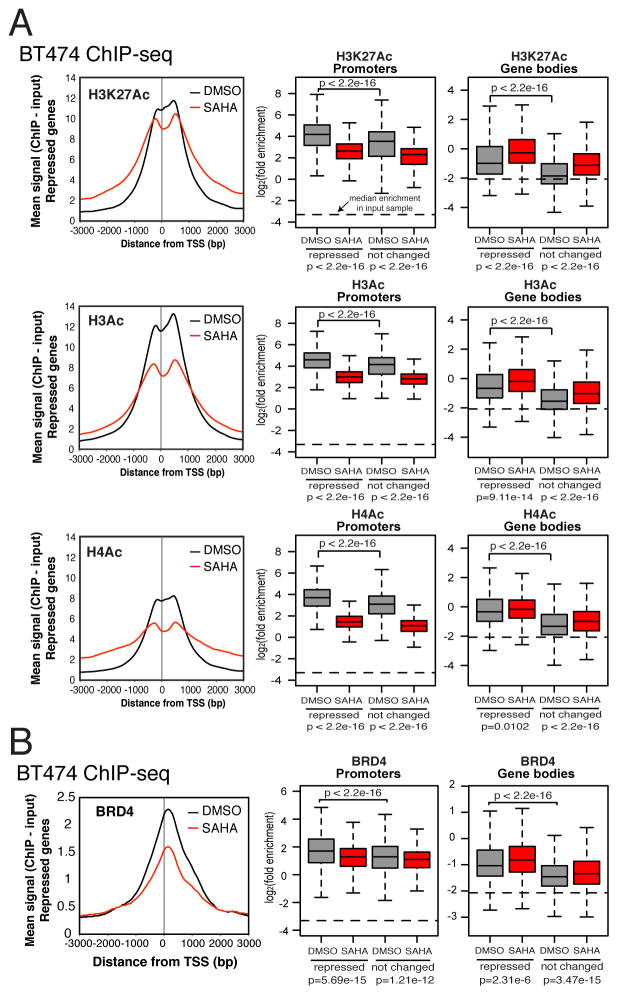
HDACI treatment globally increases acetylation, and we predicted that this results in changes in the distribution of the binding of key acetylated histone readers such as BRD4. This protein acts as a scaffold to recruit elongation factors, so its redistribution would lead to changes in the binding of other proteins. ChIP-seq shows that the acetylation of lysines in histone H3K27 (H3K27Ac), H3 (H3Ac), and H4 (H4Ac) is reduced in promoter proximal regions, while in gene bodies, acetylation increases after SAHA treatment (Figure 5A). Consistent with the changes in acetylation, SAHA treatment decreases the binding of BRD4 near TSSs, and increases its binding in gene bodies (Figure 5B, Tables S2 and S3). BRD4 is enriched in repressed genes more than not changed genes in BT474 without inhibitor treatment, suggesting their transcription is regulated by BRD4 under normal conditions, unlike the not changed genes.
Figure 5. HDACIs affect acetylation of histones and BRD4 binding.
(A) ChIP-seq profiles of acetylated histone tails around TSSs of repressed genes, and their enrichment in promoters and gene bodies. Average acetyl-ChIP-seq profiles from two biological replicates are averaged and normalized to average yield of both replicates. H3Ac antibody recognizes acetylated K9 and K14 in the H3 subunit. H4Ac antibody recognizes acetylated K5, K8, K12, and K16 in the H4 subunit.
(B) BRD4 binding at TSSs and in gene bodies of repressed genes. Quantitation of their binding in repressed and not changed genes is shown in box plots. Statistics reported are from Wilcoxon rank-sum tests. Horizontal lines on boxplot graphs represent median signal in all genes from chromatin input sample. Average of two replicates.
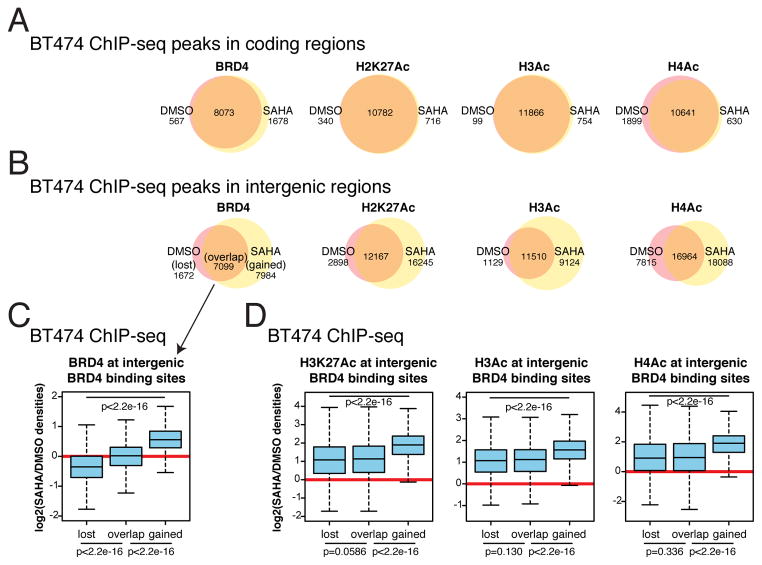
Interestingly, we noticed that in gene coding regions, while the magnitude of BRD4 binding and acetylation may differ, the locations of detected peaks did not show many changes upon SAHA treatment (Figure 6A). In intergenic regions, however, there are much more striking changes in the location of peaks, and there is an increase in the number of peaks by SAHA treatment (Figure 6B), suggesting that HDACIs may affect the global chromatin landscape far beyond regions encoding genes.
Figure 6. Intergenic changes in BRD4 binding and corollary changes in histone acetylation.
(A) Peaks and their overlaps before and after SAHA treatment in coding regions for acetylated histones and BRD4 in BT474.
(B) Peaks and their overlaps before and after SAHA treatment in intergenic regions for acetylated histones and BRD4 in BT474. Number of peaks in each category is listed. Only peaks present in both replicates were considered.
(C) Within intergenic BRD4 binding sites, those that are lost upon SAHA, stay bound, and are gained after SAHA are analyzed for intensity of BRD4 binding changes.
(D) Acetylation changes at intergenic BRD4 binding sites.
See also Figure S4 for the overlap of genes affected by TSA, SAHA, JQ1 (BRD4 inhibitor), and FLAVO treatments, and JQ1 effects on ERBB2 and MYC expression and elongation.
To investigate if changes in BRD4 binding are correlated with changes in acetylation we identified whether peaks were maintained after SAHA treatment. We identified sites where BRD4 binding peaks were lost (only present in DMSO), overlap (present in both DMSO and SAHA), and gained (only present after SAHA treatment). We determined the magnitude of change in the acetylation and binding at these sites. As expected, BRD4 binding is reduced at sites where BRD4 is lost, stays the same at overlapping sites, and increases at gained sites (Figure 6C). At these same sites, the level of acetylation at these sites maintains similar levels between at lost and overlap sites, but displays far greater increases at gained sites (Figure 6D). Therefore, BRD4 is being recruited to newly created sites of acetylation in intergenic regions. Loss of BRD4 binding at certain sites after SAHA treatment may be due to newly created acetylated sites competing for the binding of this protein.
JQ1, a BRD4 inhibitor, and HDACIs share similar gene expression change profiles (Bhadury et al., 2014). To see which genes were mutually affected by BRD4, HDAC, or P-TEFb inhibition, we defined the top 1000 most-repressed genes for each drug, and determined the percentage of genes the inhibitors were able to repress in pairwise combinations. The percentage of genes shared between JQ1 and the HDACIs is similar to the percentage shared between the two related HDACIs, TSA and SAHA. FLAVO does not have as large of an overlap with HDACIs as does JQ1, indicating more similarity between the effects of BRD4 and HDAC inhibition than P-TEFb inhibition (Figure S4A). Concordantly, JQ1 represses ERBB2 and MYC in BT474 (Figure S4B). This drug also represses elongation in a pattern similar to HDACIs in our two cell lines (Figure S4C and S4D). This indicates that JQ1 and HDACIs may have a similar mechanism of action.
eRNA transcription is repressed by HDACIs
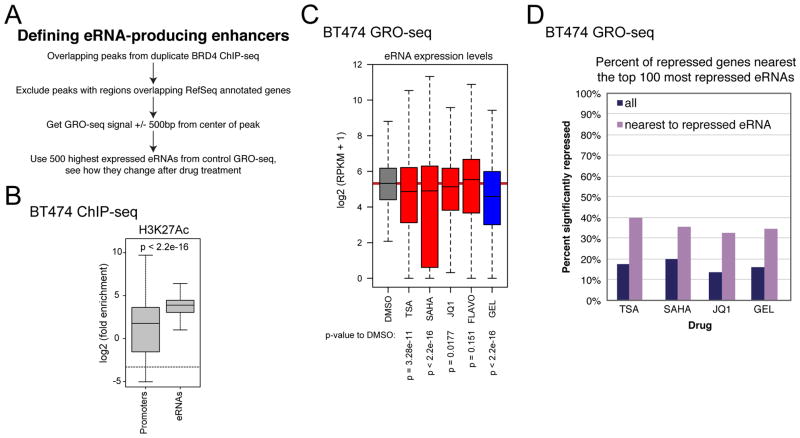
BRD4 is bound at active enhancers (Chapuy et al., 2013), and it was found recently that JQ1 reduces eRNA synthesis (Kanno et al., 2014). Since restructuring of acetylation and BRD4 binding occurs in intergenic regions, we wondered if HDACI treatment could be affecting positive regulators of transcription located in intergenic regions, namely enhancers, via loss of BRD4 at pertinent enhancer sites. We used a prediction method similar to the one defined by the Ozato group (Kanno et al., 2014) to find eRNA-generating sites by determining intergenic BRD4 peak locations in BT474 from ChIP-seq and characterized the level of eRNA synthesis around them (Figure 7A). As validation, we showed that the enrichment of H3K27Ac, a mark of active enhancers, is higher at predicted enhancer sites than at annotated promoters (Figure 7B). The expression of the most highly expressed eRNAs from a 4 hr. DMSO treatment were analyzed in comparison to their expression after different inhibitor treatments (Figure 7C). JQ1, as expected from previous reports, reduced eRNAs. HDACIs did as well, and to an even greater extent. Even after only 30 minutes of SAHA treatment, the inhibitor was able to reduce eRNAs (Figure S5A), showing that this treatment causes fast action at enhancers. Both BRD4 and HDACs are therefore positive regulators of enhancer activity, and possibly through effects of HDACIs on BRD4 binding (Figure 6, Figure S5B). In contrast, FLAVO did not decrease the median eRNA synthesis level at enhancers, suggesting that eRNA transcription is P-TEFb independent. Surprisingly, GEL treatment is a strong repressor of eRNA synthesis, indicating that HSP90 is necessary for enhancer function, and possibly linking NELF to enhancer function because of the effect HSP90 has on NELF stability. Like in the BT474 cell line, HDACI treatment reduces eRNAs in MCF10A, as do JQ1 and GEL. FLAVO again does not repress eRNAs (Figure S5C), demonstrating that the effect of HDACI on enhancers is not cancer specific. We tested if these enhancers are regulating genes nearby since, while enhancers can work from great distance, they often regulate nearby genes. We looked at the percent of genes near eRNA sites that are significantly reduced by inhibitor treatment. We found that compared the percentage of all genes that are repressed by an inhibitor, there is a higher percentage of genes repressed that are located near downregulated eRNA sites (Figure 7D, Figure S5D). This suggests that eRNA activity level affects target gene expression, and shows that enhancer activity is dependent on HDACs.
Figure 7. eRNA expression is reduced by HDACIs.
(A) BRD4 intergenic binding sites were defined, and GRO-seq reads were aligned to 1000 bp window around center of peak. Top 500 most expressed eRNAs were identified in a DMSO GRO-seq experiment and were analyzed in DMSO and the different drug conditions.
(B) Enrichment of H3K27Ac at predicted enhancer sites compared to annotated promoters.
(C) Boxplot of eRNA expression after different drug treatments. Statistics are from Wilcoxon rank-sum tests.
(D) Percent of genes that are repressed either overall or only in the genes closest to the top 100 most repressed eRNAs for each drug.
See also Figure S5 for eRNA expression changes after a 30 min. SAHA treatment, the binding changes of BRD4 at predicted enhancer sites, and the changes in eRNA production brought on by inhibitor treatments in MCF10A.
DISCUSSION
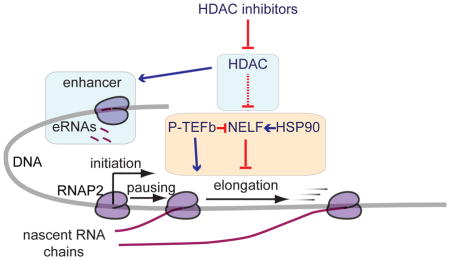
The repression of transcription by HDACIs in many genes is counterintuitive because of the well-known role HDACs play in turning off transcription. We have shown HDACIs cause a block in the elongation step of transcription by RNAP2. Some studies show evidence to support this finding. Firstly, HDACs bind to highly expressed genes more so than lowly expressed genes and heterochromatin, suggesting they play a role in active gene transcription (Wang et al., 2009). This is in line with our previous finding that HDACIs target the most highly expressed genes for repression (Kim et al., 2013). Secondly, single cell imaging experiments show that shortly after induction of transcription initiation, acetylation of histones is decreased around the time that the elongating form of RNAP2 is detected (Stasevich et al., 2014). The deacetylation caused by classical HDACs post-initiation is likely an important step in inducing gene body transcription, and may suggest that cycling of acetylation and deacetylation is important in the process of transcription elongation (Wang et al., 2009).
Based on our analysis, HDACs are required for the removal of acetylation marks in gene bodies and intergenic regions, where their levels are lower than at promoters and enhancers. We know that in yeast, the RPD3 deacetylase, related to class I HDACs in humans, acts to specifically deacetylate gene bodies. It is recruited by the H3K36me mark, which is deposited cotranscriptionally along with the elongating RNAP2 (Carrozza et al., 2005; Joshi and Struhl, 2005). When the cell is unable to deacetylate these sites due to the presence of HDACIs, the acetylation enriched near promoters and enhancers may no longer serve to demarcate these regulatory regions from the rest of the genome. BRD4, which is lost at promoters and intergenically redistributed after HDACI treatment, is important in the recruitment of elongation factors to appropriate locations to activate transcription. Indeed, HDACs are required for BRD4-inducible transcription in human cell lines (Hu et al., 2014). The reduction in BRD4 likely leads to the reduction of the factors it recruits, such as P-TEFb, at promoters after HDACI treatment. Other elongation factors may also redistribute in response to BRD4 binding to inappropriate acetylation marks when HDACs are not able to maintain a lower level of acetylation at specific sites. More work is required to determine if BRD4 binding partners are responsible for the block in elongation after applying HDACIs, or if BRD4 redistribution itself exerts this effect on elongation.
HDACIs appear to work upstream of P-TEFb and require HSP90 activity. HDAC inhibition affects NELF binding, and HDACIs cannot repress transcription in the presence of GEL. This may be through the stabilization of NELF complex as well as effects on other client proteins of HSP90 involved in elongation (Schaaf et al., 2013; Zhou et al., 2015). While there are multiple known acetylation sites on HSP90, all seem to reduce the interaction of HSP90 with client proteins (Kovacs et al., 2005; Scroggins et al., 2007; Yang et al., 2008), oppositely from what would be expected based on the results reported here. Also, HDACIs have also been shown to change the levels of reactive oxygen species in the cell, which induces some HSP90 degradation (Park et al., 2015), but this again contradicts the increase in NELF binding after HDACI treatment. So, it may be that HDACIs regulate elongation by directly affecting NELF or other elongation factors, and that these factors require HSP90 to stabilize them. Whether there are intermediate steps between HDAC function and NELF activity is unknown.
Here, we report that eRNA production is reduced when HDACs are inhibited. They are also reduced when HSP90, which is responsible for NELF stability, is repressed. This may further explain how HDACIs repress transcription, as enhancer function and elongation are possibly linked via NELF function (Schaukowitch et al., 2014). Further analysis of which genes these enhancers regulate is required to fully understand the effects HDACs and NELF have on enhancer activity.
Going forward, it will be important to identify relevant target or targets of acetylation that are necessary for transcriptional activation by HDACs is histones or other proteins since deacetylation of non-histone substrates may also be involved in promoting transcription elongation. A large amount of lysine acetylation events on non-histone substrates have been identified globally using mass spectrometry, and BRD4, several HDAC isoforms, SPT5 (a DSIF component), and NELFB all have acetylated lysines (Choudhary et al., 2009) that could have effects on elongation. Functional analysis of these sites will help determine how they affect this pathway. Also, biochemical analysis of elongation factors after HSP90 inhibition would be beneficial to elucidating how this factor affects their stability and function.
HDACIs are an effective treatment for several types of cancer (Federico and Bagella, 2011). These drugs globally increase acetylation, which is often associated with an increase in the transcription level of many genes, and they increase the expression of many important cell cycle arrest and apoptotic genes (Xu et al., 2007). In contrast, we have also found that many oncogenes are selectively targeted for repression by HDACIs through effects on elongation because of their high level of transcription (Kim et al., 2013). The elongation pathway may be a very useful therapeutic target for cancer since other elongation inhibiting drugs like FLAVO and JQ1 have also shown promise as cancer treatments (Filippakopoulos et al., 2010; Patel et al., 1998; Zuber et al., 2011). Understanding the mechanism by which HDACIs strengthen elongation blocks may facilitate the development of treatments able to more specifically target genes for therapeutic repression.
EXPERIMENTAL PROCEDURES
Cell culture
MCF10A, BT474, and SK-N-SH were obtained from American Type Culture Collection (ATCC) and cultured according to their suggested conditions.
Drug treatments
TSA and SAHA were obtained from Sigma (SAHA lot # 042M4740V), (S)-JQ1 was a gift from J. Bradner at DFCI, FLAVO was from Sigma (lot 100M4723V), and GEL was from Enzo Life Sciences (BML-EI280).
For GRO-seq, doses were 500 nM, 5 μM, 500 nM, 500 nM, and 10 μM, for TSA, SAHA, JQ1, FLAVO, and GEL, respectively. 4 hr. treatments were done for elongation inhibitors TSA, SAHA, JQ1, and FLAVO, and GEL was applied for 4 hr. and 15 min. (15 min. pre-treatment of GEL for combined GEL plus elongation inhibitor treatments). The 30 min. SAHA treatment for GRO-seq was the exception. 4 hr. treatments were used for DMSO, SAHA, and FLAVO ChIP-seq experiments, and 24 hr. treatments for RT-qPCR and western blot samples, unless otherwise stated.
Expression analysis
RT-qPCR was performed as previously described (Kim et al., 2013), except with the ImProm-II enzyme from Promega with 4.6875 mM MgCl2 in the reverse transcriptase reaction. MYC primer sequences are MYC-F 5’-CTCTGACCTTTTGCCAGGAG-3’ and MYC-R 5’-TCCTCGGATTCTCTGCTCTC-3’.
Western blot
Westerns were done with the same NELFA antibody as used in ChIP-seq or polyclonal rabbit V5 antibody from Abcam (ab9116). BT474 were treated for 24 hr. before lysis.
GRO-seq
GRO-seq was performed as previously described (Kim et al., 2013), except detergent concentration was optimized, and libraries were multiplexed to conduct high-throughput sequencing in one lane. Additional details are in the supplement.
ChIP
Chromatin was prepared and immunoprecipitated as previously described (Kim et al., 2011), except that protein A/G dynabeads (Invitrogen) were used instead of organism-specific secondary antibody bound beads. ¼ of the amount of chromatin was used for RNAP2 and acetyl ChIPs to reduce oversaturation of bead binding. Details about antibodies used are in the supplementary methods.
ChIP-seq library preparation
ThruPLEX® DNA-seq Kit from Rubicon Genomics was used for multiplexed ChIP-seq library prep of BT474 chromatin. Indexed samples were quantitated with qPCR and mixed in equimolar amounts. Input sample was prepared with an Illumina DNA-seq kit.
Sequencing and sequencing data analysis
The Yale Stem Cell Center Genomics and Bioinformatics Core Facility conducted the sequencing on an Illumina HiSeq 2000 platform. Sequencing data alignment and normalization is described in supplementary methods.
Metagenes of GRO-seq data were generated with scripts from H. Kwak (Kwak et al., 2013), and shown as 25 base pair windows considering transcript directionality. ChIP-seq metagenes were generated with our own perl scripts, which count ChIP-seq and input reads and normalize read counts by total number of mapped reads in 50 base pair sliding windows. Directionality of the gene was not considered. Boxplots and Venn diagrams were created using R version 3.1.2.
Peaks were called with MACS 1.3.7.1 (Zhang et al., 2008) with the mfold parameter set to 10. BEDTools v2.23.0 (Glass and Rosenfeld, 2000) was used to generate overlaps between duplicate samples, identify peaks and signals coming from genic and intergenic regions, and the multicov function was used to determine the amount of signal coming from a given genomic region.
In BT474, eRNA annotation was done by taking peaks called in MACS for BRD4, removing regions within 1 kilobase of Refseq annotated genes; BEDTools was used to find overlapping peaks between replicates. Partek was used to find the amount of reads coming from 500 base pairs (bp) upstream and downstream of the center of the BRD4 peaks. The highest expressed putative eRNAs in a DMSO treatment from an independently generated GRO-seq experiment, was used to sort the highest expressed eRNAs, then the DMSO from the same experiment as the other treated samples acted as the control. Genes nearest to predicted eRNA sites were determined using BEDTools closest function.
Accession numbers
All the sequencing data have been submitted to the ArrayExpress archive. The accession numbers for GRO-seq and ChIP-seq data are E-MTAB-3626 and E-MTAB-3631, respectively.
Statistical tests and categorizations
Two-tailed student’s t-tests and R2 were performed in Excel 14.4.9. Wilcoxon rank-sum tests were performed in R version 3.1.2, and the p-values reported are not corrected for multiple testing. Statistical analysis for defining repression or activation in GRO-seq was performed using the log-likelihood ratio in the Partek Genomic Suite version 6.14.0220.
GRO-seq expression level change groupings for ChIP-seq were determined by selecting significantly repressed or activated genes (log-likelihood ratio; P < 10−20) based on gene body RPKM in DMSO and SAHA treatments. Not changed genes were expressed in DMSO and SAHA conditions, not significantly changed (P > 10−20), and had less than 2-fold change in their expression, up or down. The genes common to these categorized lists from the GRO-seq prepared for this manuscript, and the GRO-seq prepared previously (Kim et al., 2013) were used to analyze ChIP-seq data (Table S1).
Additional methods and associated references are available in the supplement.
Supplementary Material
Acknowledgments
We are grateful to James Bradner (Harvard Medical School, Boston MA) for providing us with JQ1. We thank Y. Zhang and G. Greer for technical help with cloning. C.B.G. has received support from NIH Training Grant GM007324, and PhRMA foundation predoctoral fellowship. MQZ and PX acknowledge the support from NIH R01 MH102616, Cecil H. and Ida Green Endowment Chair and UTD Founder’s Fellowship. I.H.P. is partly supported by NIGMS (GM0099130-01, GM111667-01), KRIBB/KRCF research initiative program (NAP-09-1) and CT Stem Cell Grant (13-SCB-YALE-06). Grants to T.H.K. from the Rita Allen Foundation, Sidney Kimmel Foundation for Cancer Research, Yale Comprehensive Cancer Center (CA-16359), Alexander and Margaret Stewart Trust, National Institute of Allergy and Infection Disease (R21AI107067), and National Cancer Institute (R01CA140485) supported this work.
Footnotes
AUTHOR CONTRIBUTIONS
C.B.G. designed, performed, and analyzed experiments, made the figures, wrote scripts, interpreted data, and wrote the manuscript. T.H.K. conceived the project, provided overall direction and secured funding for the project. Y.T. wrote scripts for the analysis of ChIP-seq data. Y.J.K. worked with Y.T. to develop the data analysis, and oversaw sequencing library preparation. P.X. and M.Q.Z. contributed to annotation of putative eRNA sites in MCF10A. I.H.P. and T.H.K. discussed the results and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AR, Kim YJ, Kim TH. A novel virus-inducible enhancer of the interferon-beta gene with tightly linked promoter and enhancer activities. Nucleic Acids Res. 2014;42:12537–12554. doi: 10.1093/nar/gku1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadury J, Nilsson LM, Muralidharan SV, Green LC, Li Z, Gesner EM, Hansen HC, Keller UB, McLure KG, Nilsson JA. BET and HDAC inhibitors induce similar genes and biological effects and synergize to kill in Myc-induced murine lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2721–2730. doi: 10.1073/pnas.1406722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nature reviews Drug discovery. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Chou CW, Wu MS, Huang WC, Chen CC. HDAC inhibition decreases the expression of EGFR in colorectal cancer cells. PloS one. 2011;6:e18087. doi: 10.1371/journal.pone.0018087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko CG, Hyland SL, Core LJ, Martins AL, Waters CT, Lee HW, Cheung VG, Kraus WL, Lis JT, Siepel A. Identification of active transcriptional regulatory elements from GRO-seq data. Nature methods. 2015;12:433–438. doi: 10.1038/nmeth.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO reports. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico M, Bagella L. Histone deacetylase inhibitors in the treatment of hematological malignancies and solid tumors. J Biomed Biotechnol. 2011;2011:475641. doi: 10.1155/2011/475641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes & development. 2000;14:121–141. [PubMed] [Google Scholar]
- Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee GS, Chen S, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Lu X, Liu R, Ai N, Cao Z, Li Y, Liu J, Yu B, Liu K, Wang H, et al. Histone cross-talk connects protein phosphatase 1alpha (PP1alpha) and histone deacetylase (HDAC) pathways to regulate the functional transition of bromodomain-containing 4 (BRD4) for inducible gene expression. The Journal of biological chemistry. 2014;289:23154–23167. doi: 10.1074/jbc.M114.570812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Molecular cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Molecular cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR, Vahedi G, Heightman TD, Garcia BA, Reinberg D, et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nature structural & molecular biology. 2014;21:1047–1057. doi: 10.1038/nsmb.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Greer CB, Cecchini KR, Harris LN, Tuck DP, Kim TH. HDAC inhibitors induce transcriptional repression of high copy number genes in breast cancer through elongation blockade. Oncogene. 2013;32:2828–2835. doi: 10.1038/onc.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Molecular cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Molecular cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Moreau P, Hen R, Wasylyk B, Everett R, Gaub MP, Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF interacts with CBC and participates in 3' end processing of replication-dependent histone mRNAs. Molecular cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Park S, Park JA, Kim YE, Song S, Kwon HJ, Lee Y. Suberoylanilide hydroxamic acid induces ROS-mediated cleavage of HSP90 in leukemia cells. Cell stress & chaperones. 2015;20:149–157. doi: 10.1007/s12192-014-0533-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V, Senderowicz AM, Pinto D, Jr, Igishi T, Raffeld M, Quintanilla-Martinez L, Ensley JF, Sausville EA, Gutkind JS. Flavopiridol, a novel cyclin-dependent kinase inhibitor, suppresses the growth of head and neck squamous cell carcinomas by inducing apoptosis. The Journal of clinical investigation. 1998;102:1674–1681. doi: 10.1172/JCI3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawarkar R, Sievers C, Paro R. Hsp90 globally targets paused RNA polymerase to regulate gene expression in response to environmental stimuli. Cell. 2012;149:807–818. doi: 10.1016/j.cell.2012.02.061. [DOI] [PubMed] [Google Scholar]
- Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, Zhou Y, Lis JT, Dorsett D. Genome-wide control of RNA polymerase II activity by cohesin. PLoS genetics. 2013;9:e1003382. doi: 10.1371/journal.pgen.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Molecular cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott GK, Marden C, Xu F, Kirk L, Benz CC. Transcriptional repression of ErbB2 by histone deacetylase inhibitors detected by a genomically integrated ErbB2 promoter-reporting cell screen. Molecular cancer therapeutics. 2002;1:385–392. [PubMed] [Google Scholar]
- Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Molecular cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nature reviews Genetics. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, Sakata-Sogawa K, Tokunaga M, Nagase T, Nozaki N, McNally JG, et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–275. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiology and molecular biology reviews : MMBR. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Li R. Human negative elongation factor activates transcription and regulates alternative transcription initiation. The Journal of biological chemistry. 2010;285:6443–6452. doi: 10.1074/jbc.M109.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Watkins G, Blair AL, Moskaluk C, Ghosh S, Jiang WG, Li R. Deregulation of cofactor of BRCA1 expression in breast cancer cells. Journal of cellular biochemistry. 2008;103:1798–1807. doi: 10.1002/jcb.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Critical reviews in biochemistry and molecular biology. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Vidal M, Gaber RF. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Strich R, Esposito RE, Gaber RF. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991;11:6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Rodova M, Miska EA, Calvet JP, Kouzarides T. Acetylation of beta-catenin by CREB-binding protein (CBP) The Journal of biological chemistry. 2002;277:25562–25567. doi: 10.1074/jbc.M201196200. [DOI] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes & development. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Nie L, Kim SH, Sun XH. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPbeta. The EMBO journal. 2003;22:893–904. doi: 10.1093/emboj/cdg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol. 2002;22:2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yang Y, Rao R, Shen J, Tang Y, Fiskus W, Nechtman J, Atadja P, Bhalla K. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer research. 2008;68:4833–4842. doi: 10.1158/0008-5472.CAN-08-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Molecular cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Zhai S, Senderowicz AM, Sausville EA, Figg WD. Flavopiridol, a novel cyclin-dependent kinase inhibitor, in clinical development. The Annals of pharmacotherapy. 2002;36:905–911. doi: 10.1345/aph.1A162. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Fan LX, Peters DJ, Trudel M, Bradner JE, Li X. Therapeutic targeting of BET bromodomain protein, Brd4, delays cyst growth in ADPKD. Human molecular genetics. 2015 doi: 10.1093/hmg/ddv136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.